Abstract
With the advent of technologies to obtain the complete sequence of the human genome in a cost-effective manner, this decade and those to come will see an exponential increase in our understanding of the underlying genetics that lead to human disease. And where we have a deep understanding of the biochemical and biophysical basis of the machineries and pathways involved in those genetic changes, there are great hopes for the development of modern therapeutics that specifically target the actual machinery and pathways altered by individual mutations. Prime examples of such a genetic disease are those classes of hypertrophic and dilated cardiomyopathy that result from single amino-acid substitutions in one of several of the proteins that make up the cardiac sarcomere or from the truncation of myosin binding protein C. Hypertrophic cardiomyopathy alone affects ∼1 in 500 individuals, and it is the leading cause of sudden cardiac death in young adults. Here I describe approaches to understand the molecular basis of the alterations in power output that result from these mutations. Small molecules binding to the mutant sarcomeric protein complex should be able to mitigate the effects of hypertrophic and dilated cardiomyopathy mutations at their sources, leading to possible new therapeutic approaches for these genetic diseases.
Introduction
After 40 years of developing and utilizing assays to understand the molecular basis of energy transduction by the myosin family of molecular motors (1–3), all members of my laboratory are now focused on understanding the underlying biochemical and biophysical basis of hypertrophic and dilated cardiomyopathies. Hypertrophic cardiomyopathy (HCM) is most often a result of single missense mutations in one of several sarcomeric proteins, the sarcomere being the fundamental contractile unit of the cardiomyocyte. HCM has been shown to be a monogenic inherited disease (4), although the effects of secondary mutations and environmental factors on the penetrance of the disease are important areas of research (5–7).
Clinically, HCM is characterized by an asymmetric thickening of the ventricular walls and a decrease in the ventricular chamber size that occurs without a predisposing cause. In addition, fibrosis of the cardiac muscle and cardiomyocyte disarray are associated with the disease (8). Systolic performance of the heart is preserved but relaxation capacity is diminished. HCM is not a rare disease. It affects ∼1 in 500 individuals (8–10). A related genetic disease, idiopathic dilated cardiomyopathy (DCM), also results from single missense mutations in some of the same sarcomeric proteins, but leads to ventricular chamber dilation and reduced systolic performance, rather than hypertrophy (11,12). Associated with HCM and DCM worldwide are heart failure, arrhythmias, and sudden cardiac death at any age.
This review focuses on HCM and DCM resulting from single residue changes in the sarcomeric proteins. Some HCM, and approximately half of familial DCM, are associated with mutations in nonsarcomere genes, which are not discussed here. This review also does not include the syndromic myopathies like Duchenne.
The laboratories of Christine and Jon Seidman, more than 20 years ago, showed that a mutation (R403Q) in human β-cardiac myosin leads to HCM (13). Today, as a result of their work and that of others, there are reported to be >300 pathogenic mutations in β-cardiac myosin alone (14–16), and most of these are in the globular head domain (subfragment 1 of myosin, or S1) (Fig. 1 a). S1, which is the motor domain of the myosin molecule (17), has been crystallized in a wide variety of nucleotide states (18–24), and its light-chain binding region serves as a lever arm to amplify movements of the converter domain during the power-stroke of the head (20–23,25–28). Structurally, the myosin head domain consists of several subdomains that are suggested to move mainly as semirigid bodies during the chemomechanical cycle (22). A primary focus of my laboratory is on the HCM- and DCM-causing mutations in human β-cardiac myosin S1, and the discussion here relates to those studies.
Figure 1.
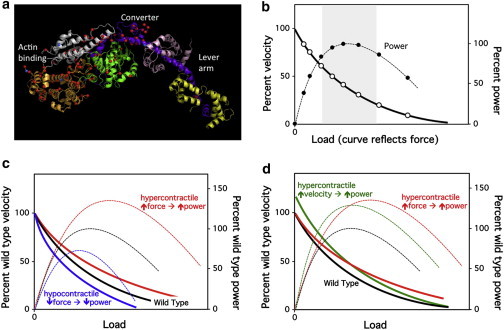
The myosin motor domain, the force-velocity curve, and power output. (a) Image of the poststroke state crystal structure of a muscle myosin motor domain (cardiac human myosin S1dC, β-isoform complexed with Mn-AMPPNP. V. A. Klenchin, J. C. Deacon, A. C. Combs, L. A. Leinwand, and I. Rayment; 4DB1 crystal structure in the RCSB database. Mutation information is from the MYOMAPR database; Buvoli 08 is the reference for those). The actin-binding, converter, and lever-arm domains are indicated. The length of the globular head from the actin-binding domain to the end of the converter is 9 nm, and from the end of the converter to the end of the lever arm is 8 nm. Representatives of reported HCM mutations (red dots) and DCM mutations (blue dots) are shown (14). (b) The hyperbolic solid line (open circles) refers to how the velocity changes as a function of the load imposed on the contractile machinery. The shape of the curve depends on how much force the contractile machinery can produce. At any velocity along the curve, the ensemble force of the system matches the equal and opposite load imposed. Power output is the force times the velocity at every point along the curve (dashed line, solid circles). (Gray zone) Region of high power output. (c) The higher force-producing hypercontractile-causing mutant forms of the contractile apparatus (solid red F-v curve; dashed red power curve) require more load to slow the contraction, and a higher power-output results. The reverse is expected for the hypocontractile-causing mutant forms (solid blue F-v curve; dashed blue power curve). (d) The force-velocity curve can be changed in two fundamental ways by hypercontractile-causing mutations: 1), The force can be increased (solid red F-v curve), resulting in increased power output (dashed red curve). 2), The velocity can be increased even in the absence of any force change (solid green curve), also resulting in increased power output (dashed green curve). Hypercontractile-causing mutations can also, of course, lead to changes in both ensemble force and velocity. To see this figure in color, go online.
HCM- and DCM-causing mutations occur in other sarcomeric proteins as well, including tropomyosin and the three troponin subunits, troponins T, I, and C, the Ca2+-regulatory proteins of the sarcomere. Mutations in these thin filament proteins have been linked to ∼5–10% of both HCM and idiopathic DCM cases (29,30). The actin, myosin, tropomyosin, troponin six-component system is the fundamental Ca2+-regulated complex of the sarcomere. There are, however, additional regulatory elements, such as myosin binding protein C and titin; mutations in myosin binding protein C are a frequent cause of HCM (4) and mutations in titin are a prevalent cause of DCM (31–33). Much of what I discuss below applies to these nonmyosin sarcomeric mutations as well as to those in human β-cardiac myosin S1.
Fundamental Parameters of the Cardiac Contractile System
Power is the product of force and velocity of contraction, and the force-velocity curve is a fundamental functional aspect of cardiac muscle function (34) (Fig. 1 b). The force axis on this curve is related to the load on the muscle; the force the muscle produces must overcome the load. Ventricular function is influenced by two types of loads that are applied to cardiac muscle. One is the preload, which is the load applied by the volume of blood in the left ventricle that results in stretching of the myofibers before the initiation of ejection. According to the Frank-Starling law, increase in ventricular filling increases contractility, and one major hypothesis is that increase in sarcomere length activates Ca2+ responsiveness in the Ca2+-regulated thin filament (35), but the Frank-Starling effect remains to be clearly understood at the molecular level. The other load that is applied to the ventricle is the pressure in the systemic circulation during contraction when the aortic valve is open, called “afterload”. The afterload is the primary load that the heart needs to overcome to eject blood out of the left ventricle, and is also the load mentioned in the F-v curves shown in Fig. 1, which depict the relationship between the velocity of the contracting muscle and the opposing load applied to the muscle. This review focuses on the biomechanical aspects of the myosin-actin interaction. Its regulation by thin filament components is also critical in determining myocardial contractility, as briefly discussed below.
Note that there is a fairly broad region of the F-v curve where the power output is near maximum (>80% power is maintained between velocities of 20–60% maximum velocity) (gray zone, Fig. 1 b). HCM patients present with clinical features of hyperdynamic ventricular function, as suggested by physical exam and echocardiographic findings (4). Thus, the HCM heart is recognized as hypercontractile (red F-v curve, Fig. 1 c), suggesting that the power output is higher than that of the normal heart. Conversely, the clinical features of DCM patients are characterized by reduced systolic function causing hypoperfusion of the body circulation. Hence, DCM is recognized as hypocontractile (blue F-v curve, Fig. 1 c), leading to lower power output than that of the normal heart. Thus, the simplest view is that there are two mechanistic buckets for changes in power output at the molecular level, mutations resulting in an increase in power (hypercontractile) and a decrease in power (hypocontractile), and small molecule therapies could be directed toward either reducing the power output or increasing it, respectively, to normalize the effect of the mutation and then allowing the heart to remodel. Life, however, is not that simple.
First of all, the relationships between the hypercontractile HCM heart and the hypocontractile DCM heart with the fundamental power outputs of the contractile molecular machinery making up the sarcomere, at the earliest stages before apparent clinical symptoms, remain to be clarified. Thus, the hypothesis that HCM is hypercontractile at the fundamental molecular level of a sarcomere whereas DCM is hypocontractile—although a common view (for reviews, see the literature (36–40))—remains to be established as a general paradigm. These mutations have not been able to be easily studied in the context of purified human β-cardiac myosin because there has not been an adequate expression system for preparing homogeneous purified human β-cardiac myosin in the laboratory. Consistent with this hypothesis, however, studies of mouse models of HCM and DCM and with purified mouse cardiac myosin suggest that HCM cardiomyopathy mutations cause hypercontractility whereas DCM cardiomyopathy mutations cause hypocontractility (41,42). However, these mutations were studied in the context of the mouse α-cardiac myosin backbone, which differs from human β-cardiac myosin by 83 amino acids in the motor domain alone, whereas myosin-based HCM and DCM are caused by a single residue change in the protein. This issue is highlighted by the findings that the same HCM-causing mutation can cause opposite effects on cardiac power output, depending on the mouse myosin isoform (α versus β) into which it is introduced (43). Recent studies of human biopsy specimens showed decreased force production in cardiomyocytes from patients carrying HCM-causing β-cardiac myosin mutations, arguing that loss of contractile function underlies the development of HCM (44). However, these samples were obtained from patients undergoing palliative surgery for severe hypertrophy and contained severely diseased myocardium with a multitude of secondary changes that make determining the primary effects of the mutations on the molecular biomechanics of the sarcomere difficult, if not impossible.
Taken together, these studies illustrate the great need to quantify the effect of HCM and DCM mutations on human cardiac myosin power generation using ensembles of highly purified and homogeneous human β-cardiac myosin. As a major advance, Srikakulam and Winkelmann (45) solved the problem of mammalian myosin expression using adenovirus expression in the mouse myogenic cell line C2C12. They have characterized the involvement of a number of chaperones involved in mammalian skeletal myosin folding, including heat shock protein 90 (Hsp90), constitutively expressed heat shock-related protein 70 (Hsc70), and Unc45b (45,46). The C2C12 expression system has been adapted to the cardiomyopathy problem by Deacon et al. (47) and Resnicow et al. (48), and it is now possible to express human β-cardiac myosin carrying HCM- and DCM-causing mutations of interest (49,50). Having the human reconstituted system now provides an opportunity to understand the detailed effects of these mutations on the power output of the fundamental unit of the human cardiac muscle, the sarcomere.
The major goal in my laboratory, as of this writing, is to understand at the molecular level, with purified reconstituted components that include human β-cardiac myosin, the detailed effects of HCM and DCM mutations on the power output by that molecular assembly. We are using a variety of assays, including loaded in vitro motility assays and single-molecule analyses with a dual-beam laser trap. Because there are currently insufficient data to conclude that HCM mutations generally cause hypercontractile sarcomeres and DCM mutations generally cause hypocontractile sarcomeres at the earliest stages before apparent clinical symptoms, I will simply refer throughout to mutations causing increased power output by the sarcomere (hypercontractile) versus those causing decreased power output (hypocontractile). Furthermore, I will focus on mutations that cause hypercontractility to explain the changes in the actin-activated myosin chemomechanical cycle that can lead to increased power output. As you will see, there are more than two mechanistic buckets, but still a limited number, probably <10. I will then discuss available assays for exploring this space, and how they can be used to search for appropriate small molecule therapies. All that I discuss below applies to the hypocontractile mutations as well, but in reverse.
Power output is the product of force and velocity and hypercontractile-causing mutations can change the F-v curve in two fundamental ways
Fig. 1 d illustrates two mechanistically distinct buckets for hypercontractile-causing mutations. An increase in power output could result from an increase in the ensemble force (red solid line) produced by the muscle, or an increase in velocity of contraction (green solid line) at various loads. A combination of change in force and change in velocity is also possible. Note that small changes in the F-v curve result in significant changes in power output. Load in Fig. 1 is the afterload on the muscle, as discussed above.
I refer to the force produced by the cardiac muscle as the ensemble force, because each myosin head is an independent force generator with its own intrinsic force, and the total force produced in the muscle is the intrinsic force times the number of heads in the ensemble that are in a force-producing state, as described in more detail below.
From a therapeutic point of view, it may be beneficial to target the increased power output caused by hypercontractile-causing mutations by developing small molecule inhibitors of the power output, regardless of whether those molecules reduce the force or the velocity. Furthermore, one could target any of the members of the fundamental six-component contractile system to do so. An agent is likely to be beneficial simply by bringing the F-v curve back down toward normal without worrying about its detailed shape. Ideal small molecule therapies, however, would normalize the F-v curve across all loads, bringing the mutant curves in line with the wild-type F-v relationship. Is this possible? Fortunately, the contractile system is one of the most thoroughly studied enzyme systems in biology, so our deep knowledge of the biochemical and biophysical parameters involved allows the creation of appropriate assays for identifying small molecules that independently modulate F or v.
Examination of the actin-activated myosin chemomechanical cycle suggests several mechanisms by which hypercontractile-causing mutations can produce increased power output
The actin-activated myosin chemomechanical cycle can be divided into two fundamental parts:
-
1.
The weakly-bound state of the myosin heads (yellow, Fig. 2; the affinity constant in this case is low and one generally refers to this state as being dissociated from the actin), with ATP or ADP.Pi bound to the active site of the motor domain; and
-
2.
The strongly-bound state that is force-producing (red, Fig. 2).
The fraction of the myosin heads in the sarcomere bound to actin in the strongly-bound state at any moment is called the “duty ratio”. In vitro assays show that under very low load, ∼5–10% of the human β-cardiac myosin heads are strongly bound to actin in a force-producing state at any moment during the chemomechanical cycle, whereas ∼90–95% are getting ready to attach to replace heads that are cycling off (49) (see also Howard (51) and Uyeda et al. (52)). I will use the value 0.1 for the duty ratio under very low load for the considerations in this review. As noted above, every head acts as an independent force generator, producing its own intrinsic force (f). Therefore, the ensemble force (Fe) produced by the contractile apparatus is the intrinsic force of each head multiplied by the total number of heads that are bound in a force-producing state,
| (1) |
where Nt is the total number of myosin heads overlapping with the actin thin filaments in the muscle.
Figure 2.
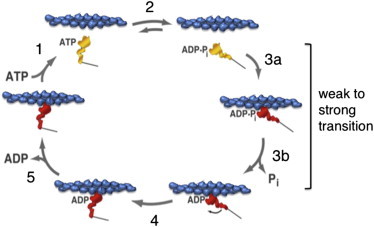
The actin-activated myosin chemomechanical cycle. Step 1: ATP binding to the strongly-bound myosin head dissociates it from the actin. Step 2: ATP hydrolysis is associated with locking the weakly-bound head into a prestroke configuration. Step 3 (a and b): Rebinding of the head to actin in a strongly-bound state is associated with Pi release from the active site. Step 4: After strong binding of the myosin to the actin, the myosin undergoes an ∼10-nm power stroke. The head is now in a strongly-bound force-producing state and cannot bind ATP until the ADP is released from the active site. Step 5: ADP is released, allowing rapid ATP binding, and the cycle continues. Modified from Lymn and Taylor (95,96). To see this figure in color, go online.
A large ΔG change occurs when ATP binds to the strongly-bound actin-myosin complex (Step 1, Fig. 2), and therefore the equilibrium of this step is very far to the right. Kinetically, ATP binding is extremely fast, as is the resultant dissociation of the myosin-ATP complex from the actin. After the myosin head dissociates from actin, hydrolysis is generally rapid (Step 2, Fig. 2), although slower in cardiac muscle than in skeletal muscle (50). There is a very small ΔG change upon hydrolysis with the ADP and Pi remaining in the active site, and therefore the equilibrium constant of this step is an important consideration (see below). For cardiac myosin, it is thought to favor the ADP.Pi state by ∼5:1 (53).
The transition from the myosin-ADP-Pi weakly-bound (to actin) state to the strongly-bound ADP state (Step 3, a and b, Fig. 2), involving concomitant release of the Pi from the active site, is a key transition in the cycle. The rate of this weak-to-strong transition is generally the rate-limiting step in the entire cycle and therefore determines the kcat, or maximum rate of the myosin ATPase activity at saturating actin concentration (although the ATP hydrolysis rate is slow enough for cardiac myosin to potentially play a role in determination of the kcat). There is a large ΔG change across this weak-to-strong transition as a result of the major increase in affinity of the myosin for actin, and therefore the equilibrium of this transition is very far to the right.
Kinetically, this bimolecular interaction of myosin with actin in the sarcomere is limited by two factors. One is the kinetic efficiency of the system or how many collisions need to happen before a productive collision occurs. The other is that in the sarcomere, the actin thin filament helical period is different from that of the myosin head arrangement along the thick filament, and therefore, at any particular amount of overlap of the thin and thick filaments, only selected myosin heads see the actin monomer to which it wants to bind in an orientation that will allow a productive collision (Fig. 3 a). In other words, the catalytic efficiencies for individual heads in the sarcomere, at any particular amount of overlap of the thin and thick filaments, are different, and are constantly changing during contraction as the two filaments slide past one another. This is in contrast to in vitro experiments with purified proteins in solution, which provide a measure of kcat for the actin-activated myosin ATPase at saturating actin concentration in which all heads behave similarly. The kcat for the actin-activated myosin ATPase in vitro at 23°C is ∼10 s−1. Thus, the mean cycle time (tc) is ∼100 ms (1/kcat). This corresponds to a tc of ∼20 ms at physiological 37°C, because the Q10 for the ATPase is between 3 and 4 (49,54). But it is worth keeping in mind that in the sarcomere, at any particular amount of overlap of the thin and thick filaments, some heads are structurally more favorably positioned for interaction with actin in contrast to in solution in vitro, limited only by the catalytic efficiency of the collision complex, whereas other heads are restricted by an unfavorable orientation for actin interaction compared to in solution in vitro.
Figure 3.
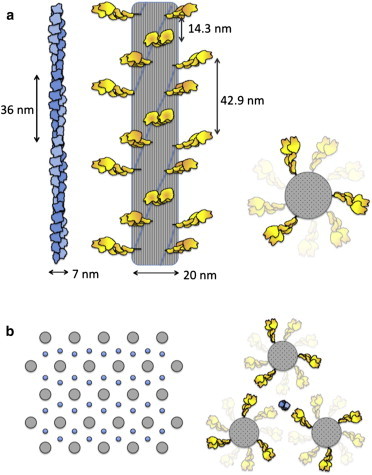
The structures of the actin filament, myosin thick filament, and their packing in the sarcomere, drawn to scale. (a) Actin is a single-start left-handed helix, more commonly described as two protofilaments with a right-handed twist with a repeat of 72 nm or pseudo-repeat of 36 nm. The myosin filament, approximately three times thicker, consists of myosin heads arranged in a three-start right-handed helix. The end-on view is shown on the right. The periodicities of the actin filament and myosin heads in the thick filament are not related. (b) Schematic representation of a cross section of a cardiac sarcomere showing a hexagonal packing of myosin thick filaments with one actin thin filament at each trigonal position (64). The packing is quite dense with heads very near the actin even in the resting state. To see this figure in color, go online.
Once the myosin head becomes strongly bound to actin, at low load the head undergoes a power stroke of ∼10 nm (Step 4, Fig. 2), producing a force in the single piconewton range (55). Note that while Fig. 2 indicates Pi release precedes the power stroke, it may occur only after the stroke occurs; this detail remains the subject of debate. The force-producing state is maintained until ADP is released from the head (Step 5, Fig. 2), a first-order process. Thus, in the presence of physiological concentrations of ATP, the rate of ADP release determines the mean time that the head remains in a strongly bound state (ts). Because the ratio of the strongly bound state time to the total cycle time (ts/tc) determines the duty ratio in the muscle, we can rewrite Eq. 1 as
| (2) |
There are only a few ways to change ensemble force
Hypercontractile-causing mutations that result in a change in Fe (solid red line, Fig. 1 d) can do so by three mechanisms: by changing the intrinsic force f, the strongly bound state time ts, or the total cycle time tc (or a combination of these). There are therefore only a few hypothetical mechanistic buckets for changing the ensemble force and thus altering the power output. However, power output can also be increased by an increase in velocity.
There are two ways to change unloaded velocity
Unloaded velocity can be thought of as the displacement caused by the myosin head stroke size (d) divided by the strongly-bound state time, or v = d/ts (51,56,57) (the velocity of the relative sliding of the actin and myosin filaments cannot be any faster than the heads can let go of the actin), although the situation is a bit more complicated due to interactions of heads with one another in an ensemble (see, for example, Walcott et al. (58)). For simplicity here, consider that hypercontractile-causing mutations that alter velocity (solid green line, Fig. 1 d) can do so by modifying the step size d or the strongly bound state time ts. Changing d or ts represent two hypothetical mechanistic buckets for affecting the velocity and thus altering the power output.
So, there are at least four hypothetical mechanistic buckets for causing an increase in power output by hypercontractile-causing mutations, changes in f, ts, tc, or d (or some combination of these). Alterations in ts are interesting because such changes have opposing effects on velocity and ensemble force. Thus, an increase in ts can lead to a decrease in velocity (decreased power) but an increase in duty ratio (increased power), and a decrease in ts can lead to the reverse. Thus, a hypercontractile-causing mutation that leads primarily to a decrease in ts would resemble the green solid line curve in Fig. 1 d, but the line would cross over the normal black curve and be below it at higher loads, due to having a diminished ensemble force to overcome load. The converse considerations apply to the same three mechanistic buckets that can lead to the reduced power output of hypocontractile-causing mutations.
How does the single cycle shown in Fig. 2 play out in the sarcomere?
How many myosin heads are in a strongly-bound state to an individual actin filament in the sarcomere at any given time? And how many times does an individual head go through its chemomechanical cycle in the course of a single contraction of the cardiac muscle (systole)? The answers surprised me and require a reasonably accurate view of the structure of the sarcomere. There are many published drawings of a sarcomere to illustrate the sliding filament model of muscle contraction, but they seldom depict the actin and myosin drawn to scale, and the conventional textbook schematic representations show a much higher extent of shortening than occurs in human cardiac sarcomeres. The cardiac sarcomere length at rest (in diastole) is variable in heart muscle, but is generally reported to be ∼2 μm (Fig. 4, top, depicts a 2-μm-long sarcomere at the beginning of its contraction). The actin filaments in cardiac muscle have been reported to be 0.8-μm-long (59) or variable in length in the 1-μm range (59–61) (Fig. 4 depicts 0.8-μm-long actin filaments, which corresponds to ∼22 pseudo-repeats (36 nm each) of the actin left-handed single-start helix, often described as a right-handed, two-stranded helical structure).
Figure 4.
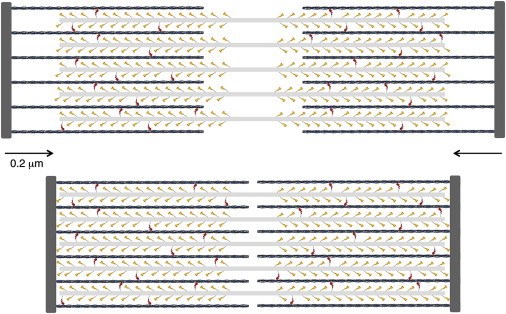
Schematic description of a cardiac sarcomere drawn to scale. (Top) A sarcomere is depicted at its resting length just beginning its contraction. The sarcomere is 2-μm long, the myosin bipolar thick filaments are 1.6-μm long, and the actin filaments are 0.8-μm long. The total number of myosin molecules that an individual actin filament sees is ∼50; only 32 are shown here because the myosin molecules from the third thick filament that is out of the plane of focus here are not shown. Note the myosin molecules are depicted as single-headed for simplicity. A duty ratio of 0.1 is depicted. Thus, ∼10% of the myosin molecules are in a force-producing state (red). (Bottom) The sarcomere has shortened by ∼20%. To see this figure in color, go online.
The cardiac myosin thick filaments are 1.6-μm long, 20-nm wide, and are bipolar, with the myosin tails packing together in an almost cylindrical backbone and the heads projecting out laterally in a helical or quasihelical fashion (62,63) (Fig. 3 a and Fig. 4). The bare zone where there are no heads is 0.2 μm. Basically, the cardiac thick filament consists of a three-start right-handed helical array of myosin heads with a ∼40° rotation and a vertical spacing of 14.3 nm between heads along the helices (Fig. 3 a). Reconstructions from electron micrographs have revealed perturbations in axial displacement, azimuthal displacement, and tilt of the heads in the cardiac myosin filament, but the radial perturbation of myosin heads is minor (62). There are three myosin molecules per 14.3 nm, or nine myosin molecules per 42.9-nm repeat.
If we simplify the thick filament helical array to a cylinder, myosin heads along one edge are spaced 42.9-nm apart. A common packing pattern has each actin filament in the sarcomere surrounded by three myosin thick filaments, which are arranged in a hexameric cross-sectional pattern (Fig. 3 b) (64). In this arrangement, 42.9-nm-spaced heads from each of three myosin filaments are available to interact with one actin filament (Fig. 3 b), which corresponds to a spacing along the actin of approximately one two-headed myosin molecule every 14.3 nm. Because the myosin thick filament in a half-sarcomere is 800-nm long, 100 nm of which is bare zone without heads, and there are three double-headed myosin molecules every 42.9 nm pointing toward a given actin filament, then (700/42.9) × 3 = ∼50 myosin molecules per half-sarcomere are pointing directly at one actin filament. Other heads that are nearby could also potentially interact, making the number of heads potentially available for interaction with one actin filament somewhat larger (Figs. 3 and 4), but for simplicity, I will consider 50 myosin molecules per half-sarcomere available for one actin filament. Note that the periodicity of heads along the thick filament is different from the periodicity of the actin filament (36-nm pseudo-repeat), insuring asynchronous binding of myosin heads to get a smooth contraction.
Although the myosin molecule is two-headed, as depicted in Fig. 3, Fig. 4 depicts only single heads projecting from the thick filament to simplify an already complex drawing. In a key study, Cooke and Franks in 1978 (65) showed that single-headed myosin generates one-half as much tension per molecule as does double-headed myosin. These experiments conclusively showed that the two heads of myosin interact independently with actin in the generation of tension, with no evidence of cooperativity between the heads. These experiments also showed that all the heads in the native two-headed myosin configuration are available for producing force because twice the force was observed with the two-headed myosin than with the single-headed myosin. Therefore, in my discussions below, I will consider all the myosin heads (two times the number of myosin molecules) as being available for force production.
Another simplification in the two-dimensional drawing of Fig. 4 is that myosin molecules from only two of the three myosin thick filaments surrounding an individual actin filament are shown, again for simplicity, which means that there are ∼30% more myosin molecules to interact with a given actin filament than illustrated in Fig. 4. Despite these simplifications, several key features become apparent from this figure, as described in more detail below:
-
1.
Even before contraction, the actin and myosin filaments in cardiac muscle are almost completely overlapping.
-
2.
At very low load, a duty ratio of 0.1 would mean that each actin would be experiencing the action of <5 myosin molecules at any moment, although this duty ratio increases as a function of load.
-
3.
An individual myosin molecule in the sarcomere may only go through one power stroke in the course of systolic contraction and that myosin does not need to even undergo one entire ATPase cycle.
The percent shortening of the cardiac sarcomere during contraction is reported to be quite small, 10–20% (66–68) (Fig. 4 depicts a 20% shortening). A 20% shortening corresponds to only 10% for each half-sarcomere, or ∼200 nm. Interestingly, even if one considers the left ventricular chamber of the heart as a simple sphere and assumes a 20% reduction of its circumference (2πr), the volume of the left ventricle (4πr3/3) would decrease to ∼50% after contraction, close to the ejection fraction of the normal human heart (∼50–70%) (69). The heart is of course not a sphere, and the effect of its complicated geometry on ejection fraction and contraction mechanics has been studied extensively (70–72).
The weakly-bound ADP-Pi-heads in the sarcomere (yellow in Figs. 2 and 4) are shown as well as the strongly-bound force-producing ADP-heads (red in Figs. 2 and 4). My surprise upon constructing this diagram was that even before contraction, the actin and myosin filaments in cardiac muscle are almost completely overlapping (top, Fig. 4). Most textbook depictions of sarcomere contraction suggest that as sliding occurs, more heads are overlapping with the actin filaments and so more of a contractile force is experienced as contraction proceeds (Eq. 2). Note from Fig. 4, however, that in cardiac muscle there may be only a modest increase in the number of myosin heads overlapping with actin filaments as contraction occurs.
The velocity of contraction of the heart once the aortic valve opens, just following isovolumetric contraction, is dictated by the relevant F-v curve (e.g., see Fig. 1 b). Assuming the system is designed for shortening of the sarcomeres to occur at maximum power output, such velocity should be in the range of 30–60% of maximum velocity at very low load (Fig. 1 b). The maximum velocity should reflect the in vitro motility velocity values, which are obtained under very low load. The velocity of cardiac muscle contraction under load at 37°C is in the range of ∼200-nm full contraction per half-sarcomere (Fig. 4) in ∼200 ms, the duration of the systolic ejection time (73–75), or ∼1 μm s−1. If we assume that the unloaded velocity should be 2–3 times that value, the unloaded in vitro motility velocity is expected to be 2–3 μm s−1 at 37°C. Mean values we have obtained for the in vitro velocity of human β-cardiac myosin are 0.8 μm s−1 at 23°C and 1.1 μm s−1 at 30°C (49). Maximum values, which I would argue more accurately reflect what occurs in the highly-ordered sarcomeres of the muscle, are just above 1 μm s−1 at 23°C (49). Thus at 37°C we expect an unloaded maximum in vitro velocity approaching 2–3 μm s−1, consistent with the prediction from the actual cardiac shortening velocity.
Systole begins when the left ventricle is full of blood and the heart undergoes an isovolumetric contraction. At this stage, the load increases rapidly while little sliding occurs. In fact, the cardiac muscle is never in a state of very low load, and load on the muscle (forces acting in the opposing direction of actin filament sliding) prevents the heads from undergoing their complete structural transition to a poststroke state (end of Step 4, Fig. 2), which slows ADP release (Step 5, Fig. 2) and prolongs ts. Thus, load changes the duty ratio (ts/tc) and more heads are bound in a force-producing state as load is increased. This is the basis of the F-v curve for muscle—as the load increases, force increases and velocity decreases. As discussed above, the velocity of contraction of the heart once the aortic valve opens is dictated by the relevant F-v curve, and the actual velocity of contraction is likely to be 30–60% of the maximum unloaded velocity. A velocity of 30% of maximum corresponds to a threefold reduction in ts and therefore a threefold increase in duty ratio. Considering a duty ratio of 0.1 at very low load, the duty ratio may increase to 0.3. A threefold increase in duty ratio corresponds to three times the number of heads bound in a strongly bound state than at very low load. As apparent from Fig. 4, which corresponds to a very low load state, this corresponds to an increase from ∼5 myosin molecules per actin filament to ∼15 myosin molecules in a strongly-bound force-producing state, still a fairly small number.
An individual myosin head may go through only one power stroke in the course of systolic contraction
Another interesting consideration illuminated by Fig. 4 relates to the number of chemomechanical cycles (Fig. 2) that a particular myosin head goes through, per systolic contraction. The fundamental unit of the myofibril is the half-sarcomere. All of the actin filaments in the half-sarcomere are linked by way of their attachment to a common Z-line (structure consisting of α-actinin and other components that bind to the (+)-end of actin filaments). Cross-sections through the myofibrils of cardiomyocytes show that sarcomere sizes in cardiac muscle are variable, ranging from ∼100 to ∼1000 actin filaments in one half-sarcomere arranged in a hexagonal array. Each actin filament is directly associated with ∼50 myosin molecules, and any movement along any individual actin filament will contribute to pulling all of the linked actin filaments toward the center of the myosin thick filaments. Considering an average of 500 actin filaments in a half-sarcomere, there are a total of ∼25,000 myosin molecules in a single average-sized half-sarcomere pointing directly at the actin filaments.
Any myosin heads that are not strongly bound to actin (Fig. 2, yellow) have already bound ATP and are in equilibrium with their ADP.Pi-bound prestroke heads. As noted above, equilibrium may favor the ADP.Pi state by as much as ∼5:1 (53). Thus, as the actin filaments encounter these heads, strong actin binding is the trigger that fires these cocked guns to undergo a step of ∼10 nm per head. If load were negligible, not many heads would be needed of the 25,000 in the half-sarcomere to complete the half-sarcomere shortening depicted in Fig. 4 of ∼200 nm; only ∼20 heads producing a stroke of 10 nm each would suffice. The muscle is not, of course, ever under negligible load. Twenty heads in the half-sarcomere would not overcome the load that exists in the system.
I already indicated that there are ∼50 double-headed myosin molecules directly poised to interact with a single actin filament in the half-sarcomere. The actual number of heads that can interact is <100 for two reasons:
-
1.
At any moment many heads are not oriented properly to bind the actin helical filament due to the different symmetries of the thin actin-containing filaments and the thick myosin-containing filaments, and must await some filament sliding to bring an actin monomer into favorable orientation.
-
2.
Any heads with ATP bound rather than ADP.Pi are unable to bind. Thus, the equilibrium constant at Step 2 of Fig. 2 is important. If the two states are distributed 1:5, then 80% of the heads are available, but if they were distributed 1:1, then only 50% of the heads would be available. An HCM or DCM mutation or small-molecule-potential drug that affects this equilibrium constant will impact force production.
Given these considerations, there are probably <50 myosin molecules available for interaction with a single actin filament in a half-sarcomere. Even if we assume all 50 molecules are available, as already discussed above, the duty ratio under load is likely to increase by a factor of ∼3, such that ∼15 myosin molecules per actin filament would be in a strongly-bound force-producing state, out of the 50 myosin molecules present. Because only ∼20 strokes of ∼10 nm each are required to traverse the 200 nm of sliding, it seems possible that no head would have to stroke more than once. This view is supported by the realization that the ATPase chemomechanical cycle time under physiological conditions of temperature and load (∼50–100 ms) is not very different from the systolic ejection time of the normal heart beat (73). Thus, there is little time for heads that have fired to recock and rebind in a strongly-bound state again before systole is finished.
These considerations also mean that an interacting head does not need to undergo one entire ATPase cycle because it would be starting in the cocked ADP.Pi state and its effect ends essentially upon ADP release, at which time ATP rapidly binds and allows the head to dissociate from the actin. Thus, a potential drug that affects the kinetics or the thermodynamics of the head may only have to apply its effect to this single partial cycle of activity during each systolic contraction.
Is the hypothesis that myosin binding to actin facilitates the Ca2+ response during contraction physiologically relevant?
My beginnings in the muscle field was 1969, when I had the great fortune to spend two years as a postdoctoral fellow at the Medical Research Council Laboratory of Molecular Biology in Cambridge, England, working with Hugh Huxley. There I reconstituted the six-component Ca2+-regulated actin-myosin-tropomyosin-troponin C,I,T system and carried out biochemical and structural studies that led us to propose the steric-blocking mechanism for tropomyosin-troponin function (76,77). In the absence of Ca2+, the troponin complex holds tropomyosin in a position on the actin filament that blocks the myosin interaction with actin (i.e., the blocked state (78)). To initiate systole, Ca2+ released from the sarcoplasmic reticulum binds to troponin C, which allosterically, through troponin I and troponin T, releases tropomyosin from its blocked state, allowing myosin to interact with actin.
It is often suggested that myosin binding to actin has a cooperative effect on moving the tropomyosin out of its inhibitory state, and that it is a combination of Ca2+ binding to troponin C and myosin binding to actin that is involved in systole (see review by Moss et al. (79)). This suggestion derives from in vitro studies with the reconstituted six-component system and with detergent-permeabilized cardiomyocytes. If one examines those data for how much myosin binding is needed to reveal such cooperativity in tropomyosin movement, it is a relatively high number—∼1 myosin head for every seven actin monomers. The considerations discussed above suggest that even under load, only ∼15 myosin molecules per 0.8-μm-long actin filament are in a strongly-bound force-producing state; that corresponds to, at most, 30 myosin heads per ∼290 actin monomers, or 1 myosin head per ∼10 actin monomers. Thus, the physiological significance for heart muscle contraction of the in vitro observed myosin-binding-induced tropomyosin movement must be questioned. Consistent with this, Sun et al. (80) present studies that suggest that although myosin binding can switch on thin filaments in rigor conditions, it does not contribute significantly under physiological conditions, and that the physiological mechanism of cooperative Ca2+ regulation of cardiac contractility must be intrinsic to the thin filaments.
Assays are Available to Assess the Fundamental Parameters that Lead to an Increase in Power Output
Fortunately, assays with purified sarcomeric proteins are available to explicitly explore the parameters f, ts, tc, and d, and see how they change in a particular HCM or DCM mutant.
Steady-state actin-activated myosin ATPase activity
The simplest and longest-standing biochemical assay is actin-activated ATPase activity, which follows Michaelis-Menten kinetics to give the maximum ATPase rate at saturating actin concentration (kcat for the cycle) and the Km or apparent affinity for myosin binding to actin. As discussed above, the mean cycle time tc falls directly out of this assay because tc = 1/kcat.
The ATPase assay extended: pCa curves to assay regulation by the tropomyosin-troponin complex
As mentioned earlier, in cardiac muscle the actin-myosin interaction is regulated through the Ca2+ sensitivity of the thin filament. Cardiac muscle is in its resting state (diastole) when Ca2+ concentration is <10−7 M (pCa < 7) and undergoes contraction when Ca2+ is released from the sarcoplasmic reticulum and binds to TnC in the sarcomere, which removes the steric block of the actin binding site for myosin. The normal operating range in cardiac muscle under normal conditions is from a pCa of ∼7 to a pCa of ∼6 (Fig. 5).
Figure 5.
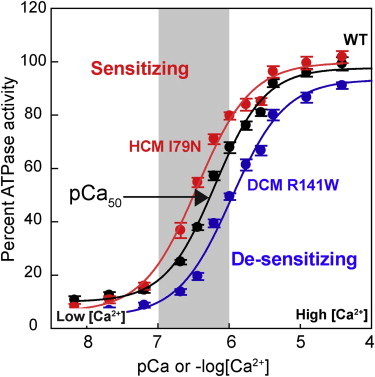
Assay for activation of myosin ATPase activity by the regulated actin thin filament as a function of free Ca2+ concentration. The in vitro generated curves mimic well the expected pCa range operating in cardiac muscle (gray zone); actually, under normal everyday conditions of behavior, the human heart seldom reaches even halfway up this curve. The two mutations shown are troponin T mutations and the assays were carried out using expressed human β-cardiac myosin S1 (data from Sommese et al. (82)). To see this figure in color, go online.
There are a variety of assays for examining the effects of HCM- and DCM-causing mutations on the Ca2+ regulation of the actin-myosin interaction. A favorite and standard assay is to measure the ATPase of the six-component system (actin, myosin, tropomyosin, and troponins T, I, and C) as a function of Ca2+ concentration, commonly referred to as a pCa curve (36,40,81,82). This assay can be applied to studies of HCM and DCM mutations in human β-cardiac myosin, but has most often been used to study the regulatory proteins carrying HCM and DCM mutations. Numerous studies show, for example, that troponin mutations shift this pCa curve to the left in the case of HCM mutations and to the right in the case of DCM mutations (36,40,81,82) (Fig. 5). Shifting the curve to the left sensitizes the system to Ca2+ and results in a higher ATPase during systole (suggesting hypercontractility) and a failure to relax completely at a pCa of 7 (a diastolic problem, consistent with the HCM clinical syndrome). Shifting the curve to the right desensitizes the system to Ca2+ and results in a lower ATPase during systole (suggesting hypocontractility, consistent with the DCM clinical syndrome).
Whereas this assay is very powerful, it, like all in vitro assays, has its limitations. It is difficult to carry out this assay in saturating concentrations of regulated thin filaments, and therefore the activity changes observed at maximal activation may result from changes in the Km of myosin binding to the thin filament, rather than changes in kcat. Km changes may not be very relevant to the function of the muscle, where the concentrations of the sarcomeric proteins are extremely high. Furthermore, steady-state ATPase only relates to changes in tc, which is only one variable in determining power output. One must use a combination of all the assays discussed here to understand changes in all the four important parameters, f, tc, ts, and d, to get the complete picture of the effects of the HCM or DCM mutation of interest.
Transient kinetic assays
Transient kinetic assays comprise a powerful set of tools in muscle research (83). There are specific assays that allow one to measure the kinetics of essentially every step in the chemomechanical cycle. From the discussion above, a critical step for cardiac myosin is the ADP release rate, because that rate determines ts, and changes in ts lead to a change in duty ratio as well as a change in velocity, both of which affect power output. One assay for the ADP release rate utilizes pyrene-labeled actin, which demonstrates an approximately twofold decrease in fluorescence upon myosin binding. If one rapidly mixes saturating ATP with a preformed actin-β-cardiac myosin S1 complex, an extremely fast dissociation rate of myosin from the actin (∼1000 s−1 at 20°C, Step 1, Fig. 2) can be measured by following the increase of fluorescence as a function of time (47). Because the ADP release rate from an actin-myosin-ADP complex is comparatively very slow (∼100 s−1 at 20°C), one can preload ADP onto an actin-myosin complex before adding saturating ATP. The ATP cannot bind to and release the myosin from the actin (Step 1, Fig. 2) until the ADP comes out of the pocket (Step 5, Fig. 2), and therefore the rate of fluorescence change becomes a readout of the rate of ADP dissociation. This is one way to estimate ts. In this experiment, the contractile system is under zero load.
For several decades, actin-activated ATPase and transient kinetic assays were the primary in vitro quantitative assays for the actin-activated myosin chemomechanical cycle, completely focused on the chemo aspect and ignoring the mechanical part of the cycle.
In vitro motility assay
In the 1980s, quantitative in vitro motility assays with purified actin and myosin were established (17,84,85) that gave a velocity readout that mimicked closely the velocities of muscle contraction (Fig. 6 a). The values obtained for velocity by this method are key, and relate to ts and d, as described above. Importantly, one can apply load to moving actin filaments by adding an actin-binding protein to the surface, which then opposes the force being applied to the filament by the myosin molecules (49,86–89). This allows one to generate a load-velocity curve by measuring the decrease of velocity as a function of increasing concentration of the load-producing protein. This is an extremely powerful tool, because with three purified proteins—actin, myosin, and the load-producing protein—one can measure the differences in the force-velocity curves shown in Fig. 1.
Figure 6.
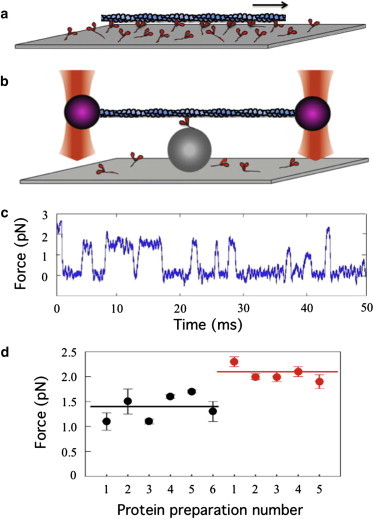
In vitro motility taken to the single-molecule level. (a) Myosin-coated surfaces drive the movement of fluorescently-labeled actin filaments at velocities comparable to those of muscle contraction (85). (b) The dual-beam laser-trap assay for measuring nanometer steps and piconewton forces of a single myosin molecule (55). (c) Force transients measured by clamping the position of the actin-bound polystyrene bead on the left (purple, in panel b) as the myosin is trying to move the actin to the right. (d) Mean intrinsic forces from multiple preparations of wild-type human β-cardiac myosin S1 (black circles) and human β-cardiac myosin S1 carrying the HCM-causing mutation R453C (red circles) (49). To see this figure in color, go online.
The single-molecule, dual-beam laser-trap assay for measuring nanometer steps and piconewton forces
None of the above assays allow direct measurement of the intrinsic force f or the step size d. Values for these parameters can be obtained using the dual-beam, single-molecule laser-trap assay developed in the 1990s (55). I like to refer to this assay as a simplified in vitro motility assay, because as is apparent in Fig. 6, the assay is the same, but simplified by examining a single surface-bound myosin molecule, perched on a polystyrene bead fixed to the surface and interacting with a single actin filament. The actin filament is suspended above the myosin molecule by virtue of being attached near its ends to two polystyrene beads held in place by two focused laser beams. The bead is being viewed by a quadrant detector that is very sensitive to even 1 nm of movement. One can change the strength by which the bead on the left of Fig. 6, for example, is held in the trap. At very low trap force compared to the intrinsic force-producing capability of the motor, the stroke size of the motor, d, can be measured by the displacement of the bead out of the laser trap.
A powerful feature of the trap is that its strength can be increased to just match the intrinsic force of the motor. A feedback circuit in the system increases the strength of the trap to hold the bead position constant when a myosin interacts with the actin. This is basically an isometric tension measurement at the single-molecule level. The trap strength can be calibrated so that one observes force transients rather than step sizes (Fig. 6 c), and the mean of many molecular events is the intrinsic force of the motor. Measurement of the intrinsic force of expressed human β-cardiac myosin motor domain (subfragment 1 of the myosin or S1; the head depicted in Figs. 2–4) carrying an HCM-causing mutation (R453C) was obtained only recently (49) and showed that the mutant myosin has a 50% increase in its intrinsic force-producing capability (Fig. 6 d).
As apparent in Fig. 6 c, one can also measure how long the myosin remains bound to the actin under the conditions used. The data in Fig. 6 c was taken at low ATP concentration and therefore the mean dwell time is a measure of the second-order rate constant for ATP binding. Such experiments carried out in saturating ATP concentrations reveal the ADP release rate and directly measure the strongly-bound state time ts under physiological conditions of ATP concentrations. Because ts is related to velocity, one can generate a F-v curve at the single-molecule level by varying the force of the trap to obtain a mean ts as a function of force.
Mutations can be Mimicked by Small Molecule Effectors that Bind to the Contractile Proteins and Alter their Structures
Just as single missense mutations slightly alter the structure of the protein of interest and therefore perturb its function, the binding of a small molecule to that protein should in many instances do the same. This is particularly true of a very allosteric enzyme such as myosin where the actin binding domain, the nucleotide binding pocket, and the converter domain are all in exquisite communication with one another.
One example of a cardiac myosin activator is omecamtiv mecarbil (OM) (90), which is being developed as a treatment for heart failure characterized primarily by systolic dysfunction (91,92). OM was discovered in a screen for actin-activated myosin ATPase activators, and its primary effect on the chemomechanical cycle shown in Fig. 2 is to increase the rate of the weak-to-strong transition, resulting in a decrease in tc. This causes an increase in duty ratio, leading to an increase in both ensemble force and power output. Understanding how OM increases the weak-to-strong transition will require a high-resolution crystal structure of the myosin-OM complex. OM is known from cross-linking studies to bind between the nucleotide-binding pocket and the converter domain of the myosin head (90), a primary communication zone between the active site and the lever arm that produces the ∼10-nm power stroke (Fig. 1). Another example of a small molecule activator of the contractile apparatus, but acting by a different mechanism, is tirasemtiv, a fast skeletal muscle activator. Tirasemtiv is being developed as a potential treatment for diseases and conditions associated with aging, muscle weakness and wasting, or neuromuscular dysfunction (93,94). In this case, the small molecule enhances contractility through allosteric interactions with the tropomyosin-troponin regulatory system.
An effort to develop small molecule therapies specifically for the inherited diseases of the heart discussed in this review is particularly attractive. In patients with HCM or DCM, we know that the disease is often caused by a single residue change in myosin or one of a number of other sarcomeric proteins, and is heritable as an autosomal dominant, monogenic Mendelian disease. Importantly, in many cases, the mutation is clearly the cause of the disease, and if one understands the effects of a particular amino-acid substitution on changes in f, ts, tc, and/or d, then it seems likely that very precise modifiers of power output can be generated to reverse the effects of that mutation, potentially interrupting, in the case of HCM, the cascade of events leading to hypertrophy, diastolic dysfunction, obstruction, and fibrosis at its source. If this hypothesis, which is eminently testable, holds true, such therapies are potentially disease modifiers, and by interrupting the pathogenic cascade, allow the diseased heart to remodel, which it is very capable of doing. Although any inhibitor of power output is likely to have a positive effect on hypercontractile-causing mutations, specific inhibitors that affect ensemble force (by altering f, ts, or tc) to reduce power and others that reduce velocity (by altering ts or d) are likely to be even better. Thus, a small number of buckets of small molecule inhibitor types married to the same mechanistic buckets of the hypercontractile-causing mutations offers an opportunity for a precision medicine approach. The same is true for hypocontractile-causing mutations, except small molecule activators for power output will be needed.
Conclusions and Future Perspectives
As I have described here, in vitro molecular studies of biochemically reconstituted sarcomeric protein complexes are critical for laying the foundation for understanding the effects of HCM- and DCM-causing mutations on power generation by the fundamental contractile apparatus of the sarcomere. With such a detailed understanding at the molecular level, one should be able to exquisitely design and screen for appropriate small molecule therapies that are desperately needed for treatment of these diseases.
What might be the best strategy for reducing the increased power output resulting from hypercontractile-causing mutations? Small molecules that significantly increase the affinity of myosin for actin should be avoided, for that will inhibit the relaxation process. The best potential drugs for hypercontractile-causing mutations may be those that park some of the heads in a state that cannot interact with actin. Thus, agents that shift the equilibrium (Step 2, Fig. 2) between myosin.ATP (which cannot bind to actin) and myosin.ADP.Pi (which can bind to actin leading to a strongly-bound force-producing state) toward the myosin.ATP state would reduce the number of myosin.ADP.Pi heads in the sarcomere available for the weak-to-strong transition. Similarly, slowing the rate of the weak-to-strong transition of myosin.ADP.Pi binding to actin and associated Pi release (Fig. 2, Step 3, a and b) will increase tc and therefore reduce the duty ratio and, therefore, the ensemble force. For those mutations that cause a decrease in the rate of ADP release, it will be important to find small molecules that can reverse that effect. This is because a reduced ADP release rate is expected to cause diastolic problems, inasmuch as the duty ratio would be increased, causing more heads to be associated with the actin filaments during systole, and it is therefore likely to take longer to remove them during diastole.
While I have focused the discussion here on hypercontractile-causing mutations in human β-cardiac myosin, hypocontractile-causing mutations are also of considerable interest. The above discussion points regarding hypercontractile-causing mutations apply to hypocontractile-causing mutations as well. Should the paradigm hold true that HCM mutations cause increased power output of the contractile apparatus at the sarcomere level, at the earliest stages before apparent clinical symptoms, whereas DCM mutations cause decreased power output, one would want small molecule inhibitors for HCM and small molecule activators for DCM. In addition to HCM- and DCM-causing mutations in the human β-cardiac myosin, mutations in other fundamental sarcomeric proteins including tropomyosin, the troponins, myosin-binding protein C, and titin also cause HCM and DCM. The regulated actin-activated myosin contractile system is one of the most thoroughly studied systems in biology, with a multitude of approaches having been applied to muscle contraction for more than seven decades. Thus, a deep understanding of the fundamental biochemical and biophysical basis of action of muscle is on hand. With this deep understanding, the development of modern therapeutics to directly and specifically target components of the sarcomeric contractile apparatus is on the horizon.
Acknowledgments
I thank Jonathan Fox and John Mercer for their insightful comments and editing in the early stages of the writing of this review. I also thank Kathy Ruppel, Ruth Sommese, Masataka Kawana, Suman Nag, Rebecca Taylor, Arjun Adhikari, Tural Aksel, Chao Liu, Jongmin Sung, Alf Månsson, Leslie Leinwand, Christine Seidman, Charles Homcy, Neil Kumar, Shaun Coughlin, Roger Cooke, Enrique De La Cruz, Donald Bers, Richard Moss, Alex Dunn, and Fady Malik for discussions and expert editing of this review. Kathy Ruppel and Masataka Kawana contributed importantly to the clinical discussions in the introduction, and Rebecca Taylor gave generous support to assembling the references.
J.A.S. is supported by National Institutes of Health grant R01 GM033289 and National Institutes of Health grant R01 HL1171138.
Footnotes
This is an Open Access article distributed under the terms of the Creative Commons-Attribution Noncommercial License (http://creativecommons.org/licenses/by-nc/2.0/), which permits unrestricted noncommercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
References
- 1.Spudich J.A. The myosin swinging cross-bridge model. Nat. Rev. Mol. Cell Biol. 2001;2:387–392. doi: 10.1038/35073086. [DOI] [PubMed] [Google Scholar]
- 2.Spudich J.A. Molecular motors: forty years of interdisciplinary research. Mol. Biol. Cell. 2011;22:3936–3939. doi: 10.1091/mbc.E11-05-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spudich J.A. One path to understanding energy transduction in biological systems. Nat. Med. 2012;18:1478–1482. doi: 10.1038/nm.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seidman C.E., Seidman J.G. Hypertrophic cardiomyopathy. In: Scriver C.R., Beaudet A.L., Valle D., Sly W.S., Childs K.W., Vogelstein B., editors. The Metabolic and Molecular Bases of Inherited Disease. McGraw-Hill; Hightstown, NJ: 2000. pp. 5532–5452. [Google Scholar]
- 5.Hilfiker-Kleiner D., Knöll R. Disease-modifying mutations in familial hypertrophic cardiomyopathy: complexity from simplicity. Circulation. 2008;117:1775–1777. doi: 10.1161/CIRCULATIONAHA.108.767657. [DOI] [PubMed] [Google Scholar]
- 6.Xu Q., Dewey S., Gomes A.V. Malignant and benign mutations in familial cardiomyopathies: insights into mutations linked to complex cardiovascular phenotypes. J. Mol. Cell. Cardiol. 2010;48:899–909. doi: 10.1016/j.yjmcc.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 7.Blauwet L., Cooper L.T. Cardiotropic viral infection in HIV-associated cardiomyopathy: pathogen or innocent bystander? Cardiovasc. J. Afr. 2013;24:199–200. [PubMed] [Google Scholar]
- 8.Harvey P.A., Leinwand L.A. The cell biology of disease: cellular mechanisms of cardiomyopathy. J. Cell Biol. 2011;194:355–365. doi: 10.1083/jcb.201101100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maron B.J. Braunwald’s Heart Disease. Elsevier; New York: 2010. Chapt. 69, Hypertrophic cardiomyopathy; pp. 1582–1594. [Google Scholar]
- 10.Maron B.J., Gardin J.M., Bild D.E. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Echocardiographic analysis of 4111 subjects in the CARDIA study. Coronary artery risk development in (young) adults. Circulation. 1995;92:785–789. doi: 10.1161/01.cir.92.4.785. [DOI] [PubMed] [Google Scholar]
- 11.Hershberger R.E., Hedges D.J., Morales A. Dilated cardiomyopathy: the complexity of a diverse genetic architecture. Nat. Rev. Cardiol. 2013;10:531–547. doi: 10.1038/nrcardio.2013.105. [DOI] [PubMed] [Google Scholar]
- 12.McNally E.M., Golbus J.R., Puckelwartz M.J. Genetic mutations and mechanisms in dilated cardiomyopathy. J. Clin. Invest. 2013;123:19–26. doi: 10.1172/JCI62862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geisterfer-Lowrance A.A., Kass S., Seidman J.G. A molecular basis for familial hypertrophic cardiomyopathy: a β-cardiac myosin heavy chain gene missense mutation. Cell. 1990;62:999–1006. doi: 10.1016/0092-8674(90)90274-i. [DOI] [PubMed] [Google Scholar]
- 14.Buvoli M., Hamady M., Knight R. Bioinformatics assessment of β-myosin mutations reveals myosin’s high sensitivity to mutations. Trends Cardiovasc. Med. 2008;18:141–149. doi: 10.1016/j.tcm.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.CardioGenomics-PGA. Genomics of Cardiovascular Development, Adaptation, and Remodeling. NHLBI Program for Genomic Applications, Harvard Medical School, Boston, MA. http://www.cardiogenomics.org [accessed Nov. 8, 2013].
- 16.Walsh R., Rutland C., Loughna S. Cardiomyopathy: a systematic review of disease-causing mutations in myosin heavy chain 7 and their phenotypic manifestations. Cardiology. 2010;115:49–60. doi: 10.1159/000252808. [DOI] [PubMed] [Google Scholar]
- 17.Toyoshima Y.Y., Kron S.J., Spudich J.A. Myosin subfragment-1 is sufficient to move actin filaments in vitro. Nature. 1987;328:536–539. doi: 10.1038/328536a0. [DOI] [PubMed] [Google Scholar]
- 18.Dominguez R., Freyzon Y., Cohen C. Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: visualization of the pre-power stroke state. Cell. 1998;94:559–571. doi: 10.1016/s0092-8674(00)81598-6. [DOI] [PubMed] [Google Scholar]
- 19.Rayment I., Rypniewski W.R., Holden H.M. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 20.Holmes K.C. The molecular basis of cross-bridge function. Adv. Exp. Med. Biol. 2005;565:13–23. doi: 10.1007/0-387-24990-7_2. 359–369. [DOI] [PubMed] [Google Scholar]
- 21.Preller M., Holmes K.C. The myosin start-of-power stroke state and how actin binding drives the power stroke. Cytoskeleton (Hoboken) 2013;70:651–660. doi: 10.1002/cm.21125. [DOI] [PubMed] [Google Scholar]
- 22.Houdusse A., Kalabokis V.N., Cohen C. Atomic structure of scallop myosin subfragment S1 complexed with MgADP: a novel conformation of the myosin head. Cell. 1999;97:459–470. doi: 10.1016/s0092-8674(00)80756-4. [DOI] [PubMed] [Google Scholar]
- 23.Houdusse A., Szent-Gyorgyi A.G., Cohen C. Three conformational states of scallop myosin S1. Proc. Natl. Acad. Sci. USA. 2000;97:11238–11243. doi: 10.1073/pnas.200376897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sweeney H.L., Houdusse A. Structural and functional insights into the myosin motor mechanism. Ann. Rev. Biophys. 2010;39:539–557. doi: 10.1146/annurev.biophys.050708.133751. [DOI] [PubMed] [Google Scholar]
- 25.Whittaker M., Wilson-Kubalek E.M., Sweeney H.L. A 35-Å movement of smooth muscle myosin on ADP release. Nature. 1995;378:748–751. doi: 10.1038/378748a0. [DOI] [PubMed] [Google Scholar]
- 26.Uyeda T.Q., Abramson P.D., Spudich J.A. The neck region of the myosin motor domain acts as a lever arm to generate movement. Proc. Natl. Acad. Sci. USA. 1996;93:4459–4464. doi: 10.1073/pnas.93.9.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shih W.M., Gryczynski Z., Spudich J.A. A FRET-based sensor reveals large ATP hydrolysis-induced conformational changes and three distinct states of the molecular motor myosin. Cell. 2000;102:683–694. doi: 10.1016/s0092-8674(00)00090-8. [DOI] [PubMed] [Google Scholar]
- 28.Geeves M.A., Holmes K.C. The molecular mechanism of muscle contraction. Adv. Protein Chem. 2005;71:161–193. doi: 10.1016/S0065-3233(04)71005-0. [DOI] [PubMed] [Google Scholar]
- 29.Lakdawala N.K., Funke B.H., Ho C.Y. Genetic testing for dilated cardiomyopathy in clinical practice. J. Card. Fail. 2012;18:296–303. doi: 10.1016/j.cardfail.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richard P., Charron P., Komajda M. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107:2227–2232. doi: 10.1161/01.CIR.0000066323.15244.54. [DOI] [PubMed] [Google Scholar]
- 31.Harris S.P., Lyons R.G., Bezold K.L. In the thick of it: HCM-causing mutations in myosin binding proteins of the thick filament. Circ. Res. 2011;108:751–764. doi: 10.1161/CIRCRESAHA.110.231670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herman D.S., Lam L., Seidman C.E. Truncations of titin causing dilated cardiomyopathy. N. Engl. J. Med. 2012;366:619–628. doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Driest S.L., Ommen S.R., Ackerman M.J. Sarcomeric genotyping in hypertrophic cardiomyopathy. Mayo Clin. Proc. 2005;80:463–469. doi: 10.1016/S0025-6196(11)63196-0. [DOI] [PubMed] [Google Scholar]
- 34.Opie L.H. 4th Ed. Lippincott Williams & Wilkins; Philadelphia, PA: 2004. Heart Physiology: From Cell to Circulation. [Google Scholar]
- 35.de Tombe P.P., Mateja R.D., Irving T.C. Myofilament length dependent activation. J. Mol. Cell. Cardiol. 2010;48:851–858. doi: 10.1016/j.yjmcc.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willott R.H., Gomes A.V., Potter J.D. Mutations in troponin that cause HCM, DCM AND RCM: what can we learn about thin filament function? J. Mol. Cell. Cardiol. 2010;48:882–892. doi: 10.1016/j.yjmcc.2009.10.031. [DOI] [PubMed] [Google Scholar]
- 37.Moore J.R., Leinwand L., Warshaw D.M. Understanding cardiomyopathy phenotypes based on the functional impact of mutations in the myosin motor. Circ. Res. 2012;111:375–385. doi: 10.1161/CIRCRESAHA.110.223842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tardiff J.C. Thin filament mutations: developing an integrative approach to a complex disorder. Circ. Res. 2011;108:765–782. doi: 10.1161/CIRCRESAHA.110.224170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alcalai R., Seidman J.G., Seidman C.E. Genetic basis of hypertrophic cardiomyopathy: from bench to the clinics. J. Cardiovasc. Electrophysiol. 2008;19:104–110. doi: 10.1111/j.1540-8167.2007.00965.x. [DOI] [PubMed] [Google Scholar]
- 40.Chang A.N., Potter J.D. Sarcomeric protein mutations in dilated cardiomyopathy. Heart Fail. Rev. 2005;10:225–235. doi: 10.1007/s10741-005-5252-6. [DOI] [PubMed] [Google Scholar]
- 41.Debold E.P., Schmitt J.P., Warshaw D.M. Hypertrophic and dilated cardiomyopathy mutations differentially affect the molecular force generation of mouse α-cardiac myosin in the laser trap assay. Am. J. Physiol. Heart Circ. Physiol. 2007;293:H284–H291. doi: 10.1152/ajpheart.00128.2007. [DOI] [PubMed] [Google Scholar]
- 42.Chuan P., Sivaramakrishnan S., Spudich J.A. Cell-intrinsic functional effects of the α-cardiac myosin Arg-403-Gln mutation in familial hypertrophic cardiomyopathy. Biophys. J. 2012;102:2782–2790. doi: 10.1016/j.bpj.2012.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lowey S., Lesko L.M., Robbins J. Functional effects of the hypertrophic cardiomyopathy R403Q mutation are different in an α- or β-myosin heavy chain backbone. J. Biol. Chem. 2008;283:20579–20589. doi: 10.1074/jbc.M800554200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Witjas-Paalberends E.R., Piroddi N., van der Velden J. Mutations in MYH7 reduce the force generating capacity of sarcomeres in human familial hypertrophic cardiomyopathy. Cardiovasc. Res. 2013;99:432–441. doi: 10.1093/cvr/cvt119. [DOI] [PubMed] [Google Scholar]
- 45.Srikakulam R., Winkelmann D.A. Chaperone-mediated folding and assembly of myosin in striated muscle. J. Cell Sci. 2004;117:641–652. doi: 10.1242/jcs.00899. [DOI] [PubMed] [Google Scholar]
- 46.Liu L., Srikakulam R., Winkelmann D.A. Unc45 activates Hsp90-dependent folding of the myosin motor domain. J. Biol. Chem. 2008;283:13185–13193. doi: 10.1074/jbc.M800757200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deacon J.C., Bloemink M.J., Leinwand L.A. Identification of functional differences between recombinant human α and β cardiac myosin motors. Cell. Mol. Life Sci. 2012;69:2261–2277. doi: 10.1007/s00018-012-0927-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Resnicow D.I., Deacon J.C., Leinwand L.A. Functional diversity among a family of human skeletal muscle myosin motors. Proc. Natl. Acad. Sci. USA. 2010;107:1053–1058. doi: 10.1073/pnas.0913527107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sommese R.F., Sung J., Spudich J.A. Molecular consequences of the R453C hypertrophic cardiomyopathy mutation on human β-cardiac myosin motor function. Proc. Natl. Acad. Sci. USA. 2013;110:12607–12612. doi: 10.1073/pnas.1309493110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bloemink M., Deacon J., Geeves M.A. The hypertrophic cardiomyopathy myosin mutation R453C alters ATP-binding and hydrolysis of human cardiac β-myosin. J. Biol. Chem. 2013 doi: 10.1074/jbc.M113.511204. http://www.jbc.org/content/early/2013/12/16/jbc.M113.511204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Howard J. Sinauer; Sunderland, MA: 2001. Mechanics of Motor Proteins and the Cytoskeleton. [Google Scholar]
- 52.Uyeda T.Q., Kron S.J., Spudich J.A. Myosin step size. Estimation from slow sliding movement of actin over low densities of heavy meromyosin. J. Mol. Biol. 1990;214:699–710. doi: 10.1016/0022-2836(90)90287-V. [DOI] [PubMed] [Google Scholar]
- 53.Bloemink M.J., Adamek N., Geeves M.A. Kinetic analysis of the slow skeletal myosin MHC-1 isoform from bovine masseter muscle. J. Mol. Biol. 2007;373:1184–1197. doi: 10.1016/j.jmb.2007.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yengo C.M., Takagi Y., Sellers J.R. Temperature dependent measurements reveal similarities between muscle and non-muscle myosin motility. J. Muscle Res. Cell Motil. 2012;33:385–394. doi: 10.1007/s10974-012-9316-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Finer J.T., Simmons R.M., Spudich J.A. Single myosin molecule mechanics: piconewton forces and nanometer steps. Nature. 1994;368:113–119. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- 56.Spudich J.A. Optical trapping: motor molecules in motion. Nature. 1990;348:284–285. doi: 10.1038/348284a0. [DOI] [PubMed] [Google Scholar]
- 57.Spudich J.A. How molecular motors work. Nature. 1994;372:515–518. doi: 10.1038/372515a0. [DOI] [PubMed] [Google Scholar]
- 58.Walcott S., Warshaw D.M., Debold E.P. Mechanical coupling between myosin molecules causes differences between ensemble and single-molecule measurements. Biophys. J. 2012;103:501–510. doi: 10.1016/j.bpj.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Littlefield R., Fowler V.M. Measurement of thin filament lengths by distributed deconvolution analysis of fluorescence images. Biophys. J. 2002;82:2548–2564. doi: 10.1016/S0006-3495(02)75598-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Burgoyne T., Muhamad F., Luther P.K. Visualization of cardiac muscle thin filaments and measurement of their lengths by electron tomography. Cardiovasc. Res. 2008;77:707–712. doi: 10.1093/cvr/cvm117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robinson T.F., Winegrad S. Variation of thin filament length in heart muscles. Nature. 1977;267:74–75. doi: 10.1038/267074a0. [DOI] [PubMed] [Google Scholar]
- 62.Al-Khayat H.A., Kensler R.W., Morris E.P. Atomic model of the human cardiac muscle myosin filament. Proc. Natl. Acad. Sci. USA. 2013;110:318–323. doi: 10.1073/pnas.1212708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kensler R.W., Harris S.P. The structure of isolated cardiac myosin thick filaments from cardiac myosin binding protein-C knockout mice. Biophys. J. 2008;94:1707–1718. doi: 10.1529/biophysj.107.115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ovalle W.K., Nahimey P. 2nd Ed. Saunders; Philadelphia, PA: 2013. Netter’s Essential Histology. [Google Scholar]
- 65.Cooke R., Franks K.E. Generation of force by single-headed myosin. J. Mol. Biol. 1978;120:361–373. doi: 10.1016/0022-2836(78)90424-2. [DOI] [PubMed] [Google Scholar]
- 66.Botcherby E.J., Corbett A., Bub G. Fast measurement of sarcomere length and cell orientation in Langendorff-perfused hearts using remote focusing microscopy. Circ. Res. 2013;113:863–870. doi: 10.1161/CIRCRESAHA.113.301704. [DOI] [PubMed] [Google Scholar]
- 67.Laks M.M., Nisenson M.J., Swan H.J.C. Myocardial cell and sarcomere lengths in the normal dog heart. Circ. Res. 1967;21:671–678. doi: 10.1161/01.res.21.5.671. [DOI] [PubMed] [Google Scholar]
- 68.Rodriguez E.K., Hunter W.C., Weisman H.F. A method to reconstruct myocardial sarcomere lengths and orientations at transmural sites in beating canine hearts. Am. J. Physiol. 1992;263:H293–H306. doi: 10.1152/ajpheart.1992.263.1.H293. [DOI] [PubMed] [Google Scholar]
- 69.Lang R.M., Bierig M., Stewart W.J. Recommendations for chamber quantification: a report from the American Society of Echocardiography’s Guidelines and Standards Committee and the Chamber Quantification Writing Group, developed in conjunction with the European Association of Echocardiography, a branch of the European Society of Cardiology. J. Am. Soc. Echocardiogr. 2005;18:1440–1463. doi: 10.1016/j.echo.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 70.Bovendeerd P.H.M., Arts T., Reneman R.S. Dependence of local left ventricular wall mechanics on myocardial fiber orientation: a model study. J. Biomech. 1992;25:1129–1140. doi: 10.1016/0021-9290(92)90069-d. [DOI] [PubMed] [Google Scholar]
- 71.Eriksson T., Prassl A., Holzapfel G. Influence of myocardial fiber/sheet orientations on left ventricular mechanical contraction. Math. Mech. Solids. 2013;18:592–606. [Google Scholar]
- 72.Huisman R.M., Sipkema P., Elzinga G. Comparison of models used to calculate left ventricular wall force. Med. Biol. Eng. Comput. 1980;18:133–144. doi: 10.1007/BF02443288. [DOI] [PubMed] [Google Scholar]
- 73.Wallace A.G., Mitchell J.H., Sarnoff S.J. Duration of the phases of left ventricular systole. Circ. Res. 1963;12:611–619. doi: 10.1161/01.res.12.6.611. [DOI] [PubMed] [Google Scholar]
- 74.Leddet P., Couppié P., Hanssen M. Value of cardiac MRI for intraventricular thrombi’s diagnosis. Ann. Cardiol. Angeiol. (Paris) 2010;59:285–293. doi: 10.1016/j.ancard.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 75.Hanssen H., Keithahn A., Halle M. Magnetic resonance imaging of myocardial injury and ventricular torsion after marathon running. Clin. Sci. 2011;120:143–152. doi: 10.1042/CS20100206. [DOI] [PubMed] [Google Scholar]
- 76.Spudich J.A., Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- 77.Spudich J.A., Huxley H.E., Finch J.T. Regulation of skeletal muscle contraction. II. Structural studies of the interaction of the tropomyosin-troponin complex with actin. J. Mol. Biol. 1972;72:619–632. doi: 10.1016/0022-2836(72)90180-5. [DOI] [PubMed] [Google Scholar]
- 78.McKillop D.F., Geeves M.A. Regulation of the interaction between actin and myosin subfragment 1: evidence for three states of the thin filament. Biophys. J. 1993;65:693–701. doi: 10.1016/S0006-3495(93)81110-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Moss R.L., Razumova M., Fitzsimons D.P. Myosin crossbridge activation of cardiac thin filaments: implications for myocardial function in health and disease. Circ. Res. 2004;94:1290–1300. doi: 10.1161/01.RES.0000127125.61647.4F. [DOI] [PubMed] [Google Scholar]
- 80.Sun Y.B., Lou F., Irving M. Calcium- and myosin-dependent changes in troponin structure during activation of heart muscle. J. Physiol. 2009;587:155–163. doi: 10.1113/jphysiol.2008.164707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gomes A.V., Barnes J.A., Potter J.D. Role of troponin T in disease. Mol. Cell. Biochem. 2004;263:115–129. doi: 10.1023/B:MCBI.0000041853.20588.a0. [DOI] [PubMed] [Google Scholar]
- 82.Sommese R.F., Nag S., Ruppel K.M. Effects of troponin T cardiomyopathy mutations on the calcium sensitivity of the regulated thin filament and the actomyosin cross-bridge kinetics of human β-cardiac myosin. PLoS ONE. 2013;8:e83403. doi: 10.1371/journal.pone.0083403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.De La Cruz E.M., Ostap E.M. Kinetic and equilibrium analysis of the myosin ATPase. Methods Enzymol. 2009;455:157–192. doi: 10.1016/S0076-6879(08)04206-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Spudich J.A., Kron S.J., Sheetz M.P. Movement of myosin-coated beads on oriented filaments reconstituted from purified actin. Nature. 1985;315:584–586. doi: 10.1038/315584a0. [DOI] [PubMed] [Google Scholar]
- 85.Kron S.J., Spudich J.A. Fluorescent actin filaments move on myosin fixed to a glass surface. Proc. Natl. Acad. Sci. USA. 1986;83:6272–6276. doi: 10.1073/pnas.83.17.6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Greenberg M.J., Moore J.R. The molecular basis of frictional loads in the in vitro motility assay with applications to the study of the loaded mechanochemistry of molecular motors. Cytoskeleton (Hoboken) 2010;67:273–285. doi: 10.1002/cm.20441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Warshaw D.M., Desrosiers J.M., Trybus K.M. Smooth muscle myosin cross-bridge interactions modulate actin filament sliding velocity in vitro. J. Cell Biol. 1990;111:453–463. doi: 10.1083/jcb.111.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Haeberle J.R. Calponin decreases the rate of cross-bridge cycling and increases maximum force production by smooth muscle myosin in an in vitro motility assay. J. Biol. Chem. 1994;269:12424–12431. [PubMed] [Google Scholar]
- 89.Bing W., Knott A., Marston S.B. A simple method for measuring the relative force exerted by myosin on actin filaments in the in vitro motility assay: evidence that tropomyosin and troponin increase force in single thin filaments. Biochem. J. 2000;350:693–699. [PMC free article] [PubMed] [Google Scholar]
- 90.Malik F.I., Hartman J.J., Morgans D.J. Cardiac myosin activation: a potential therapeutic approach for systolic heart failure. Science. 2011;331:1439–1443. doi: 10.1126/science.1200113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cleland J.G., Teerlink J.R., Malik F.I. The effects of the cardiac myosin activator, omecamtiv mecarbil, on cardiac function in systolic heart failure: a double-blind, placebo-controlled, crossover, dose-ranging phase 2 trial. Lancet. 2011;378:676–683. doi: 10.1016/S0140-6736(11)61126-4. [DOI] [PubMed] [Google Scholar]
- 92.Teerlink J.R., Clarke C.P., Wolff A.A. Dose-dependent augmentation of cardiac systolic function with the selective cardiac myosin activator, omecamtiv mecarbil: a first-in-man study. Lancet. 2011;378:667–675. doi: 10.1016/S0140-6736(11)61219-1. [DOI] [PubMed] [Google Scholar]
- 93.Shefner J.M., Watson M.L., Wolff A.A. A study to evaluate safety and tolerability of repeated doses of tirasemtiv in patients with amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. Frontotemporal Degener. 2013;14:574–581. doi: 10.3109/21678421.2013.822517. [DOI] [PubMed] [Google Scholar]
- 94.Shefner J.M., Wolff A.A., Meng L. The relationship between tirasemtiv serum concentration and functional outcomes in patients with ALS. Amyotroph. Lateral Scler. Frontotemporal Degener. 2013;14:582–585. doi: 10.3109/21678421.2013.817587. [DOI] [PubMed] [Google Scholar]
- 95.Lymn R.W., Taylor E.W. Transient state phosphate production in the hydrolysis of nucleoside triphosphates by myosin. Biochemistry. 1970;9:2975–2983. doi: 10.1021/bi00817a007. [DOI] [PubMed] [Google Scholar]
- 96.Lymn R.W., Taylor E.W. Mechanism of adenosine triphosphate hydrolysis by actomyosin. Biochemistry. 1971;10:4617–4624. doi: 10.1021/bi00801a004. [DOI] [PubMed] [Google Scholar]


