Abstract
Homologous recombination (HR)-mediated instability of the repetitively organized ribosomal DNA (rDNA) has been proposed as a mediator of cell senescence in yeast triggering the DNA damage response. High individual variability in the content of human rDNA suggests that this genomic region remained relatively unstable throughout evolution. Therefore, quantitative real time PCR was used to determine the genomic content of rDNA in post mortem samples of parietal cortex from 14 young- and 9 elderly individuals with no diagnosis of a chronic neurodegenerative/neurological disease. In addition, rDNA content in that brain region was compared between 10 age-matched control individuals and 10 patients with dementia with Lewy bodies (DLB) which involves neurodegeneration of the cerebral cortex. Probing rRNA-coding regions of rDNA revealed no effects of aging on the rDNA content. Elevated rDNA content was observed in DLB. Conversely, in the DLB pathology-free cerebellum, lower genomic content of rDNA was present in the DLB group. In the parietal cortex, such a DLB-associated instability of rDNA was not accompanied by any major changes of CpG methylation of the rDNA promoter. As increased cerebro-cortical rDNA content was previously reported in Alzheimer's diseases, neurodegeneration appears to be associated with instability of rDNA. The hypothetical origins and consequences of this phenomenon are discussed including possibilities that the DNA damage-induced recombination destabilizes rDNA and that differential content of rDNA affects heterochromatin formation, gene expression and/or DNA damage response.
Keywords: Nucleolus, Genomic Instability, Dementia with Lewy Bodies, Neurodegeneration, Aging
1. Introduction
Aging-associated disorders including proliferative senescence, cancer and neurodegeneration have been proposed to originate from accumulation of DNA damage leading to genomic instability [1, 2]. Thus, loss of telomeres, as well as de-regulated homologous recombination (HR)1 have been associated with aging and/or aging-related disorders [2, 3]. Telomere shortening that in proliferating cells triggers DNA damage response and cell cycle arrest has been observed in leukocytes from patients with Alzheimer's disease (AD) and dementia with Lewy bodies (DLB) [4-6]. In these conditions, dementia is believed to originate from degenerative changes in the hippocampus and/or the cerebral cortex. In AD, telomere shortening may have complex effects including neuroprotective reduction of inflammation but also increased neuronal dysfunction [7]. While the role of HR malfunction in proliferative senescence or cancer is well established [2, 3], its contribution to neurodegeneration is unclear.
The ribosome is a critical component of translation machinery. Hence, complex mechanisms evolved to adjust ribosomal production to cellular needs. For instance, in Eukaryotes, genes that encode rRNA, the key component of ribosomes were amplified [8-10]. Such amplification resulted in approximately 300 copies of nucleolar 47S rRNA genes (rDNA) per haploid human genome. These genes are organized in five clusters of tandem repeats on short arms of acrocentric chromosomes 13, 14, 15, 21 and 22. It is believed that the primary force driving rDNA amplification was a need to produce large number of ribosomes during periods of rapid proliferative growth. However, under normal growth conditions, only about half of rDNA units participate in ribosomal biogenesis while the other half remains transcriptionally inactive and epigenetically silenced [8]. Hence, possessing multiple inactive copies of rRNA genes may have enabled cells to tolerate loss of some of the active copies of these critical genes. Finally, rDNA acquired additional functions regulating chromatin structure, gene expression and stress response [11-14]. These activities do not appear to be directly related to ribosomal biogenesis.
Repetitive nature of rDNA poses a challenge for genome maintenance. HR of rDNA units within one cluster or between clusters that are located on different chromosomes may lead to genomic instability including loss of rDNA, its amplification and chromosomal translocations [3, 15]. In 1972, Bernard Strehler proposed a hypothesis that aging-related loss of rDNA in post-mitotic cells, including muscles and neurons, leads to aging associated dysfunction simply by insufficient ribosome supply and translational failure [16]. However, subsequent studies including those in post mortem samples of the human cerebral cortex did not confirm early observations of aging-associated changes in the genomic rDNA content [17, 18]. Likewise, no disruption of rDNA stability has been found in cell culture models of replicative senescence [19, 20].
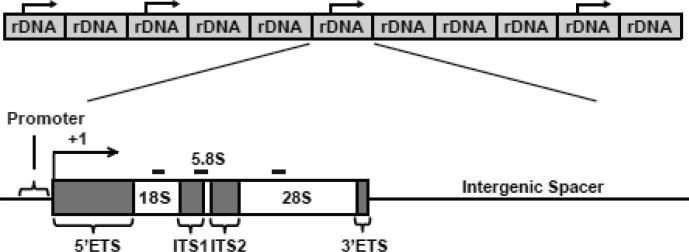
However, the negative reports on effects of aging on rDNA copy number in vertebrates used hybridization methods that did not allow for a detailed, high resolution insight into stability of this region. Each human rDNA unit is 43 kb long and contains the 34 kb-long intergenic spacer and the 47S pre-rRNA gene including a promoter and exons that correspond to 18S-, 5.8S-, and 28S rRNAs [8] (Fig. 1). Indeed, more recent work that employed qPCR technique to selectively probe the 18S-, 5.8S- and 28S- rRNA-coding portions of rDNA suggested age-dependent decrease of rDNA content in human adipose tissue that was limited to 5.8S and 28Ss regions [21]. However, in this study, the age effects were modest despite investigating a relatively large population (n=120). Conversely, the in situ hybridization-based approach of molecular combing revealed that in cells derived from normal individuals about 30% of rDNA units are organized as palindromic rather than tandem repeats [22]. In cells from WS cases, frequency of such non-canonical repeats increased to 50%. Moreover, high dynamics of human rDNA is suggested by high individual variability in length of the entire rDNA clusters as revealed by pulsed field gel electrophoresis studies [23]. Such an effect was attributed to a high rate of meiotic rDNA recombination which was estimated at at least 10% per meiosis per each cluster. Mitotic rDNA recombination was also detected in normal individuals. Interestingly, increased mitotic instability of rDNA has been found in at least 50% of human colon- or lung cancer samples [24]. Finally, genomic instability disorders including ataxia telangiectasia and Bloom syndrome have been linked to very high rates of mitotic rDNA cluster length instability [25]. Hence, in higher Eukaryotes including humans, rDNA is unstable. However, the role of human rDNA instability in aging or aging-associated diseases remains unresolved.
Figure 1. The qPCR-based assay to determine the genomic content of rDNA.
The rDNA copies are organized as long tandem repeats located on five acrocentric chromosomes. Each copy consists of the rRNA gene and the intergenic spacer (IGS). Each rRNA gene includes a Pol1-dependent promoter and exons that correspond to 18S-, 5.8S- and 28S rRNAs. They are separated by introns (5’ETS, ITS1, ITS2 and 3’ETS). The positions of the analyzed rDNA amplicons are indicated by the thick black lines. The schematics are not drawn in scale.
In contrast, there is strong evidence that replicative senescence in yeast is caused by rDNA instability [26, 27]. DNA damage is presumed to trigger such instability. Initially, it has been proposed that the pro-senescence effector of rDNA instability in yeast was accumulation of a toxic byproduct of rDNA HR, the extrachromosomal rDNA circles [28]. However, current data better fit to a model in which rDNA loss by itself triggers senescence by activating the DNA damage response (DDR) in a ribosomal biogenesis-independent manner [13, 29]. Such a model implies that the extra rDNA copies that are not active in ribosome production are required to suppress the DDR [26]. Thus, Kobayashi has proposed that rDNA copy number could work as a sensor of environmental exposures to genotoxic agents complementing the genome integrity sensing function of telomeres which measure the number of cell divisions [26]. However, it remains to be tested whether rDNA content affects DDR in higher Eukaryotes.
In yeast, HR underlies rDNA instability including rDNA loss and rDNA expansion [15, 30]. These mechanisms operate during DNA replication and involve unequal sister chromatid exchange. The rDNA loss and expansion appear to balance each other in non-senescent yeast cells [26]. Likewise, HR appears to play an important role in human rDNA stability as the excessive HR in cells from Bloom syndrome patients is associated with a pronounced increase in mitotic rDNA instability [25]. In yeast- and mouse cells, maintaining a pool of inactive, epigenetically silenced copies of rDNA is critical for rDNA stability [26, 31].
Previously we have used qPCR to demonstrate increased genomic content of 18S rDNA region in the parietal cortex of Alzheimer's disease (AD) patients [32]. Moreover, the increased copy number was associated with an increase in rDNA promoter methylation which is a signature of epigenetic silencing of rDNA. Hence, a question could be raised whether in the brain, aging involves segmental instability of rDNA that is prevented by the AD-associated epigenetic silencing of rDNA. Alternatively, rDNA instability including its amplification may be associated with neurodegeneration. To address these possibilities, we have compared rDNA content in the parietal cortex from young and old individuals. In addition, we have determined rDNA content and its methylation in the cerebral cortex and the cerebellum of DLB patients.
2. Methods
2.1 Subjects and sample preparation
For aging studies fresh flash frozen samples of parietal cortex (Brodmann areas 39 and 40) were obtained from the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland, Baltimore, MD. The young donor group consisted of 7 men and 7 women (1-25 years old, median age: 20); the old donor group consisted of 4 men and 5 women (73-90 years old, median age: 79). The donors had no prior history of a chronic neurodegenerative- or neurological disease. The post mortem intervals (PMIs) were comparable for both groups averaging 7.5 h (PMIs were always less than 14 h). For DLB studies, the donors were participants of the IRB-approved University of Kentucky Alzheimer's Disease Center cohort and were followed for at least 2 years before death [33]. The follow up included annual Minimental State Examination (MMSE) as well as neurological and physical examinations. The donors had no history of substance abuse, head injury, encephalitis, meningitis, epilepsy, or stroke/transient ischemic attack. The MMSE score closest to death was used as an indicator of overall cognitive status. During autopsy (usually 5 or less hours after death), tissue samples including parietal cortex and cerebellum were processed for neuropathological evaluations or flash-frozen in liquid nitrogen and stored at −80°C, as described previously [33, 34]. All included DLB subjects met the clinical and histopathological criteria for diagnosis of DLB [35]. The control subjects received MMSE scores ≥ 23 with Braak staging at ≤ 2. Detailed donor information is presented in Tables 1 and 2.
Table 1.
Young and old donors of the parietal cortex samples that were used for the aging studies.
| Group | Age (years) | PMI (h)1 | Sex |
|---|---|---|---|
| Young | 2 | 10 | Male |
| 15 | 9 | Female | |
| 16 | 7 | Male | |
| 18 | 8 | Female | |
| 19 | 7 | Female | |
| 20 | 6 | Male | |
| 20 | 5 | Male | |
| 21 | 7 | Male | |
| 22 | 10 | Female | |
| 22 | 7 | Male | |
| 24 | 9 | Female | |
| 24 | 7 | Female | |
| 24 | 9 | Male | |
| 25 | 4 | Female | |
| Old | 73 | 13 | Female |
| 76 | 3 | Female | |
| 76 | 3 | Male | |
| 77 | 8 | Female | |
| 79 | 5 | Male | |
| 79 | 10 | Male | |
| 88 | 8 | Female | |
| 89 | 14 | Male | |
| 90 | 4 | Female |
PMI, post mortem interval
Table 2.
Control and DLB donors of the parietal cortex and the cerebellum samples that were used for the DLB studies.
| Group | Age (years) | PMI (h)1 | Sex | Parietal Cortex | Cerebellum |
|---|---|---|---|---|---|
| Control | 77 | 3.5 | Male | + | unavailable |
| 81 | 2 | Male | + | unavailable | |
| 87 | 2.4 | Male | + | unavailable | |
| 82 | 2.1 | Male | + | unavailable | |
| 85 | 2.5 | Female | + | + | |
| 90 | 2 | Female | + | + | |
| 79 | 1.8 | Female | + | + | |
| 78 | 1.2 | Male | + | + | |
| 81 | 2.8 | Male | + | + | |
| 96 | 2.1 | Female | + | + | |
| 86 | 2.5 | Female | not included | + | |
| 90 | 3.8 | Female | not included | + | |
| 80 | 2.5 | Female | not included | + | |
| 95 | 2.8 | Female | not included | + | |
| DLB | 78 | 2.5 | Male | + | + |
| 78 | 3.8 | Male | + | + | |
| 81 | 2.5 | Male | + | + | |
| 72 | 2.8 | Male | + | + | |
| 65 | 9.5 | Male | + | + | |
| 87 | 2 | Female | + | + | |
| 68 | 3.8 | Male | + | + | |
| 90 | 2.5 | Female | + | + | |
| 82 | 2.3 | Male | + | + | |
| 91 | 10.5 | Female | + | + |
PMI, post mortem interval
2.2 Analysis of genomic rDNA content
Genomic qPCR using standard curve based analysis was utilized to determine rDNA content. Amplicons targeting 18S, 5.8S, and the 28S coding regions of rDNA were used (Fig 1). For normalization, amplicons corresponding to the coding regions of the tRNAK-CTT gene or the albumin gene (ALB) were amplified. Primer sequences were as follows, 18S- forward agcctgagaaacggctacca and reverse ggtcgggagtgggtaatttgc, 5.8S- forward gaggcaaccccctctcctctt and reverse gagccgagtgatccaccgcta, 28S- forward gacctcagatcagaggtggcga and reverse ttcactcgccgttactgagggaat, tRNA-KCTT forward ctagctcagtcggtagagcatg and reverse ccaacgtggggctcgaac, and ALB forward cggcggcgggcggcgcgggctgggcggaaatgctgcacagaatccttg and reverse gcccggcccgccgcgcccgtcccgccggaaaagcatggtcgcctgtt.
2.3 Bisulfite mapping of rDNA promoter methylation was performed as described previously [32].
Briefly, two hundred ng of DNA/sample was treated with bisulfite to convert unmethylated cytosines to uracil. The rDNA promoter region was PCR-amplified and cloned into the pGEM-T vector. Following bacterial transformation, individual clones were isolated and sequenced. Only clones with unique methylation patterns were included in the analysis to avoid potential PCR/cloning artifacts. Clones with incomplete bisulfate conversion were also discarded. Thus, twenty fully converted, and unique clones were analyzed for each individual.
2.4 DNA methylation analysis using the methyl-CpG sensitive restriction enzyme HpaII
Methylation of the HpaII site located at position -9 relative to the transcription start site (CpG #23 of the human rDNA promoter) was analyzed with a quantitative real-time PCR of the HpaII-digested genomic DNA as described previously [32] with modifications. Importantly, as a reference, the HpaII-digested DNA was used as template to determine the content of an amplicon adjacent to the one with the HpaII site. Such a normalization resulted in reproducible methylation values as opposed to using a non-digested DNA with the HpaII amplicon. QPCR primer sequences were as follows: HpaII forward: gtatatctttcgctccgagtcg ; HpaII reverse: acaggtcgccagaggacag; reference forward: acggacgttcgtggcga; reference reverse: ggagaggagagacgagggg.
2.5 Evaluation of methylation effects on qPCR efficiency
An unmethylated DNA template for the 5.8S amplicon was obtained by PCR on genomic DNA (primer sequences: forward cccgtggtgtgaaacctt, reverse agctagctgcgttcttcatc). The PCR product was then in vitro methylated in a buffer containing 60ng DNA/μL, 0.16 mM S-adenosylmethionine and 0.8 unit M.SssI/μL (New England Biolabs) at 37°C for 2 h followed by M.SssI inactivation at 65°C for 20’. Methylated and unmethylated templates were used for qPCR with the 5.8S amplicon primers. To verify the extent of methylation, both methylated and unmethylated templates were incubated with the methyl sensitive restriction enzyme HpaII. Template resistance to HpaII was determined by qPCR.
2.6 Statistical analysis
was performed using the non-parametric Kruskal-Wallis one way ANOVA and linear regression fitting. In addition, comparisons of individual rDNA methylation sites were accomplished using a modified significance analysis of microarrays (SAM), as reported previously [32].
3. Results
3.1 Stability of the cerebro-cortical rDNA copy number in aging
To assess the effects of aging on rDNA copy number in the brain, genomic DNA was isolated from post mortem parietal cortex samples that were collected from two groups of donors. The young donor group consisted of 14 individuals with a median age of 20±1.6 (range 1-25, Table 1). The old donor group included 9 individuals with a median age of 79±2.1 (range 73-90, Table 1). None of the donors were known to suffer from a chronic neurodegenerative and/or neurological disease. The genomic content of 18S- and 28S rRNA coding regions of rDNA was analyzed by a qPCR assay (Fig. 1). For normalization, a coding region of the multi-copy gene tRNAK-ctt was used. There are 17 almost identical copies of this gene in the haploid human genome. However, in contrast to rDNA, they are not clustered together [36] and therefore, are less likely than rDNA to undergo recombination-associated instability.
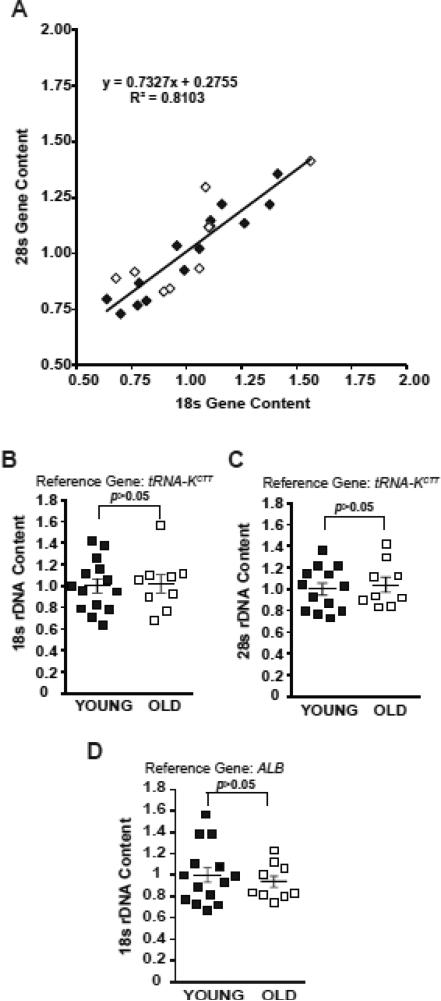
As the investigated rDNA regions are physically linked, a direct correlation between their genomic content for each analyzed sample is expected (Fig. 1). Indeed, the individual values obtained for the 18S- and the 28S amplicons were strongly correlated fitting to a linear model of a direct correlation (Fig. 2A, a=0.7327, R2=0.8103). These results support accuracy of the applied qPCR methodology for determination of the genomic rDNA content.
Figure 2. Effects of age on the cerebro-cortical content of rDNA.
Genomic rDNA content was quantified in post mortem samples of the parietal cortex from young- and old individuals without any neurodegenerative conditions (n=14, median age 20, and n=9, median age 79, respectively). The 18S- and 28S amplicons were used for rDNA content determinations; the reference genes are indicated. For each amplicon, the rDNA content was normalized to average value of young individuals. A, Regression analysis of individual values of rDNA content as determined with 18S- or 28S probes. As expected for a close physical linkage between these genomic templates, a direct correlation is present supporting validity of the qPCR assay. Filled or opened diamonds indicate young or old individuals, respectively. B-D, Content of rDNA is similar in young- and old individuals. Individual values are depicted by squares; mean values are indicated by the lines intersecting the error bars (SEM); p values of the Kruskal-Wallis ANOVA are shown.
However, genomic content of 18S- and 28S- regions of rDNA did not significantly differ between the young- and the old subjects (Fig. 2B, C). Usage of a coding region of the albumin gene (ALB) as an alternative genomic reference produced similar results (Fig. 2D). Therefore, in the cerebral cortex, genomic content of rDNA appears to be stable throughout the lifespan.
3.2 DLB-associated instability of brain rDNA
To assess whether aging-related neurodegeneration affects genomic rDNA content in the brain, post mortem parietal cortex samples from ten patients who were diagnosed with DLB were analyzed (Table 2). The control group included ten age-matched individuals who died of non-neurological diseases and whose brains did not display any evidence of neurodegeneration (Table 2). Similarly to the midbrain-focused Parkinson's disease (PD), DLB is a form of a synucleinopathy [37, 38]. Their common characteristic is the presence of intracytoplasmic α-synuclein-containing inclusions, the Lewy bodies. In addition, DLB is associated with neuronal atrophy, neuronal death and reactive gliosis in the cerebral cortex including the parietal region [37]. Oxidative damage of macromolecules including DNA is observed in both conditions [39, 40]. Hence, DLB and PD may represent a similar pathological process that affects different areas of the brain.
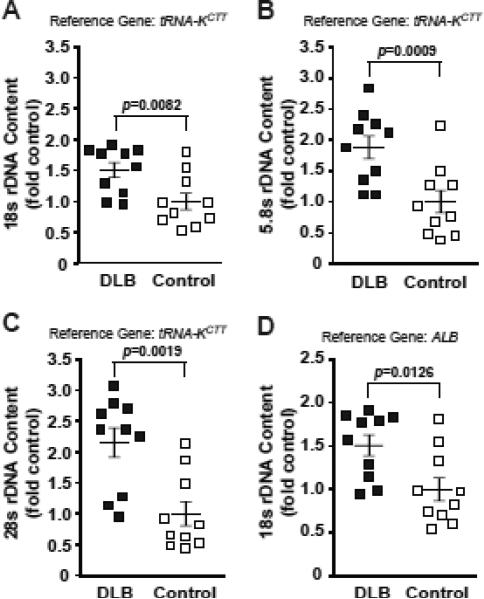
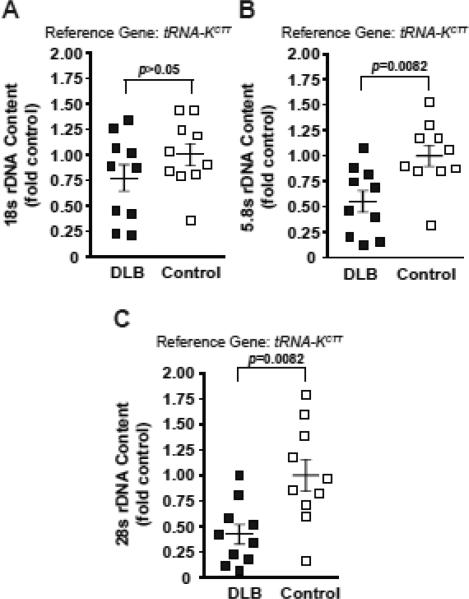
Three rDNA coding regions were analyzed including probes corresponding to the 18S-, 5.8S-, and 28S exons. As expected for a tight physical linkage, the DLB- and the control group-derived individual values obtained with these probes correlated with each other fitting a linear model of direct correlation (28S(18S), y=1.7416*x-0.6165, R2=0.8495; 5.8S(18S), y=1.4776*x-0.4126, R2=0.8957; 28S(5.8S), y=1.1008*x-0.0133, R2=0.8274). Interestingly, genomic content of rDNA was higher in DLB. In this group, the 18S-, 5.8S-, and 28S probes revealed rDNA content that was 1.6-, 2.0-, and 2.3 fold controls respectively (Kruskal-Wallis ANOVA, p<0.01, Fig. 3A-C). Similar results were obtained when ALB was used as an alternative genomic reference (Fig. 3D). To determine whether this DLB-associated effect on rDNA content was directly related to pathological changes in the cortex, samples from the DLB pathology-free cerebellum were analyzed. In samples from this structure, direct correlations between individual values for various rDNA probes were observed confirming accuracy of our determinations (data not shown). Surprisingly, in the cerebellum, rDNA content was lower in the DLB group. Significant decreases of 0.55- and 0.42 fold control were observed for the 5.8S- and 28S probes, respectively (Kruskal-Wallis ANOVA, p<0.01, Fig. 4). While the 18S probe did not detect significant differences between the two groups, the genomic 18S content also showed a downward trend in DLB (Fig. 4). Therefore, DLB appears to be associated with instability of brain rDNA.
Figure 3. Increased rDNA content in the DLB parietal cortex.
Genomic rDNA content was quantified in post mortem samples of the parietal cortex from DLB- and age-matched control individuals without any neurodegenerative conditions (n=10 each). The 18S-, 5.8S- and 28S amplicons were used for the determinations; the reference genes are indicated. Regression analysis of individual values for pairs of rDNA amplicons revealed strong correlations similar to those in Fig. 2A (see the Results section for more details). In the DLB group, significant increases of rDNA content were detected using the 18S/tRNAK-ctt (A), 5.8S/tRNAK-ctt (B), 28S/tRNAK-ctt (C) and 18S/ALB (D) ratios. Individual values are depicted by squares; mean values are indicated by the lines intersecting the error bars (SEM); p values of the Kruskal-Wallis ANOVA are shown.
Figure 4. Decreased rDNA content in the DLB cerebellum.
Genomic rDNA content was quantified in post mortem samples of the cerebellum from 10 DLB- and 10 aged-matched control individuals. Regression analysis of individual values for pairs of rDNA amplicons revealed strong correlations similar to those in Fig. 2A (data not shown). While a downward trend was observed for the 18S amplicon (A), the 5.8S- and the 28S probes revealed significant decreases of rDNA content in DLB samples (B-C). Individual values are depicted by squares; mean values are indicated by the lines intersecting the error bars (SEM); p values of the Kruskal-Wallis ANOVA are shown.
3.3 Effects of DLB on methylation of the rDNA promoter region
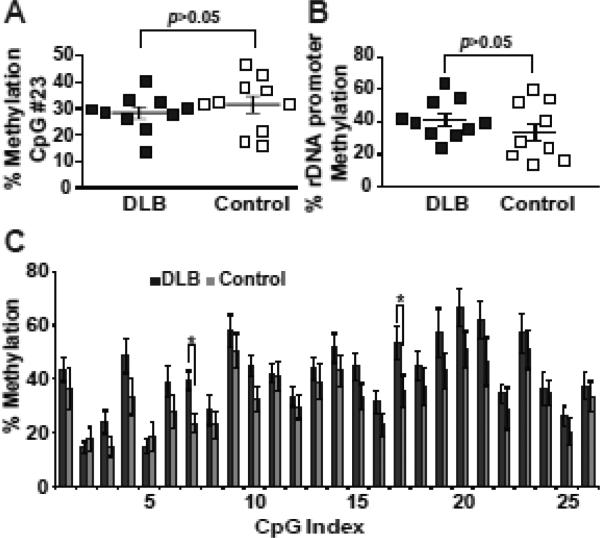
At least in cancer cell lines, amplification of rDNA may be associated with CpG hypermethylation [41]. As CpG methylation of the rDNA is associated with epigenetic silencing of rDNA copies a combination of amplification and hypermethylation may help to keep the number of active rDNA copies constant [8]. Moreover, epigenetic silencing of rDNA may stabilize the inactive genes preventing recombination; conversely, de-silencing may have an opposite effect [26, 31]. Thus, methylation of the rDNA promoter region was analyzed in the parietal cortex of the DLB- and control groups using the CpG methylation-sensitive restriction endonuclease HpaII or bisulfite sequencing. Methylation of the rDNA promoter was similar in the DLB- and the control group (Fig. 5). Therefore, in the DLB pathology-affected cerebral cortex, rDNA amplification is not associated with changes in CpG methylation of the rDNA promoter.
Figure 5. Similar methyl-cytosine content of the rDNA promoter region in the control- and DLB parietal cortex.
A, Methylation of the rDNA promoter CpG#23 (position -9 from the transcription start site) content was determined using a methylation-sensitive enzyme HpaII and qPCR. B-C, Methylation of all 26 CpG sites within the human rDNA promoter region (positions -186 to +26) was investigated using bisulfate sequencing. B, Average CpG methylation in the rDNA promoter. C, Effect of DLB on distribution of CpG methylation across the rDNA promoter. The data represent averages ±SEM from 10 DLB- and 10 control individuals. Individual values are depicted by squares; mean values are indicated by the lines intersecting the error bars. In A and B, p values of the Kruskal-Wallis ANOVA are shown; in C, *, p<0.05 (SAM statistics). Although CpG#7 and 17 appeared hypermethylated in DLB, the overall trend of CpG methylation across the 26 rDNA promoter CpGs was not significantly affected by DLB (local regression analysis, data not shown).
3.4 Effects of template methylation on qPCR efficiency
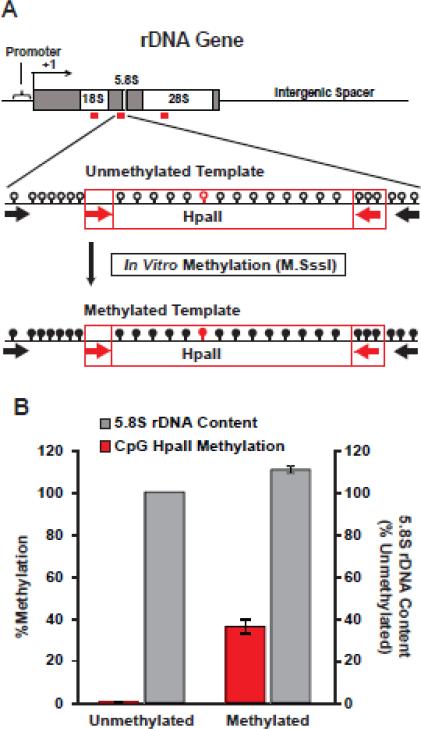
Methylation of rDNA is observed not only in the promoter region but also in the rRNA-coding exons that have been targeted for qPCR-based rDNA quantification. Therefore a possibility exists that the apparent DLB-associated changes in rDNA content are due to altered methyl-CpG content in those regions if methylation affects qPCR amplification efficiency. To exclude such a possibility, nonmethylated rDNA fragment containing the 5.8S amplicon template was produced by PCR. As the 5.8S amplicon contains 19 CpG sites including 3 in the reverse primer target sequence methylated template was generated in vitro using a non-selective CpG DNA methyltransferase M.SssI (Fig. 6). As compared to the unmethylated template, methylation only slightly increased qPCR efficiency resulting in about 10% overestimation of the template content. Therefore, even dramatic changes in CpG methylation of the rDNA templates are unlikely to explain up to 130%- increases or decreases of rDNA content that were observed between control- and DLB samples.
Figure 6. Effects of template methylation on qPCR efficiency.
A, PCR on genomic DNA was used to produce an unmethylated DNA fragment including a template for the 5.8S qPCR amplicon that revealed rDNA instability in DLB (red box, qPCR primers indicated by red arrows, PCR primers indicated by black arrows). DNA was then in vitro methylated with M.SssI DNA methyltransferase that indiscriminately targets all CpG sites. Note presence of multiple CpGs in the 5.8S amplicon (opened and filled circles correspond to CpGs or methyl-CpGs, respectively; one of the CpGs is a part of an HpaII site). B, Methylation of a CpG that was part of an HpaII site was confirmed using HpaII-qPCR assay. When methylated DNA fragment was used as a template for 5.8S qPCR, rDNA content was overestimated by just 10% as compared to unmethylated template. Hence, differential methylation of rDNA templates of the qPCR amplicons is unlikely to account for DLB-associated changes in rDNA content. Data represent two independent experiments; error bars are SDs.
Discussion
In this study, the qPCR- based analysis of the genomic rDNA content in the cerebral cortex revealed no major differences between young- and old individuals. However, in DLB- and age-matched control brains rDNA content differed significantly. Increased rDNA levels were observed in the neurodegeneration-affected parietal cortex and decreased rDNA levels were found in the DLB pathology-free cerebellum. Therefore, rDNA instability in the brain appears to be associated with this form of synucleinopathy. One might speculate that similar instability accompanies other synucleinopathies including PD.
In concert with our findings, no aging effects on rDNA content in the human cerebral cortex were found using a hybridization approach [42]. Conversely, we have reported an apparent increase in the genomic rDNA levels in the parietal- and frontal cortex of individuals with advanced AD or mild cognitive impairment (MCI), which often represents early stage AD [32]. In the cerebellum, rDNA content did not seem to be affected by AD. As both the parietal- and frontal cortex but not the cerebellum display AD-related pathology including plaques, tangles, neuronal atrophy, neuronal loss and reactive gliosis, the AD-associated elevations of the cerebro-cortical rDNA content appear to be associated with neurodegeneration. A similar conclusion applies to the current study of DLB cases. However, unlike AD, DLB seems to be associated with reduced rDNA content in the neurodegeneration-free cerebellum. Thus, at least in DLB, rDNA becomes unstable in neurodegeneration-affected and unaffected brain regions. In the degenerating areas, the rDNA expansions would be the predominant product of such a destabilization; in the non-degenerating areas, rDNA instability would result in rDNA loss. While causes for such different outcomes of rDNA instability are unclear, similar divergence has been reported for another unstable region of the genome, the telomere. Telomere shortening has been observed in the peripheral blood leukocytes in AD, and DLB [4-6]. However, at least in AD, telomere expansion has been reported in the AD pathology-affected hippocampus [4].
Although somatic rDNA instability appears to be the most probable source of the observed differences between the DLB- and the control group one can not formally exclude a possibility that in addition to rDNA instability, our results are also affected by germ line-derived rDNA copy number polymorphisms that are associated with an increased risk of DLB.
HR has been identified as a major mechanism of rDNA instability [15, 26, 30]. Hence, our current as well as previously published results suggest that in DLB or AD HR becomes activated in the brain. As this process is thought to occur during or after DNA replication peaking in the S phase of the cell cycle [43, 44], the observed rDNA instability may be localized to reactive glia that underwent divisions in response to neurodegeneration [45, 46]. Alternatively, as neuronal cell cycle re-entry that has been documented in AD-affected brain regions, reactivation of HR in normally post-mitotic neurons may also be possible [47-49]. Finally, as rDNA is present in 5 clusters on 5 distinct chromosomes that all reside in close physical proximity within the nucleolus and the nucleolus-associated heterochromatin, HR of rDNA may, at least theoretically, occur in the absence of the replication-generated sister chromatids. Such a “non-replicative” recombination could engage homologous rDNA units within the same chromatid or from different chromosomes [3]. Interestingly, recent reports have documented HR-like activity of non-mitotic human somatic cells in G0 [50, 51]. As that activity required the participating DNA to be transcribed, such a mechanism may be possible at the active rDNA units.
As rDNA instability may have serious adverse consequence for the cell including induction of the DDR, HR of rDNA is under tight control [15, 26, 30]. For instance, in yeast, the critical component of the HR machinery, Rad52 is generally excluded from the nucleolus where the rDNA is located [52]. In addition, rDNA double strand breaks associate with Rad52 only after a transient exit of the affected rDNA region from the nucleolus. These restrictions ensure rDNA stability and are at least in part mediated by sumolylation of Rad52. Interestingly, in mouse G0 and S cells Rad52 is enriched in the nucleolus suggesting it may also participate in HR of rDNA in mammals [53]. While mediators of HR such as Rad52 appear to have a restricted access and/or activity at rDNA, anti-recombinogenic regulators have been found in proximity of rDNA including yeast DNA helicase Srs2 [52]. Such an anti-recombinogenic activity also appears to stabilize human rDNA as its remarkable destabilization has been reported in cell lines from patients with the defective human DNA helicase BLM [25]. Like Srs2, BLM inhibits HR. While most components of the HR machinery are not expressed in normal mature mammalian brain (http://www.brain-map.org/), there are no data on the activity of these components in neurodegeneration. However, our results indicate that at least in AD and DLB such a pathway may become re-activated. This conclusion is in agreement with the reported expansion of telomeres in the AD hippocampus as HR is presumed to be a major positive regulator of telomere length in most if not all somatic cells where telomerase is not expressed [4, 43, 44].
Rates of HR in rDNA may be affected by changes in chromatin structure and Pol1 activity. For instance, loss of rDNA-condensin interactions or reduced repressive chromatin modifications including DNA- and histone- methylation as well as histone de-acetylation increase rDNA recombination and reduce its stability in yeast-, fly- and murine cells [54, 55] [31, 56, 57]. In yeast, loss of Pol1 activity due to deletions of its critical subunits or associated co-factors promotes rDNA instability [58, 59]. Such effects may be due to structural disruption of the nucleolus in response to Pol1 inhibition and subsequent changes to rDNA chromatin. Thus, in the neurodegeneration-affected human brain tissue, altered epigenetic status of rDNA and/or changes to Pol1 activity may also contribute to increased recombination of rDNA.
HR including that of rDNA is initiated by DNA damage [15, 30, 44]. DNA double strand breaks that are the major recombinogenic form of DNA damage have been recently reported in normal mouse forebrain neurons following periods of increased physiological neuronal activity [60]. Moreover, their repair appeared to be impaired in a mouse model of AD-like amyloidosis. Evidence for double strand break-associated phosphorylation of the histone variant H2AX has also been found in astrocytes from AD brain [61]. In addition, HR may be activated by bulky DNA adducts and single strand DNA gaps that are generated by the adduct removal via the nucleotide excision repair [44, 62]. Oxidative DNA damage is well documented in both AD and DLB [40, 63] and oxidative bulky DNA adducts have been observed in the brain [64]. Hence, in the degenerating brain, HR and the subsequent destabilization of rDNA may be a result of excessive DNA damage.
At this stage one can only speculate about possible consequences of rDNA destabilization in human cells including those in the brain. The reported changes in rDNA content are unlikely to affect ribosomal biogenesis as only a fraction of rDNA units is required for efficient ribosome supply even during accelerated growth in development [65, 66]. Conversely, in the fly, the heterochromatin content appeared to be directly influenced by the number of rDNA copies independently of ribosomal biogenesis suggesting that rDNA instability may affect heterochromatin maintenance in mammalian cells [11]. In support of such a notion, in a mouse cell line, loss of heterochromatin coincided with loss of rDNA [31]. This occurred in response to a deficiency in rDNA silencing by the nucleolar repressive complex (NoRC) [31]. These observations indicate that the silenced, inactive copies of rDNA play an important role in maintenance of the heterochromatin. In turn, the rDNA-linked changes in heterochromatin content could affect expression of nearby genes and alter global genomic stability [67]. Finally, structural and functional connections between the heterochromatin and rDNA are suggested by physical interactions between these two regions in interphase nuclei and by a direct involvement of NoRC in heterochromatin formation [68, 69].
Interestingly, flies with variation in rDNA copy number also have functional changes to their euchromatin including relatively small but widespread effects on gene expression [70]. Such changes were not only observed in the engineered mutants of rDNA but were also found in flies with natural variation in rDNA copy number. Interestingly, genes involved in mitochondrial function and lipid metabolism were particularly affected by rDNA copy number. Such effects would fit well with the AD- and DLB-associated dysfunction of the mitochondria and/or dysregulation of brain lipid homeostasis [71-74]
Last but not least, yeast studies led to a proposition that the inactive copies of rDNA sequester mediators of the cytotoxic DDR pathway [13, 26]. Hence, loss of rDNA copies could result in activation of such a pathway. Conversely, expansion of rDNA could increase its activation threshold making cells more resistant to DNA damage. In support of this possibility, yeast sensitivity to a genotoxic chemical, methyl methanesulfonate (MMS) was inversely proportional to the genomic rDNA content [13]. Of note, in mammalian cells, stress-induced sequestration of various proteins have been demonstrated by the rRNA non-coding regions of rDNA [14]. Therefore, changes in rDNA copy number may affect cellular stress response including that to DNA damage.
To firmly establish and characterize the association of rDNA instability with neurodegeneration, our findings should be repeated on bigger cohorts of control-, DLB-, and AD individuals and sample more brain regions and non-neuronal tissues. Presence of rDNA instability in other neurodegenerative diseases should also be examined. As rDNA instability is documented not only in neurodegeneration but also in cancer, identifying its causes and consequences deserves attention from the research community. In the meantime, a working model can be proposed where the apparent instability of rDNA in the degenerating brain is a result of DNA damage-induced HR (Fig. 7). The altered number of rDNA copies would then affect genomic heterochromatin levels, expression of euchromatin-located genes and/or the DDR. If the rDNA content increases, heterochromatin levels go up, the rDNA influence over expressed genes is stronger and DDR resistance is higher (Fig. 7). The opposite may be true if rDNA content declines (Fig.7). Thus the observed increases of rDNA in the degeneration-affected cerebral cortex may contribute to compensation for the cellular damage/distress and promote cell survival. Such an effect could explain the apparent discrepancy between the cerebellar- and the cerebro-cortical rDNA content in DLB. Namely, as in DLB, the cerebellum has no obvious pathology/cell loss, there would be no selection factor against “inferior” genomic arrangements and the major end result of rDNA instability will be overall reduction of rDNA content. Conversely, in the cortex, cell death would produce enrichment of most resistant cells with expanded rDNA. Likewise, one could speculate that rDNA copy number variability may be an important factor increasing the ability of cancer cells to adapt to changes in cellular environment including therapy resistance.
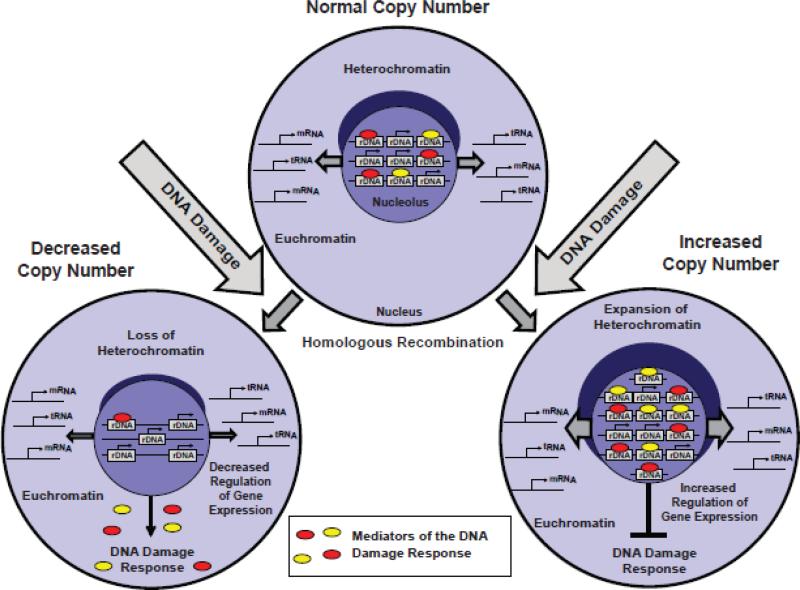
Figure 7. A hypothetical model summarizing possible causes and consequences of rDNA instability.
The DNA damage-stimulated homologous recombination (HR) is a likely cause of rDNA instability in the degenerating brain. The moderate changes in rDNA copy number as those reported here are not expected to significantly affect ribosomal biogenesis. Instead, ribosomal biogenesis-independent functions of rDNA may be affected including control of the DNA damage response (DDR), maintenance of heterochromatin, and expression regulation of the non-rRNA genes that are located in the euchromatin. One can speculate that higher content of rDNA may support cell survival by suppressing the DDR, stabilizing chromatin and stronger regulatory control of the euchromatic genes. Conversely, lower rDNA content may have opposite effects on cell sensitivity to DNA damage as well as other forms of stress by enhancing the DDR, reducing control over euchromatin gene expression and destabilizing the chromatin architecture. Thus, rDNA amplification in the degenerating regions of the DLB- or AD brain may be a consequence of increased genotoxic stress. In turn, cells with the higher genomic content of rDNA may become overrepresented due to lower sensitivity to subsequent injuries.
In summary, we did not find any indication for brain rDNA instability in aging. Conversely, we have obtained an initial evidence of DLB-associated rDNA instability in the brain including an apparent rDNA expansion in the neurodegeneration-affected cerebral cortex. While these findings raise many questions on the origin and the role of rDNA instability in aging-associated neurodegenerative diseases, they also suggest that HR may become activated in these conditions. Hence, the reported rDNA instability may be a component of a wider shift towards genomic disorganization with hyper-recombination of homologous sequences being one of its causes.
Highlights.
Brain rDNA instability is shown in Dementia with Lewy Bodies but not in aging.
These data suggest activation of homologous recombination in neurodegeneration.
Methylation of the rDNA promoter is not affected by Dementia with Lewy Bodies.
Causes and consequences of rDNA instability are discussed.
Acknowledgments
We are grateful for the research volunteers that contributed to this work. This work was supported by NIH (NS073584-01 and 8P30GM103507 to MH; CA152158 to GR), NSF (IOS1021860 to MH; DMS1318886 to GR), The Commonwealth of Kentucky Challenge for Excellence, and the Kentucky Spinal Cord and Head Injury Research Trust. The authors wish to thank Ms. Jing-Juan Zheng for excellent technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations- ALB- Albumin; AD- Alzheimer's Disease; CpG- cytosine-phosphate-guanine; DLB- Dementia with Lewy Bodies; DDR- DNA Damage Response; HR- Homologous recombination; tRNAK-ctt- Lysine-ctt tRNA; MCI- Mild Cognitive Impairment; MMSE- Minimental State Examination; NoRC- nucleolar repressive complex; PD- Parkinson's Disease; PMI- Post Mortem Interval; rDNA- Ribosomal DNA; rRNA- Ribosomal RNA; qPCR- quantitative real time polymerase chain reaction; SAM- significance analysis of microarrays.
References
- 1.Katyal S, McKinnon PJ. DNA strand breaks, neurodegeneration and aging in the brain. Mech. Ageing Dev. 2008;129:483–491. doi: 10.1016/j.mad.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lombard DB, Chua KF, Mostoslavsky R, Franco S, Gostissa M, Alt FW. DNA repair, genome stability, and aging. Cell. 2005;120:497–512. doi: 10.1016/j.cell.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 3.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas P, NJ OC, Fenech M. Telomere length in white blood cells, buccal cells and brain tissue and its variation with ageing and Alzheimer's disease. Mech. Ageing Dev. 2008;129:183–190. doi: 10.1016/j.mad.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Kume K, Kikukawa M, Hanyu H, Takata Y, Umahara T, Sakurai H, Kanetaka H, Ohyashiki K, Ohyashiki JH, Iwamoto T. Telomere length shortening in patients with dementia with Lewy bodies. Eur. J. Neurol. 2012;19:905–910. doi: 10.1111/j.1468-1331.2011.03655.x. [DOI] [PubMed] [Google Scholar]
- 6.Cai Z, Yan LJ, Ratka A. Telomere shortening and Alzheimer's disease. Neuromolecular Med. 2013;15:25–48. doi: 10.1007/s12017-012-8207-9. [DOI] [PubMed] [Google Scholar]
- 7.Rolyan H, Scheffold A, Heinrich A, Begus-Nahrmann Y, Langkopf BH, Holter SM, Vogt-Weisenhorn DM, Liss B, Wurst W, Lie DC, Thal DR, Biber K, Rudolph KL. Telomere shortening reduces Alzheimer's disease amyloid pathology in mice. Brain. 2011;134:2044–2056. doi: 10.1093/brain/awr133. [DOI] [PubMed] [Google Scholar]
- 8.McStay B, Grummt I. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu. Rev. Cell Dev. Biol. 2008;24:131–157. doi: 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- 9.Sirri V, Urcuqui-Inchima S, Roussel P, Hernandez-Verdun D. Nucleolus: the fascinating nuclear body. Histochem Cell Biol. 2008;129:13–31. doi: 10.1007/s00418-007-0359-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drygin D, Rice WG, Grummt I. The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Annu. Rev. Pharmacol. Toxicol. 2010;50:131–156. doi: 10.1146/annurev.pharmtox.010909.105844. [DOI] [PubMed] [Google Scholar]
- 11.Paredes S, Maggert KA. Ribosomal DNA contributes to global chromatin regulation. Proc. Natl. Acad. Sci. U. S. A. 2009;106:17829–17834. doi: 10.1073/pnas.0906811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santoro R, Schmitz KM, Sandoval J, Grummt I. Intergenic transcripts originating from a subclass of ribosomal DNA repeats silence ribosomal RNA genes in trans. EMBO Rep. 2010;11:52–58. doi: 10.1038/embor.2009.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ide S, Miyazaki T, Maki H, Kobayashi T. Abundance of ribosomal RNA gene copies maintains genome integrity. Science. 2010;327:693–696. doi: 10.1126/science.1179044. [DOI] [PubMed] [Google Scholar]
- 14.Audas TE, Jacob MD, Lee S. Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol. Cell. 2012;45:147–157. doi: 10.1016/j.molcel.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 15.Tsang E, Carr AM. Replication fork arrest, recombination and the maintenance of ribosomal DNA stability. DNA Repair (Amst) 2008;7:1613–1623. doi: 10.1016/j.dnarep.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 16.Johnson R, Strehler BL. Loss of genes coding for ribosomal RNA in ageing brain cells. Nature. 1972;240:412–414. doi: 10.1038/240412a0. [DOI] [PubMed] [Google Scholar]
- 17.Gaubatz J, Prashad N, Cutler RG. Ribosomal RNA gene dosage as a function of tissue and age for mouse and human. Biochim. Biophys. Acta. 1976;418:358–375. doi: 10.1016/0005-2787(76)90297-5. [DOI] [PubMed] [Google Scholar]
- 18.Peterson CR, Cryar JR, Gaubatz JW. Constancy of ribosomal RNA genes during aging of mouse heart cells and during serial passage of WI-38 cells. Arch. Gerontol. Geriatr. 1984;3:115–125. doi: 10.1016/0167-4943(84)90004-9. [DOI] [PubMed] [Google Scholar]
- 19.Halle JP, Muller S, Simm A, Adam G. Copy number, epigenetic state and expression of the rRNA genes in young and senescent rat embryo fibroblasts. Eur. J. Cell Biol. 1997;74:281–288. [PubMed] [Google Scholar]
- 20.Machwe A, Orren DK, Bohr VA. Accelerated methylation of ribosomal RNA genes during the cellular senescence of Werner syndrome fibroblasts. Faseb J. 2000;14:1715–1724. doi: 10.1096/fj.99-0926com. [DOI] [PubMed] [Google Scholar]
- 21.Zafiropoulos A, Tsentelierou E, Linardakis M, Kafatos A, Spandidos DA. Preferential loss of 5S and 28S rDNA genes in human adipose tissue during ageing. Int. J. Biochem. Cell Biol. 2005;37:409–415. doi: 10.1016/j.biocel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 22.Caburet S, Conti C, Schurra C, Lebofsky R, Edelstein SJ, Bensimon A. Human ribosomal RNA gene arrays display a broad range of palindromic structures. Genome Res. 2005;15:1079–1085. doi: 10.1101/gr.3970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stults DM, Killen MW, Pierce HH, Pierce AJ. Genomic architecture and inheritance of human ribosomal RNA gene clusters. Genome Res. 2008;18:13–18. doi: 10.1101/gr.6858507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stults DM, Killen MW, Williamson EP, Hourigan JS, Vargas HD, Arnold SM, Moscow JA, Pierce AJ. Human rRNA gene clusters are recombinational hotspots in cancer. Cancer Res. 2009;69:9096–9104. doi: 10.1158/0008-5472.CAN-09-2680. [DOI] [PubMed] [Google Scholar]
- 25.Killen MW, Stults DM, Adachi N, Hanakahi L, Pierce AJ. Loss of Bloom syndrome protein destabilizes human gene cluster architecture. Hum Mol Genet. 2009;18:3417–3428. doi: 10.1093/hmg/ddp282. Epub 2009 Jun 3419. [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi T. How does genome instability affect lifespan?: roles of rDNA and telomeres. Genes Cells. 2011;16:617–624. doi: 10.1111/j.1365-2443.2011.01519.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobayashi T. Regulation of ribosomal RNA gene copy number and its role in modulating genome integrity and evolutionary adaptability in yeast. Cell. Mol. Life Sci. 2011;68:1395–1403. doi: 10.1007/s00018-010-0613-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kennedy BK, Gotta M, Sinclair DA, Mills K, McNabb DS, Murthy M, Pak SM, Laroche T, Gasser SM, Guarente L. Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S-cerevisiae. Cell. 1997;89:381–391. doi: 10.1016/s0092-8674(00)80219-6. [DOI] [PubMed] [Google Scholar]
- 29.Ganley AR, Ide S, Saka K, Kobayashi T. The effect of replication initiation on gene amplification in the rDNA and its relationship to aging. Mol. Cell. 2009;35:683–693. doi: 10.1016/j.molcel.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 30.Eckert-Boulet N, Lisby M. Regulation of rDNA stability by sumoylation. DNA Repair (Amst) 2009;8:507–516. doi: 10.1016/j.dnarep.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 31.Guetg C, Lienemann P, Sirri V, Grummt I, Hernandez-Verdun D, Hottiger MO, Fussenegger M, Santoro R. The NoRC complex mediates the heterochromatin formation and stability of silent rRNA genes and centromeric repeats. EMBO J. 2010;29:2135–2146. doi: 10.1038/emboj.2010.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pietrzak M, Rempala G, Nelson PT, Zheng JJ, Hetman M. Epigenetic silencing of nucleolar rRNA genes in Alzheimer's disease. PLoS One. 2011;6:e22585. doi: 10.1371/journal.pone.0022585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nelson PT, Jicha GA, Schmitt FA, Liu H, Davis DG, Mendiondo MS, Abner EL, Markesbery WR. Clinicopathologic correlations in a large Alzheimer disease center autopsy cohort: neuritic plaques and neurofibrillary tangles “do count” when staging disease severity. J. Neuropathol. Exp. Neurol. 2007;66:1136–1146. doi: 10.1097/nen.0b013e31815c5efb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abdul HM, Sama MA, Furman JL, Mathis DM, Beckett TL, Weidner AM, Patel ES, Baig I, Murphy MP, LeVine H, 3rd, Kraner SD, Norris CM. Cognitive decline in Alzheimer's disease is associated with selective changes in calcineurin/NFAT signaling. J. Neurosci. 2009;29:12957–12969. doi: 10.1523/JNEUROSCI.1064-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nelson PT, Kryscio RJ, Jicha GA, Abner EL, Schmitt FA, Xu LO, Cooper G, Smith CD, Markesbery WR. Relative preservation of MMSE scores in autopsy-proven dementia with Lewy bodies. Neurology. 2009;73:1127–1133. doi: 10.1212/WNL.0b013e3181bacf9e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKeith I, Mintzer J, Aarsland D, Burn D, Chiu H, Cohen-Mansfield J, Dickson D, Dubois B, Duda JE, Feldman H, Gauthier S, Halliday G, Lawlor B, Lippa C, Lopez OL, Carlos Machado J, O'Brien J, Playfer J, Reid W. D.L.B. International Psychogeriatric Association Expert Meeting on, Dementia with Lewy bodies. Lancet Neurol. 2004;3:19–28. doi: 10.1016/s1474-4422(03)00619-7. [DOI] [PubMed] [Google Scholar]
- 38.Spillantini MG, Crowther RA, Jakes R, Hasegawa M, Goedert M. alpha-Synuclein in filamentous inclusions of Lewy bodies from Parkinson's disease and dementia with lewy bodies. Proc. Natl. Acad. Sci. U. S. A. 1998;95:6469–6473. doi: 10.1073/pnas.95.11.6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alam ZI, Jenner A, Daniel SE, Lees AJ, Cairns N, Marsden CD, Jenner P, Halliwell B. Oxidative DNA damage in the parkinsonian brain: an apparent selective increase in 8-hydroxyguanine levels in substantia nigra. J. Neurochem. 1997;69:1196–1203. doi: 10.1046/j.1471-4159.1997.69031196.x. [DOI] [PubMed] [Google Scholar]
- 40.Lyras L, Perry RH, Perry EK, Ince PG, Jenner A, Jenner P, Halliwell B. Oxidative damage to proteins, lipids, and DNA in cortical brain regions from patients with dementia with Lewy bodies. J. Neurochem. 1998;71:302–312. doi: 10.1046/j.1471-4159.1998.71010302.x. [DOI] [PubMed] [Google Scholar]
- 41.Tantravahi U, Guntaka RV, Erlanger BF, Miller OJ. Amplified ribosomal RNA genes in a rat hepatoma cell line are enriched in 5-methylcytosine. Proc. Natl. Acad. Sci. U. S. A. 1981;78:489–493. doi: 10.1073/pnas.78.1.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gaubatz JW, Cutler RG. Age-related differences in the number of ribosomal RNA genes of mouse tissues. Gerontology. 1978;24:179–207. doi: 10.1159/000212250. [DOI] [PubMed] [Google Scholar]
- 43.Mao Z, Bozzella M, Seluanov A, Gorbunova V. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle. 2008;7:2902–2906. doi: 10.4161/cc.7.18.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krejci L, Altmannova V, Spirek M, Zhao X. Homologous recombination and its regulation. Nucleic Acids Res. 2012;40:5795–5818. doi: 10.1093/nar/gks270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boekhoorn K, Joels M, Lucassen PJ. Increased proliferation reflects glial and vascular-associated changes, but not neurogenesis in the presenile Alzheimer hippocampus. Neurobiol. Dis. 2006;24:1–14. doi: 10.1016/j.nbd.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 46.Robel S, Berninger B, Gotz M. The stem cell potential of glia: lessons from reactive gliosis. Nat. Rev. Neurosci. 2011;12:88–104. doi: 10.1038/nrn2978. [DOI] [PubMed] [Google Scholar]
- 47.Yang Y, Mufson EJ, Herrup K. Neuronal cell death is preceded by cell cycle events at all stages of Alzheimer's disease. J. Neurosci. 2003;23:2557–2563. doi: 10.1523/JNEUROSCI.23-07-02557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Y, Geldmacher DS, Herrup K. DNA replication precedes neuronal cell death in Alzheimer's disease. J. Neurosci. 2001;21:2661–2668. doi: 10.1523/JNEUROSCI.21-08-02661.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Busser J, Geldmacher DS, Herrup K. Ectopic cell cycle proteins predict the sites of neuronal cell death in Alzheimer's disease brain. J. Neurosci. 1998;18:2801–2807. doi: 10.1523/JNEUROSCI.18-08-02801.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gandhi M, Evdokimova VN, Cuenco KT, Bakkenist CJ, Nikiforov YE. Homologous chromosomes move and rapidly initiate contact at the sites of double-strand breaks in genes in G(0)-phase human cells. Cell Cycle. 2013;12:547–552. doi: 10.4161/cc.23754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gandhi M, Evdokimova VN, K TC, Nikiforova MN, Kelly LM, Stringer JR, Bakkenist CJ, Nikiforov YE. Homologous chromosomes make contact at the sites of double-strand breaks in genes in somatic G0/G1-phase human cells. Proc. Natl. Acad. Sci. U. S. A. 2012;109:9454–9459. doi: 10.1073/pnas.1205759109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Torres-Rosell J, Sunjevaric I, De Piccoli G, Sacher M, Eckert-Boulet N, Reid R, Jentsch S, Rothstein R, Aragon L, Lisby M. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat. Cell Biol. 2007;9:923–931. doi: 10.1038/ncb1619. [DOI] [PubMed] [Google Scholar]
- 53.Liu Y, Li M, Lee EY, Maizels N. Localization and dynamic relocalization of mammalian Rad52 during the cell cycle and in response to DNA damage. Curr. Biol. 1999;9:975–978. doi: 10.1016/s0960-9822(99)80427-8. [DOI] [PubMed] [Google Scholar]
- 54.Kobayashi T, Ganley AR. Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science. 2005;309:1581–1584. doi: 10.1126/science.1116102. [DOI] [PubMed] [Google Scholar]
- 55.Mekhail K, Seebacher J, Gygi SP, Moazed D. Role for perinuclear chromosome tethering in maintenance of genome stability. Nature. 2008;456:667–670. doi: 10.1038/nature07460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peng JC, Karpen GH. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat. Cell Biol. 2007;9:25–35. doi: 10.1038/ncb1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Larson K, Yan SJ, Tsurumi A, Liu J, Zhou J, Gaur K, Guo D, Eickbush TH, Li WX. Heterochromatin formation promotes longevity and represses ribosomal RNA synthesis. PLoS Genet. 2012;8:e1002473. doi: 10.1371/journal.pgen.1002473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kobayashi T, Heck DJ, Nomura M, Horiuchi T. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 1998;12:3821–3830. doi: 10.1101/gad.12.24.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsang CK, Zheng XF. Opposing role of condensin and radiation-sensitive gene RAD52 in ribosomal DNA stability regulation. J. Biol. Chem. 2009;284:21908–21919. doi: 10.1074/jbc.M109.031302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suberbielle E, Sanchez PE, Kravitz AV, Wang X, Ho K, Eilertson K, Devidze N, Kreitzer AC, Mucke L. Physiologic brain activity causes DNA double-strand breaks in neurons, with exacerbation by amyloid-beta. Nat. Neurosci. 2013;16:613–621. doi: 10.1038/nn.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Myung NH, Zhu X, Kruman II, Castellani RJ, Petersen RB, Siedlak SL, Perry G, Smith MA, Lee HG. Evidence of DNA damage in Alzheimer disease: phosphorylation of histone H2AX in astrocytes. Age (Dordr) 2008;30:209–215. doi: 10.1007/s11357-008-9050-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ma W, Westmoreland JW, Resnick MA. Homologous recombination rescues ssDNA gaps generated by nucleotide excision repair and reduced translesion DNA synthesis in yeast G2 cells. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E2895–2904. doi: 10.1073/pnas.1301676110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Markesbery WR, Lovell MA. DNA oxidation in Alzheimer's disease. Antioxid Redox Signal. 2006;8:2039–2045. doi: 10.1089/ars.2006.8.2039. [DOI] [PubMed] [Google Scholar]
- 64.Brooks PJ. The 8,5′-cyclopurine-2′-deoxynucleosides: candidate neurodegenerative DNA lesions in xeroderma pigmentosum, and unique probes of transcription and nucleotide excision repair. DNA Repair (Amst) 2008;7:1168–1179. doi: 10.1016/j.dnarep.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Delany ME, Muscarella DE, Bloom SE. Effects of rRNA gene copy number and nucleolar variation on early development: inhibition of gastrulation in rDNA-deficient chick embryos. J. Hered. 1994;85:211–217. doi: 10.1093/oxfordjournals.jhered.a111437. [DOI] [PubMed] [Google Scholar]
- 66.Paredes S, Maggert KA. Expression of I-CreI endonuclease generates deletions within the rDNA of Drosophila. Genetics. 2009;181:1661–1671. doi: 10.1534/genetics.108.099093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- 68.Nemeth A, Conesa A, Santoyo-Lopez J, Medina I, Montaner D, Peterfia B, Solovei I, Cremer T, Dopazo J, Langst G. Initial genomics of the human nucleolus. PLoS Genet. 2010;6:e1000889. doi: 10.1371/journal.pgen.1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Postepska-Igielska A, Krunic D, Schmitt N, Greulich-Bode KM, Boukamp P, Grummt I. The chromatin remodelling complex NoRC safeguards genome stability by heterochromatin formation at telomeres and centromeres. EMBO Rep. 2013;14:704–710. doi: 10.1038/embor.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Paredes S, Branco AT, Hartl DL, Maggert KA, Lemos B. Ribosomal DNA deletions modulate genome-wide gene expression: “rDNA-sensitive” genes and natural variation. PLoS Genet. 2011;7:e1001376. doi: 10.1371/journal.pgen.1001376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cutler RG, Kelly J, Storie K, Pedersen WA, Tammara A, Hatanpaa K, Troncoso JC, Mattson MP. Involvement of oxidative stress-induced abnormalities in ceramide and cholesterol metabolism in brain aging and Alzheimer's disease. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2070–2075. doi: 10.1073/pnas.0305799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reddy PH, Beal MF. Are mitochondria critical in the pathogenesis of Alzheimer's disease? Brain Res. Brain Res. Rev. 2005;49:618–632. doi: 10.1016/j.brainresrev.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 73.Bosco DA, Fowler DM, Zhang Q, Nieva J, Powers ET, Wentworth P, Jr., Lerner RA, Kelly JW. Elevated levels of oxidized cholesterol metabolites in Lewy body disease brains accelerate alpha-synuclein fibrilization. Nat. Chem. Biol. 2006;2:249–253. doi: 10.1038/nchembio782. [DOI] [PubMed] [Google Scholar]
- 74.Navarro A, Boveris A, Bandez MJ, Sanchez-Pino MJ, Gomez C, Muntane G, Ferrer I. Human brain cortex: mitochondrial oxidative damage and adaptive response in Parkinson disease and in dementia with Lewy bodies. Free Radic. Biol. Med. 2009;46:1574–1580. doi: 10.1016/j.freeradbiomed.2009.03.007. [DOI] [PubMed] [Google Scholar]