Abstract
The recent discovery of mutations in metabolic enzymes has rekindled interest in harnessing the altered metabolism of cancer cells for cancer therapy. One potential drug target is isocitrate dehydrogenase 1 (IDH1), which is mutated in multiple human cancers. Here, we examine the role of mutant IDH1 in fully transformed cells with endogenous IDH1 mutations. A selective R132H-IDH1 inhibitor (AGI-5198) identified through a high-throughput screen blocked, in a dose-dependent manner, the ability of the mutant enzyme (mIDH1) to produce R-2-hydroxyglutarate (R-2HG). Under conditions of near-complete R-2HG inhibition, the mIDH1 inhibitor induced demethylation of histone H3K9me3 and expression of genes associated with gliogenic differentiation. Blockade of mIDH1 impaired the growth of IDH1-mutant—but not IDH1–wild-type—glioma cells without appreciable changes in genome-wide DNA methylation. These data suggest that mIDH1 may promote glioma growth through mechanisms beyond its well-characterized epigenetic effects.
Somatic mutations in the metabolic enzyme isocitrate dehydrogenase (IDH) have recently been identified in multiple human cancers, including glioma (1, 2), sarcoma (3, 4), acute myeloid leukemia (5, 6), and others. All mutations map to arginine residues in the catalytic pockets of IDH1 (R132) or IDH2 (R140 and R172) and confer on the enzymes a new activity: catalysis of alpha-ketoglutarate (2-OG) to the (R)-enantiomer of 2-hydroxyglutarate (R-2HG) (7, 8). R-2HG is structurally similar to 2-OG and, due to its accumulation to millimolar concentrations in IDH1-mutant tumors, competitively inhibits 2-OG–dependent dioxygenases (9).
The mechanism by which mutant IDH1 contributes to the pathogenesis of human glioma remains incompletely understood. Mutations in IDH1 are found in 50 to 80% of human low-grade (WHO grade II) glioma, a disease that progresses to fatal WHO grade III (anaplastic glioma) and WHO grade IV (glioblastoma) tumors over the course of 3 to 15 years. IDH1 mutations appear to precede the occurrence of other mutations (10) and are associated with a distinctive gene-expression profile (“proneural” signature), DNA hypermethylation [CpG island methylator phenotype (CIMP)], and certain clinicopathological features (11–13). When ectopically expressed in immortalized human astrocytes, R132H-IDH1 promotes the growth of these cells in soft agar (14) and induces epigenetic alterations found in IDH1-mutant human gliomas (15, 16). However, no tumor formation was observed when R132H-IDH1 was expressed from the endogenous IDH1 locus in several cell types of the murine central nervous system (17).
To explore the role of mutant IDH1 in tumor maintenance, we used a compound that was identified in a high-throughput screen for compounds that inhibit the IDH1-R132H mutant homodimer (fig. S1 and supplementary materials) (18). This compound, subsequently referred to as AGI-5198 (Fig. 1A), potently inhibited mutant IDH1 [R132H-IDH1; half-maximal inhibitory concentration (IC50), 0.07 µM) but not wild-type IDH1 (IC50 > 100 µM) or any of the examined IDH2 isoforms (IC50 > 100 µM) (Fig. 1B). We observed no induction of nonspecific cell death at the highest examined concentration of AGI-5198 (20 µM).
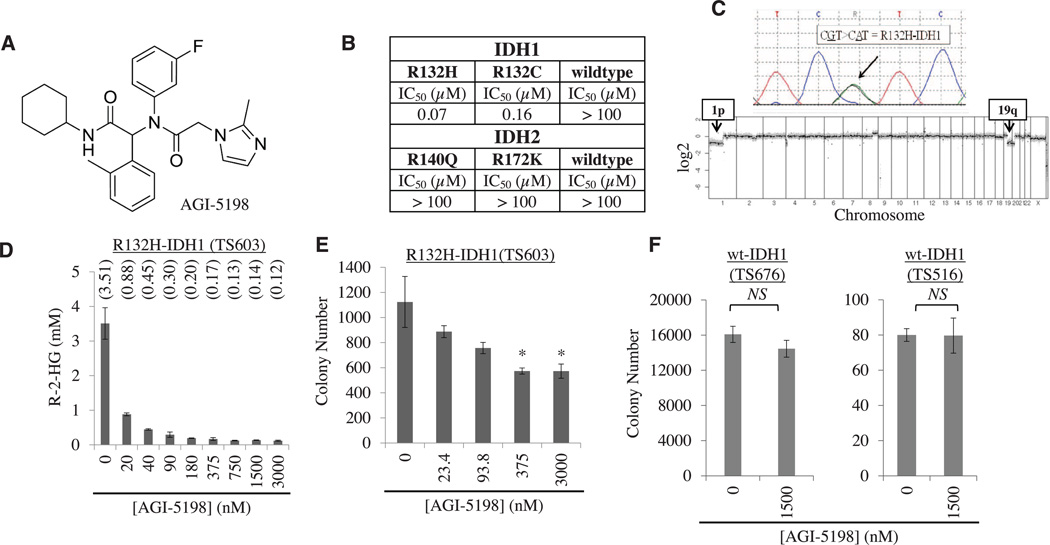
Fig. 1. An R132H-IDH1 inhibitor blocks R-2HG production and soft-agar growth of IDH1-mutant glioma cells.
(A) Chemical structure of AGI-5198. (B) IC50 of AGI-5198 against different isoforms of IDH1 and IDH2, measured in vitro. (C) Sanger sequencing chromatogram (top) and comparative genomic hybridization profile array (bottom) of TS603 glioma cells. (D) AGI-5198 inhibits R-2HG production in R132H-IDH1 mutant TS603 glioma cells. Cells were treated for 2 days with AGI-5198, and R-2HG was measured in cell pellets. R-2HG concentrations are indicated above each bar (in mM). Error bars, mean ± SEM of triplicates. (E and F) AGI-5198 impairs soft-agar colony formation of (E) IDH1-mutant TS603 glioma cells [*P < 0.05, one-way analysis of variance (ANOVA)] but not (F) IDH1–wild-type glioma cell lines (TS676 and TS516). Error bars, mean ± SEM of triplicates.
We next explored the activity of AGI-5198 in TS603 glioma cells with an endogenous heterozygous R132H-IDH1 mutation, the most common IDH mutation in glioma (2). TS603 cells were derived from a patient with anaplastic oligodendroglioma (WHO grade III) and harbor another pathognomomic lesion for this glioma subtype, namely co-deletion of the short arm of chromosome 1 (1p) and the long arm of chromosome 19 (19q) (19) (Fig. 1C). Measurements of R-2HG concentrations in pellets of TS603 glioma cells demonstrated dose-dependent inhibition of the mutant IDH1 enzyme by AGI-5198 (Fig. 1D). When added to TS603 glioma cells growing in soft agar, AGI-5198 inhibited colony formation by 40 to 60% (Fig. 1E). AGI-5198 did not impair colony formation of two patient-derived glioma lines that express only the wild-type IDH1 allele (TS676 and TS516) (Fig. 1F), further supporting the selectivity of AGI-5198.
After exploratory pharmacokinetic studies in mice (fig. S2), we examined the effects of orally administered AGI-5198 on the growth of human glioma xenografts. When given daily to mice with established R132H-IDH1 glioma xenografts, AGI-5198 [450 mg per kg of weight (mg/kg) per os] caused 50 to 60% growth inhibition (Fig. 2A). Treatment was tolerated well with no signs of toxicity during 3 weeks of daily treatment (fig. S3). Tumors from AGI-5198– treated mice showed reduced staining with an antibody against the Ki-67 protein, a marker used for quantification of tumor cell proliferation in human brain tumors. In contrast, staining with an antibody against cleaved caspase-3 showed no differences between tumors from vehicle and AGI-5198–treated mice (fig. S4), suggesting that the growth-inhibitory effects of AGI-5198 were primarily due to impaired tumor cell proliferation rather than induction of apoptotic cell death. AGI-5198 did not affect the growth of IDH1 wild-type glioma xenografts (Fig. 2B).
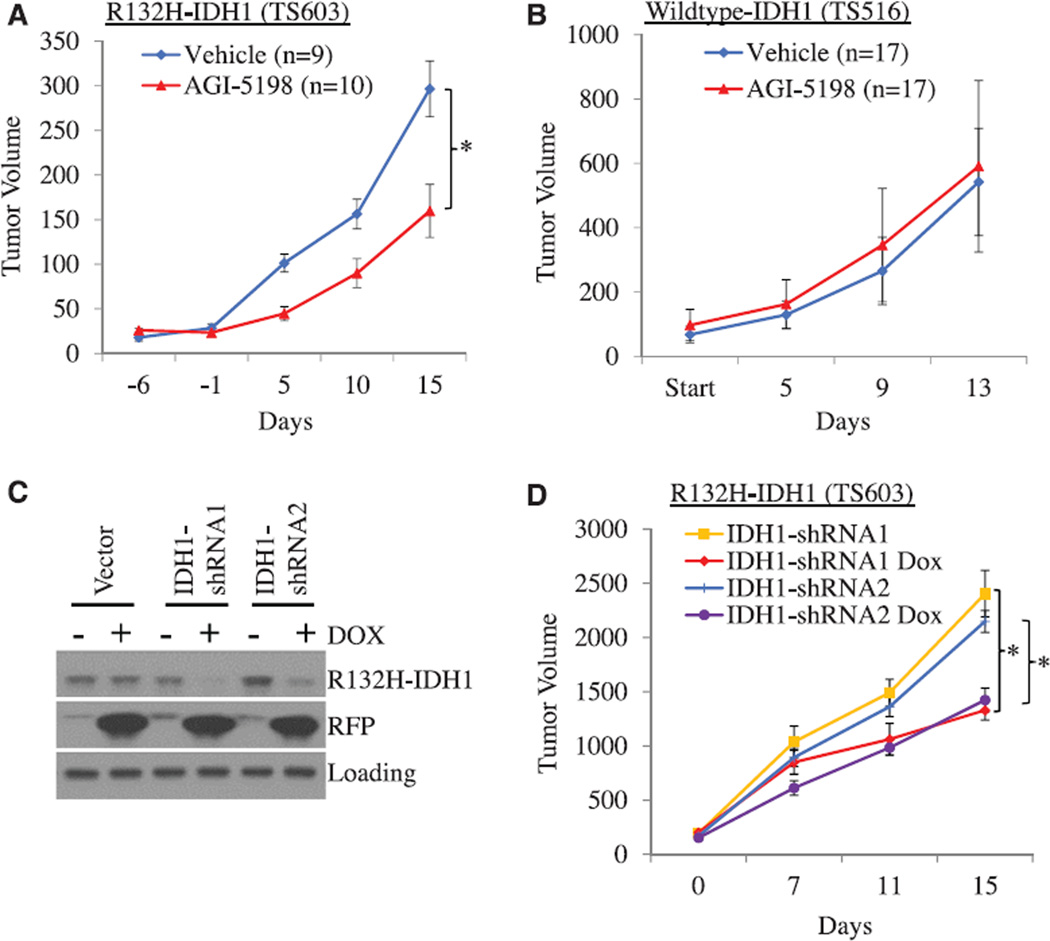
Fig. 2. AGI-5198 impairs growth of IDH1-mutant glioma xenografts in mice.
(A) AGI-5198 impairs growth of IDH1±mutant TS603 glioma xenografts (*P = 0.015, two-tailed t test). Error bars, mean ± SEM. (B) AGI-5198 does not impair the growth of IDH1–wild-type TS516 glioma xenografts. Treatment with AGI-5198 (450 mg/kg per day via gavage) was started after subcutaneous xenografts were established. Error bars, mean ± SEM. (C) Inducible knockdown of IDH1 in IDH1-mutant glioma cells. Shown are Western blots from TS603 cells engineered to express either empty vector or IDH1-shRNAs from a doxycycline-inducible cassette and treated with doxycycline (2.5 µg/mL) for 6 days before lysis. (D) IDH1 knockdown impairs growth of IDH1-mutant glioma xenografts. Mice were randomized to vehicle versus doxycycline after subcutaneous tumors were established (*P < 0.05, two-tailed t test; n = 15 mice per cohort). Error bars, mean ± SEM.
Given the likely prominent role of R-2HG in the pathogenesis of IDH-mutant human cancers, we investigated whether intratumoral depletion of this metabolite would have similar growth inhibitory effects on R132H-IDH1-mutant glioma cells as AGI-5198. We engineered TS603 sublines in which IDH1–short hairpin RNA (shRNA) targeting sequences were expressed from a doxycycline-inducible cassette. Doxycycline had no effect on IDH1 protein levels in cells expressing the vector control but depleted IDH1 protein levels by 60 to 80% in cells infected with IDH1-shRNA targeting sequences (Fig. 2C). We next injected these cells into the flanks of mice with severe combined immunodeficiency and, after establishment of subcutaneous tumors, randomized the mice to receive either regular chow or doxycycline-containing chow. As predicted from our experiments with AGI-5198, doxycycline impaired the growth of TS603 glioma cells expressing inducible IDH1-shRNAs in soft agar (fig. S5) and in vivo (Fig. 2D) but had no effect on the growth of tumors expressing the vector control (fig. S6). Immunohistochemistry (IHC) with a mutant-specific R132H-IDH1 antibody confirmed depletion of the mutant IDH1 protein in IDH1-shRNA tumors treated with doxycycline. This was associated with an 80 to 90% reduction in intratumoral R-2HG levels, similar to the levels observed in TS603 glioma xenografts treated with AGI-5198 (fig. S7). Knockdown of the IDH1 protein in R132C-IDH1-mutant HT1080 sarcoma cells similarly impaired the growth of these cells in vitro and in vivo (fig. S8).
To explore candidate molecular mechanisms of tumor growth inhibition by AGI-5198, we isolated RNA from the glioma xenografts in vehicle- and AGI-5198–treated mice, hybridized it to Affymetrix U133 plus 2.0 gene-expression arrays, and queried the RNA expression data for genes with a ≥2.0-fold increase or decrease in RNA expression in response to AGI-5198 (Fig. 3A) (table S1). The list of genes induced by AGI-5198 included several genes that are associated with astrocytic (aquaporin-4-AQP4; adenosine triphosphatase, Na+/K+ transporting, alpha 2 polypeptide-ATP1A2) (20, 21) and oligodendrocytic differentiation [prostaglandin D2 synthase 21 kD (brain)-PTGDS] in the central nervous system (21). AGI-5198 also induced the expression of zinc finger and BTB domain– containing protein 16 (ZBTB16) [also known as promyelocytic leukemia zinc finger (PLZF)], a transcriptional repressor protein that is located on chromosome 11q23 and has been shown to promote glial differentiation in the central nervous system (22).
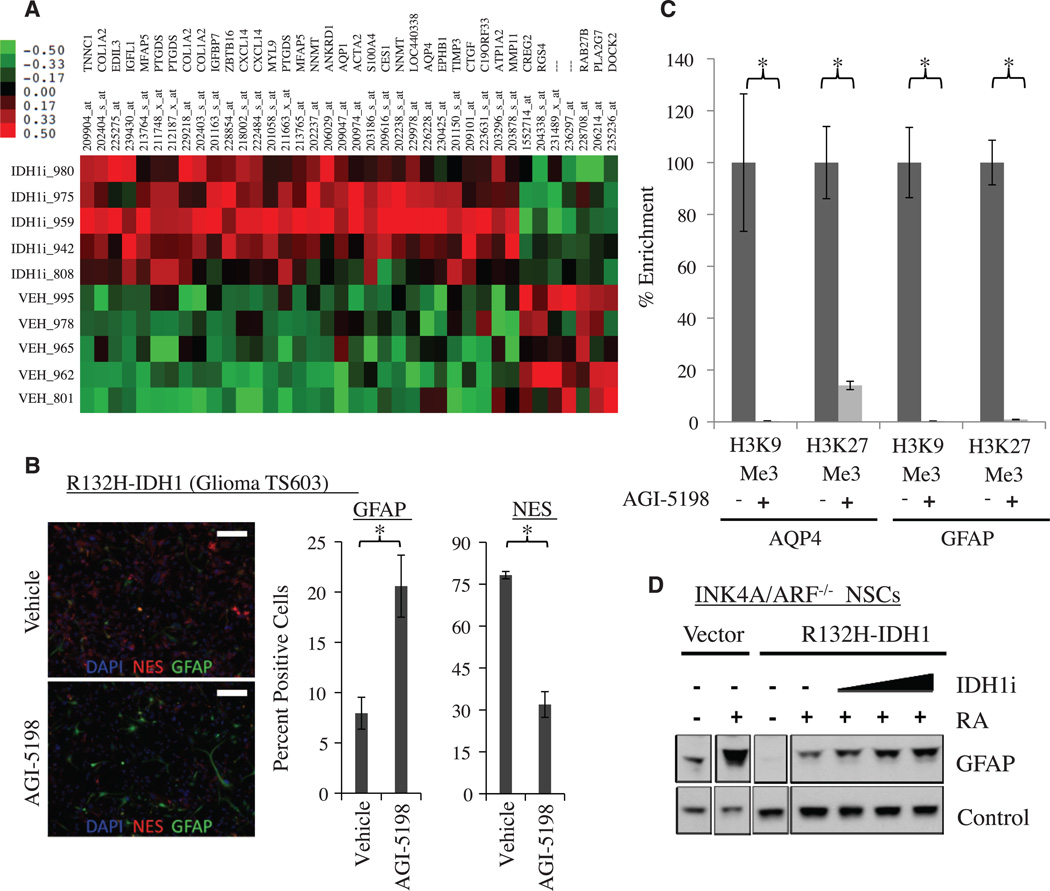
Fig. 3. AGI-5198 promotes astroglial differentiation in R132H-IDH1 mutant cells.
(A) Heat map of genes that are up- or down-regulated in TS603 glioma xenografts treated with AGI-5198 (more than twofold up or down). (B) Increased expression of GFAP (green) and decreased expression of NES (red) in TS603 cells treated in vitro with AGI-5198. Shown are immunofluorescence images of cells incubated in 1% fetal bovine serum (FBS) and 1 µM retinoic acid for 7 days in the presence of vehicle (top) or 1.5 µM AGI-5198 (bottom). Scale bar, 200 µM. The bar graph on the right represents a quantification of GFAP and NES staining (*P < 0.05, two-tailed t test). DAPI (4´,6-diamidino-2-phenylindole) staining in blue. Error bars, mean ± SEM of triplicates. (C) AGI-5198 promotes removal of repressive H3K9me3 and H3K27me3 marks at the GFAP and AQP4 promoters. Shown is ChIP (percent enrichment normalized to vehicle) of TS603 cells grown for 7 days in FBS (1%) and retinoic acid (1 µM) in the presence of vehicle or 1.5 µM AGI-5198 (*P < 0.05, two-tailed t test). Error bars, mean ± SEM for four repeats. (D) Blockade of mIDH1 restores the ability of R132HIDH1 mutant Ink4a/Arf−/− murine neuroprogenitor cells (NPCs) to express GFAP in response to retinoic acid. Shown is a Western blot of parental (vector) and R132H-IDH1 expressing Ink4a/Arf−/− NPCs treated with 1 µM retinoic acid (RA) in the presence of vehicle or mIDH1 inhibitor.
The gene-expression data suggested that treatment of IDH1-mutant glioma xenografts with AGI-5198 promotes a gene-expression program akin to gliogenic (i.e., astrocytic and oligodendrocytic) differentiation. To examine this question further, we treated TS603 glioma cells ex vivo with AGI-5198 and performed immunofluorescence for glial fibrillary acidic protein (GFAP) and nestin (NES) as markers for astrocytes and undifferentiated neuroprogenitor cells, respectively. Compared with vehicle, AGI-5198 treatment markedly increased the fraction of GFAP-positive cells and reduced the fraction of NES-positive cells (Fig. 3B). Chromatin-immunoprecipitation (ChIP) experiments showed that induction of the astrocytic markers GFAP and AQP4 in response to AGI-5198 was associated with marked reduction in repressive H3K9me3 and H3K27me3 marks on the promoters of these genes (Fig. 3C). Previous work showed that R132H-IDH1 expression in INK4A/ARF−/− murine neuroprogenitor cells (NPCs) restricts the ability of these cells to express GFAP in response to the differentiation cue retinoic acid (23). We investigated whether blockade of mutant IDH1 could restore this ability, and this was indeed the case (Fig. 3D). These results indicate that mIDH1 plays an active role in restricting cellular differentiation potential, and this defect is acutely reversible by blockade of the mutant enzyme.
In the developing central nervous system, gliogenic differentiation is regulated through changes in DNA and histone methylation (24). Mutant IDH1 can affect both epigenetic processes through R-2HG mediated suppression of TET (ten-eleven translocation) methyl cytosine hydroxylases and Jumonji-C domain histone demethylases (JHDMs). We therefore sought to define the epigenetic changes that were associated with the acute growth-inhibitory effects of AGI-5198 in vivo. We included a lower dose of AGI-5198 (150 mg/kg) in these experiments because of the differential sensitivity of 2-OG–dependent dioxygenases to competitive inhibition by R-2HG (25). We first examined intratumoral R-2HG concentrations after 2 weeks of treatment. Tumors from vehicle-treated mice showed R-2HG concentrations in the 4- to 6-mM range, similar to the concentrations of this metabolite in IDH1-mutant gliomas (7). Treatment of mice with AGI-5198 resulted in dose-dependent reduction of intratumoral R-2HG with partial R-2HG reduction at the 150 mg/kg dose (0.85 ± 0.22 mM) and near-complete reduction at the 450 mg/kg dose (0.13 ± 0.03 mM) (Fig. 4A).
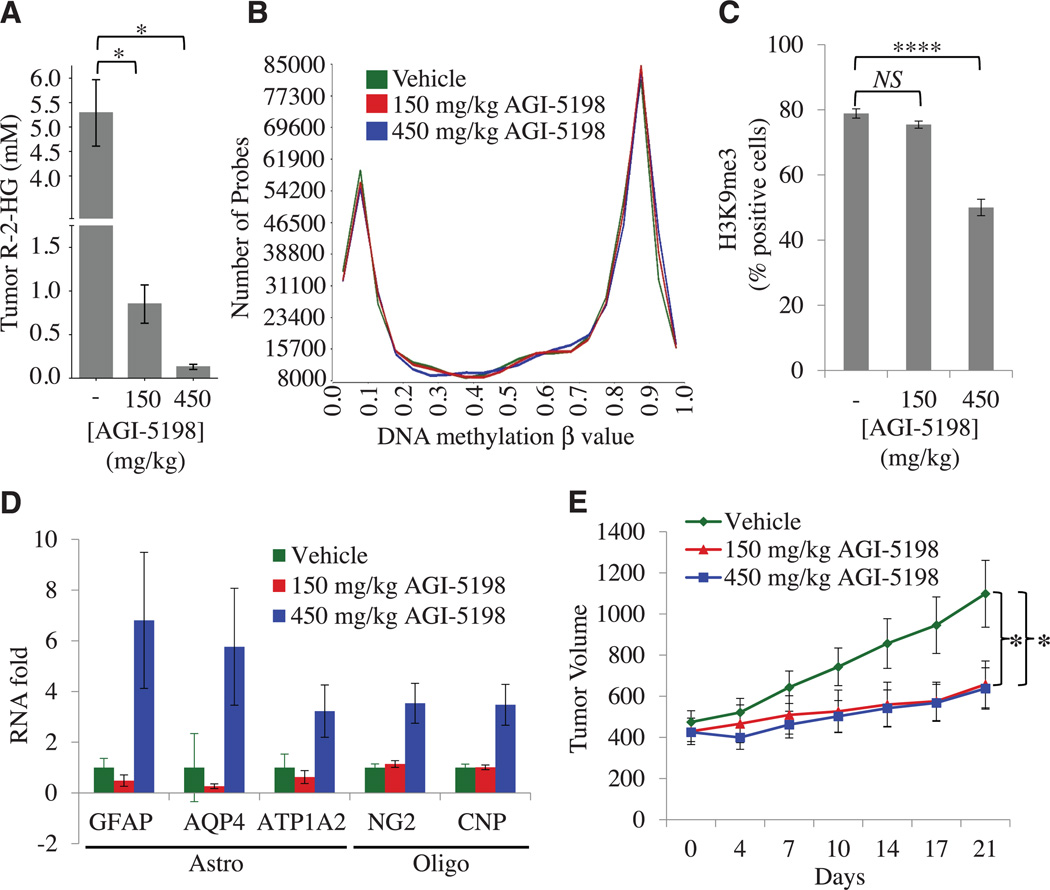
Fig. 4. Dose-dependent inhibition of histone methylation in IDH1-mutant gliomas after shortterm treatment with AGI-5198.
(A) Intratumoral concentrations of R-2HG in TS603 xenografts treated for 2 weeks with vehicle (n = 10 mice per cohort), 150 mg/kg AGI-5198 (n = 10 mice per cohort), or 450 mg/kg AGI-5198 (n = 8 mice per cohort) (*P < 0.05, two-tailed t test). Error bars, mean ± SEM. (B) Genome-wide distribution of DNA methylation in TS603 glioma xenografts treated for 2 weeks with vehicle, 150 mg/kg AGI-5198, or 450 mg/kg AGI-5198. (C) Effect of AGI-5198 on H3K9 trimethylation in TS603 glioma xenografts. Shown is the quantification of IHC results (****P < 0.00001) (see also fig. S11). Error bars, mean ± SEM of triplicates. (D) RNA levels of astrocytic (GFAP, AQP4, and ATP1A2) and oligodendrocytic (NG2 and CNP) differentiation markers in vehicle-versus AGI-5198– treated TS603 xenografts (P = 4 × 10−8, ANOVA). RNA expression was measured by RT-PCR. Error bars, mean ± SEM for 7 or 8 repeats (7 for vehicle, 8 for 150 mg/kg, and 7 for 450 mg/kg). (E) Tumor volumes of TS603 xenografts in mice treated with vehicle, 150 mg/kg AGI-5198, or 450 mg/kg AGI-5198 (*P < 0.05, two-tailed t test); n = 10 mice per cohort. Error bars, mean ± SEM.
We next examined whether acute pharmacological blockade of the mutant IDH1 enzyme reversed the CIMP, which is strongly associated with IDH1-mutant human gliomas (12). This CIMP phenotype is readily appreciated in untreated R132H-IDH1 TS603 glioma xenografts using genome-wide DNA methylation arrays (Illumina Infinium Human Methylation 450 Arrays) by their increased number of highly methylated probes (β value > 0.7) (fig. S9) and by their unsupervised clustering with IDH-mutant human glioblastomas (fig. S10A) and intermediate-grade gliomas (fig. S10B). On a genome-wide scale, we observed no statistically significant change in the distribution of β values between AGI-5198– and vehicle-treated tumors (Fig. 4B) (supplementary materials).
We next examined the kinetics of histone demethylation after inhibition of the mutant IDH1 enzyme. The histone demethylases JMJD2A and JMJD2C, which remove bi- and trimethyl marks from H3K9, are significantly more sensitive to inhibition by the R-2HG oncometabolite than other 2-OG–dependent oxygenases (8, 9, 14, 25). Restoring their enzymatic activity in IDH1-mutant cancer cells would thus be expected to require near-complete inhibition of R-2HG production. Consistent with this prediction, tumors from the 450 mg/kg AGI-5198 cohort showed a marked decrease in H3K9me3 staining, but there was no decrease in H3K9me3 staining in tumors from the 150 mg/kg AGI-5198 cohort (Fig. 4C) (fig. S11). Of note, AGI-5198 did not decrease H3K9 trimethylation in IDH1–wild-type glioma xenografts (fig. S12A) or in normal astrocytes (fig. S12B), demonstrating that the effect of AGI-5198 on histone methylation was not only dose-dependent but also IDH1-mutant selective.
Because the inability to erase repressive H3K9 methylation can be sufficient to impair cellular differentiation of nontransformed cells (16), we examined the TS603 xenograft tumors for changes in the RNA expression of astrocytic (GFAP, AQP4, and ATP1A2) and oligodendrocytic (CNP and NG2) differentiation markers by real-time polymerase chain reaction (RT-PCR). Compared with vehicletreated tumors, we observed an increase in the expression of astroglial differentiation genes only in tumors treated with 450 mg/kg AGI-5198 (Fig. 4D).
Despite its inability to reverse H3K9 trimethylation and induce gliogenic differentiation markers, the lower dose of AGI-5198 (150 mg/kg) resulted in a similar tumor growth inhibition as the higher dose of AGI-5198 (Fig. 4E) (fig. S13). Induction of the differentiation gene-expression program was thus associated with H3K9me3 demethylation but not required for tumor growth inhibition by AGI-5198, suggesting that mutant IDH regulated proliferation and differentiation in glioma through distinct effector programs with differential sensitivity, kinetic response, or reversibility to the R-2HG oncometabolite.
To identify pathways that are associated with the growth-inhibitory effects of AGI-5198, we ran Affymetrix RNA expression arrays. Many of the genes that showed significant changes in expression in both AGI-5198–treated cohorts relate to cardiovascular system development and tissue morphology (table S2) (supplementary materials). Interestingly, vascular abnormalities and disturbed collagen maturation were recently reported as the most prominent phenotype in mice with brain-specific expression of R132H-IDH1 (17).
When viewed on a genome-wide scale, the 150 mg/kg dose of AGI-5198, which was sufficient for maximal growth inhibition, showed only small effects on RNA expression patterns because 150 mg/kg-treated xenografts clustered with the untreated tumors (fig. S14). Furthermore, an integrated analysis of the DNA methylation and RNA expression data showed no correlation between changes in RNA expression and changes in DNA methylation (fig. S15). Together, these data suggest that mutant IDH1 may promote glioma growth through transcriptional and nontranscriptional mechanisms that are independent of its epigenetic effects.
In summary, we describe a tool compound (AGI-5198) that impairs the growth of R132H-IDH1-mutant, but not IDH1 wild-type, glioma cells. This data demonstrates an important role of mutant IDH1 in tumor maintenance, in addition to its ability to promote transformation in certain cellular contexts (14, 26). Effector pathways of mutant IDH remain incompletely understood and may differ between tumor types, reflecting clinical differences between these disorders. Although much attention has been directed toward TET-family methyl cytosine hydroxylases and Jumonji-C domain histone demethylases, the family of 2-OG–dependent dioxygenases includes more than 50 members with diverse functions in collagen maturation, hypoxic sensing, lipid biosynthesis/metabolism, and regulation of gene expression (27). Our study suggests that a broader investigation of the role of these enzymes in IDH1-mutant glioma may be warranted.
Supplementary Material
Acknowledgments
We thank W. G. Kaelin Jr., for sharing unpublished results. This work was supported by the National Brain Tumor Society (T.G.G., K.E.Y., and I.K.M.), the National Institutes of Health (1R01NS080944-01 and U54CA143798), the Leon Levy Foundation (I.K.M), and the James S. McDonnell Foundation (I.K.M.). C.G. was supported through an American Brain Tumor Association Basic Research Fellowship Award and the Lymphoma Research Foundation. T.G.G. is the recipient of a Research Scholar Award from the American Cancer Society. I.K.M. is the recipient of an Advanced Clinical Research Award in Glioma from the American Society of Clinical Oncology. Agios Pharmaceuticals has filed a patent application on AGI-5198. Microarray data are deposited in the Gene Expression Omnibus (GEO accession no. GSE45200).
Footnotes
This work is based on equal contributions from the laboratories of T.G.G., K.E.Y., and I.K.M.
Supplementary Materials
www.sciencemag.org/cgi/content/full/science.1236062/DC1
Materials and Methods
Supplementary Text
Figs. S1 to S16
Tables S1 and S2
References
References and Notes
- 1.Parsons DW, et al. Science. 2008;321:1807. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yan H, et al. N. Engl. J. Med. 2009;360:765. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amary MF, et al. J. Pathol. 2011;224:334. doi: 10.1002/path.2913. [DOI] [PubMed] [Google Scholar]
- 4.Borger DR, et al. Oncologist. 2012;17:72. doi: 10.1634/theoncologist.2011-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paschka P, et al. J. Clin. Oncol. 2010;28:3636. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
- 6.Mardis ER, et al. N. Engl. J. Med. 2009;361:1058. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dang L, et al. Nature. 2010;465:966. doi: 10.1038/nature09132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Figueroa ME, et al. Cancer Cell. 2010;18:553. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu W, et al. Cancer Cell. 2011;19:17. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. Am. J. Pathol. 2009;174:1149. doi: 10.2353/ajpath.2009.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verhaak RG, et al. Cancer Genome Atlas Research Network. Cancer Cell. 2010;17:98. [Google Scholar]
- 12.Noushmehr H, et al. Cancer Genome Atlas Research Network. Cancer Cell. 2010;17:510. [Google Scholar]
- 13.Lai A, et al. J. Clin. Oncol. 2011;29:4482. doi: 10.1200/JCO.2010.33.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koivunen P, et al. Nature. 2012;483:484. doi: 10.1038/nature10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turcan S, et al. Nature. 2012;483:479. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu C, et al. Nature. 2012;483:474. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sasaki M, et al. Genes Dev. 2012;26:2038. doi: 10.1101/gad.198200.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Popovici-Muller J, et al. ACS Medicinal Chem. Lett. 2012;3:850. doi: 10.1021/ml300225h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Louis DN, et al. Acta Neuropathol. 2007;114:97. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cahoy JD, et al. J. Neurosci. 2008;28:264. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lein ES, et al. Nature. 2007;445:168. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 22.Sobieszczuk DF, Poliakov A, Xu Q, Wilkinson DG. Genes Dev. 2010;24:206. doi: 10.1101/gad.554510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bachoo RM, et al. Cancer Cell. 2002;1:269. doi: 10.1016/s1535-6108(02)00046-6. [DOI] [PubMed] [Google Scholar]
- 24.Ma DK, et al. Nat. Neurosci. 2010;13:1338. doi: 10.1038/nn.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chowdhury R, et al. EMBO Rep. 2011;12:463. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sasaki M, et al. Nature. 2012;488:656. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDonough MA, Loenarz C, Chowdhury R, Clifton IJ, Schofield CJ. Curr. Opin. Struct. Biol. 2010;20:659. doi: 10.1016/j.sbi.2010.08.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.