Abstract
HNO has broad biological effects and pharmacological activities. Direct HNO probes for in vivo applications were recently reported, which are CuII-based complexes having fluorescence reporters with reaction to HNO resulting in CuI systems and the release of NO. Their coordination environments are similar to that in Cu,Zn-superoxide dismutase (SOD), which plays a significant role in cellular HNO/NO conversion. However, none of these conversion mechanisms are known. A quantum chemical investigation was performed here to provide structural, energetic, and electronic profiles of HNO/NO conversion pathways via the first CuII-based direct HNO probe. Results not only are consistent with experimental observations but also provide numerous structural and mechanistic details unknown before. Results also suggest the first HNO/NO conversion mechanism for Cu,Zn-SOD, as well as useful guidelines for future design of metal-based HNO probes. These results shall facilitate development of direct HNO probes and studies of HNO/NO conversions via metal complexes and metalloproteins.
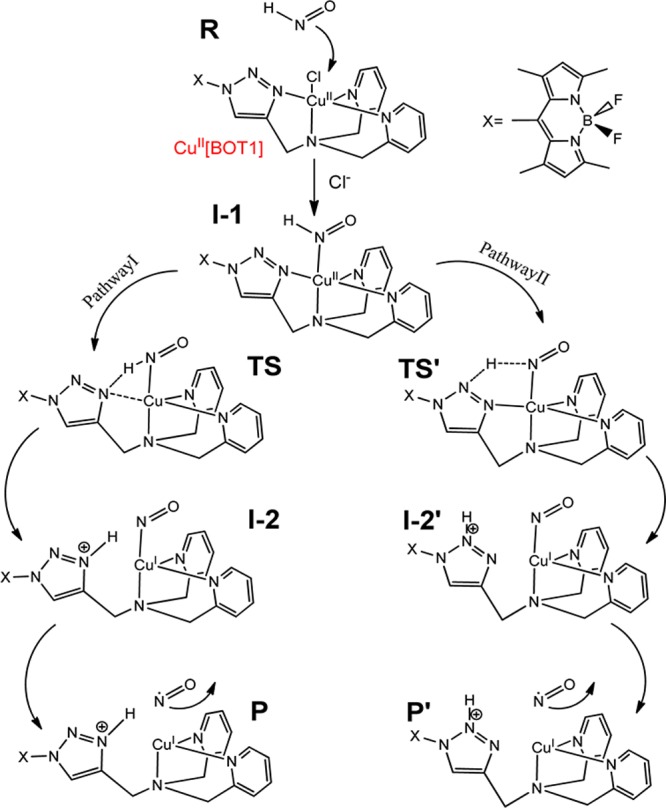
HNO plays significant roles in many biological processes, such as vascular relaxation, enzyme activity regulation, and neurological function regulation.1−3 It has also been found to possess unique pharmacological effects.2,4−6 For instance, it has a more favorable vasodilative effect than the well-known signaling molecule NO and an increased contractility effect, which enables HNO donors as a promising new class of vasodialtors and heart failure treatment.7 HNO has also been reported to inhibit GAPDH, allowing it to be an effective anticancer agent.8 However, most HNO detection methods, though useful in many scientific studies, are indirect or inconvenient for in vivo uses.1,4,9−18 Recently, intriguing breakthroughs were reported regarding direct HNO probes for in vivo applications,19−21 which are Cu-based complexes coupled with fluorescence reporters. The first complex with high selectivity against other reactive nitrogen and oxygen species including NO is CuII[BOT1].19 As shown in Figure 1, it has a BODIPY-based fluorescence reporter (X) and a tripodal receptor coupled via a triazole bridge. Upon reaction with HNO, the CuII center is reduced to CuI to trigger strong “turn-on” type fluorescence in the visible light range, together with the release of NO. More recently, a new complex, CuII[COT1], with excellent in vivo HNO imaging function and highly selective HNO detection was reported, which has a different fluorophore (coumarin-based) but the same CuII coordination site compared to CuII[BOT1].20 Its reaction with HNO also results in a CuI complex. These results indicate the importance of this Cu center in these excellent Cu-based HNO probes. Interestingly, the Cu coordination environment of four nitrogen-containing ligands including the nitrogen-containing bridging ligand in these systems is essentially the same as that found in Cu,Zn-superoxide dismutase (SOD),22,23 which was found to play a significant role in the cellular interconversion between HNO and NO.1,4,12 However, the conversion mechanisms in these Cu-based systems, identities of important species in the conversion pathways, and key structural and electronic factors responsible for reactivities remain to be elucidated.
Figure 1.
HNO/NO conversion pathways via CuII[BOT1].
Previous quantum chemical investigations have provided useful information to understand structural, spectroscopic, and mechanistic properties of HNO involvement in metalloproteins and related models.24−32 Here, we present results from a quantum chemical investigation of the HNO/NO conversion mechanisms via CuII[BOT1]. This first mechanistic information of HNO/NO conversion via copper-containing systems shall facilitate the development of additional HNO probes and studies of HNO reactions with other copper systems,12,13,16,20,21 particularly Cu,Zn-SOD.12,22,23
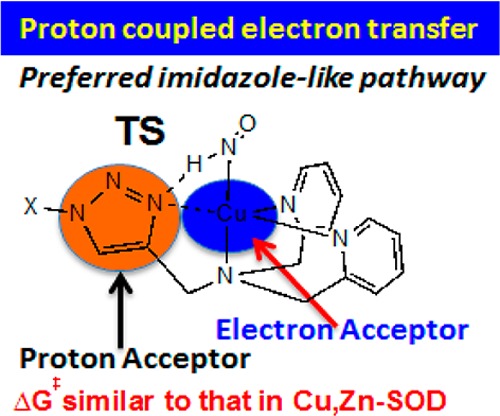
As shown in Figure 1, based on the experimental results,19 HNO/NO conversion via CuII[BOT1] starts after HNO replaces the chloride ligand in the first step (R → I-1). Then, the bound HNO is converted to bound NO, which is followed by the release of NO from the Cu complex. The HNO conversion to NO involves both proton transfer and electron transfer, which is similar to the superoxide reduction step by CuII,Zn-SOD.33,34 CuII[BOT1] also has four nitrogen-containing ligands around the redox center like CuII,Zn-SOD, including a bridging ligand, triazole, which is known as an efficient proton-transfer mediator like the bridging histidine ligand in CuII,Zn-SOD. However, because triazole can accept the proton via the terminal and middle nitrogen atoms in the triazole ring, respectively,35−39 called the imidazole-like pathway (I) and the pyrazole-like pathway (II), both pathways as illustrated in Figure 1 were investigated. This process involves transition states (TS/TS′) between the HNO-bound CuII complex (I-1) and the NO-bound CuI complex (I-2/I-2′) and is completed with the formation of corresponding products (P/P′) and the concomitant release of NO.
In order to obtain the appropriate first HNO/NO conversion mechanistic information, we first performed a methodological investigation of a number of density functional theory (DFT) methods, including the frequently used hybrid method B3LYP40 and another hybrid functional mPW1PW9141 as well as the more recently developed functionals M0642 and ωB97XD43 recommended previously for reaction energy calculations,44−48 together with several basis sets of different sizes and relativistic effects for metal centers. On the basis of the detailed comparisons in the Supporting Information, the mPW1PW91 method with the LanL2DZ basis for Cu, the 6-311++G(2d,2p) basis for core atoms including HNO, all copper coordinated atoms, and all heavy atoms in the triazole ligand, plus the 6-31G(d) basis for the rest part of molecules, were used in subsequent studies. Solvent effects are included in all geometry optimizations and subsequent frequency and energy calculations via the PCM model (see the Supporting Information). Because the complete molecules including the exact fluorophores in the experimental systems were used here, the best method was then used to investigate the conformational effect of the fluorophore on the reaction pathways, which was found to be basically negligible; see the Supporting Information. This is consistent with the fact that it is separated from the metal reaction center by the bridging ligand, as well as the experimental results that both CuII[BOT1] and CuII[COT1] with the same reaction site and only different by the fluorophore part are excellent HNO probes.19,20 Therefore, the relatively more stable fluorophore conformation was used in the following investigations, as shown by the optimized structures in Figure 2.
Figure 2.
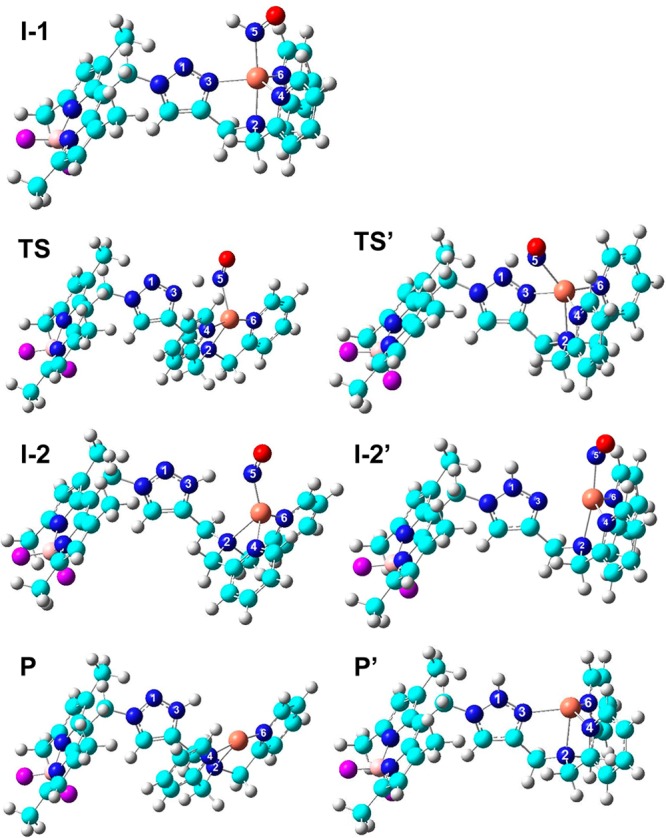
Optimized structures of the studied species in HNO/NO conversion pathways. Atom color scheme: N, blue; O, red; C, cyan; H, gray; Cu, orange; B, pink; F, purple.
As seen from Table 1, for the imidazole-like pathway I, the relative electronic energies with respect to I-1 (ΔE’s) show that after the uphill TS, the NO-bound I-2 state is actually more stable than the HNO-bound I-1 state. The NO binding with the Cu center in I-2 is also favorable as the release of NO consumes 7.11 kcal/mol of electronic energy. The inclusion of the quantum zero-point energy contribution (ΔEZPE) reduces the reaction barrier (I-1 → TS) by 2.44 kcal/mol. This barrier is further reduced at the enthalpy level (ΔH). However, after the inclusion of entropic contributions, the Gibbs free energy reaction barrier (ΔG‡) goes up by 2.63 kcal/mol. However, the formation of P in the NO release step becomes thermodynamically favorable due to a negative Gibbs free energy change from I-2 to P + NO. Because the enthalpy change in this step is positive, this thermodynamically favorable step is entropy-driven. The qualitative energetic trends were found to be the same for the pyrazole-like pathway II; see Table 1. However, the imidazole-like pathway I was found to be the dominant mechanism compared to the pyrazole-like pathway II here due to both kinetically lower ΔG‡ by 6.76 kcal/mol and thermodynamically lower overall reaction ΔG by 7.44 kcal/mol. The preference of the imidazole-like pathway is consistent with previous mechanistic studies of triazole systems in other reactions.35,37−39 For this preferred imidazole-like pathway, the small reaction barrier (12.12 kcal/mol) coupled with a favorable overall Gibbs free energy change (−12.80 kcal/mol) is in good accord with the experimental observation that CuII[BOT1] is an excellent HNO detection system with facile reactivity.19 These results also suggest that for Cu,Zn-SOD with basically the same four nitrogen-containing ligands including an imidazole-like histidine bridging ligand, the HNO conversion to NO reaction may proceed in a similar imidazole-like mechanism as described here. The HNO to NO conversion via Cu,Zn-SOD may also be driven by both the more favorable enthalpy of the NO-bound system than the HNO-bound system (similar to I-2 versus I-1) and the favorable entropy change of NO release as in I-2 → P + NO. Interestingly, the HNO/NO conversion barrier of 12.12 kcal/mol for CuII[BOT1] is also close to the experimental HNO/NO conversion barrier in CuII,Zn-SOD (10.55 kcal/mol) calculated from the experimental rate constant of 1.0 × 105 M–1 s–1 from the Eyring–Polanyi equation.15 The relatively more favorable ΔG‡ in Cu,Zn-SOD could be due to the protein environment effect, which is under investigation in our lab now.
Table 1. Relative Energies of Key Species in HNO/NO Conversion Pathways (unit: kcal/mol).
| pathway | state | ΔE | ΔEZPE | ΔH | ΔG |
|---|---|---|---|---|---|
| I | I-1 | 0.00 | 0.00 | 0.00 | 0.00 |
| TS | 12.55 | 10.11 | 9.49 | 12.12 | |
| I-2 | –10.03 | –9.27 | –9.19 | –9.14 | |
| P + NO | –2.92 | –3.23 | –1.68 | –12.80 | |
| II | I-1 | 0.00 | 0.00 | 0.00 | 0.00 |
| TS′ | 19.98 | 17.05 | 16.55 | 18.88 | |
| I-2′ | –1.47 | –1.68 | –1.33 | –2.07 | |
| P′ + NO | 5.50 | 4.23 | 4.60 | –5.36 |
In I-1, the middle N1 atom (see Figure 2 for atom numbering) involved in the pyrazole-like pathway is actually in a better position to form hydrogen bonding with the H in HNO than the terminal N3 atom involved in the imidazole-like pathway; ∠N1···H–N of 109.7° is larger than ∠N3···H–N of 84.5°, and the N1···H distance is ∼0.02 Å shorter than that of N3···H. However, the above data show that the imidazole-like pathway is more favorable. To help understand this result, a molecular orbital analysis of the HNO/NO conversion precursor I-1 was performed. As illustrated in Figure 3, in the highest occupied molecular orbital (HOMO), N3 is the only atom in the triazole ring with dominant contribution, while the contribution from the middle N1 is negligible. In addition, the lobe of N3′s p orbital that is close to H of HNO was also found to be in-phase with this hydrogen atom in the HOMO (see Figure 3) by the same green colors, which may help the formation of the new N3–H bond in I-2 after proton transfer. In contrast, the first occupied orbital with a reasonably large contribution from the middle N1 atom, the proton acceptor in the pyrazole-like pathway, is the HOMO–3, which is of lower energy than the HOMO and thus harder to be accessed. Therefore, these results suggest that the preference of the imidazole-like pathway is due to relatively easier access of N3 than N1 as the proton acceptor.
Figure 3.
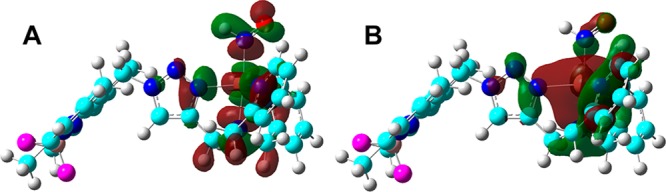
Isosurface representations of the (A) HOMO and (B) HOMO–3 in I-1. Contour values = ±0.02 au.
Further analysis of the electronic and geometric results (full data in Tables S7–S9 (Supporting Information) and selected data in Table 2) shows that the proton transfer and electron transfer are coupled during HNO/NO conversion pathways I and II, similar to the reduction step of CuII,Zn-SOD by superoxide.22,23 For instance, for the preferred imidazole-like pathway I, the N5–H bond in HNO is elongated by ∼0.2 Å from I-1 to TS and is further elongated by ∼1.0 Å at I-2. These results indicate that the proton transfer is partially done in TS. From I-1 to I-2, as shown in Table S7 (Supporting Information), the most significant geometric change in the Cu coordination environment occurs with the Cu–N3 bond, associated with the proton-acceptor site in the bridging triazole ligand. As shown in Table 2, this bond is already broken in TS, and its change is basically done at I-2 because the RCu–N3 difference between I-2 and P is only 0.021 Å. This kind of bridging ligand change is also similar to the reduction step of CuII,Zn-SOD by superoxide.22,23 As seen from Table 2, the NO moiety’s spin density (ραβNO) increases from 0.099 e in I-1 to 0.258 e in TS and subsequently changes to 0.946 e in I-2, which is basically the same as 1.000 e observed as released NO in the experimental products.19 Accordingly, copper’s spin density (ραβCu) decreases by 22% from I-1 to TS and then further reduces by 74% to be 0.024 e at I-2, which indicates that the metal center is essentially CuI, as in P observed experimentally.19 These data show that similar to the proton-transfer process, the electron transfer from the NO moiety of HNO to the Cu center is also partially done in TS and mostly finished at I-2, indicating that it is coupled with the proton transfer. Additional calculations of initial setups with either proton transfer preceding electron transfer or vice versa (i.e., they are not coupled), however, all end up with the same results as those described above, which suggests that the proton-transfer and electron-transfer processes in HNO/NO conversion have a strong tendency to be coupled. In addition, these results indicate that during HNO conversion to NO, CuII and triazole are the electron and proton acceptors, respectively. Therefore, the following approaches may help build metal-based HNO probes with enhanced reactivities: (1) a metal center with stronger oxidizing power to facilitate electron transfer; (2) the proton-accepting ligand with higher proton affinity and/or with electron-rich substituents to facilitate proton transfer; (3) the other metal ligand(s) in the system with electron-deficient substituents to facilitate electron transfer to the metal center. Other efforts such as using the ligand with strong fluorescence response and strong metal binding capability may also help.
Table 2. Key Geometric and Electronic Properties in HNO/NO Conversion Pathways.
| pathway | state | RN5–H (Å) | RCu–N3 (Å) | ραβCu (e) | ραβNO (e) |
|---|---|---|---|---|---|
| I | I-1 | 1.043 | 2.085 | 0.549 | 0.099 |
| TS | 1.191 | 3.161 | 0.430 | 0.258 | |
| I-2 | 2.238 | 3.561 | 0.024 | 0.946 | |
| P + NO | 3.540 | 0.000 | 1.000 | ||
| II | I-1 | 1.043 | 2.085 | 0.549 | 0.099 |
| TS′ | 1.346 | 2.022 | 0.339 | 0.436 | |
| I-2′ | 3.716 | 2.411 | –0.122 | 0.758 | |
| P′ + NO | 2.144 | 0.000 | 1.000 |
In summary, this work provides the first structural, energetic, and electronic profiles of HNO/NO conversion pathways in the metal-based systems. Results not only are consistent with experimental observations but also provide numerous structural and mechanistic details unknown before. Results also suggest a HNO/NO conversion mechanism for Cu,Zn-SOD, which has the same coordination sphere and a similar bridging ligand with a similar HNO/NO conversion barrier. In addition, key structural and electronic factors responsible for observed reactivities were revealed for the first time, and specific guidelines for future design of metal-based HNO probes with enhanced reactivities were reported. These first time HNO/NO mechanistic results shall facilitate development of additional direct HNO probes and investigations of HNO/NO conversions via other metal complexes and metalloproteins.
Acknowledgments
This work was supported by the NIH Grant GM085774 and the NSF Grant CHE-1300912 to Y.Z.
Supporting Information Available
Computational details and optimized coordinates (Tables S1–S24) are available. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Miranda K. M. The Chemistry of Nitroxyl (HNO) and Implications in Biology. Coord. Chem. Rev. 2005, 249, 433–455. [Google Scholar]
- Nagasawa H. T.; Demaster E. G.; Redfern B.; Shirota F. N.; Goon J. W. Evidence for Nitroxyl in the Catalase-Mediated Bioactivation of the Alcohol Deterrent Agent Cyanamide. J. Med. Chem. 1990, 33, 3120–3122. [DOI] [PubMed] [Google Scholar]
- Sidorkina O.; Espey M. G.; Miranda K. M.; Wink D. A.; Laval J. Inhibition of Poly(ADP-ribose) Polymerase (PARP) by Nitric Oxide and Reactive Nitrogen Oxide Species. Free Radical Biol. Med. 2003, 35, 1431–1438. [DOI] [PubMed] [Google Scholar]
- Fukuto J. M.; Bartberger M. D.; Dutton A. S.; Paolocci N.; Wink D. A.; Houk K. N. The Physiological Chemistry and Biological Activity of Nitroxyl (HNO): The Neglected, Misunderstood, and Enigmatic Nitrogen Oxide. Chem. Res. Toxicol. 2005, 18, 790–801. [DOI] [PubMed] [Google Scholar]
- Boje K. M. K.; Lakhman S. S. Nitric Oxide Redox Species Exert Differential Permeability Effects on the Blood–Brain Barrier. J. Pharmacol. Exp. Ther. 2000, 293, 545–550. [PubMed] [Google Scholar]
- Booth B. P.; Tabrizi-Fard M. A.; Fung H. L. Calcitonin Gene-Related Peptide-Dependent Vascular Relaxation of Rat Aorta — An Additional Mechanism for Nitroglycerin. Biochem. Pharmacol. 2000, 59, 1603–1609. [DOI] [PubMed] [Google Scholar]
- Feelisch M. Nitroxyl Gets to the Heart of the Matter. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 4978–4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Switzer C. H.; Flores-Santana W.; Mancardi D.; Donzelli S.; Basudhar D.; Ridnour L. A.; Miranda K. M.; Fukuto J. M.; Paolocci N.; Wink D. A. The Emergence of Nitroxyl (HNO) as a Pharmacological Agent. Biochim. Biophys. Acta 2009, 1787, 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough P. N.; Thrush B. A.; Ramsay D. A.; Stamper J. G. Vibrational Frequencies of HNO. Chem. Phys. Lett. 1973, 23, 155–156. [Google Scholar]
- Butkovskaya N. I.; Muravyov A. A.; Setser D. W. Infrared Chemiluminescence from the NO + HCO Reaction: Observation of the 2ν1–ν1 Hot Band of HNO. Chem. Phys. Lett. 1997, 266, 223–226. [Google Scholar]
- Cline M. R.; Tu C.; Silverman D. N.; Toscano J. P. Detection of Nitroxyl (HNO) by Membrane Inlet Mass Spectrometry. Free Radical Biol. Med. 2011, 50, 1274–1279. [DOI] [PubMed] [Google Scholar]
- Murphy M. E.; Sies H. Reversible Conversion of Nitroxyl Anion to Nitric-Oxide by Superoxide-Dismutase. Proc. Natl. Acad. Sci. U.S.A. 1991, 88, 10860–10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuto J. M.; Hobbs A. J.; Ignarro L. J. Conversion of Nitroxyl (HNO) to Nitric Oxide (NO) in Biological Systems: The Role of Physiological Oxidants and Relevance to the Biological Activity of HNO. Biochem. Biophys. Res. Commun. 1993, 196, 707–713. [DOI] [PubMed] [Google Scholar]
- Suarez S. A.; Marti M. A.; De Biase P. M.; Estrin D. A.; Bari S. E.; Doctorovich F. HNO Trapping and Assisted Decomposition of Nitroxyl Donors by Ferric Hemes. Polyhedron 2007, 26, 4673–4679. [Google Scholar]
- Liochev S. I.; Fridovich I. Nitroxyl (NO−): A Substrate for Superoxide Dismutase. Arch. Biochem. Biophys. 2002, 402, 166–171. [DOI] [PubMed] [Google Scholar]
- Nelli S.; Hillen M.; Buyukafsar K.; Martin W. Oxidation of Nitroxyl Anion to Nitric Oxide by Copper Ions. Br. J. Pharmacol. 2000, 131, 356–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisz J. A.; Zink C. N.; King S. B. Rapid and Selective Nitroxyl (HNO) Trapping by Phosphines: Kinetics and New Aqueous Ligations for HNO Detection and Quantitation. J. Am. Chem. Soc. 2011, 133, 11675–11685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline M. R.; Toscano J. P. Detection of Nitroxyl (HNO) by a Prefluorescent Probe. J. Phys. Org. Chem. 2011, 24, 993–998. [Google Scholar]
- Rosenthal J.; Lippard S. J. Direct Detection of Nitroxyl in Aqueous Solution Using a Tripodal Copper(II) BODIPY Complex. J. Am. Chem. Soc. 2010, 132, 5536–5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y.; Liu K.; Li J. Y.; Fang Y.; Zhao T. C.; Yao C. Visualization of Nitroxyl in Living Cells by a Chelated Copper(II) Coumarin Complex. Org. Lett. 2011, 13, 1290–1293. [DOI] [PubMed] [Google Scholar]
- Apfel U.-P.; Buccella D.; Wilson J. J.; Lippard S. J. Detection of Nitric Oxide and Nitroxyl with Benzoresorufin-Based Fluorescent Sensors. Inorg. Chem. 2013, 52, 3285–3294. [DOI] [PubMed] [Google Scholar]
- Schmidt H.; Hofmann H.; Schindler U.; Shutenko Z. S.; Cunningham D. D.; Feelisch M. No •NO from NO Synthase. Proc. Natl. Acad. Sci. U.S.A. 1996, 93, 14492–14497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hough M. A.; Hasnain S. S. Crystallographic Structures of Bovine Copper–Zinc Superoxide Dismutase Reveal Asymmetry in Two Subunits: Functionally Important Three and Five Coordinate Copper Sites Captured in the Same Crystal. J. Mol. Biol. 1999, 287, 579–592. [DOI] [PubMed] [Google Scholar]
- Silaghi-Dumitrescu R. Nitric Oxide Reduction by Heme-thiolate enzymes (P450nor): A Reevaluation of the Mechanism. Eur. J. Inorg. Chem. 2003, 1048–1052. [Google Scholar]
- Lehnert N.; Praneeth V. K. K.; Paulat F. Electronic Structure of Iron(II)–Porphyrin Nitroxyl Complexes: Molecular Mechanism of Fungal Nitric Oxide Reductase (P450nor). J. Comput. Chem. 2006, 27, 1338–1351. [DOI] [PubMed] [Google Scholar]
- Marti M. A.; Bari S. E.; Estrin D. A.; Doctorovich F. Discrimination of Nitroxyl and Nitric Oxide by Water-Soluble Mn(III) Porphyrins. J. Am. Chem. Soc. 2005, 127, 4680–4684. [DOI] [PubMed] [Google Scholar]
- Einsle O.; Messerschmidt A.; Huber R.; Kroneck P. M. H.; Neese F. Mechanism of the Six-Electron Reduction of Nitrite to Ammonia by Cytochrome c Nitrite Reductase. J. Am. Chem. Soc. 2002, 124, 11737–11745. [DOI] [PubMed] [Google Scholar]
- Ling Y.; Mills C.; Weber R.; Yang L.; Zhang Y. NMR, IR/Raman, and Structural Properties in HNO and RNO (R = Alkyl and Aryl) Metalloporphyrins with Implication for the HNO–Myoglobin Complex. J. Am. Chem. Soc. 2010, 132, 1583–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L.; Ling Y.; Zhang Y. HNO Binding in a Heme Protein: Structures, Spectroscopic Properties, and Stabilities. J. Am. Chem. Soc. 2011, 133, 13814–13817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramos B.; Menyhard D. K.; Olah J. Direct Hydride Shift Mechanism and Stereoselectivity of P450nor Confirmed by QM/MM Calculations. J. Phys. Chem. B 2012, 116, 872–885. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Computational Investigations of HNO in Biology. J. Inorg. Biochem. 2013, 118, 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L.; Fang W. H.; Zhang Y. Metal Centre Effects on HNO Binding in Porphyrins and the Electronic Origin: Metal’s Electronic Configuration, Position in the Periodic Table, and Oxidation State. Chem. Commun. 2012, 48, 3842–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konecny R.; Li J.; Fisher C. L.; Dillet V.; Bashford D.; Noodleman L. CuZn Superoxide Dismutase Geometry Optimization, Energetics, and Redox Potential Calculations by Density Functional and Electrostatic Methods. Inorg. Chem. 1999, 38, 940–950. [DOI] [PubMed] [Google Scholar]
- Pelmenschikov V.; Siegbahn P. E. M. Copper–Zinc Superoxide Dismutase: Theoretical Insights into the Catalytic Mechanism. Inorg. Chem. 2005, 44, 3311–3320. [DOI] [PubMed] [Google Scholar]
- Zhou Z.; Liu R.; Wang J.; Li S.; Liu M.; Bredas J.-L. Intra- and Intermolecular Proton Transfer in 1H(2H)-1,2,3-Triazole Based Systems. J. Phys. Chem. A 2006, 110, 2322–2324. [DOI] [PubMed] [Google Scholar]
- Subbaraman R.; Ghassemi H.; Zawodzinski T. A. Jr. 4,5-Dicyano-1H-[1,2,3]-Triazole as a Proton Transport Facilitator for Polymer Electrolyte Membrane Fuel Cells. J. Am. Chem. Soc. 2007, 129, 2238–2239. [DOI] [PubMed] [Google Scholar]
- Subbaraman R.; Ghassemi H.; Zawodzinski T. A. Jr. Triazole and Triazole Derivatives as Proton Transport Facilitators in Polymer Electrolyte Membrane Fuel Cells. Solid State Ionics 2009, 180, 1143–1150. [Google Scholar]
- Nagamani C.; Versek C.; Thorn M.; Tuominen M. T.; Thayumanavan S. Proton Conduction in 1H-1,2,3-Triazole Polymers: Imidazole-Like or Pyrazole-Like?. J. Polym. Sci. A: Polym. Chem. 2010, 48, 1851–1858. [Google Scholar]
- Li A.; Yan T.; Shen P. Exploring Proton Transfer in 1,2,3-Triazole-Triazolium Dimer with Ab Initio Method. J. Power Sources 2011, 196, 905–910. [Google Scholar]
- Becke A. D. Density-Functional Thermochemistry. 3. the Role of Exact Exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar]
- Adamo C.; Barone V. Exchange Functionals with Improved Long-Range Behavior and Adiabatic Connection Methods without Adjustable Parameters: The mPW and mPW1PW models. J. Chem. Phys. 1998, 108, 664–675. [Google Scholar]
- Truhlar D. G.; Zhao Y. The M06 Suite of Density Functionals for Main Group Thermochemistry, Thermochemical Kinetics, Noncovalent Interactions, Excited States, and Transition Elements: Two New Functionals and Systematic Testing of Four M06-Class Functionals and 12 Other Functionals. Theor. Chem. Acc. 2008, 120, 215–241. [Google Scholar]
- Chai J.-D.; Head-Gordon M. Long-Range Corrected Hybrid Density Functionals with Damped Atom–Atom Dispersion Corrections. Phys. Chem. Chem. Phys. 2008, 10, 6615–6620. [DOI] [PubMed] [Google Scholar]
- Siegbahn P. E. M. The Performance of Hybrid DFT for Mechanisms Involving Transition Metal Complexes in Enzymes. J. Biol. Inorg. Chem. 2006, 11, 695–701. [DOI] [PubMed] [Google Scholar]
- Sousa S. F.; Fernandes P. A.; Ramos M. J. General Performance of Density Functionals. J. Phys. Chem. A 2007, 111, 10439–10452. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Guo Z. J.; You X. Z. Hydrolysis Theory for Cisplatin and Its Analogues Based on Density Functional Studies. J. Am. Chem. Soc. 2001, 123, 9378–9387. [DOI] [PubMed] [Google Scholar]
- Yang K.; Zheng J.; Zhao Y.; Truhlar D. G. Tests of the RPBE, revPBE, tHCTHhyb, wB97X-D, and MOHLYP Density Functional Approximations and 29 Others against Representative Databases for Diverse Bond Energies and Barrier Heights in Catalysis. J. Chem. Phys. 2010, 132, 164117. [DOI] [PubMed] [Google Scholar]
- Zhao Y.; Truhlar D. G. Density Functional Theory for Reaction Energies: Test of Meta and Hybrid Meta Functionals, Range-Separated Functionals, and Other High-Performance Functionals. J. Chem. Theory Comput. 2011, 132, 164117. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


