Abstract
Solar ultraviolet (UV) radiation, an ubiquitous environmental carcinogen, is classified depending on the wave-length, into three regions; short-wave UVC (200–280 nm), mid-wave UVB (280–320 nm), and long-wave UVA (320–400 nm). The human skin, constantly exposed to UV radiation, particularly the UVB and UVA components, is vulnerable to its various deleterious effects such as erythema, photoaging, immunosuppression and cancer. To counteract these and for the maintenance of genomic integrity, cells have developed several protective mechanisms including DNA repair, cell-cycle arrest and apoptosis. The network of damage sensors, signal transducers, mediators, and various effector proteins is regulated through changes in gene expression. MicroRNAs (miRNAs), a group of small non-coding RNAs, act as post-transcriptional regulators through binding to complementary sequences in the 3′-untranslated region of their target genes, resulting in either translational repression or target degradation. Recent studies show that miRNAs add an additional layer of complexity to the intricately controlled cellular responses to UV radiation. This review summarizes our current knowledge of the role of miRNAs in the regulation of the human skin response upon exposure to UV radiation.
Keywords: MicroRNAs, skin, UV
1. INTRODUCTION
1.1. Structure of the Human Skin
The skin is the largest organ of the body which acts as an interface with the outside world [1]. In addition to possessing unique biomechanical properties that allow it to protect and conform to the body, its functions include maintenance of water equilibrium, protection against ultraviolet (UV) radiation, immunologic surveillance, and regulation of thermal exchange [1]. There are three structural layers to the skin: the epidermis, the dermis and the hypodermis. The epidermis is the cell-rich superficial layer composed mainly of keratinocytes, along with pigment producing melanocytes, and antigen presenting Langerhans cells. The keratinocytes forming the muti-layered epidermis possess different characteristics [2]. In the innermost basal cell layer of the epidermis, keratinocytes are undifferentiated and able to proliferate. This layer contains stem cells as well as the transient amplifying cells. Over this, several spinous cell layers reside, characterized by a high number of desmosomes. Next is the granulosum layer, that comprises of 3–5 cell layers of cells containing lamellar bodies as well as keratohyalin granules. Finally, the uppermost layer of the epidermis is the stratum corneum, which consists of dead cells or corneocytes and intercellular lipids and forms an integral part of the skin barrier [3]. The dermis is a thick layer of fibrous and elastic tissue, made mostly of collagen, elastin, and fibrillin, that gives the skin its flexibility and strength. The dermis contains nerve endings, sweat and sebaceous glands, hair follicles, and blood vessels. Beneath the dermis lies the hypodermis or the subcutaneous tissue [3].
1.2. Ultraviolet Radiation
Solar UV radiation, an important environmental carcinogen is divided, depending on the wavelength, into three regions; short-wave UVC (200–280 nm), mid-wave UVB (280–320 nm), and long-wave UVA (320–400 nm) [4]. Most of the UV to which the human skin is exposed to, is absorbed by the epidermis, with transmission of only the longer wavelengths into the dermis. UV acts by initiating a cascade of events in the skin which starts with the absorption of UV by chromophores such as DNA or urocanic acid, followed by membrane damage, induction of cytoplasmic transcription factors, DNA damage and isomerization of urocanic acid [5].
Exposure to UV may induce genomic lesions in the nuclear and mitochondrial DNA directly as well as through the generation of reactive oxygen species. The degree and type of UV-induced DNA damage depends heavily on the wavelength [4]. For a more detailed understanding of UV induced DNA damage, we recommend literature reviews by Cadet et al. and others where they have provided in depth analysis of the various mechanisms involved [6, 7]. Briefly, specific UV induced DNA damage includes formation of cyclobutane pyrimidine dimers (CPDs), pyrimidine pyrimidone photoproducts (6-4PPs) and their Dewar valence isomers. In addition, UV through the production of reactive oxygen species can produce oxidative base damage such as 8-hydroxydeoxyguanosine (8-OHdG) and thymine glycol in DNA or can cause single or double strand breaks [8]. The heterogeneous distribution of the UV-induced photolesions in the DNA depends on the sequences that facilitate DNA bending as well as chromatin modulation through binding of the specific protein [8, 9]. These lesions, if not repaired in a timely manner, can cause severe structural distortions in the DNA molecule, thereby affecting important cellular processes such as DNA replication and transcription, and compromising cellular viability and functional integrity [9].
1.3. Skin Response to Ultraviolet Radiation
To cope with the detrimental effects of UV exposure, the cellular machinery responds by mounting a rapid, inducible, transient response often termed the ‘UV stress response’. This response developed by the eukaryotic cells for the maintenance of genomic integrity is ensured by a repertoire of DNA repair systems and cell cycle checkpoints [10]. Failure of this response in the human skin may lead to immune suppression, inflammation, photoaging, and skin carcinogenesis [10].
The DNA repair system is selected based on the nature of the lesion. Similar lesions may be dealt with differently depending on whether they occur in a quiescent or a dividing cell [11]. Regardless of the repair mechanism, a mandatory step of the DNA damage response in proliferating cells is cell cycle arrest, mediated via the checkpoint cascade that results in the inhibition of cyclin-dependent kinases (Cdks), the enzymes responsible for driving cell division. DNA lesions are recognized by a network of sensor and mediator factors that result in the rapid recruitment of proteins involved in DNA damage repair such as ataxia telangiectasia mutated (ATM) and ATM-Rad3 related (ATR), which then activate the checkpoint proteins Chk1 and Chk2 with subsequent arrest of the cell cycle [12]. Checkpoint-arrested cells resume cell-cycle progression once damage has been repaired, whereas cells with irreparable DNA lesions undergo permanent cell-cycle arrest or apoptosis [13]. For the many different types of lesions that can occur, several repair pathways have evolved which include base excision repair (BER), nucleotide excision repair (NER), mismatch repair (MMR), and double strand break repair. The NER is a highly conserved strategy for repairing a variety of bulky DNA damages, such as CPDs and 6-4PPs whereas base change, such as 8-OHdG, is repaired by the BER system [14].
1.4. MicroRNAs
MicroRNAs (miRNAs) are a group of endogenous small noncoding RNAs (~23 nucleotides) that negatively regulate gene expression at the post-transcriptional level, mainly via binding to the 3′-untranslated region (3′-UTR) of the target gene [15]. The binding of the miRNA with target mRNA may lead to blockage of protein translation as well as reduced mRNA stability, however the latter seems to be the predominant mechanism in miRNA-dependent gene repression [16].
The ~70-nucleotide long primary miRNA gene transcripts, termed primary miRNAs, are recognized by RNase III endonuclease Drosha-DGCR8 microprocessor complex in the nucleus and cleaved into precursor miRNAs, which are then exported to the cytoplasm by exportin 5, where another RNase III endonuclease, Dicer, further processes them to yield mature miRNAs. These are then loaded onto RNA-induced silencing complex along with Argonaute (Ago) proteins for targeting mRNAs through interactions with sites of imperfect complementarity [16]. The binding of the seed region, located between nucleotides 2 to 8 of the mature miRNA, to the 3′-UTR of the target mRNA directs posttranscriptional repression [17]. A remarkable feature of miRNA-mediated gene regulation is that each gene may be regulated by multiple miRNAs. It is thought that this mechanism is in place for more efficient gene inhibition. To add further to the complexity, one miRNA can have several functional targets within a cell type where each gene carries the complementary sequence of the miRNA seeds in its 3′-UTR. This may be especially true of genes with long 3′-UTRs [17]. For this reason, the mechanism of action of miRNAs has been compared by some to that of transcription factors in terms of their pleiotropic effects though the effect of each is mediated at different levels in the flow of genetic information.
The relative lack of attention to miRNAs in the previous years has changed, as scientists realize that genes may have an additional layer of regulation never touched upon. Thus miRNAs are evolving as a whole new area of research. The diversity of miRNA targets demonstrates that they are involved in various cellular networks. Indeed, miRNAs have been shown to regulate many biological processes including embryonal development, cell differentiation, apoptosis, and proliferation [18]. The variations in the expression of miRNAs have been linked to a wide range of human diseases, especially cancer [16]. Several tools have been developed to control the function of individual miRNAs and have been applied to study their biogenesis, biological role, and therapeutic potential. Light-activated miRNA antagomirs have been developed through the site-specific installation of caging groups where miRNA-inhibitory activity is restored upon decaging through a brief UV exposure [19].
2. MicroRNAs and UV-Exposed Skin
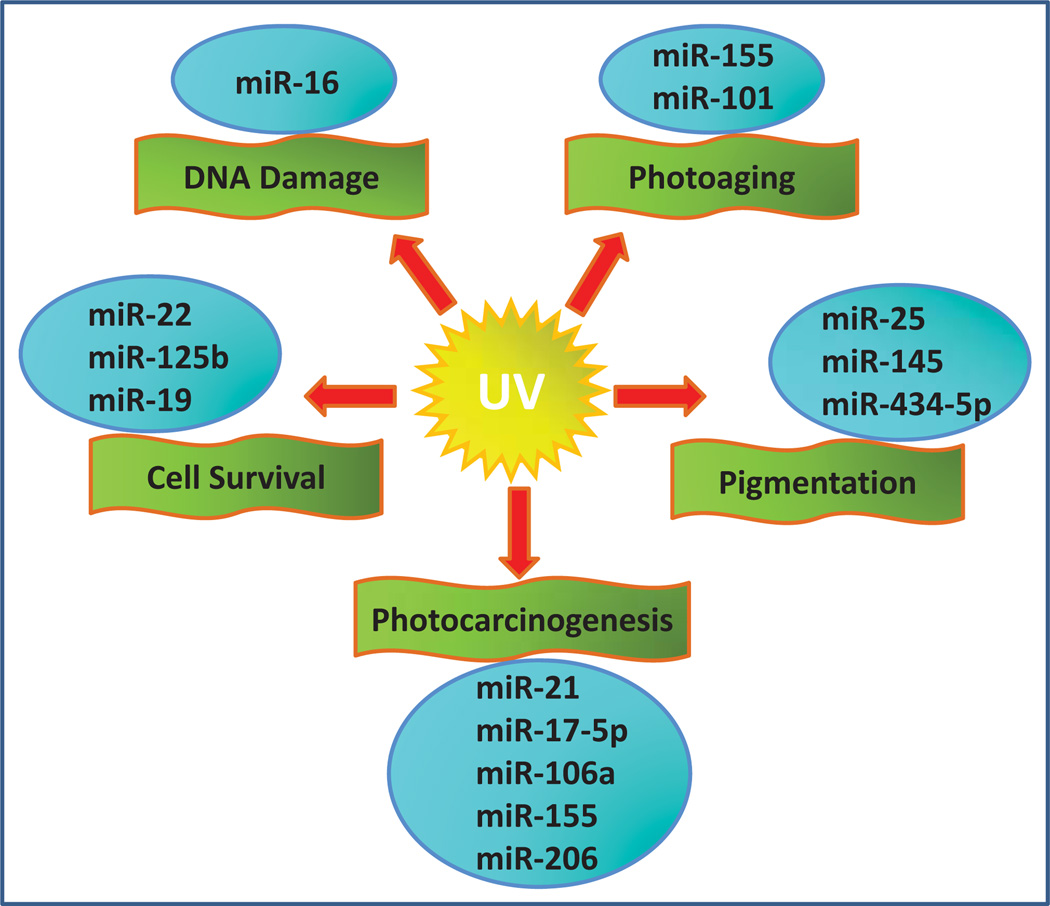
Studies of miRNAs in the field of dermatology have predominantly concentrated on their role in wound healing, skin differentiation and cancer. In spite of the rapidly growing interest in miRNAs in cutaneous physiology and pathology, surprisingly little is known about the precise mechanism(s) through which miRNAs regulate the response of normal human skin exposed to UV radiation. Recent findings that link epigenetic regulation of skin cancer to miRNAs have been the subject of several well written detailed reviews [20–23]. These analyses have mostly focused on studies that have examined miRNAs and their targets in melanoma and non-melanoma skin cancers. In this review, we have limited our scrutiny to studies that have used UV radiation in their experimental work and/or used human skin or human skin derived cells, examining a direct link between UV induced changes in miRNA expression (Fig. 1). Here we provide a brief summary of the available literature, in an attempt to gain some insight into the mechanistic aspects of miRNA-mediated regulation in the context of UV induced skin response.
Fig. (1).
Depicts selected miRNAs reportedly involved in UV-induced skin response.
2.1. MicroRNA Profile in UV-Exposed Normal Skin
The specific patterns of miRNA expression in response to UVB irradiation have been studied to some extent in keratinocytes and fibroblasts [24, 25]. Primary cultures of normal human keratinocytes 4 and 24 h post UVB irradiation (30 or 60 mJ/cm2) were analyzed using miRNA microarray and real-time PCR platform. The results showed more than two fold change in the expression of 44 miRNAs compared with non-irradiated keratinocytes [24]. Subsequent unsupervised hierarchical clustering analysis of miRNA expression based on the time of exposure (4 vs 24 h), and not on the radiation dose, revealed distinct patterns of miRNA expression change, such that some miRNAs were found modulated only at 4 h while others showed consistent change at both time points. Importantly, this pattern was similar to miRNA expression observed in human thyroid cells following exposure to ionizing radiation and in HeLa cells after UVC irradiation (8 J/m2) suggesting that these time-dependent patterns represent a common paradigm of miRNA response to radiation exposure [24, 26, 27].
The miRNAs expression in primary human fibroblasts, 4 and 24 h post UV exposure was comparable and divided into three main classes as a short-term early regulation, and a longer-lasting regulation that started early or late after UV irradiation. Interestingly, most of the miRNAs in the short term were upregulated, indicating that repression of gene expression by upregulation of UV-responsive miRNAs, rather than upregulation of gene expression by miRNA repression, is the main function of miRNA-mediated gene silencing during the first hours after UV damage [27]. Using similar methods, the differential expression of miRNAs was profiled in mouse fibroblasts after exposure to UVB irradiation (50 J/m2). In this study, UVB irradiation was found to result in robust changes in the miRNA expression profiles in a time-dependent manner. The early UVB response (at 4 h) involved upregulation of 14 miRNAs, whereas the change at 12 h was relatively small with 4 miRNAs that were upregulated and 2 that were downregulated. The authors inferred that their findings have the potential to be extrapolated to the human situation as miRNA sequences are well conserved in mice, chickens and humans. The increase in mmu-miR-365 and mmu-miR-21 in mouse fibroblasts upon UV exposure suggest that these miRNAs may be involved in the regulation of cell growth and apoptosis in response to UV-induced DNA damage in the human skin [25].
2.2. MicroRNAs and UV-Induced DNA Damage
The cellular response elicited as a consequence of DNA damage to UV radiation, is accompanied by profound changes in gene expression. Recent studies indicate that in addition to being regulated at the transcriptional and post-translational levels this response is controlled at the post-transcriptional level by miRNAs [27, 28]. Using a combination of fluorescence microscopy, miRNA profiling and reverse genetics, it was demonstrated that Ago2 and miRNA expression regulate several aspects of the DNA damage response, eventually leading to increased survival after UV irradiation [27]. UV damage triggered a cell-cycle-dependent relocalization of Ago2 into stress granules and various miRNA expression changes. It was shown that the Ago2 relocalization required Cdk activity, but was independent of ATM/ATR checkpoint signaling. Both miRNA expression changes and stress-granule formation were most pronounced within the first hours after genotoxic stress, suggesting that miRNA-mediated gene regulation operates earlier than most transcriptional responses [27]. Knocking down essential components of the miRNA processing pathway, Dicer and Ago2 reduced miRNA-mediated gene-silencing inhibition and compromised the survival and checkpoint response of the cells post UV damage. The functionality of the miRNA response in UV exposed cells was further manifested by miR-16-dependent CDC25a regulation studies [27]. CDC25a has an essential role in the cell cycle and its downregulation leads to an immediate cell-cycle stop. Gene silencing and over expression studies indicated that CDC25a mRNA is directly downregulated by miR-16 and provided evidence that miRNA-mediated gene silencing contributes to the cellular responses to UV stress in a timed manner. It was suggested that miRNA mediated regulation of UV response operates at an intermediate time point between fast protein modifications that occurs within minutes and the much slower transcriptional reprogramming that takes several hours to days to develop [27, 28].
2.3. MicroRNAs and UV-Induced Pigmentation
The skin, through its pigments, provides a unique defense system against UV radiation. The transfer of melanosomes from melanocytes to keratinocytes plays a critical role in reducing UV-induced DNA damage in the human epidermis [29]. Very few studies have described the involvement of miRNAs in the pigmentation process. A functional role for miR-25 in the regulation of pigmentation in Alpaca (Lama pacos) skin melanocytes through the suppression of micropthalmia-associated transcription factor MITF has been demonstrated [30].
Dynoodt et al. sought to identify the miRNAs involved in the regulation of melanogenesis in the human skin [31]. For their preliminary studies, they selected normal pigment-producing melan-a mouse melanocytes. Compared with human primary melanocytes, these cells are known to be significantly more responsive to physiological pigmentation factors, such as α-melanocyte-stimulating hormone and UV radiation. The authors contended that by using the mouse melan-a cells, a less complex in vitro cell culture model was introduced, in contrast to human primary melanocytes that are typically characterized by donor-specific variability. miRNA profiling was performed on melan-a cells after three consecutive treatments of a low solar-simulated UV dose (60 mJ/ cm2) and forskolin (20 µM), a known stimulator of the cAMP pathway. The rationale behind this treatment regimen was that it would induce pigmentation, whereby the phenotypic appearance of the cells could be monitored by bright-field microscopy. Increased proliferation of the treated cells with additional production of melanin, including an increased processing/movement of the melanosomes toward the periphery of the melanocytes and into the dendritic tips, was observed. Subsequently, the expression of 540 miRNAs was studied where the cutoff value for considering a miRNA as deregulated was set at 1.5-fold between treated and control samples. This filtering resulted in a miRNA signature identifying 16 differentially expressed miRNAs, of which the majority were uniformly downregulated and included miR-125b, miR-139-5p, miR-145, miR-155, miR-193*, miR-206, miR-218, miR-221, miR-222, miR-28, miR-335*, miR-365, and miR-455. In contrast, miR-130b, miR-182, and miR-9 were observed to be upregulated in the treated cells. Remarkably, a 15-fold downregulation of miR-145 was detected. A repeat of these experiments in human primary melanocytes showed that overexpression or downregulation of miR-145 reduced and increased the expression of Myo5a, Sox9, Mitf, Tyr, Trp1, Rab27a, and Fscn1, respectively [31]. Furthermore, direct targeting of Myo5a by miR-145 was demonstrated. Immunofluorescence tagging of melanosomes in miR-145-transfected human melanocytes displayed perinuclear accumulation of melanosomes with additional hypopigmentation of harvested cell pellets. Given the changes in major pigmentation genes, upon modulation of miR-145 expression, a key role for miR-145 in regulating melanogenesis was suggested. Since the current treatments for hyperpigmentation are not as effective and are often associated with adverse side effects, the authors further speculated that miRNA-based treatments by specifically targeting key genes in melanogenesis may be an attractive alternative [31]. In this context, Wu et al. designed a miR-434-5p homologue targeting Tyrosinase, the rate-limiting enzyme of melanin biosynthesis in human and mouse melanocytes and used it to demonstrate the feasibility of miRNA-mediated skin whitening and lightening in vitro and in vivo [32].
2.4. MicroRNAs and Photoaging
Photoaging is a process of aging of skin attributed to continuous, long-term exposure to UV radiation, natural or synthetic. Studies have shown that UV radiation, through activation of growth factor receptors and protein kinase cascades, up-regulates the expression of c-Jun and c-Fos, integral components of the AP-1 transcription factor complex. AP-1, in turn not only stimulates transcription of matrix metalloproteinases to induce degradation of extracellular matrix proteins, but also negatively regulates the collagen synthesis pathway [2]. Song et al. studied the mechanism through which UVA irradiation up-regulates c-Jun expression in the human skin [33]. They showed that increased c-Jun protein and mRNA levels correlated with markedly reduced miR-155 expressions in dermal fibroblasts. Luciferase reporter assays further assays further identified c-Jun as a target of miR-155. Transfection of miR-155 mimic decreased c-Jun protein levels in both UVA-exposed and unexposed fibroblasts while miR-155 inhibitor increased c-Jun protein levels. Interestingly, neither had any effect on c-Jun mRNA expression, suggesting that miR-155-induced c-Jun inhibition occurred at the post-transcriptional level [33]. The study provided new insights into the pathogenesis of UVA-induced photoaging and indicated that miR-155 might function as a protective miRNA and can serve as a potential target for the treatment of photoaging.
Skin aging has further been linked to UV irradiation mediated cellular senescence. The molecular mechanisms underlying UVB induced senescence were investigated in human diploid fibroblasts [34]. Using genome-wide transcriptome analysis, a transcriptional signature of UVB-induced senescence was established. In parallel, miRNAs screening identified five miRNAs, miR-15a, miR-20a, miR-20b, miR-93 and miR-101, modulated during UVB-induced senescence. Subsequent analysis, focused on miR-101, identified Ezh2 as a target gene of miR-101. Interestingly, downregulation of miR-101 was not sufficient to block the phenotype of UVB-induced senescence, suggesting that other UVB-induced processes may induce the senescence response in a pathway redundant with upregulation of miR-101 [34]. Moreover, An et al. studying the protective activity of a titrated extract of Centella asiatica against photoaging showed that the extract may serve as a potential natural chemoprotective agent against UVB-mediated damage through inducing changes in the expression of specific miRNAs in dermal fibroblasts. Using functional bioinformatic tools, it was determined that miRNAs that showed alteration in expression after treatment with the extract were functionally related with inhibition of apoptosis, positive regulation of cell proliferation and activation of MAP Kinases [35].
The miRNA expression pattern of the photodamaged skin has also been determined in human subjects. A total of 16 human biopsies were collected from eight healthy volunteers with no prior history of skin disease. All subjects had grade 2 photoaging on the outer arm and no signs of photoaging on the inner arm. Hence the biopsies taken from the inner side of the arm (sun-protected area) served as control to the outer side (sun-exposed) photoaged skin. The analysis of 936 human miRNAs, registered in the miR-base 13.0 database, did not show any significant differences in miRNA expression between sun protected and photodamaged skin. The study however validated the previously reported expression pattern of mi-RNAs present in the human skin [36].
2.5. MicroRNAs and UV-Induced Cell Survival
In an effort to explore the impact of UV radiation on miRNA regulation in relation to cell survival, the expression of a group of selected miRNAs was examined employing qPCR in cells exposed to UVC treatment. MiR-22 was found to be significantly upregulated upon exposure to UV radiation, in HaCaT human keratinocytes and HEK293 human embryonic kidney cells [37]. UV-induced increase in miR-22 expression appeared to be dependent on the activation of the DNA damage responsive ATM kinase, as UV treatment induced a significant increase of miR-22 expression in wild-type mouse embryonic fibroblasts (MEF) cells, but not in ATM-deficient MEF cells. It was proposed that the increased miR-22 expression may result from enhanced miR-22 maturation in cells exposed to UV radiation. An interesting link between miR-22 and the tumor suppressor gene phosphatase and tensin homolog (PTEN) was observed. It was found that PTEN expression inversely correlated with miR-22 induction in UV exposed cells [37]. Furthermore, UV-induced PTEN repression was attenuated by overexpression of a miR-22 inhibitor. Increased miR-22 expression significantly inhibited the activation of the proapoptotic caspase signaling cascade [37]. The study indicated that miR-22 is an important player in the cellular stress response to UV radiation, which promotes cell survival via the repression of PTEN expression thereby providing a window in time for cells to repair the DNA damage and mitigate the detrimental impact of UV radiation on cells. Remarkably, overexpression or a long-lasting high miR-22 level has been shown to contribute to tumorigenesis of skin cancers, such as melanoma, where solar UV radiation is one of the major risk factors for the disease [36]. In this context, comparative microarray analysis of miRNA expression profiles showed miR-22 along with miR-130b, miR-146b-5p, miR-223, miR-301a, miR-484, miR-663, miR-720, miR-1260, miR-1274a, miR-1274b, miR-3663-3p, miR-4281, and miR-4286 to be significantly upregulated in the melanoma tissue when compared to benign melanocytic nevi [36]. PTEN loss has been show to promote carcinogenesis of skin cancer upon UV radiation and miR-22-mediated PTEN repression may contribute to this pathological process [38].
Another miRNA that has been studied in connection with the cellular response to UV radiation is miR-125b. It was found that the protein level of p38α MAPK substantially decreased in HaCaT keratinocytes and HEK293 cells after UV exposure. In depth studies indicated that the p38α gene 3′-UTR is subjected to miR-125b -dependent repression, resulting in down-regulation of p38α expression in response to UV treatment. It was shown that increase of miR-125b depended on UV-induced NF-κB activation, which enhanced miR-125b gene transcription. Moreover, ATM appeared to be essential for UV-induced NF-κB activation, at least in HaCaT and HEK293 cells. UV-induced p38α activation was substantially inhibited by ATM inhibitor, suggesting the involvement of ATM in p38α activation however the precise mechanisms by which ATM regulates p38α activation is not yet fully understood. Collectively, these studies indicate that NF-κB-dependent up-regulation of miR-125b, forms a negative feedback loop to repress p38α activation and promote cell survival upon UV radiation [39].
Human antigen R (HuR) is a ubiquitous RNA-binding protein that shuttles between the nucleus and cytoplasm and regulates several aspects of the post-transcriptional control of gene expression. Numerous mRNAs that encode stress response proteins involved in apoptotic control are regulated through HuR binding at specific sequences in their 3′-UTRs [40]. The Ras homolog B (RhoB), a member of the Rho family of small GTPases, is an important player in UV induced cellular response. Downregulation of RhoB potentiates UV-induced apoptosis, whereas its overexpression protects human keratinocytes against UV-induced apoptosis. Moreover, RhoB binds to HuR on three distinct locations, at its 3′-UTR end [41]. Glorian et al. studied the functional consequence of interactions between RhoB mRNA, HuR and Ago2, following UV exposure. It was shown that miR-19, an oncogenic component of the miR-17-92/Oncomir-1 miRNA polycistron regulates the expression of RhoB in UV exposed keratinocytes. Interestingly, there was no evidence of deregulated expression of miR-19 post UV exposure. However, miR-19-mediated regulation of antiapoptotic RhoB expression required the binding of HuR, to the 3′-UTR of the RhoB mRNA. It was proposed that the loss of the interdependent binding between HuR and miR-19 to the RhoB mRNA upon UV exposure relieves this mRNA from miR-19-dependent inhibition of translation and contributes to the apoptotic response [42].
2.6. MicroRNAs and Photocarcinogenesis
The most common nonmelanoma skin cancers are basal cell carcinoma and squamous cell carcinoma (SCC), which start in either the basal or squamous cells located at the base of the epidermis. Although there are many risk factors involved, it is well established that exposure to UV radiation plays a major role in the causation of these cancers. Interestingly, the incidence of SCC is increased more than 60 fold in organ transplant recipients, making it the most common malignancy in these patients. To examine if UV mediated changes in miRNA expression can be related to the development of SCC, the miRNAs of the SCC skin tissue were analyzed and compared to the miRNA profile of immunocompetent patients and normal skin samples obtained from healthy individuals. Real-time RT-PCR analysis showed significantly increased expression of miR-21, extending the list of tumors that show upregulated miR-21 expression. In addition, miR-184, barely detectable in the normal epidermis, was considerably increased in SCC samples, suggesting an essential role of these miRNAs in the development or maintenance of SCC of the skin. For proof of principle, the authors further studied these miRNAs in monolayer keratinocyte cultures after exposing them to UV radiation. It was observed that UVA radiation increased the expression of miR-21, miR-203, and miR-205. UVB radiation, however, had no effect on miR-21, but significantly decreased the expression of miR-205 whereas miR-203 increased slightly. The finding that UVA, but not UVB, increased the expression of miR-21, was particularly interesting, as UVA radiation more than UVB has been implicated in the pathogenesis of SCC. Both UVA and UVB induced miR-203 expression, which is possibly in line with the differentiation and aging of keratinocytes after solar irradiation. The study recognized a role for miRNAs in SCC formation, through affecting important processes such as differentiation and apoptosis. The diverging effects of UVA and UVB radiation on miRNA expression, underlined the different mechanisms of cell damage mediated by these wavelengths and may explain the differential influence of these two wavelengths on SCC formation [43].
It has been known for some time that individuals suffering from the genetic disease Xeroderma Pigmentosum, with mutation in the NER repair enzymes are predisposed to the development of skin cancer, even with exposure to very modest levels of UV radiation. Transgenic partially repair-deficient Xpc+/− mice modeling the spectrum of UV vulnerability and increased susceptibility to cancer have been developed to better understand the possible mechanisms of photocarcinogenesis [44]. The miRNA expression profile of Xpc heterozygous and WT repair-proficient mice were compared, using a mammalian miRNA microarray comprising whole mice mature and precursor miRNA sequences [45]. MiRBase and GO analysis employed for the prediction of miRNA targets revealed that a total of 20 miRNAs were differentially expressed in the transgenic group of which 13 miRNAs were down-regulated and 7 miRNAs up-regulated when compared to the WT counterparts. Differentially expressed miRNAs were predicted to have links with several signaling pathway in skin cells, of which regulation of epidermal growth factor receptor signaling pathway and transforming growth factor beta receptor signaling pathway were notable [45].
A plethora of evidence suggests that cutaneous human papillomavirus (HPV) infection may be an additional risk factor for SCC. A recently concluded study determined that the association between cutaneous HPVs and skin cancers appears to be specific to SCC and to genus β HPVs in a general US population [46]. The involvement of miRNAs in the causation of SCC, in the context of β HPV and UV radiation was examined, using the transgenic mouse model HPV8-CER [47]. Skin tissues of UV-irradiated wild-type and HPV8-CER were analyzed. The carcinogenic potential of HPV8 was evident, as a single UVA/B-dose induced oncogene expression and led to papilloma growth transgenic mouse model within three weeks. Early steps in skin tumor formation in HPV8-CER mice were associated with an upregulation of the oncogenic miR-17-5p, miR-21 and miR-106a and a downregulation of the tumor-suppressive miR-155 and miR-206. In addition, PTEN, PDCD4 and Rb, respective targets of miR-21 and miR -106a with their pivotal role in cell cycle regulation, apoptosis and proliferation were affected. The study demonstrated that deregulations of miRNA expression are closely related to the UV-induced upregulation of HPV8 oncogene levels and subsequent tumorigenesis [47].
Recent studies have explored and established associations between malignant melanoma and varying risk factors, such as nevi, hair and eye color, history of skin cancer, childhood cancers, Parkinson's Disease, hormone exposure etc. However, UV radiation exposure remains the most important modifiable risk factor for the disease [48]. The melanoma cells are expected to bear mutations due to the typical UV-induced DNA damage. Based on whole genome sequencing of a melanoma tumor, using three different computational algorithms, it was proposed that the melanoma somatic mutations globally reduce binding of miRNAs to the mutated 3'-UTRs. It was shown that seed regions are enriched with guanine, thus rendering miRNAs prone to reduced binding to UV-mutated 3'-UTRs. The reduced binding is melanoma-specific as it is mainly a consequence of the type of mutation rather than its exact position [49]. Accordingly, mutation patterns in non UV-induced malignancies e.g. lung cancer and leukemia do not yield similar predictions. The importance of GC nucleotides and their thermodynamic advantage in conferring thermodynamic stability to DNA structures in response to UV-induced damage has been suggested. Interestingly, a significant difference in the GC/AU ratio in the 3′-UTR SNPs between dark- and light-skinned human populations was noted which implies that GC nucleotides may act as facilitators of miRNA binding and thereby global cell regulation. It was speculated that this might be an evolutionary strategy to minimize UV radiation effects and reduce risk for development of skin cancer. The observed differences in GC content between SNPs falling within predicted target sites of miRNAs highly expressed in melanoma and target sites of other miRNAs strongly supports this speculation [49].
CONCLUSION
The transcriptional regulation and posttranslational modification of proteins in response to UV radiation has been well studied. It is only recently that the involvement of miRNAs in the regulation of gene translation is being recognized. Alteration of miRNAs has been found in cells exposed to UV, both in plant and human study systems; however the precise role of miRNAs in the UV stress response of human skin has not yet been fully elucidated. Although a number of UV-responsive miRNAs have been shown to be modulated in skin cancer, the mechanistic details through which miRNAs play a role in the pathogenesis of UV-induced skin cancer are still unknown. From the data generated so far, it is clear that miRNAs contribute to regulation of cell cycle checkpoints and apoptosis in UV exposed skin. It has further been suggested that miRNA mediated regulation of gene expression in response to UV induced DNA damage operates somewhere between fast protein modifications that occurs within minutes and the much slower transcriptional reprogramming which may take several hours to days to develop. The information that we have acquired so far has answered some initial questions yet we are nowhere close to understanding the delicate interplay of gene regulation at the miRNA level in the UV-exposed skin. The functions of specific miRNAs in cell cycle checkpoints, apoptosis, differentiation and other aspects of the cellular responses to UV irradiation require detailed studies. Comparative studies in human skin defining similarities and differences between various types of DNA damaging agents on miRNA responses are needed. It is hoped that the ongoing research in this field will increase our understanding of the correlation of UV induced miRNAs with skin tumorigenesis.
ACKNOWLEDGEMENTS
Declared none.
Footnotes
CONFLICT OF INTEREST
The authors confirm that this article content has no conflicts of interest.
REFERENCES
- 1.Hussain SH, Limthongkul B, TR H, et al. The biomechanical properties of the skin. Dermatologic surgery : official publication for American Society for Dermatologic Surgery. 2013;39:193–203. doi: 10.1111/dsu.12095. [DOI] [PubMed] [Google Scholar]
- 2.Rittie L, Fisher GJ. UV-light-induced signal cascades and skin aging. Ageing research reviews. 2002;1:705–720. doi: 10.1016/s1568-1637(02)00024-7. [DOI] [PubMed] [Google Scholar]
- 3.Kirschner N, Brandner JM. Barriers and more: functions of tight junction proteins in the skin. Annals of the New York Academy of Sciences. 2002;1257:158–156. doi: 10.1111/j.1749-6632.2012.06554.x. [DOI] [PubMed] [Google Scholar]
- 4.Afaq F, Mukhtar H. Botanical antioxidants in the prevention of photocarcinogenesis and photoaging. Experimental dermatology. 2006;15:678–684. doi: 10.1111/j.1600-0625.2006.00466.x. [DOI] [PubMed] [Google Scholar]
- 5.Duthie MS, Kimber I, Norval M. The effects of ultraviolet radiation on the human immune system. The British journal of dermatology. 1999;140:995–1009. doi: 10.1046/j.1365-2133.1999.02898.x. [DOI] [PubMed] [Google Scholar]
- 6.Cadet J, Sage E, Douki T. Ultraviolet radiation-mediated damage to cellular DNA. Mutation research. 2005;571:3–17. doi: 10.1016/j.mrfmmm.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 7.Schuch AP, Garcia CC, Makita K, et al. DNA damage as a biological sensor for environmental sunlight. Photochemical & photobiological sciences: Official journal of the European Photochemistry Association and the European Society for Photobiology. 2013 doi: 10.1039/c3pp00004d. [DOI] [PubMed] [Google Scholar]
- 8.Ikehata H, Ono T. The mechanisms of UV mutagenesis. Journal of radiation research. 2011;52:115–125. doi: 10.1269/jrr.10175. [DOI] [PubMed] [Google Scholar]
- 9.Rastogi RP, Richa KA, Tyagi MB, et al. Molecular mechanisms of ultraviolet radiation-induced DNA damage and repair. Journal of nucleic acids. 2010;59:2980. doi: 10.4061/2010/592980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mailand N, Falck J, Lukas C, et al. Rapid destruction of human Cdc25A in response to DNA damage. Science. 2000;288:1425–1429. doi: 10.1126/science.288.5470.1425. [DOI] [PubMed] [Google Scholar]
- 11.Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nature reviews. Molecular cell biology. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- 12.Matsuoka S, Ballif BA, Smogorzewska A, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 13.Cerqueira A, Santamaria D, Martinez-Pastor B, et al. Overall Cdk activity modulates the DNA damage response in mammalian cells. The Journal of cell biology. 2009;187:773–780. doi: 10.1083/jcb.200903033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ichihashi M, Ueda M, Budiyanto A, et al. UV-induced skin damage. Toxicology. 2003;189:21–39. doi: 10.1016/s0300-483x(03)00150-1. [DOI] [PubMed] [Google Scholar]
- 15.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 16.Baer C, Claus R, Plass C. Genome-wide epigenetic regulation of miRNAs in cancer. Cancer research. 2013;73:473–477. doi: 10.1158/0008-5472.CAN-12-3731. [DOI] [PubMed] [Google Scholar]
- 17.Sonkoly E, Pivarcsi A. MicroRNAs in inflammation and response to injuries induced by environmental pollution. Mutation research. 2011;717:46–53. doi: 10.1016/j.mrfmmm.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 18.Lee D, Shin C. MicroRNA-target interactions: new insights from genome-wide approaches. Annals of the New York Academy of Sciences. 2012;1271:118–128. doi: 10.1111/j.1749-6632.2012.06745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Connelly CM, Uprety R, Hemphill J, Deiters A. Spatiotemporal control of microRNA function using light-activated antagomirs. Molecular BioSystems. 2012;8:2987–2993. doi: 10.1039/c2mb25175b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Segura MF, Greenwald HS, Hanniford D, et al. MicroRNA and cutaneous melanoma: from discovery to prognosis and therapy. Carcinogenesis. 2012;33:1823–1832. doi: 10.1093/carcin/bgs205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ning MS, Andl T. Control by a hair's breadth: the role of microRNAs in the skin. Cellular and molecular life sciences : CMLS. 2013;70:1149–1169. doi: 10.1007/s00018-012-1117-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sand M, Sand D, Altmeyer P, et al. MicroRNA in non-melanoma skin cancer. Cancer biomarkers : section A of Disease markers. 2012;11:253–257. doi: 10.3233/CBM-2012-0274. [DOI] [PubMed] [Google Scholar]
- 23.Bonazzi VF, Stark MS, Hayward NK. MicroRNA regulation of melanoma progression. Melanoma research. 2012;22:101–113. doi: 10.1097/CMR.0b013e32834f6fbb. [DOI] [PubMed] [Google Scholar]
- 24.Zhou BR, Xu Y, Permatasari F, et al. Characterization of the miRNA profile in UVB-irradiated normal human keratinocytes. Experimental Dermatology. 2012;21:317–319. doi: 10.1111/j.1600-0625.2012.01465.x. [DOI] [PubMed] [Google Scholar]
- 25.Guo L, Huang ZX, Chen XW, et al. Differential expression profiles of microRNAs in NIH3T3 cells in response to UVB irradiation. Photochemistry and photobiology. 2009;85:765–773. doi: 10.1111/j.1751-1097.2008.00482.x. [DOI] [PubMed] [Google Scholar]
- 26.Nikiforova MN, Gandhi M, Kelly L, et al. MicroRNA dysregulation in human thyroid cells following exposure to ionizing radiation. Thyroid. 2011;21:261–266. doi: 10.1089/thy.2010.0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pothof J, Verkaik NS, van IW, et al. MicroRNA-mediated gene silencing modulates the UV-induced DNA-damage response. The EMBO J. 2009;28:2090–2099. doi: 10.1038/emboj.2009.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pothof J, Verkaik NS, Hoeijmakers JH, et al. MicroRNA responses and stress granule formation modulate the DNA damage response. Cell cycle. 2009;8:3462–3468. doi: 10.4161/cc.8.21.9835. [DOI] [PubMed] [Google Scholar]
- 29.Costin GE, Hearing VJ. Human skin pigmentation: melanocytes modulate skin color in response to stress. FASEB. 2007;21:976–994. doi: 10.1096/fj.06-6649rev. [DOI] [PubMed] [Google Scholar]
- 30.Zhu Z, He J, Jia X, et al. MicroRNA-25 functions in regulation of pigmentation by targeting the transcription factor MITF in Alpaca (Lama pacos) skin melanocytes. Domestic animal endocrinology. 2010;38:200–209. doi: 10.1016/j.domaniend.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 31.Dynoodt P, Mestdagh P, Van Peer G, et al. Identification of miR-145 as a key regulator of the pigmentary process. The Journal of investigative dermatology. 2013;133:201–209. doi: 10.1038/jid.2012.266. [DOI] [PubMed] [Google Scholar]
- 32.Wu D, Chen JS, Chang DC, et al. Mir-434-5p mediates skin whitening and lightening. Clinical, cosmetic and investigational dermatology. 2008;1:19–35. doi: 10.2147/ccid.s4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song J, Liu P, Yang Z, et al. MiR-155 negatively regulates c-Jun expression at the post-transcriptional level in human dermal fibroblasts in vitro: implications in UVA irradiation-induced photoaging. Cellular physiology and biochemistry. 2012;29:331–340. doi: 10.1159/000338488. [DOI] [PubMed] [Google Scholar]
- 34.Greussing R, Hackl M, Charoentong P, et al. Identification of microRNA-mRNA functional interactions in UVB-induced senescence of human diploid fibroblasts. BMC genomics. 2013;14:224. doi: 10.1186/1471-2164-14-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.An IS, An S, Kang SM, et al. Titrated extract of Centella asiatica provides a UVB protective effect by altering microRNA expression profiles in human dermal fibroblasts. International journal of molecular medicine. 2012;30:1194–1192. doi: 10.3892/ijmm.2012.1117. [DOI] [PubMed] [Google Scholar]
- 36.Sand M, Skrygan M, Sand D, et al. Comparative microarray analysis of microRNA expression profiles in primary cutaneous malignant melanoma, cutaneous malignant melanoma metastases, and benign melanocytic nevi. Cell and tissue research. 2013;351:85–98. doi: 10.1007/s00441-012-1514-5. [DOI] [PubMed] [Google Scholar]
- 37.Tan G, Shi Y, Wu ZH. MicroRNA-22 promotes cell survival upon UV radiation by repressing PTEN. Biochemical and biophysical research communications. 2012;417:546–551. doi: 10.1016/j.bbrc.2011.11.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ming M, He YY. PTEN: new insights into its regulation and function in skin cancer. The Journal of investigative dermatology. 2009;129:2109–2112. doi: 10.1038/jid.2009.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan G, Niu J, Shi Y, et al. NF-kappaB-dependent microRNA-125b up-regulation promotes cell survival by targeting p38alpha upon ultraviolet radiation. The Journal of biological chemistry. 2012;287:33036–33047. doi: 10.1074/jbc.M112.383273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang W, Furneaux H, Cheng H, et al. HuR regulates p21 mRNA stabilization by UV light. Molecular and cellular biology. 2000;20:760–769. doi: 10.1128/mcb.20.3.760-769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Westmark CJ, Bartleson VB, Malter JS. RhoB mRNA is stabilized by HuR after UV light. Oncogene. 2005;24:502–501. doi: 10.1038/sj.onc.1208224. [DOI] [PubMed] [Google Scholar]
- 42.Glorian V, Maillot G, Poles S, et al. HuR-dependent loading of miRNA RISC to the mRNA encoding the Ras-related small GTPase RhoB controls its translation during UV-induced apoptosis. Cell death and differentiation. 2011;18:1692–1701. doi: 10.1038/cdd.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dziunycz P, Iotzova-Weiss G, Eloranta JJ, et al. Squamous cell carcinoma of the skin shows a distinct microRNA profile modulated by UV radiation. The Journal of investigative dermatology. 2010;130:2686–2689. doi: 10.1038/jid.2010.169. [DOI] [PubMed] [Google Scholar]
- 44.Nahari D, McDaniel LD, Task LB, et al. Mutations in the Trp53 gene of UV-irradiated Xpc mutant mice suggest a novel Xpc-dependent DNA repair process. DNA repair. 2004;3:379–376. doi: 10.1016/j.dnarep.2003.03.001. [DOI] [PubMed] [Google Scholar]
- 45.Zhou B, Wu H, Li W, et al. Microarray analysis of microRNA expression in skin of Xpc(+)/(−) mice and wild-type mice. Irish journal of medical science. 2011;180:721–726. doi: 10.1007/s11845-010-0609-9. [DOI] [PubMed] [Google Scholar]
- 46.Farzan SF, Waterboer T, Gui J, et al. Cutaneous alpha, beta and gamma human papillomaviruses in relation to squamous cell carcinoma of the skin: A population-based study. Int J Cancer. 2013;133(7):1713–1720. doi: 10.1002/ijc.28176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hufbauer M, Lazic D, Reinartz M, et al. Skin tumor formation in human papillomavirus 8 transgenic mice is associated with a deregulation of oncogenic miRNAs and their tumor suppressive targets. Journal of dermatological science. 2011;64:7–15. doi: 10.1016/j.jdermsci.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 48.Chen ST, Geller AC, Tsao H. Update on the Epidemiology of Melanoma. Current dermatology reports. 2013;2:24–34. doi: 10.1007/s13671-012-0035-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Greenberg E, Rechavi G, Amariglio N, et al. Mutagen-specific mutation signature determines global microRNA binding. PloS one. 2011;6:e27400. doi: 10.1371/journal.pone.0027400. [DOI] [PMC free article] [PubMed] [Google Scholar]