Abstract
Over 900 genes have been annotated within duplicated regions of the human genome, yet their functions and potential roles in disease remain largely unknown. One major obstacle has been our inability to accurately and comprehensively assay genetic variation for these genes in a high-throughput manner. We developed a sequencing-based method for rapid and high-throughput genotyping of duplicated genes using molecular inversion probes designed to unique paralogous sequence variants. We apply this method to genotype all members of two gene families, SRGAP2 and RH, among a diversity panel of 1,056 humans. The approach can accurately distinguish copy number in paralogs having up to ∼99.6% sequence identity, identify small gene-disruptive deletions, detect single nucleotide variants, define breakpoints of unequal crossover, and discover regions of interlocus gene conversion. Our analysis of SRGAP2 suggests that nonreciprocal genetic exchange akin to interlocus gene conversion can occur over long distances (> 80 Mbp) between paralogs. The ability to rapidly and accurately genotype multiple gene families in thousands of individuals at low cost enables the development of genome-wide gene conversion maps and unlocks many duplicated genes for association with human traits.
Introduction
Duplicated genes are important contributors to genetic variation1-4, evolutionary adaptation5-8, and human disease9-12. Despite this, most individual duplicated genes remain poorly characterized at the genetic level13 due to high sequence identity13-14, extensive copy number polymorphism1-4, missing sequencing data13, and low correlation with flanking single nucleotide polymorphisms2, 15-16. As a result, these genes and regions have often been excluded from genetic analyses17-18, or contradictory associations with disease have been reported19-20.
Several different technologies have been applied to assay copy number for such genes21. Both qPCR and the paralog ratio test22, which utilizes PCR product specificity to distinguish copies, are labor-intensive, requiring the design and testing of multiple primers. Multiplex ligation-dependent probe amplification (MLPA)23 and multiplex amplification and probe hybridization (MAPH)24 allow for copy number analysis at up to 50 loci simultaneously, but cannot be applied to genotype many gene families at high spatial resolution in a single reaction. Array comparative genomic hybridization (CGH) lacks paralog-specificity and can only access a fraction of duplicated genes, typically where the number of duplicated copies is low2, 16. Finally, mapping whole-genome sequence (WGS) data to singly unique nucleotide (SUN) identifiers that tag a particular paralog and analyzing read-depth1, 25 has yielded genome-wide paralog-specific copy number estimates. The sensitivity of this approach depends on high genome sequencing coverage—still a costly proposition that cannot be applied to thousands of samples in a laboratory setting.
Here we report a new approach for genotyping duplicated genes using molecular inversion probes (MIPs), short oligonucleotides designed to capture targeted genomic regions26-29, together with massively parallel DNA sequencing. We evaluate this method by examining SRGAP2 and RH genetic variation in 1,056 individuals and explore its potential application to the discovery of novel interlocus gene conversion events in humans. The method scales well to thousands of samples and yields accurate, paralog-specific sequence and copy number genotypes at a very low cost.
Results
Genotyping strategy
Our approach leverages SUN variants, fixed paralogous sequence variants that uniquely tag a specific paralog, distinguishing it from all other copies1. We systematically design MIP assays targeting SUNs across the length of the duplicated segment (Fig. 1a and Methods) and additional MIP assays targeting exons. We designed MIPs to hybridize to sequence that is identical between paralogs (Fig. 1b), such that the probability of an individual MIP capturing sequence (Fig. 1c) from a particular paralog is a function of its copy number relative to the copy number of related paralogs. Massively parallel sequencing of amplified capture products allows simultaneous quantification of sequences derived from each paralog (Fig. 1d) and detection of sequence-level genetic variation. We selected two gene families to demonstrate proof-of-principle of our approach and to assess its power to discover novel genetic variation in duplicated regions: SRGAP213—a highly identical (> 99%) human-specific gene family and RH—a clinically relevant blood antigen gene family that has been extensively characterized for common copy number polymorphism30, rearrangement breakpoints31, and interlocus gene conversion32 in the human population.
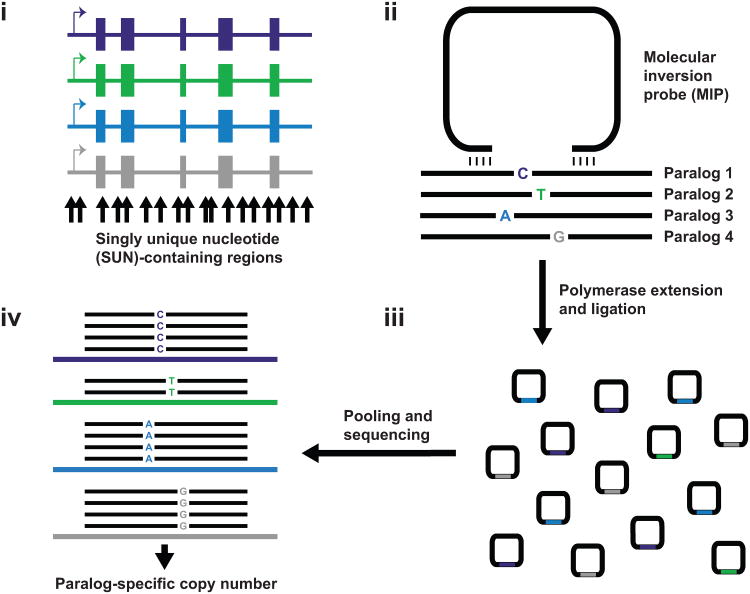
Figure 1. MIP copy number genotyping assay for duplicated genes.
a) 112 nt regions (black arrows) containing sequence variants which uniquely distinguish one paralog (potential SUNs) are identified based on alignment of genomic sequence. b) 70 nt MIPs used for copy number genotyping have 16-24 nt hybridization arms complementary to sequence flanking SUN-containing regions. Several such MIPs are designed collectively spanning the spatial extent of duplicated genic sequence. c) DNA polymerase extension and ligation incorporates SUN-containing sequences into covalently closed circular molecules, which are then barcoded, pooled, and sequenced. d) Reads are mapped to reference sequences for each paralog and paralog-specific read counts for each MIP quantified. A genotyping program is used to infer paralog-specific copy number from these counts. The schematic shows counts consistent with a deletion of paralog 2 (red). Incorporating data from all MIPs overwhelms noisy signals from individual poorly performing MIPs. The dynamic range response and sequence specificity of this method result in accurate copy number genotyping for each paralog with high spatial resolution.
Copy number and sequence genotyping
For SRGAP2, we designed a total of 142 MIPs to sites corresponding to potential SUNs (Supplementary Tables 1 and 2, Methods) that could reliably differentiate SRGAP2 paralogs. Forty of these MIP targets harbor nucleotide differences that distinguish all four SRGAP2 paralogs from one another, 28 distinguish two SRGAP2 paralogs from the other two paralogs, and the remaining 74 distinguish a single SRGAP2 paralog from the remaining three. We initially employed these MIPs to genotype 48 individuals where orthogonal SRGAP2 copy number data were generated or were available from WGS data (1000 Genomes Project (1KG)), array CGH, and/or fluorescent in situ hybridization (FISH). All captured sequences from a given DNA sample were barcoded, pooled with those from other samples, and sequenced using Illumina HiSeq or MiSeq to an approximate coverage of 350 reads per MIP per individual. For each individual, paralog-specific read counts served as a proxy for copy number for each SRGAP2 gene. We developed a maximum-likelihood approach using paralog-specific read count data to generate SRGAP2 paralog-specific copy number calls across the spatial extent of duplicated SRGAP2 sequence (Methods, Software). Figure 2 shows MIP data for 90 high-performing copy number MIPs (Methods and Supplementary Figs. 1 and 2) alongside FISH data for three representative individuals, highlighting the precision with which MIP genotyping detected known duplications and deletions of SRGAP2B and SRGAP2D.
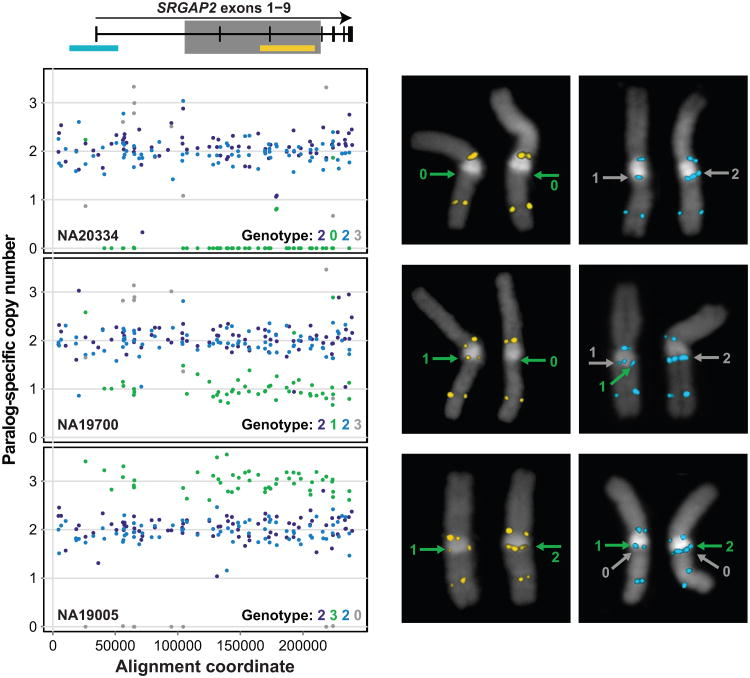
Figure 2. Accuracy of paralog-specific copy number genotyping.
142 MIPs and FISH were used for genotyping SRGAP2 copy number in the HapMap individuals NA20334, NA19700, and NA19005. SRGAP2 exons 1-9 are shared between SRGAP2A, B, C, and D, with the exception of exons 2-3, which are deleted from SRGAP2D (gray box indicates region deleted in SRGAP2D). SRGAP2 paralogs exhibit > 99% sequence identity. Exon locations are plotted relative to the FISH probes (cyan and yellow rectangles) and MIP data below. Paralog-specific copy number estimates are shown for 90 high-performing MIPs across ∼240 kbp of aligned SRGAP2 genomic sequence. Each point indicates a paralog-specific copy number estimate, calculated as the product of the paralog-specific read count frequency for a particular MIP and the aggregate estimated SRGAP2 copy number at the corresponding locus. Analysis of the MIP data using an automated paralog-specific copy number caller successfully identified known homozygous and heterozygous deletions and a duplication of SRGAP2B (red) and duplications and a homozygous deletion of SRGAP2D (gray). FISH validates the MIP-based genotypes for these individuals. The FISH data for NA20334 and NA19700 are somewhat ambiguous regarding SRGAP2D, consistent with either two or three diploid copies of this paralog in these individuals. Array CGH data for NA19700 supports the MIP-based genotype for SRGAP2D. All HapMap genotypes were independently confirmed by WGS data.
We found that 97.2% (35/36) of copy number calls were concordant with FISH, 8/8 were consistent with array CGH data and 91.5% (150/164) agreed with estimates made from WGS data (Supplementary Table 3). All inconsistencies involved genotyping results for the SRGAP2D pseudogene. This paralog is the shortest and most recently duplicated segment having ∼99.6% identity to SRGAP2B. Low WGS coverage from the 1KG together with the paucity of SRGAP2D SUNs likely confounded sequencing-based copy number estimates for SRGAP2D. To explore this possibility, we generated aggregate SRGAP2 copy number estimates from WGS data. These aggregate estimates are more accurate than corresponding paralog-specific estimates1 because all reads mapping to SRGAP2 (rather than just those mapping to SUN identifiers) inform this analysis. Strikingly, 13/14 aggregate SRGAP2 copy number estimates were consistent with MIP-based paralog-specific estimates rather than corresponding WGS-based estimates in cases where these results disagreed. We extended our analysis to include 1,056 HapMap individuals, 73 of which we genotyped more than once using MIPs to examine the reproducibility of our approach. We found 99.5% (390/392) of replicate SRGAP2 paralog-specific copy number genotypes were concordant with initial genotypes (Supplementary Table 4).
Our data allowed us to estimate allele frequencies for SRGAP2 duplications and deletions in nine human populations (Supplementary Table 5). As expected based on a previous analysis of WGS data from the 1KG13, SRGAP2B and SRGAP2D showed evidence of complete loss or gain, ranging in copy number from 0-4 in the human population. In contrast, complete duplication or deletion of SRGAP2A or SRGAP2C was not observed, consistent with the notion that these two paralogs are functional copies. Interestingly, our analysis of SRGAP2B and SRGAP2D copy number variation suggests population stratification. Deletion of SRGAP2B, for example, is more common in populations of African descent in contrast to deletions of SRGAP2D, which have risen in frequency in several Out-of-Africa populations.
Unlike most other copy number genotyping assays, MIPs also provide information on the sequence content of targeted regions28-29, 33. We reasoned that in some cases, linkage of discovered single nucleotide variants (SNVs) to a nearby paralog-distinguishing SUN would allow inference of paralog-of-origin. We evaluated whether our method could accurately genotype such SNVs by comparing MIP sequence data (Methods) with fosmid clone end-sequence data34 and WGS data for NA18507, an individual previously sequenced to high coverage35. The WGS data validated 93.8% (15/16) of our genotype calls (Supplementary Table 6), including a heterozygous nonsynonymous variant. Fosmid end-sequence data including a putative variant site were only available in three cases, but each validated the SNV identified from MIP data. Thus, our method can successfully detect SNVs within highly identical duplicated sequence and in some cases accurately assign them to specific paralogs.
Internal SRGAP2 deletion and duplication discovery
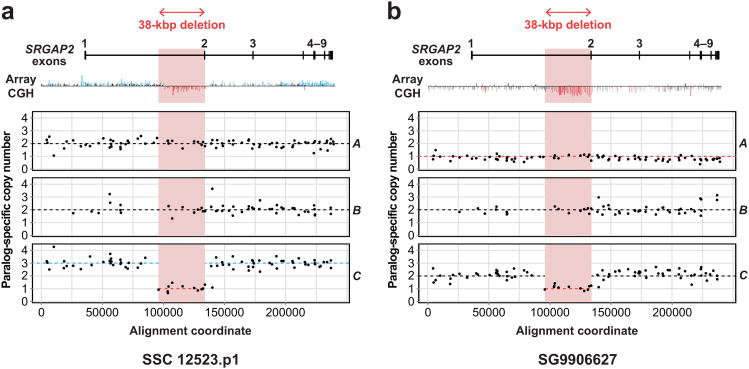
We applied our MIP-based method to two individuals having array CGH profiles showing complex structural variation in SRGAP213. MIP genotyping (Fig. 3) correctly identified large SRGAP2C and SRGAP2A events discovered via array CGH and resolved the internal deletions as specifically affecting SRGAP2C, removing exon 2 and inducing a frameshift. MIP-based genotyping of 1,056 HapMap individuals indicates this deletion is segregating at low frequency (< 3%) exclusively in populations with some European ancestry. In addition to this SRGAP2C deletion, we identified seven other additional internal deletion and duplication events in HapMap individuals ranging in size from 1.5 kbp to 144 kbp and assigned them to specific SRGAP2 paralogs (Supplementary Table 5, Supplementary Fig. 3). These structural variants include three distinct exon-overlapping events in SRGAP2B and an intronic duplication in SRGAP2A.
Figure 3. Resolution of complex structural variation in SRGAP2.
a) The array CGH profile for SRGAP2 loci predicts a gain and an interstitial loss for a patient with autism but cannot distinguish which paralogs the variation affects. The MIP copy number assay predicts copy number two for A, B, and D (not shown), but duplication of a copy of C having an ∼38 kbp internal deletion containing exon 2. Dashed lines indicate paralog-specific copy number calls from the automated caller. b) Similar analysis of a patient with developmental delay shows that the individual is diploid for B and D (not shown), but has lost a copy of A (the ancestral locus) and carries the internal deletion for C.
RH gene conversion, copy number, and NAHR breakpoint resolution
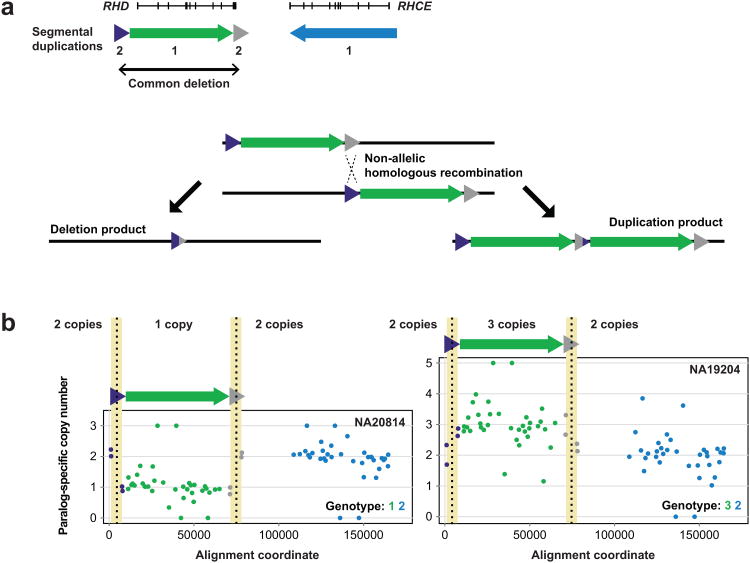
To assess the applicability of our method to assaying interlocus gene conversion and resolving breakpoints associated with non-allelic homologous recombination (NAHR), we applied our MIP genotyping method to RHD and RHCE—sites of known gene conversion and unequal crossover with clinical relevance for Rh antigen presentation. We reasoned that these two forms of mutation would generate characteristic sequence signatures with respect to SUN copy number. In the case of gene conversion, we would expect to observe a reciprocal copy number shift at a pocket of homology with no difference in copy number of flanking regions. Gains would correspond to donors and losses to acceptors of gene conversion, allowing inference of the directionality of the event. In contrast, at a site of unequal crossover, a reciprocal SUN copy number transition should be observed around the NAHR breakpoint.
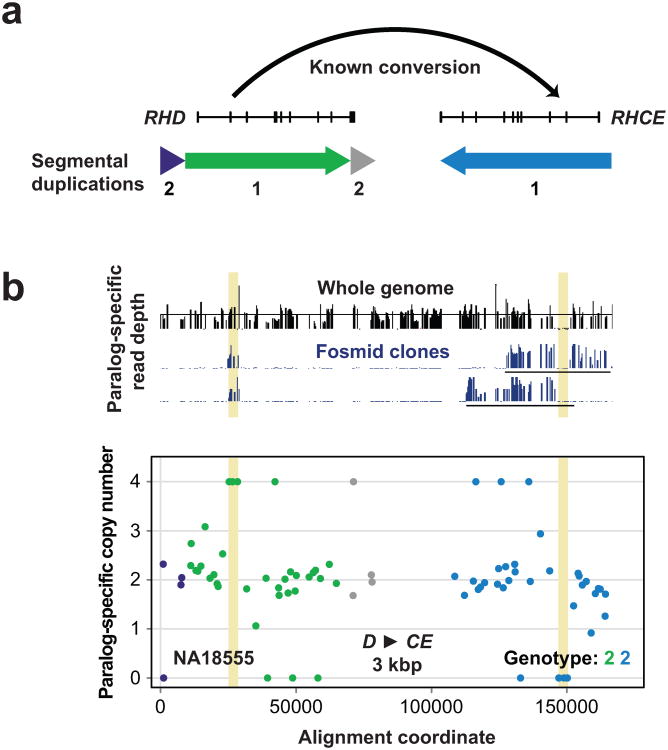
We designed 39 MIPs targeting RH paralogs and flanking regions (Fig. 4a) and included them in the same capture reactions as SRGAP2 MIPs, allowing us to simultaneously genotype the same individuals described above for RH. Searching for reciprocal copy number shifts, we observed 7 distinct putative RH gene conversion events, ranging in length from 1709 bp to ∼39 kbp (Supplementary Table 5, Supplementary Fig. 4). Although we denote these events as gene conversions, other mutational mechanisms36-38 may be responsible for the signatures we observed. Four events involved a transfer of genetic information from RHCE to RHD, four corresponded to polymorphic variants reported in the Blood Group Antigen Gene Mutation Database (dbRBC at NCBI), and four were supported by at least one observed instance of transmission from parent to child. The most common involved sequence transfer from RHD to RHCE at a known gene conversion site including RHD exon 239 and was confirmed by whole-genome and fosmid clone sequencing data1 from an individual predicted from MIP data to be homozygous for this event (Fig. 4b).
Figure 4. Detection of gene conversion at the RH locus.
a) RHD and RHCE lie within an ∼60 kbp segmental duplication (red and blue arrows) and frequently undergo interlocus gene conversion events. A common known RH gene conversion affects exon 2 and serves as the primary genetic basis for the RhC phenotype. b) 39 MIPs were used for genotyping paralog-specific RH copy number in the HapMap individual NA18555. MIP data is plotted relative to locations of RH exons and associated segmental duplications in (a). Colors correspond to segmental duplications shown in (a). The MIP data show a strong signature of homozygous gene conversion (highlighted in yellow) from RHD to RHCE spanning at least ∼3 kbp at the common conversion site including exon 2. Mapping whole-genome and fosmid clone short-read sequence data from this individual to SUN identifiers and examining paralog-specific read depth validates this homozygous conversion.
Using our copy number genotyping strategy, we identified known deletions and duplications in RHD associated with unequal crossover between flanking segmental duplications (Fig. 5a). We found 97.6% (80/82) of our RH paralog-specific copy number estimates agreed with those from WGS data (Supplementary Table 3). Reproducibility was lower for RH copy number genotyping than for SRGAP2 genotyping (Supplementary Table 4), as only 91.8% (180/196) of replicate MIP-based RH paralog-specific copy number genotypes were concordant with initial genotypes. We calculated logarithm of odds confidence scores for each of these genotypes (Methods) and observed that discordancies' scores fell at the low end of the score distribution (Supplementary Table 7, Supplementary Fig. 5), suggesting potential errors can be readily distinguished from high-confidence genotype calls. To attempt to refine NAHR-associated breakpoints, we looked for instances of a reciprocal paralog-specific copy number transition within the segmental duplications flanking RHD. This approach allowed us to narrow breakpoint locations to within ∼6 kbp windows (Fig. 5b), regions previously found to contain RHD deletion breakpoints based on Sanger sequencing of spanning PCR products31.
Figure 5. Resolution of NAHR-associated RHD deletion and duplication breakpoints.
a) An ∼9 kbp segmental duplication (green and gray triangles) flanks RHD. NAHR between these flanking sequences frequently results in deletion (and more rarely, duplication) of RHD. Homozygous deletion of RHD results in the Rhd phenotype, most common in individuals of European ancestry. b) Data from 39 RH MIPs for HapMap individuals NA20814 and NA19204 reveal copy number variation at RHD. Note the signatures of NAHR in the four MIP data points corresponding to the RHD-flanking segmental duplications. These data refine the NAHR-associated breakpoints to ∼6 kbp homologous genomic regions (highlighted in yellow) where RHD deletion breakpoints have been previously reported.
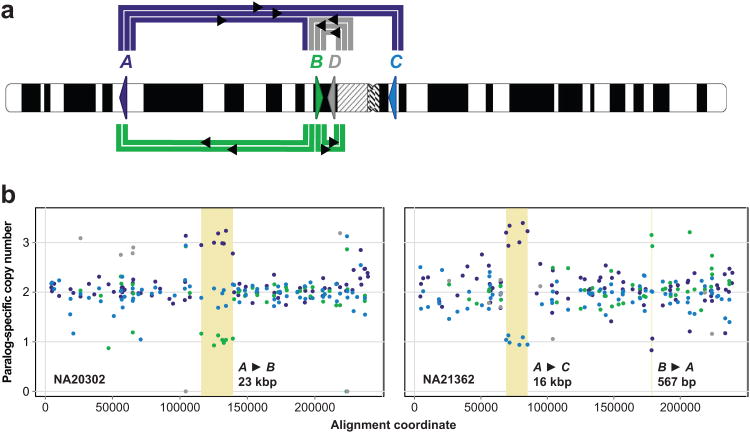
Discovery of novel sites of interlocus gene conversion in SRGAP2
Given the > 99% sequence identity between SRGAP2 paralogs, we reasoned that interlocus gene conversion or other mechanisms of nonreciprocal sequence transfer may have occurred at these loci and left signatures detectable using our MIP genotyping method. Analysis of the 1,056 HapMap individuals revealed 10 such events ranging in size from 416 bp to 23 kbp (Supplementary Table 5, Supplementary Fig. 6), collectively involving all four SRGAP2 paralogs (Fig. 6a). All paralogs except SRGAP2C were observed as putative gene conversion donors. Unlike RHD/CE, these putative conversion events appear to have occurred over large genetic distances. For example, two distinct nonreciprocal exchanges of genetic information occur across the centromere between SRGAP2A and SRGAP2C—paralogs over 80 Mbp apart on chromosome 113 (Fig. 6b, Supplementary Fig. 6).
Figure 6. Extensive interlocus gene conversion between SRGAP2 paralogs.
a) Schematic denotes location and orientation (triangles) of SRGAP2 paralogs on human chromosome 1. Thick colored lines connect SRGAP2 paralogs exhibiting signatures of interlocus gene conversion in the MIP data. Line colors correspond to conversion donors, and each line corresponds to a distinct conversion event. b) Examples of different interlocus gene conversion events (highlighted in yellow). Reported sizes indicate the minimum length of the conversion event based on the MIP data, assuming a single conversion event underlies the conversion signature. All events shown, except for the B to A conversion revealed by two MIPs, were detected by the automated caller.
To corroborate these findings, we examined inheritance for putative gene conversion events detected in members of HapMap trios. We observed at least one instance of transmission from parent to child for six distinct putative gene conversions, and no such events were inferred as de novo. We also validated one putative conversion using paralog-specific qPCR and array CGH. MIP data suggested a complete SRGAP2D duplication and a gene conversion resulting in replacement of SRGAP2C sequence with paralogous sequence in a patient with intellectual disability (Supplementary Fig. 7). If the MIP genotyping were accurate, SRGAP2C-specific qPCR using primers in the putative conversion region would be expected to signal a loss in SRGAP2C copy number, but array CGH would be expected to signal a slight gain in aggregate SRGAP2 copy number over SRGAP2 sequence shared with SRGAP2D. Performing the qPCR and array CGH experiments yielded precisely these results, providing additional support for the accuracy of our method and its applicability to detect novel signatures of interlocus gene conversion.
Discussion
We developed a method for genotyping duplicated genes that is well-suited for large-scale studies of genetic variation. Hallmarks of our approach include: (i) extreme scalability—2,400 MIPs can be combined in a single reaction to simultaneously assay many genes, and reactions for several hundred individuals can be performed within a week, with DNA input requirements as low as 100 ng per sample and a cost of potentially less than $1 per gene per individual33(Methods); (ii) broad scope—a single experiment yields copy number and sequence information, allowing several forms of genetic variation to be studied simultaneously; (iii) high accuracy—with spatial resolution to delineate structural variation breakpoints; and (iv) paralog-specificity—with a demonstrated ability to distinguish genes having up to ∼99.6% sequence identity.
What would be required to obtain the same volume of genotype information for an arbitrary gene family comparable to SRGAP2 or RH using existing approaches? Whole-genome sequencing offers great potential given its comprehensive nature1, 25, but remains prohibitively expensive for genotyping projects of even moderate size, especially given that accuracy demands high coverage. More scalable available targeted methods, on the other hand, provide limited genotyping power. PCR-based strategies for copy number genotyping query, at most, a few sites per reaction because PCR multiplexes poorly40-41. MLPA and MAPH allow for the simultaneous analysis of up to 50 loci, but even this greater scale of multiplexing cannot match the ability of our method to assay many gene families each at high spatial resolution. None of the targeted methods above provide exonic sequence information, and none have been successfully applied in large-scale studies of gene conversion. As our analyses demonstrate, genetic variation in duplicated genes exhibits considerable complexity. Any method for genotyping such genes should be developed with this consideration in mind.
Although we focused on SRGAP2 and RH, our method will be useful for studying other duplicated genes that have proven very difficult to genotype accurately, including CCL3L119-20, beta-defensins42-43, and C444 (Methods). We provide programs to obtain genotypes with confidence scores from MIP-sequence data and to assist in the identification of informative sites from aligned sequences (Software). We also provide a complete list of ∼3.8 million SUNs based on the current human reference genome (GRCb37) for use with other duplicated regions and gene families (Methods, Supplementary Table 8). While higher copy number and more polymorphic gene families will pose additional challenges, generating high coverage sequence data precisely over the most informative sites promises to significantly improve our understanding of genetic variation of these complex regions of the genome.
Successful application of our method to a particular gene family of interest depends on several factors, including availability of accurate sequence, the number of paralogs, their sequence identity, their G+C content31-32, their copy number ranges, and their sizes. First, optimal MIP design requires high-quality reference sequences for all family members, so gene families lacking complete sequence characterization (Methods, Supplementary Table 9) will be at least partially inaccessible using MIP-based genotyping. Second, some genetic variation must distinguish different paralogs from one another—our method cannot determine copy number if copies are identical at the genomic level (< 1% of all paralogous sequences). Third, gene families with high numbers of paralogs, or with paralogs at high copy numbers showing a range of copy number variation, pose several challenges for MIP-based genotyping. In general, as the number of distinct paralogs increases, fewer potential target regions will contain SUNs allowing discrimination of all paralogs, and thus, more MIPs will need to be designed for copy number genotyping. Furthermore, paralog-specific read count frequencies become more difficult to confidently distinguish as aggregate copy number for a gene family increases. This particular issue could be mitigated somewhat via the use of single molecule MIPs to quantify individual capture events45. Accurate sequence genotyping also becomes more difficult as aggregate copy number increases and the number of possible assignments of sequences to paralog copies grows.
Our method will facilitate efforts to map NAHR-associated structural variation breakpoints, which often occur in complex regions of segmental duplication. Identifying SUNs which discriminate the high-identity paralogs followed by MIP genotyping will provide sequence-level precision to determine the effect of such rearrangements on the genes embedded within such complex regions46. We anticipate MIP-based genotyping will also be very valuable for studies of interlocus gene conversion, providing an experimental platform for surveying the most highly identical paralogs where this mechanism frequently operates47. In this study, we provide evidence of conversion-like events between paralogs separated by more than 80 Mbp—a somewhat surprising finding given that conversion is thought to occur most frequently between high-identity segments in close proximity48-50. Most significantly, our MIP-based method will encourage the inclusion of many previously intractable duplicated genes in future genetic analyses of human phenotypes. With accurate, scalable genotyping, we will be well-positioned to assess the impacts of hundreds of these genes on human traits and disease.
Online Methods
MIP design
SRGAP2 exon-targeting MIPs were designed as previously described33. MIPs employed for paralog-specific copy number inference were designed in a similar fashion, with the following additional considerations. Careful selection of paralogous regions to target for MIP capture is critical for applying our copy number genotyping method to any particular gene family of interest. Suitable target regions contain genetic variation between paralogs that has fixed in the human species. Obtaining paralog-specific read counts from a targeted region requires that the region contain genetic variation such that at least one paralog can be distinguished from all others. Ensuring that these counts reflect underlying relative paralog-specific copy numbers demands that variants used for distinguishing paralogs have very low levels of polymorphism.
To identify regions containing paralog-distinguishing variation, we aligned SRGAP2 sequences13, RH sequences (GRCb37/hg19, chr1:25594516-25655519 and chr1:25688914-25751819), and RHD flanking segmental duplications (GRCb37/hg19, chr1:25585374-25594516 and chr1:25655517-25664845) using Clustal 2.151. Regions of the alignments where sequences identical between all paralogs (20 bp each side) flanked a 112 bp region where at least a single paralog had a distinct sequence were selected as potential targets and input to the MIP design pipeline33. This pipeline attempted to design MIPs to capture each of these potential target regions, outputting MIP oligonucleotides, information about their corresponding arm hybridization sequences and their capture targets, and scores corresponding to their predicted capture performances. We eliminated from consideration any MIPs determined to have arm hybridization sequences with copy counts in the genome (GRCb37 augmented with SRGAP2contig sequences) > 8 to avoid capturing repeat sequences and restrict MIP hybridization to SRGAP2 and RH loci. We also ensured that all MIP arm hybridization sequences were complementary to sequences identical between all paralogs of interest—any MIPs not meeting this criterion were eliminated from further consideration. Finally, we eliminated from consideration all MIPs with the lowest design score (-1) and most MIPs having a target region with < 35% or > 55% G+C content.
For remaining MIPs under consideration for design, we analyzed polymorphism at potential SUNs within the corresponding capture target regions. Briefly, potential SUNs distinguishing each paralog were extracted from the alignment and scored with regard to likely fixation status via analysis of 12 high-coverage genomes (Supplementary Table 1). For each SRGAP2 and RHparalog, we computed all 30-mer sequences found within that paralog and absent from the rest of the genome (singly-unique nucleotide k-mers1 (SUNKs)). We then mapped 12 unrelated high-coverage genomes to SRGAP2 and RHparalog sequences (masked using RepeatMasker52 and Tandem Repeats Finder53) using mrsFAST54 and parsed mapping output to assess the presence of each SUNK in each genome analyzed. A SUNK was considered present if observed in at least a single read mapped with no mismatches. Using only high-coverage genomes for this analysis minimizes the possibility of simply not having sequenced SUNKs that are truly present in a genome. Because each potential SUN typically contributes to (as a single base in the sequence of) many 30-mer SUNKs, the presence or absence of such SUNKs can serve as a proxy for the presence or absence of each potential SUN. Thus, a score from 0-12 was calculated for each potential SUN corresponding to the average number of high-coverage genomes supporting a potential SUN's presence (Supplementary Fig. 8). For example, if a particular potential SUN contributed to four different 30-mer SUNKs, and these SUNKs were determined to be present in 11, 9, 11, and 12 high-coverage genomes, respectively, the score for that potential SUN would be 10.75 ((11+9+11+12)/4). True SUNs are paralog-distinguishing single nucleotide variants that have fixed in the human population. We defined potential SUNs having scores greater than or equal to 11 (for SRGAP2A, SRGAP2B, and SRGAP2C) or 8 (for SRGAP2D, RHD, and RHCE) as true SUNs. (The threshold is lower for these latter paralogs due to a paucity of higher-scoring potential SUNs for these paralogs across the spatial extent of duplicated sequence, reflecting in part heterozygous SRGAP2D and RHD deletions in some of the individuals sequenced to high coverage). Biologically, these defined true SUNs are most likely to be fixed in a particular paralog in the human species and thus most useful for copy number genotyping. Given the observation that the majority (84.8%) of putative autosomal SUNs genome-wide were present in 12/12 high-coverage genomes previously analyzed1, however, SUN scoring, though useful, is not necessary for successful application of our method.
MIPs used for copy number genotyping were selected from remaining MIPs under consideration based on the paralog-specificity, SUN content, and relative genic location of their corresponding target regions. SRGAP2A and SRGAP2C were prioritized in the SRGAP2 copy number genotyping MIP design based on the likely pseudogenicity of SRGAP2B and SRGAP2D13. All MIPs were ordered from Integrated DNA Technologies as previously described33. Supplementary Table 2 provides specific details regarding MIPs designed for this study and their pooling.
MIP pooling, 5′ phosphorylation, and multiplex capture of targeted sequences
MIPs were pooled (Supplementary Table 2), phosphorylated, and used to capture targeted sequences as previously described33, with the following modifications. Initial capture reactions used in the 48-individual experiment were performed with genomic DNA input levels of 50 ng and 100 ng, with subsequent reactions involving HapMap samples using 200 ng DNA input and the reaction involving sample Troina2665 using 100 ng DNA input. MIPs were added to capture reactions at a ratio of 800 MIP copies per haploid genome copy. Incubation of capture reactions at 60°C was performed for 23-24 h, and incubation of exonuclease reactions at 37°C was performed for 45 min. Supplementary Table 10 summarizes MIP capture experiments performed for this study and details sample sets assayed.
Amplification, barcoding, pooling, cleanup, and sequencing of captured sequences
Captured sequences were amplified, barcoded, pooled, and purified as previously described33, with the following specifications. PCR was performed in a 25 μL reaction. Libraries with excessive off-target captures were not observed, and thus, the standard Agencourt purification protocol was followed. Final library DNA concentrations were quantified using the QubitdsDNA HS assay (Life Technologies). Sequencing pools of capture reactions was performed using either a MiSeq or a HiSeq 2000, depending on the number of individual capture reactions included in the pool for sequencing and the number of MIPs used in each individual capture reaction. Supplementary Table 10 provides specific details regarding sequencing performed for different MIP capture experiments in this study.
Paralog-specific copy number genotyping
Initial 151 bp reads (MiSeq) or 101 bp reads (HiSeq 2000) were trimmed from their 3′ ends to 76 bp to eliminate low quality data from the ends of reads while ensuring coverage of each targeted base in nearly all cases. All MIPs are designed such that a 152 bp region (target sequence plus hybridization arms) is sequenced. With 151 bp reads, all bases except the first and last base in this 152 bp region are sequenced during both the forward and reverse reads. Thus, retaining only the first 76 bp from each read eliminates low quality data from the ends of reads while ensuring coverage of each targeted base in all cases except those in which there is a net insertion. Trimmed reads were mapped to SRGAP2 and RHparalog sequences using mrFAST 2.555 in paired-end mode with the maximum allowed edit distance set to 4 and the minimum and maximum inferred distances allowed between paired-end sequences set to 144 and 160, respectively.
Mapping output was parsed to yield counts of reads mapping to each paralog for each MIP for each individual. The following stringent filters were applied to ensure accuracy: the mapping location of a read pair was required to be within 4 bp of the expected mapping location, the strandedness of reads had to be consistent with expectation based on MIP design, the inferred insert size had to be within 2 bp of its expected value (152 bp), any bases covered by forward and reverse trimmed reads had to have the same base call, the quality scores at all base positions showing variation between paralogs (base positions that affect mapping paralog-specificity) had to be at least Q30, no mismatches could occur at likely fixed true SUN positions, and reported barcode sequences had to perfectly match a known barcode sequence. Read pairs violating any of these filters were not included in final counts. For SRGAP2paralog-specific copy number analysis, final counts served as input to a genotyping program which generated SRGAP2paralog-specific copy number calls for each individual across the spatial extent of duplicated SRGAP2 sequences. For RHparalog-specific copy number analysis, genotyping calls were made in a similar automated fashion, except no copy number state transitions were allowed. Thus, all internal RH gene conversion events were called based on manual visual inspection of paralog-specific count frequency plots.
The SRGAP2 genotyping program generates paralog-specific copy number calls using a maximum likelihood approach together with dynamic programming. For each individual, log-likelihoods of observing the paralog-specific read count data for each MIP are calculated under 400 different possible hidden underlying SRGAP2paralog-specific copy number states, where SRGAP2A and SRGAP2C can have copy numbers from 0-3 and SRGAP2B and SRGAP2D can have copy numbers from 0-4 (4*5*4*5 = 400 combinations)13. For each paralog-specific copy number state, log-likelihoods were calculated as logarithms of multinomial probabilities. Specifically, for each paralog-specific copy number state, a multinomial probability of the observed data was computed for each MIP, with the number of trials equal to the total number of mapped reads for that MIP and the vector of outcome probabilities equal to the copy numbers of specific paralogs over the aggregate copy number for the gene family given the paralog-specific copy number state. (An outcome in this case is observing a read coming from a particular paralog.) The SRGAP2D internal deletion is built into the log-likelihood calculations: all copy number states for MIPs in this region have SRGAP2D copy number set to 0. Log-likelihood values below -30 are set to -30 to limit the ability of count data from a single MIP to potentially single-handedly invalidate a particular SRGAP2paralog-specific copy number state as possibly underlying the count data.
Next, for each individual, log-likelihoods are used to construct a weighted directed acyclic graph, with prior probabilities based on observed SRGAP2 copy number genotype data from previous experiments13 incorporated into the log-likelihoods for the first (most 5′ with respect to SRGAP2) MIP. The graph is constructed by iteratively considering log-likelihoods for the next MIP and tracking the highest scoring paths ending at each copy number state allowing 0, 1, and 2 transitions between copy number states as well as the values of the corresponding log-likelihoods of these paths until the graph spans all MIPs. Allowed transitions between copy number states are restricted to copy number gains or losses affecting a single paralog or cases where the copy numbers of two paralogs change, but the total number of SRGAP2 copies remains constant. All transitions meeting these criteria and transition probabilities associated with remaining in the same state have probability 1; all other transitions have probability 0. Three highest-scoring paths through the likelihood graph are calculated: one for 0 allowed total transitions between copy number states, one for 1 allowed transition between copy number states, and one for 2 allowed transitions between copy number states. Restricting the nature of allowed copy number state transitions reflects the fact that true biological events should fall into one of two categories (single-paralog-affecting duplication/deletion or interlocus gene conversion). Restricting the number of transitions reflects the fact that a single individual is most likely to have, at most, a single duplication, deletion, or interlocus gene conversion restricted to within SRGAP2. If an individual were to have multiple events restricted to within SRGAP2, the program would still flag this individual as having a complex SRGAP2paralog-specific copy number genotype, and the second internal event would be apparent upon subsequent visual inspection of paralog-specific count frequency plots.The program ultimately identifies the highest-scoring paths through the likelihood graph (most likely paralog-specific copy number states across the spatial extent of duplicated SRGAP2 sequence) allowing 0, 1, and 2 transitions and their corresponding log-likelihood scores. Heuristics (Supplementary Table 11) are used to assess increases in likelihood of the 1-transition and 2-transition paths compared to the 0-transition path and determine whether they signal an event within SRGAP2 and warrant calling the paralog-specific copy number genotype for an individual as complex. In most cases, the scores of the 1-transition and 2-transition paths will not be substantially higher than that of the 0-transition path, and the genotype for an individual will be called as simple (a single copy number state across the entirety of duplicated SRGAP2 sequence).
The program also calculates a logarithm of odds confidence score associated with the simple (0-transition) genotype call for each individual. Specifically, this score is equal to the log-likelihood of the chosen 0-transition path minus the highest log-likelihood for a 0-transition path having a distinct set of associated multinomial probabilities. For example, if an individual was called as having two copies of each SRGAP2paralog, the confidence score would be the log-likelihood of the 0-transition path for this copy number state minus the highest log-likelihood of a 0-transition path among all other copy number states except those having equal copy numbers for each SRGAP2paralog. The logic behind this requirement is that likelihoods of 0-transition paths with the same set of associated multinomial probabilities will differ only because they have distinct prior probabilities—that is, the paralog-specific read count frequency data, independent of any prior knowledge, support each such path equally well. Confidence scores should be interpreted relative to confidence scores for other individuals genotyped for the same gene family using the same set of MIPs (Supplementary Table 7, Supplementary Fig. 5) rather than in an absolute sense.
The RH genotyping program works the same way as the SRGAP2 genotyping program, except that there are 25 different possible RHparalog-specific copy number states (each of RHD and RHCE is allowed to vary in copy number from 0-4), prior probabilities used were based on our estimates of RHparalog-specific copy number from the 1KG, and no transitions between copy number states were allowed, such that all RH genotypes are called as simple (a single copy number state across the entirety of duplicated RH sequence).
Fluorescent in situ hybridization
Metaphase spreads were obtained from lymphoblast and fibroblast cell lines from human HapMap individuals NA19700, NA19703, NA19901, NA20127, NA20334, NA19005, NA19190, NA19201, and NA12878 (Coriell Cell Repository, Camden, NJ). FISH experiments were performed using fosmid clones (WIBR2-2926C23_G248P88292B12 and WIBR2-3738J10_G248P802587E5)13 directly labeled by nick-translation with Cy3-dUTP (Perkin-Elmer), Cy5-dUTP (Perkin-Elmer), and fluorescein-dUTP (Enzo) as described previously56 with minor modifications. Briefly: 300 ng of labeled probe were used for the FISH experiments; hybridization was performed at 37°C in 2×SSC, 50% (v/v) formamide, 10% (w/v) dextran sulphate, and 3 μgsonicated salmon sperm DNA, in a volume of 10 μL. Posthybridization washing was at 60°C in 0.1×SSC (three times, high stringency). Nuclei were simultaneously DAPI stained. Digital images were obtained using a Leica DMRXA2 epifluorescence microscope equipped with a cooled CCD camera (Princeton Instruments). DAPI, Cy3, Cy5 and fluorescein fluorescence signals, detected with specific filters, were recorded separately as gray-scale images. Pseudocoloring and merging of images were performed using Adobe Photoshop software.
Other orthogonal validations
Array CGH data, qPCR data, and whole-genome shotgun sequence data from the 1000 Genomes Project used for validation purposes were collected and processed as previously described1,13.
Paralog-specific SNV genotyping
Initial reads were trimmed from their 3′ ends to 100 bp to eliminate some low quality data from the ends of reads while ensuring coverage of each targeted base. Trimmed reads were mapped separately to individual SRGAP2paralog sequences using the Burrows-Wheeler Aligner57 (version 0.5.9, paired-end mapping) with the following options: -e 50 -l 17 -q 20 -d 5 -i 5 -I. Mapping output, combined with each individual's SRGAP2paralog-specific copy number genotype determined as described above, was parsed to yield sequence genotypes for each copy of each paralog for each MIP for each individual. The Hungarian method58 was employed to optimally assign distinct sequences to copies of different SRGAP2paralogs based on observed counts of distinct sequences, treating paralog-specific mapping edit distances as the costs of sequence assignments to copies of different paralogs. In cases where equally optimal but biologically distinct sets of assignments could be made, each assignment set and its corresponding paralog-specific sequence genotypes was reported and flagged as having some ambiguity. All detected single nucleotide variants were annotated with regard to location and likely functional impact. Reported single nucleotide variants in Supplementary Table 6 have the following properties: (i) they were called based on MIP sequence data from NA18507, (ii) they occur at alignment positions where all paralogs share the same nucleotide, (iii) they occur in close proximity to a SUN or on a paralog-distinguishing haplotype such that their paralog-of origin can be accurately inferred, (iv) they were unambiguously assigned to a particular paralog, and (v) all copies of the paralog they were assigned to have no ambiguity in sequence at the variant site.
Preparation of final SRGAP2 MIP pool
Data from the 48-individual genotyping experiment revealed that most MIPs in the initial pool captured their corresponding targets well (Supplementary Fig. 9). Considering only the capture reactions using 100 ng DNA input, the mean and median mapped read counts per MIP were 18,447 and 15,809, respectively. On average, this translates to approximately 350 mapped reads per MIP per individual. To optimize our MIP pool before extending our genotyping efforts to thousands of samples, we rebalanced exon-targeting MIPs that failed to efficiently capture their corresponding targets and removed SRGAP2 copy number genotyping MIPs that did not meet a high performance standard.
Specifically, we increased the amount of any exon-targeting MIPs having a total mapped read count fewer than 2,500 times the number of paralogs that include the targeted exon. For example, if a MIP targeted exon 1, shared between all four SRGAP2paralogs, but had fewer than 10,000 reads from the initial capture reactions using 100 ng DNA input, we rebalanced it. Rebalancing was performed such that this count threshold would be achieved if mapped read count per MIP increases proportionally with the amount of MIP added to the pool, that is, for example, if doubling the amount of MIP added to the pool results in twice the number of corresponding mapped reads. We thus added 7 exon-targeting MIPs to be at a relative amount of 2× in the final pool and another 5 such MIPs to be at a relative amount of 5×. Eleven MIPs, however, would still fail to meet the count threshold even if their corresponding mapped read counts increased fivefold. These worst-performing exonic MIPs were added to be at a relative amount of 50× in the final MIP pool to maximize their chances for successful capture.
To evaluate the performance of SRGAP2 copy number genotyping MIPs, we compared observed paralog-specific count frequencies for each MIP with corresponding expected frequencies for 31 genomes from the 48-individual experiment (Supplementary Table 3). These genomes were selected because we had very high confidence in their true paralog-specific copy number genotypes—genotyping results were concordant between all methods used for 30 of these genomes, and FISH results supported MIP results in the remaining case. For each SRGAP2 copy number genotyping MIP, we calculated the mean and standard deviation of per-genome error in paralog-specific count frequencies (Supplementary Fig. 1). We removed MIPs having mean per-genome errors ≥ 0.125 or corresponding standard deviations ≥ 0.25 from our final set, with a few exceptions. For example, we retained some MIPs in the SRGAP2C deletion region having mean errors or standard deviations slightly above these values because we wanted to maximize our power to genotype this event. Reducing the number of MIPs used for genotyping SRGAP2 in our final pool in this manner increases our capacity to assay additional genes of interest and larger numbers of individuals in the same experiment while ensuring SRGAP2 genotyping remains highly accurate.Selection of a high-performing final MIP set from all initial MIPs tested, though useful for increasing multiplexing potential, is not necessary for successful application of our method (Supplementary Fig. 2).
Cost estimation
The approximate cost per gene per individual associated with using MIPs can be estimated as follows. Each MIP is 70 bp, and each synthesized base costs $0.09. Thus, each MIP costs $6.30. We usually multiplex ∼2000 MIPs in a single MIP pool, so the cost of generating a typical MIP pool is $12,600. Because a very small amount of the MIP pool is used in each capture reaction, a single order of oligos can be used to assay tens of thousands of samples, and thus we assume this cost is effectively fixed (i.e. independent of the number of samples tested). The oligo cost per sample thus depends on the number of samples tested—assuming we assay 4000 samples, for example, the per sample oligo cost is $3.15. The cost of reagents associated with the experimental protocol is $2.57 per sample. The cost of a lane of sequencing using the HiSeq is $1,388. We have found that up to 192 samples can be multiplexed per lane to obtain high coverage per MIP per sample assuming the MIP pool used in capture experiments contained 2000 MIPs. Thus, the sequencing cost per sample is approximately $7.23. Adding these results, we obtain a cost of $12.95 per sample. Assuming each gene can be effectively assayed by 50 MIPs, on average, each MIP pool covers 40 genes. Thus, the final cost per gene per sample in this scenario is ∼$0.32. If we eventually assay 10,000 samples using this same MIP pool, the final cost per gene per sample works out to $0.28. Even if we were to only assay 1000 samples, the final cost per gene per sample would still be less than $1 (∼$0.56).
Internal SRGAP2 deletion and duplication genotyping using WGS data
We leveraged data from the 1KG to evaluate WGS-based discovery of novel structural variation within duplicated genes and compare these results with our MIP data. Specifically, we genotyped SRGAP2B copy number in an individual genotyped by MIPs as having two copies of SRGAP2B with an 83 kbp internal SRGAP2B duplication and SRGAP2C copy number in seven individuals genotyped by MIPs as having two copies of SRGAP2C, one harboring the 38 kbp internal deletion. All WGS-based paralog-specific copy number estimates1 for these individuals were 2; however, specifying the regions affected by these events prior to genotyping allowed for successful identification of the internal events in 7/8 cases (Supplementary Table 12). These data provide additional support for the internal duplications and deletions called by our MIP-based method and suggest that naïve paralog-specific copy number genotyping using low-coverage WGS data cannot reliably discover them.
Application of our method to other gene families of interest
To use our method to study a gene family of interest other than SRGAP2 or RH, one would first need to obtain accurate genomic sequences for as many paralogs as possible. Having reliable sequence data for all paralogs allows MIP design to be optimized to achieve complete paralog-specificity and maximize genotyping power. Second, one would align paralogous sequences and identify SUN-containing regions to guide MIP design. We provide a program (Software) to identify such regions from aligned sequences. Third, one would attempt to design MIPs to all such regions as well as exons using the publicly available MIP design software33, select a final set of MIPs based on criteria detailed above, and order them from a commercial oligo provider. Fourth, one would perform the MIP experiments and analyses described (Software) to obtain genotypes for each duplicated segment. If possible, we recommend testing every new MIP set on a panel of genomes having known paralog-specific copy numbers to ensure accuracy and reproducibility.
Interestingly, we found RH more difficult to genotype for copy number than SRGAP2 using our approach. Two factors likely contribute to this observation. First, RHparalog-specific copy number genotyping included data from only 35 MIPs, while that for SRGAP2 incorporated data from either 90 or 142 MIPs. Second, fewer independent MIP capture events occur per genome for RH than SRGAP2 because there are fewer total genomic copies of RH than SRGAP2. Thus, there are effectively fewer experimental trials for RH than SRGAP2, resulting in increased sampling error in RHparalog-specific read count data. Designing more MIPs targeting RH and increasing DNA input would mitigate these issues and improve future RH genotyping performance. These issues warrant consideration in applying our method to other gene families of interest.
CCL3L119-20, 59-60, beta-defensins42-43, and C444 present a few novel challenges for our method: (i) CCL3L1 and beta-defensins are much smaller than SRGAP2 and RH, such that only a few MIPs may be able to interrogate SUN-containing regions within them; (ii) unlike SRGAP2 and RH, beta-defensins and C4 have no obvious family member fixed or nearly fixed at diploid copy number two in the human population, making copy number determination based on relative counts more ambiguous. For these gene families, it will be necessary to perform absolute in addition to relative read depth analysis, perhaps via singular value decomposition analysis as has been done to normalize exome capture variability from a large number of samples61. Another possible strategy would be to use some MIPs as MLPA probes (Supplementary Fig. 10) targeting genes of interest and regions of known invariant diploid copy number to calibrate aggregate or paralog-specific copy number estimates based on absolute read depth data. In addition, genotyping copy number of the blocks of duplicated sequence containing CCL3L1 and beta-defensins should provide accurate copy number genotypes for these genes, as common copy number variation at these loci occurs at the level of such blocks rather than affecting individual genes within them47. This approach leverages the much larger sample of SUNs these blocks contain compared to the genes themselves.
Identification of missing paralogous sequences in GRCb37
We genotyped regions in GRCb37 that had been previously described as missing paralogous sequence in GRCb361to identify regionsof the reference genome still lackingcomplete sequence characterization. We successfully lifted over 326 of the original 333 regions from GRCb36 to GRCb37 and calculated paralog-specific copy numbers for each region with 885 individuals from the 1KG. A region was considered "missing" from GRCb37 if the paralog-specific copy number for that region was greater than 2 for at least 90% of the individuals we genotyped. Based on this definition, we found 21 regions that are still missing paralogous sequence in GRCb37. Comparing these regions with public NCBI patches to GRCb37 reveals that 7 of the 21 regions are completely covered by a fix patch and will likely be resolved in GRCb38.
Identification of SUNs from GRCb37
We used previously calculated SUNKs and segmental duplications for GRCb37 to calculate the set of all SUNs that uniquely identify individual segmental duplications. For each pair of segmental duplications, we globally aligned the corresponding sequences and identified all mismatches, insertions, and deletions. We identified the diagnostic differences between related duplications by intersecting the coordinates of all differences with coordinates for SUNKs across GRCb37. With this approach we identified ∼4 million SUNs. After filtering out any of these SUNs that were within 36 bp of repeats identified by RepeatMasker52 or TandemRepeats Finder53, we identified ∼3.8 million SUNs.
Supplementary Material
Acknowledgments
We thank P. Sudmant, E. Karakoc, F. Hormozdiari, B. Dumont, and O. Penn for thoughtful discussion, L. Vives, K. Mohajeri, and C. Lee for technical assistance, and T. Brown for assistance with manuscript preparation. X.N. is supported by a National Science Foundation Graduate Research Fellowship under Grant No. DGE-1256082. This work was supported by NIH Grant HG004120 to E.E.E. E.E.E. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Software: Associated software, documentation, and an example dataset are freely available via GitHub at https://github.com/xnuttle/mips_cnv_typer.
Author Contributions: X.N., J.S., and E.E.E. designed the study. X.N. and B.J.O. designed the MIPs. X.N. performed capture experiments, wrote analysis software, and analyzed data. F.A. performed FISH experiments. J.H. contributed to the analysis software, prepared it for public access, and identified SUNs from the reference genome. M.F. and C.R. contributed to sample collection. X.N. and E.E.E. wrote the paper, with input and approval from all coauthors.
Competing Financial Interests: E.E.E. is on the scientific advisory boards for Pacific Biosciences, Inc., SynapDx Corp., and DNAnexus, Inc.
References
- 1.Sudmant PH, et al. Diversity of human copy number variation and multicopy genes. Science. 2010;330:641–646. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campbell CD, et al. Population-genetic properties of differentiated human copy number polymorphisms. Am J Hum Genet. 2011;88:317–332. doi: 10.1016/j.ajhg.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sebat J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 5.Ohno S. Evolution by Gene Duplication. New York: Springer-Verlag; 1970. [Google Scholar]
- 6.Semple CA, Rolfe M, Dorin JR. Duplication and selection in the evolution of primate beta-defensin genes. Genome Biol. 2003;4:R31. doi: 10.1186/gb-2003-4-5-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han MV, Demuth JP, McGrath CL, Casola C, Hahn MW. Adaptive evolution of young gene duplicates in mammals. Genome Res. 2009;19:859–867. doi: 10.1101/gr.085951.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bailey JA, Eichler EE. Primate segmental duplications: crucibles of evolution, diversity and disease. Nat Rev Genet. 2006;7:552–564. doi: 10.1038/nrg1895. [DOI] [PubMed] [Google Scholar]
- 9.Lefebvre S, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 10.Olbrich H, et al. Recessive HYDIN mutations cause primary ciliary dyskinesia without randomization of left-right body asymmetry. Am J Hum Genet. 2012;91:672–684. doi: 10.1016/j.ajhg.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bunge S, et al. Homologous nonallelic recombinations between the iduronate-sulfatase gene and pseudogene cause various intragenic deletions and inversions in patients with mucopolysaccharidosis type II. Eur J Hum Genet. 1998;6:492–500. doi: 10.1038/sj.ejhg.5200213. [DOI] [PubMed] [Google Scholar]
- 12.Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–422. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- 13.Dennis MY, et al. Evolution of human-specific neural SRGAP2 genes by incomplete segmental duplication. Cell. 2012;149:912–922. doi: 10.1016/j.cell.2012.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doggett NA, et al. A 360-kb interchromosomal duplication of the human HYDIN locus. Genomics. 2006;88:762–771. doi: 10.1016/j.ygeno.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 15.Locke DP, et al. Linkage disequilibrium and heritability of copy number polymorphisms within duplicated regions of the human genome. Am J Hum Genet. 2006;79:275–290. doi: 10.1086/505653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCarroll SA, Altshuler DM. copy number variation and association studies of human disease. Nat Genet. 2007;39:S37–S42. doi: 10.1038/ng2080. [DOI] [PubMed] [Google Scholar]
- 17.Eichler EE, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11:446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez E, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 20.Bhattacharya T, et al. CCL3L1 and HIV/AIDS susceptibility. Nat Med. 2009;15:1112–1115. doi: 10.1038/nm1009-1112. [DOI] [PubMed] [Google Scholar]
- 21.Cantsilieris S, Baird PN, White SJ. Molecular methods for genotyping complex copy number polymorphisms. Genomics. 2013;101:86–93. doi: 10.1016/j.ygeno.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 22.Armour JAL, et al. Accurate, high-throughput typing of copy number variation using paralogue ratios from dispersed repeats. Nucleic Acids Res. 2007;35:e19. doi: 10.1093/nar/gkl1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schouten JP, et al. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Armour JA, Sismani C, Patsalis PC, Cross G. Measurement of locus copy number by hybridisation with amplifiable probes. Nucleic Acids Res. 2000;28:605–609. doi: 10.1093/nar/28.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waszak SM, et al. Systematic inference of copy number genotypes from personal genome sequencing data reveals extensive olfactory receptor gene content diversity. PLoS Comput Biol. 2010;6:e1000988. doi: 10.1371/journal.pcbi.1000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hardenbol P, et al. Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat Biotechnol. 2003;21:673–678. doi: 10.1038/nbt821. [DOI] [PubMed] [Google Scholar]
- 27.Hardenbol P, et al. Highly multiplexed molecular inversion probe genotyping: over 10,000 targeted SNPs genotyped in a single tube assay. Genome Res. 2005;15:269–275. doi: 10.1101/gr.3185605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Porreca GJ, et al. Multiplex amplification of large sets of human exons. Nat Methods. 2007;4:931–936. doi: 10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
- 29.Turner EH, et al. Massively parallel exon capture and library-free resequencing across 16 genomes. Nat Methods. 2009;6:315–316. doi: 10.1038/nmeth.f.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Colin Y, et al. Genetic basis of the RhD-positive and RhD-negative blood group polymorphism as determined by Southern analysis. Blood. 1991;78:2747–2752. [PubMed] [Google Scholar]
- 31.Wagner FF, Flegel WA. RHD gene deletion occurred in the Rhesus box. Blood. 2000;95:3662–3668. [PubMed] [Google Scholar]
- 32.Kitano T, Saitou N. Evolution of Rh blood group genes have experienced gene conversions and positive selection. J Mol Evol. 1999;49:615–626. doi: 10.1007/pl00006583. [DOI] [PubMed] [Google Scholar]
- 33.O'Roak BJ, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kidd JM, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 37.Zhang F, et al. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat Genet. 2009;41:849–853. doi: 10.1038/ng.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fledel-Alon A, et al. Broad-scale recombination patterns underlying proper disjunction in humans. PLoS Genet. 2009;5:e1000658. doi: 10.1371/journal.pgen.1000658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carritt B, Kemp TJ, Poulter M. Evolution of the human RH (rhesus) blood group genes: a 50 year old prediction (partially) fulfilled. Hum Mol Genet. 1997;6:843–850. doi: 10.1093/hmg/6.6.843. [DOI] [PubMed] [Google Scholar]
- 40.Edwards MC, Gibbs RA. Multiplex PCR: advantages, development, and applications. PCR Methods Appl. 1994;3:S65–S75. doi: 10.1101/gr.3.4.s65. [DOI] [PubMed] [Google Scholar]
- 41.Markoulatos P, Siafakas N, Moncany M. Multiplex polymerase chain reaction: a practical approach. J Clin Lab Anal. 2002;16:47–51. doi: 10.1002/jcla.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Groth M, et al. High-resolution mapping of the 8p23.1 beta-defensin cluster reveals strictly concordant copy number variation of all genes. Hum Mutat. 2008;29:1247–1254. doi: 10.1002/humu.20751. [DOI] [PubMed] [Google Scholar]
- 43.Aldhous MC, et al. Measurement methods and accuracy in copy number variation: failure to replicate associations of beta-defensin copy number with Crohn's disease. Hum Mol Genet. 2010;19:4930–4938. doi: 10.1093/hmg/ddq411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fernando MM, et al. Assessment of complement C4 gene copy number using the paralog ratio test. Hum Mutat. 2010;31:866–874. doi: 10.1002/humu.21259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hiatt JB, et al. Single molecule molecular inversion probes for targeted, high accuracy detection of low frequency variation. Genome Res. 2013;23:843–854. doi: 10.1101/gr.147686.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Itsara A, et al. Resolving the breakpoints of the 17q21.31 microdeletion syndrome with next-generation sequencing. Am J Hum Genet. 2012;90:599–613. doi: 10.1016/j.ajhg.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jackson MS, et al. Evidence for widespread reticulate evolution within human duplicons. Am J Hum Genet. 2005;77:824–840. doi: 10.1086/497704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schildkraut E, Miller CA, Nickoloff JA. Gene conversion and deletion frequencies during double-strand break repair in human cells are controlled by the distance between direct repeats. Nucleic Acids Res. 2005;33:1574–1580. doi: 10.1093/nar/gki295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ezawa K, et al. Proceedings of the SMBE Tri-National Young Investigators' Workshop 2005 Genome-wide search of gene conversions in duplicated genes of mouse and rat. Mol Biol Evol. 2006;23:927–940. doi: 10.1093/molbev/msj093. [DOI] [PubMed] [Google Scholar]
- 50.Chen JM, et al. Gene conversion: mechanisms, evolution and human disease. Nat Rev Genet. 2007;8:762–775. doi: 10.1038/nrg2193. [DOI] [PubMed] [Google Scholar]
- 51.Thompson JD, Gibson TJ, Higgins DG. Current Protocols in Bioinformatics. New York: John Wiley & Sons; 2002. Multiple sequence alignment using ClustalW and ClustalX. [DOI] [PubMed] [Google Scholar]
- 52.Tarailo-Graovac M, Chen N. Current Protocols in Bioinformatics. New York: John Wiley & Sons; 2009. Using RepeatMasker to identify repetitive elements in genomic sequences. [DOI] [PubMed] [Google Scholar]
- 53.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hach F, et al. mrsFAST: a cache-oblivious algorithm for short-read mapping. Nat Methods. 2010;7:576–577. doi: 10.1038/nmeth0810-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alkan C, et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet. 2009;41:1061–1067. doi: 10.1038/ng.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Antonacci F, et al. A large and complex structural polymorphism at 16p12.1 underlies microdeletion disease risk. Nat Genet. 2010;42:745–750. doi: 10.1038/ng.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuhn HW. The Hungarian Method for the assignment problem. Nav Res Logist Q. 1955;2:83–97. [Google Scholar]
- 59.Carpenter D, Walker S, Prescott N, Schalkwijk J, Armour JA. Accuracy and differential bias in copy number measurement of CCL3L1 in association studies with three auto-immune disorders. BMC Genomics. 2011;12:418. doi: 10.1186/1471-2164-12-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nordang GB, et al. Association analysis of the CCL3L1 copy number locus by paralogue ratio test in Norwegian rheumatoid arthritis patients and healthy controls. Genes Immun. 2012;13:579–682. doi: 10.1038/gene.2012.30. [DOI] [PubMed] [Google Scholar]
- 61.Krumm N, et al. Copy number variation detection and genotyping from exome sequence data. Genome Res. 2012;22:1525–1532. doi: 10.1101/gr.138115.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang J, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park H, et al. Discovery of common Asian copy number variants using integrated high-resolution array CGH and massively parallel DNA sequencing. Nat Genet. 2010;42:400–405. doi: 10.1038/ng.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ahn SM, et al. The first Korean genome sequence and analysis: full genome sequencing for a socio-ethnic group. Genome Res. 2009;19:1622–1629. doi: 10.1101/gr.092197.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schuster SC, et al. Complete Khoisan and Bantu genomes from southern Africa. Nature. 2010;463:943–947. doi: 10.1038/nature08795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.1000 Genomes Project Consortium et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fischbach GD, Lord C. The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron. 2010;68:192–195. doi: 10.1016/j.neuron.2010.10.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.