Abstract
The σ subunit of bacterial RNA polymerase (RNAP) has been implicated in all steps of transcription initiation, including promoter recognition and opening, priming of RNA synthesis, abortive initiation and promoter escape. The post-promoter-recognition σ functions were proposed to depend on its conserved region σ3.2 that directly contacts promoter DNA immediately upstream of the RNAP active centre and occupies the RNA exit path. Analysis of the transcription effects of substitutions and deletions in this region in Escherichia coli σ70 subunit, performed in this work, suggests that (i) individual residues in the σ3.2 finger collectively contribute to RNA priming by RNAP, likely by the positioning of the template DNA strand in the active centre, but are not critical to promoter escape; (ii) the physical presence of σ3.2 in the RNA exit channel is important for promoter escape; (iii) σ3.2 promotes σ dissociation during initiation and suppresses σ-dependent promoter-proximal pausing; (iv) σ3.2 contributes to allosteric inhibition of the initiating NTP binding by rifamycins. Thus, region σ3.2 performs distinct functions in transcription initiation and its inhibition by antibiotics. The B-reader element of eukaryotic factor TFIIB likely plays similar roles in RNAPII transcription, revealing common principles in transcription initiation in various domains of life.
INTRODUCTION
In contrast to DNA polymerases that require a primer and are recruited to the replisome sequence-nonspecifically, RNA polymerases (RNAPs) start gene transcription from specific promoter sites and begin RNA synthesis from NTPs de novo. The process of transcription initiation involves specific recognition of promoter sequences, DNA melting around the starting point of transcription, priming of RNA synthesis and promoter escape that requires breaking of the RNAP-promoter contacts (1–3). All cellular multisubunit RNAPs rely on specialized factors for transcription initiation. The principal factor of transcription initiation in bacteria, the σ subunit of RNAP, is involved in all steps of initiation. The σ subunit binds the catalytic core enzyme of RNAP to form holoenzyme capable of promoter recognition. During promoter recognition, σ directly binds specific promoter elements [in the case of the primary σ70 factor, the −10 (TATAAT), −35 (TTGACA), TG and discriminator (GGGA) motifs] and participates in DNA melting (3–7). Conserved σ subunit domains σ2 and σ4, involved in the recognition of the −10 and −35 elements, respectively, are separated by a flexible linker formed by conserved region σ3.2 (7–10). In holoenzyme RNAP, region σ3.2 forms a loop (‘σ3.2 finger’) that approaches the RNAP active centre and partially occupies the path for RNA exit (Figure 1). Based on its position in the initiating complex, region σ3.2 was proposed to play important roles at post-promoter-recognition steps of transcription initiation (7,8,11). In particular, it was demonstrated to stimulate the binding of the initiating NTPs (iNTPs), participate in abortive initiation and facilitate promoter escape by RNAP, likely by competing with the nascent RNA near the active site and in the RNA exit channel (11,12). The involvement of region σ3.2 in the RNA priming conforms to the large stimulatory effect of the σ subunit on the iNTP binding that was first observed many years ago (13,14). Similarly, region σ3.2 was implicated in iNTP binding and stabilization of short RNA primers during transcription initiation on phage single-stranded replication origins, whose recognition may not require specific σ-DNA interactions, suggesting that the RNA priming σ3.2 functions are universal for various types of transcription templates (15).
Figure 1.
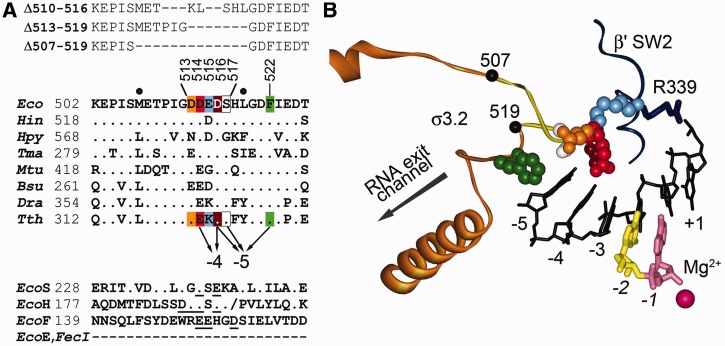
The structure of region σ3.2 and its contacts in the open promoter complex. (A) Sequence alignment of region σ3.2 in σ factors from various bacteria and positions of analyzed σ3.2 mutations. Eco, E. coli; Hin, Haemophilus influenzae; Hpy, Helicobacter pylori; Tma, Thermotoga maritima; Mtu, Mycobacterium tuberculosis; Bsu, Bacillus subtilis; Dra, Deinococcus radiodurans; Tth, T. thermophilus. Interactions of the T. thermophilus σA residues with the −4 and −5 nucleotides of the template DNA (corresponding to promoter positions p-3 and p-4) are indicated with arrows. Positions of alanine substitutions in the E. coli σ70 subunit studied in this work are highlighted; positions of the σ3.2 deletions are shown above the alignment. Alignment of regions σ3.2 in E. coli alternative σ factors is shown at the bottom of the figure. EcoS, EcoH, EcoF and EcoE correspond to σS, σH, FliA and σE, respectively; σE and FecI completely lack region σ3.2. The slash in the RpoH sequence corresponds to an insertion (QPMA) at this position. Negatively charged amino acid residues on the tip of the σ3.2 loop are underlined. (B) The structure of region σ3.2 in the T. thermophilus RNAP–promoter complex [4G7O, (7)]. The template DNA strand is black, the catalytic magnesium ion is shown as a sphere. The dinucleotide primer (yellow/pink) corresponding to promoter positions p − 1/p + 1 is bound just upstream of the +1 site of the RNAP active centre. Region σ3.2 is shown in orange, the 507–519 segment is yellow, residues substituted in the analyzed σ3.2 mutants are shown as CPK models (the colour code corresponds to panel A). The β′ switch2 region (SW2), interacting with σ3.2, is shown in dark blue, residue R339 (corresponding to R615 in T. thermophilus RNAP) is shown as a stick model. The direction of RNA exit is indicated with an arrow.
Within primary σ factors, region σ3.2 is highly conserved and contains several negatively charged amino acid residues at the tip of the σ3.2 finger (Figure 1A and B). While the sequences of this region significantly differ in alternative σs (16), it also contains similarly placed negatively charged residues, as illustrated in Figure 1A for Escherichia coli σS, σH and FliA. Intriguingly, extracytoplasmic functions (ECF) σ factors (such as E. coli σE and FecI) completely lack region 3, which is replaced by a short linker of unrelated sequence that connects regions σ2 and σ4 (16). However, structural modelling suggests that the σ2–σ4 linker may be similarly placed within RNAP holoenzyme and, probably, functionally replace region 3.2 (17). Curiously, E. coli heat-shock σH with a deletion of most of region σ3.2 (Δ178–201, see Figure 1A for amino acid numbering) could still support expression of heat-shock promoters, although it had a reduced affinity for core RNAP (18). Besides this observation, no functional studies of the σ3.2 region (or the σ2–σ4 linker) in alternative σ factors have been performed to date, and the exact roles of these regions in transcription initiation by alternative holoenzymes are unknown. At the same time, the σ3.2 functions seem to be conserved in evolution, as parts of archaeal factor TFB and eukaryotic general transcription factor TFIIB occupy similar places in the initiation complexes and participate in the first steps of RNA synthesis and promoter escape by their cognate RNAPs (19,20).
Recent structural analysis of a transcription initiation complex of Thermus thermophilus RNAP holoenzyme assembled on a synthetic promoter scaffold revealed direct contacts of the region σ3.2 finger with the template DNA strand and suggested that it positions the DNA template in the active centre for priming of RNA synthesis (Figure 1B) (7). In particular, residues D514, D516, S517 and F522 (E. coli residue numbers are used throughout) interact with DNA bases at promoter positions p-3 and p-4 (corresponding to positions −4 and −5 relative to the +1 site of the RNAP active centre, Figure 1B), suggesting their specific role(s) in DNA positioning and/or promoter escape. However, substitutions of individual amino acid residues in region σ3.2 led to only a subtle decrease in RNAP activity, putting roles of these contacts into question (7). In this study, we dissect the roles of individual σ3.2 residues and the region as a whole at different steps of transcription initiation and discuss structural and functional parallels between transcription initiation by bacterial RNAP and eukaryotic RNAPII.
MATERIALS AND METHODS
RNAPs and promoters
Wild-type and mutant R339A E. coli core RNAPs were purified from E. coli BL21(DE3) cells overproducing RNAP subunits from plasmids pVS10 and pIA830, respectively, as previously described (21). Plasmids encoding mutant variants of the σ70 subunit with deletions Δ510–516 and Δ509–519 were generously provided by L. Minakhin and I. Artsimovitch. Other mutant variants of the σ70 subunit were obtained by polymerase chain reaction (PCR) mutagenesis of plasmid pET28rpoD encoding wild-type σ70 subunit with an N-terminal hexahistidine tag (11). Wild-type and mutant σ70 subunits were expressed and purified as described (11). The DNA fragment containing wild-type T7A1 promoter followed by λtR2 terminator was obtained as in (22). The λPR promoter (followed by hisT terminator) and galP1 promoter fragments were obtained by PCR from plasmids pIA226 and pTZ19galP1, respectively (provided by I. Artsimovitch and L. Minakhin). The T7A1cons, T7A1_σP+6, T7A1_σP+6mut and rrnB P1 promoters were obtained by PCR from synthetic oligonucleotides. All promoter sequences are presented in Supplementary Figure S1.
In vitro transcription
Holoenzyme RNAPs were prepared by incubating core RNAP (50 nM final concentration) with either wild-type or mutant σ70 subunits (500 nM) in transcription buffer (40 mM Tris–HCl, pH 7.9, 40 mM KCl and 10 mM MgCl2) for 5 min at 37°C. The DNA template was added (10 nM) and the samples were further incubated for 5 min at 37°C. For full-length RNA synthesis, all four NTPs were added (10 µM ATP, CTP, GTP and 5 µM UTP with addition of α-[32P]-UTP in most experiments, unless otherwise indicated), either in the absence or in the presence of RNA primers (25 µM CpA, ApU or CpApU). The reactions were stopped after 7 min by addition of an equal volume of stop buffer containing 8 M urea and 20 mM EDTA. RNA products were separated by denaturing polyacrylamide gel electrophoresis (PAGE; 15% in Figures 2 and 3; 20% and 30% in Figure 4; 20% in Figure 5) and analyzed with Typhoon 9500 scanner (GE Healthcare).
Figure 2.
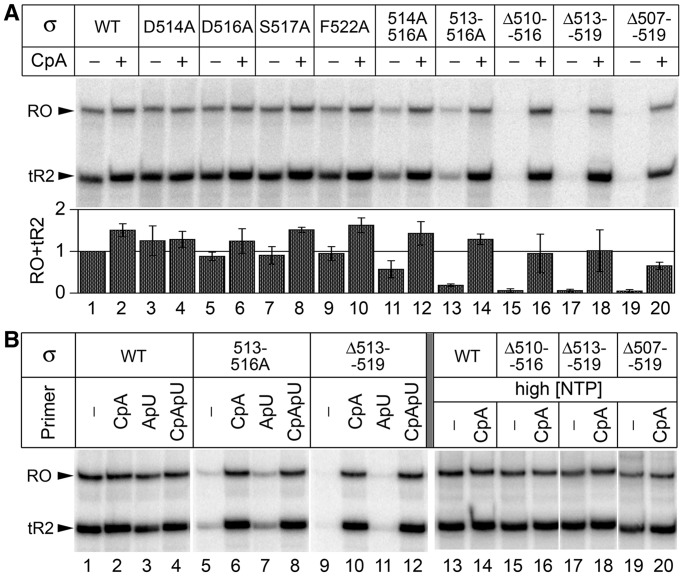
Transcription activity of RNAPs containing mutant σ70 subunits on the T7A1 promoter. (A) Full-length RNA synthesis on the T7A1 promoter template. Positions of the terminated (tR2) and full-length run-off (RO) transcripts are indicated. Reactions contained 10 µM ATP, CTP, GTP and 5 µM UTP; RNA primer CpA was added when indicated. Relative activities of wild-type and mutant RNAPs, calculated as a sum of tR2 and RO transcripts, are shown below the gel (averages and standard deviations from three independent experiments). (B) RNAP activities on the T7A1 promoter template in the presence of various initiating primers (lanes 1–12) and at high NTP concentrations (200 µM ATP, CTP, GTP and 10 µM UTP) (lanes 13–20). Positions of the tR2 and RO transcripts are indicated.
Figure 3.
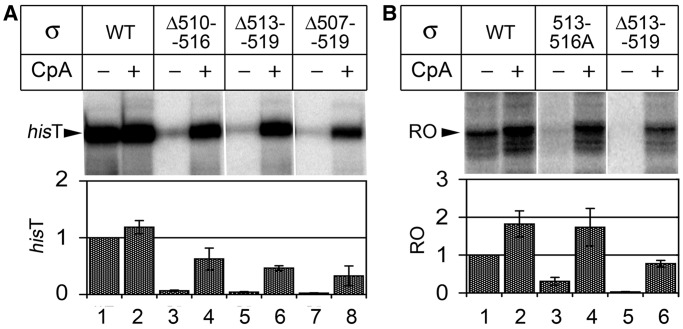
Transcription activities of mutant RNAPs on the λPR and rrnB P1 promoters. (A) RNA synthesis on the λPR promoter template; position of the major terminated product (hisT) is indicated. (B) RNAP activities on the rrnB P1 promoter template; position of the full-length run-off (RO) transcript is indicated. Reactions contained 25 µM ATP, GTP, CTP, 10 µM UTP and the CpA primer, when indicated. Relative RNAP activities are shown below the gels (averages and standard deviations from two to three independent experiments).
Figure 4.
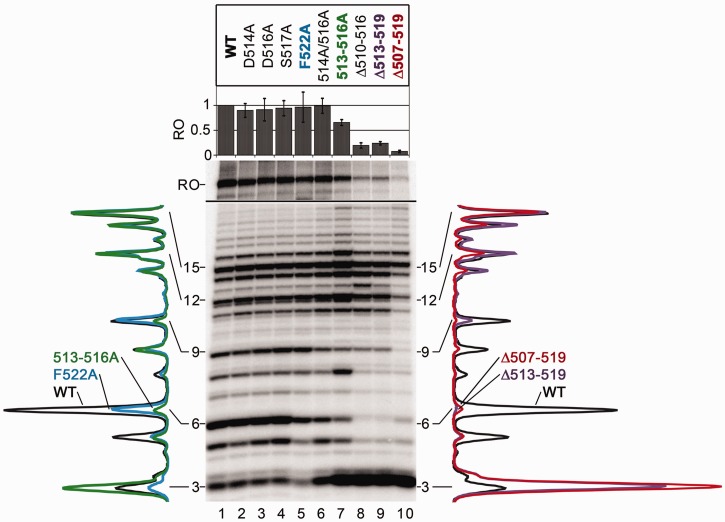
Abortive initiation and promoter escape by RNAPs with mutations in region σ3.2. Transcription was performed on the T7A1cons-promoter template in the presence of the CpA primer, 25 µM ATP, GTP, CTP and 5 µM UTP; heparin was added to 10 µg/ml together with NTPs to prevent re-initiation. Positions of abortive and run-off (RO) RNA products are indicated; to resolve abortive and full-length RNAs, the samples were separated on gels of various concentrations (see ‘Materials and Methods’ section for details). The relative efficiencies of the run-off RNA synthesis are shown above the gel (averages and standard deviations from three independent experiments). Scanned profiles of abortive products (normalized by the amounts of 15 nt RNAs) are shown on the sides of the gel. Left, profiles for wild-type (WT), 513–516A and 522A σ70 subunits; right, profiles for WT, Δ513–519 and Δ507–519 σ70 subunits.
Figure 5.
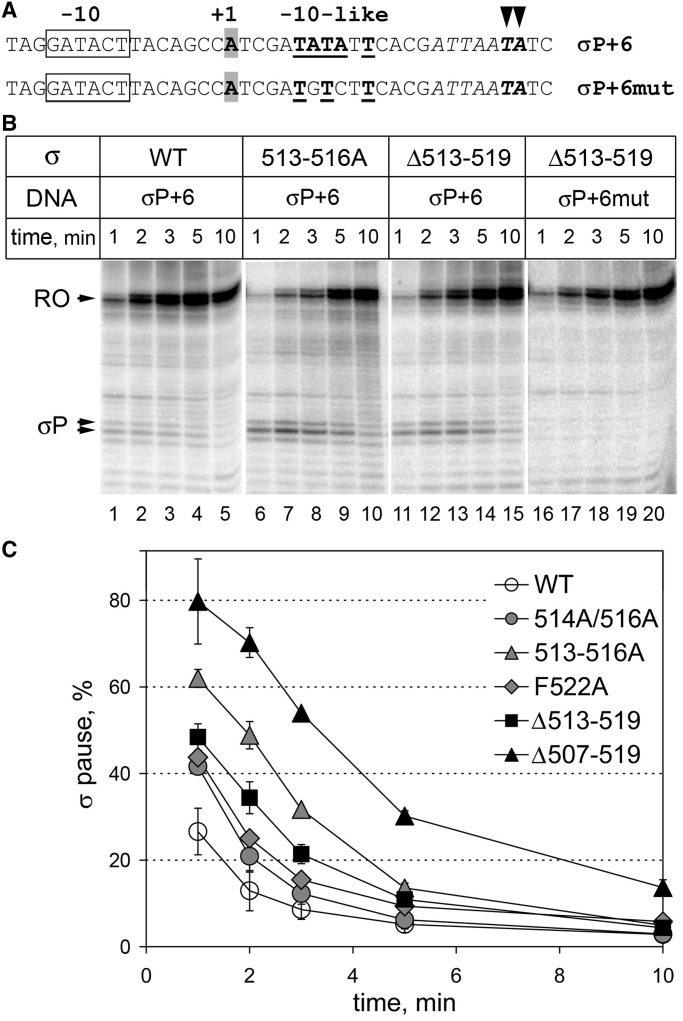
Effects of the σ3.2 mutations on σ70-dependent transcription pausing. (A) Initially transcribed sequences of the T7A1-promoter templates containing the −10-like pause-inducing signal (bold underlined, σP+6) and a mutant variant of this signal (σP+6mut). Major pausing positions are indicated with arrowheads; the A/T-rich sequence at the site of pausing is italicized. (B) Analysis of σ70-dependent pausing. Transcription was performed in the presence of the CpA primer. Positions of the paused and run-off (RO) transcripts are indicated. (C) Kinetics of pausing induced by wild-type and mutant σ70 variants. The efficiency of σ-pausing was calculated as a ratio of the intensities of paused RNAs to the sum of the paused and full-length transcripts. Averages and standard deviations from three independent experiments are shown.
Apparent KMs for iNTPs were measured on the wild-type T7A1 promoter in reactions containing ATP and UTP, or CpA and UTP substrates. One of the two substrates was taken at fixed concentration (1 mM for ATP or UTP, and 100 µM for CpA), while the concentration of the other was varied (from 1 µM to 6 mM for ATP or UTP; from 1 µM to 1 mM for CpA). Either α-[32P]-UTP or γ-[32P]-ATP was added to the reactions to label the RNA products. When required, rifapentin was added to 10 µg/ml 5 min before NTPs. The reactions were stopped after 1 min at 37°C, and the samples were analyzed by 30% PAGE (20:3, acrylamide:bisacrylamide) to separate reaction products and mononucleotides. To calculate apparent KM values, the data were fit to the Michaelis–Menten equation A = Amax × [NTP]/(KM + [NTP]), where A is the amount of the synthesized RNA product and Amax is the amount of RNA product at saturation, using GraFit software (Erithacus Software).
Analysis of σ-dependent pausing in transcription complexes assembled on synthetic oligonucleotide scaffold (Supplementary Figure S3) was performed as described in (23). Either wild-type or mutant σ subunits were added at various concentrations (from 50 to 2000 nM) to pre-assembled transcription complexes containing 5′-labeled 20-nt RNA. After 5-min incubation at 37°C, NTPs were added to 100 µM. The reactions were terminated after 1.5 min by addition of the stop buffer and the RNA products were analyzed by 15% PAGE. The data were fit to the hyperbolic equation P = Pmax × [σ]/(Kd + [σ]), where P is the pause efficiency, Pmax is the pause efficiency at saturating concentration of σ and Kd is apparent dissociation constant for σ binding to the elongation complex.
RESULTS
RNAPs with mutations in region σ3.2 require a primer for transcription initiation
To highlight the functions of region σ3.2 in transcription initiation, we analyzed the effects of amino acid substitutions and deletions in region σ3.2 in the E. coli σ70 subunit, including (i) individual alanine substitutions of residues D514, D516, S517 and F522; (ii) combinations of several substitutions (double substitution 514A/516A and quadruple substitution 513–516A at the tip of the σ3.2 loop); (iii) deletions of varying lengths (Δ510–516, residues 510–516 substituted with KL; Δ513–519; Δ507–519) (Figure 1A).
RNA synthesis by RNAPs containing mutant σ70 subunits was first analyzed on a DNA fragment containing T7A1 promoter followed by λtR2 terminator (Figure 2A). At low NTP concentrations (10 µM), wild-type RNAP and RNAPs with single amino acid substitutions efficiently synthesized full-length RNA products, and their activity was only slightly stimulated by the addition of a dinucleotide primer CpA, corresponding to positions p − 1/p + 1 of the promoter (Figure 2A, lanes 1–10; see promoter sequences in Supplementary Figure S1). In contrast, RNAPs with double and quadruple substitutions in the σ3.2 loop displayed significantly reduced activity (lanes 11, 13), and all three RNAPs with σ3.2 deletions were essentially inactive (lanes 15, 17, 19) in the absence of the primer. The activity of these RNAPs was restored on addition of the initiating primer CpA (lanes 12, 14, 16, 18 and 20). Furthermore, the defects of the mutant RNAPs in the full-length RNA synthesis were compensated at increased NTP concentrations (200 µM, Figure 2B, lanes 13–20). This result implied that amino acid substitutions and deletions in region σ3.2 affect the first RNA bond formation, probably owing to the impaired binding of the initiating nucleotides (iNTPs), as was previously proposed for the Δ513–519 deletion (11).
The location of the σ3.2 loop several nucleotides upstream of the active centre makes it unlikely that this region directly participates in the iNTP binding (Figure 1B). Rather, mutations in σ3.2 might affect the template DNA strand positioning, in turn resulting in the impaired iNTP binding. The primers may then help to stabilize the template strand in the active centre and improve the iNTP binding. The CpA primer used in the previous experiment is optimally placed just upstream of the +1 NTP-binding site of the active centre (which corresponds to promoter position p + 2 in the open complex, see Figure 1B). To establish whether the stimulating effect of the primer was position-specific, we analyzed RNA synthesis by wild-type, 513–516A and Δ513–519 RNAPs in the presence of primers ApU (corresponds to promoter positions p + 1/p + 2) and CpApU (positions p − 1/p + 1/p + 2) (Figure 2B). Notably, the mutant RNAPs could not initiate transcription in the presence of the ApU primer, which is shifted 1-nt downstream relative to the CpA primer (lanes 7 and 11). This was not a result of a suboptimal positioning of the primer 3′-end because the CpApU primer with the identical 3′-end fully restored the RNAP activities (lanes 8 and 12). Thus, the activation of the mutant RNAPs seemingly requires primer positioning upstream of the starting point of transcription. The primer may thereby compensate for the absence of the upstream stabilizing contacts of region σ3.2 with the template.
Mutant RNAPs possess similar defects in transcription initiation on various promoters
To determine whether the observed defects of the mutant RNAPs in transcription initiation are universal for various promoters, we analyzed RNAP activities on the λPR (for the three deletant RNAPs) and rrnB P1 (for the 513–516A and Δ513–519 RNAPs) promoter templates (Figure 3). In comparison with T7A1, λPR forms more stable complexes, while rrnB P1 forms highly unstable complexes with RNAP (21,24). In both cases, the mutant RNAPs were unable to synthesize full-length RNA in the presence of mononucleotide substrates and were reactivated on addition of the CpA primer (which also corresponds to positions p − 1/p + 1 in these promoters, see Supplementary Figure S1). In the case of the rrnB P1 promoter, the primer also stimulated the activity of wild-type RNAP (∼2-fold; Figure 3B, compare lanes 1 and 2). This likely reflects inefficient transcription initiation on this promoter under the reaction conditions because it requires high iNTP concentrations for full activity (25). At the same time, the initiating primer could still at least partially compensate for the transcription initiation defects of the mutant RNAPs, suggesting that the region σ3.2 mutations have basically the same effects on the first steps of RNA synthesis on various promoters.
σ3.2 mutations increase apparent KMs for iNTPs
To highlight possible effects of the σ3.2 mutations on the iNTP binding, we measured apparent KM values for the 5′- and 3′-iNTPs in the reaction of dinucleotide synthesis on the T7A1 promoter. In this reaction, the dinucleotide products are released from the complex, thus allowing reiterative abortive RNA synthesis. The overall rate of the reaction greatly depends not only on the iNTP binding and the catalytic rate but also on the rate of product release. We did not compare the reaction rates for the mutant RNAPs because their interpretation may be complicated, and limited our analysis to comparison of the KM values for iNTPs, which can be used as a measure of their binding in the active centre (26).
As we reported previously (11,21), the Δ513–519 deletion slightly increased (3.7-fold) apparent KM value for the 5′-ATP and dramatically increased (54-fold) KM for the 3′-UTP substrate (Table 1). RNAP 513–516A with four amino acid substitutions in the σ3.2 loop had comparable defects in the iNTP binding, resulting in 5.1- and 11.3-fold changes in the ATP and UTP KMs, respectively. RNAP 514A/516A containing two substitutions in the σ3.2 loop had much weaker effects on the iNTP binding (∼1.9- and 4.4-fold increase in the ATP and UTP KMs), and RNAP F522A with a single substitution of the phenylalanine residue located further upstream from the active site only modestly affected the iNTP KMs.
Table 1.
Apparent KM values for the initiating substrates on the T7A1 promoter for wild-type (WT) and mutant RNAPs
| RNAP | ATP+UTP → pppApU |
|||
|---|---|---|---|---|
|
KM, ATP (µM) |
KM, UTP (µM) |
|||
| −Rif | +Rif | −Rif | +Rif | |
| WT | 190 ± 4a | 850 ± 150 | 9.2 ± 0.5a | 170 ± 35 |
| 1 | 4.5 | 1 | 18.5 | |
| σ70 Δ513–519 | 700 ± 28a | 965 ± 105 | 490 ± 30a | 815 ± 36 |
| 3.7 | 5.1 | 53.7 | 89.1 | |
| σ70 513–516A | 960 ± 60 | 1920 ± 170 | 105 ± 6.1 | 605 ± 78 |
| 5.1 | 10.1 | 11.3 | 66.1 | |
| σ70 514A,516A | 360 ± 81 | ndb | 40.0 ± 5.7 | nd |
| 1.9 | 4.4 | |||
| σ70 F522A | 240 ± 23 | nd | 32 ± 7.1 | nd |
| 1.3 | 3.5 | |||
| β′ R339A | 670 ± 120 | 1700 ± 550 | 59.3 ± 7.0 | 385 ± 77 |
| 3.5 | 9.0 | 6.5 | 41.9 | |
| CpA+UTP → CpApU |
||||
|---|---|---|---|---|
| KM, CpA (µM) | KM, UTP (µM) | |||
| WT | 66.3 ± 26.4 | nd | 5.2 ± 1.0 | nd |
| 1 | 1 | |||
| σ70 Δ513–519 | 125 ± 17.2 | nd | 15.4 ± 6.9 | nd |
| 1.9 | 2.9 | |||
The numbers in bold show changes in KM relative to wild-type RNAP in the absence of Rif.
aData from ref. (21).
bnd, not determined.
We then measured the apparent KMs for the wild-type and Δ513–519 RNAPs in the reaction of trinucleotide synthesis with the CpA and UTP substrates. The UTP KMs in this reaction can be directly compared with the corresponding values in the reaction of dinucleotide synthesis (Table 1). As can be seen, the presence of the CpA primer had slightly stimulating effect on the UTP binding by wild-type RNAP but dramatically (>30-fold) decreased the UTP KM for the Δ513–519 RNAP, thus greatly decreasing the difference in the UTP binding between the two RNAPs (2.9-fold as compared with the 54-fold difference for the dinucleotide reaction). There was also ∼2-fold difference in the CpA KM values, demonstrating that the σ3.2 deletion somewhat impairs the dinucleotide binding (Table 1).
The observed defects in the iNTP binding likely explain the inability of the mutant RNAPs to initiate transcription in the absence of primers. The stronger transcription defects of RNAPs with multiple substitutions in σ3.2 suggest that individual amino acids from this region collectively contribute to the iNTP binding, probably by the positioning of the DNA template strand in the active centre. The mutations in σ3.2 primarily affect the 3′-iNTP binding, consistently with the view that the 5′-iNTP site is preformed in the RNAP core enzyme and the 5′-iNTP binding does not require the σ subunit and the DNA template (13,14,27).
Rifamycin affects iNTP binding through region σ3.2
Rifamycin (Rif) antibiotics were demonstrated to directly interact with σ region 3.2 in T. thermophilus and E. coli RNAPs, with possible implications in RNAP inhibition (10,28). In particular, it was suggested that Rif may affect the first phosphodiester bond formation through changes in the position of the template DNA strand in the active centre, mediated by changes in the conformation of region σ3.2 (10). Similar to the σ3.2 mutations, Rif had been shown to slightly increase KMs for iNTPs during initiation (29); in contrast, Rif did not inhibit dinucleotide synthesis by core RNAP (14). We therefore measured the effects of a semi-synthetic Rif, rifapentin, on the iNTP KMs on the T7A1 promoter for wild-type and mutant RNAPs. The effects of Rif on the iNTP binding by wild-type RNAP were significantly stronger than previously reported (29), with ∼4.5- and 18.5-fold increase in KMs for ATP and UTP, respectively (Table 1). The absolute iNTP KM values measured in the Rif presence were in the sub-millimolar range, suggesting that the allosteric effects of Rif on the iNTP binding may significantly contribute to transcription inhibition (both in vitro and in vivo), before its steric effects on the RNA extension (30,31) (although the steric effects would be likely sufficient for complete RNAP inhibition even in the absence of the allosteric component). The inhibitory effect of Rif on the first bond synthesis is usually masked in the in vitro transcription reactions because the steric blocking of RNA extension results in the trapping of RNAP in the process of abortive transcription and in a huge increase in the amounts of short abortive products [see, for example, (30,32)].
Strikingly, Rif had much weaker effects on the iNTP binding by RNAPs with σ3.2 mutations, which by themselves impaired the iNTPs binding. In particular, addition of Rif resulted in ∼1.5-fold increase in KMs for both iNTP in the case of Δ513–519 RNAP and in ∼2- and 6-fold increase in KMs for ATP and UTP, respectively, in the case of 513–516A RNAP (Table 1).
Previously, we proposed that the β′ subunit region switch2, which interacts with both region σ3.2 and the template DNA strand (Figure 1B), may cooperate with the σ subunit in the positioning of the template DNA in the active centre. In particular, substitutions of a conserved E. coli switch2 R339 residue were also shown to result in a significant increase in the apparent KM values for iNTPs (21). We confirmed this result and showed that the R339A substitution increased KM values for initiating ATP and UTP on the T7A1 promoter 3.5- and 6.5-fold, respectively (Tale 1). Similar to the σ3.2 mutant RNAPs, Rif had weaker effects on the iNTP KMs in the case of the R339A RNAP, which were increased ∼2.5- and 6.5-fold in comparison with the Rif-less reaction (Table 1). Thus, Rif affects iNTP binding in the same way as changes in regions σ3.2 and β′ switch2, probably by altering the template DNA positioning through region σ3.2, as suggested by structural analysis (10).
Deletions in region σ3.2 impair promoter escape by RNAP
Positioning of region σ3.2 in the RNA exit channel immediately suggested that it may be involved in promoter clearance by RNAP (7–9). Previously we demonstrated that deletion Δ513–519 in the σ3.2 loop impaired promoter escape by E. coli RNAP (11). To dissect the role of contacts of region σ3.2 with DNA, we compared the effects of amino acid substitutions and deletions in σ3.2 on transcription from the T7A1cons promoter, a consensus variant of the T7A1 promoter (Supplementary Figure S1), which forms strong interactions with RNAP and is characterized by a high efficiency of abortive synthesis (11,21). The transcription was performed in the presence of the CpA primer, to suppress the priming defects of RNAP variants. The reactions contained labeled α-[32P]-UTP, which allowed visualization of all abortive products staring from trinucleotide RNA (see promoter sequence in Supplementary Figure S1). On this template, wild-type RNAP synthesized large amounts of abortive transcripts varying from 3 to 16 nt in length (Figure 4, lane 1). Point amino acid substitutions of residues D514, D516, S517 and F522 in the σ3.2 loop did not significantly affect the full-length RNA synthesis (90–100% of the wild-type level, lanes 2–4). The first three substitutions also did not change the pattern of abortive RNA products. At the same time, the F522A substitution slightly decreased the amounts of short 3–6 nt abortive RNAs, without affecting the synthesis of longer abortive RNAs (lane 5). Combinations of substitutions in the 516A/516A and 513–516A RNAPs increased the synthesis of trinucleotide products (CpApU in this reaction) but did not dramatically affect longer abortive products (lanes 6 and 7). The double 516A/516A substitution also did not change the efficiency of run-off RNA synthesis, while the quadruple 513–516A substitution slightly decreased the level of full-length RNA (65% of wild-type RNAP).
Much more pronounced effects on the pattern of RNA products were observed for RNAPs with deletions in region σ3.2. First, all three deletions significantly increased the amounts of the trinucleotide RNA, similar to the 516A/516A and 513–516A RNAPs (Figure 4, lanes 8–10). This effect is likely explained by destabilization of the trinucleotide binding in the active centre because of the altered template DNA positioning, resulting in its faster release and increasing product turnover. The Δ513–519 deletion increases apparent KMs not only for iNTPs but also for the CpA primer (see Table 1), suggesting that it may similarly affect the trinucleotide binding. The latter effect seemingly outweighs the moderate unfavorable increase in the KMs for CpA and UTP, resulting in the increase in the overall rate of trinucleotide synthesis. No increase in the synthesis of longer products was observed, suggesting that region σ3.2 is most important for stabilization of the binding of iNTPs and very short 2–3 nt RNAs. Second, the deletions resulted in the disappearance of middle-size abortive RNAs (5–9 nt), likely as a result of the removal of region σ3.2 from the RNA exit path, thus allowing further RNA extension. In support of this, the σ3.2 deletions only slightly affected the longest 15–16 nt abortive products, which appear as a result of clashing of the extended RNA with region σ4 bound to the β flap domain in the RNA exit channel (33). Furthermore, the deletions differed in the ranges of affected middle-size abortive RNAs: the largest (Δ507–519) deletion decreased the amounts of up to 14 nt long RNAs (lane 10), while the shorter deletions (Δ510–516 and Δ513–519) decreased the synthesis of ≤9 nt RNAs (lanes 8 and 9). This suggests that the physical presence of region σ3.2 in the RNA exit channel is important for abortive initiation (see ‘Discussion’ section). Third, all deletions dramatically decreased the efficiency of full-length RNA synthesis (20, 24 and 7% of wild-type RNAP activity for Δ510–516, Δ513–519 and Δ507–519 RNAPs, respectively), indicative of serious problems with promoter escape.
Similar effects of the region σ 3.2 mutations on abortive synthesis and promoter escape were observed in the case of the consensus galP1 promoter, which has an extended −10 element (Supplementary Figures S1 and S2). This promoter forms a distinct set of interactions with RNAP, involving region σ3.0, and was also shown to form highly stable complexes with RNAP (34,35). Thus, deletions in region σ3.2 impair promoter escape and full-length RNA synthesis by RNAP on promoters that form strong specific interactions with RNAP.
Mutations in region σ3.2 stimulate σ-dependent promoter-proximal pausing
The classical view of transcription initiation implied that the σ subunit dissociates from core RNAP on completion of abortive synthesis and transition to processive elongation. During transcription initiation, region σ3.2 (and, subsequently, region σ4) should be ejected from the RNA exit channel to allow RNA extension and transition to processive elongation (Figure 1B) (8,9). However, recent studies suggested that σ may nevertheless remain bound to the transcription elongation complex and induce pausing through interactions between region σ2 and −10-like elements present in the initially transcribed sequences in a number of phage and bacterial genes (23,36–42). The interconnection between promoter escape, σ dissociation and σ-dependent pausing, and a possible role of region σ3.2 in the pausing remain poorly understood.
We proposed that σ3.2 alterations that affect promoter escape by RNAP may affect σ dissociation during initiation by impairing its displacement from the RNA exit channel, in turn resulting in enhanced promoter-proximal pausing. To test this hypothesis, we used a variant of the T7A1 promoter template with a −10-like pause-inducing signal located 6 nt downstream of the transcription start site (σP+6) and a control template that contained three point substitutions in the −10-like element (σP + 6mut; Figure 5A). We also introduced an A/T-rich sequence at the expected pausing site, to stimulate RNAP backtracking that was shown to be essential for pausing (43). Transcription was performed in the presence of the CpA primer; as we showed above, all mutant RNAPs can efficiently initiate transcription and escape to elongation on the T7A1 promoter under these conditions (see Figure 2). Paused RNA products of the expected lengths were observed with wild-type RNAP and disappeared during the course of the reaction (Figure 5B, lanes 1–5 and Figure 5C). Remarkably, the pause efficiency was significantly increased in the case of RNAPs containing amino acid substitutions (double 514/516A, quadruple 513–516A and single F522A substitutions) or deletions in region σ3.2 (Figure 5B lanes 6–15 and Figure 5C). Substitutions in the pause-inducing −10-like element completely abolished pausing (Figure 5B, lanes 16–20). The strongest stimulating effect on the pausing was observed in the case of the largest deletion Δ507–519. This correlates with its strongest effect on promoter escape (see Figure 4). It should be noted that the increased pausing by the mutant RNAPs cannot be explained by their slower escape to elongation, which could affect the pausing kinetics but not the efficiency of pausing in elongation complexes that had already leaved the promoter.
The enhanced pausing by the mutant RNAPs might result not only from impaired σ dissociation during promoter escape but also from changes in the σ-dependent pausing per se, such as changes in σ-RNAP and σ-DNA interactions or in the efficiency of RNAP backtracking at the pause site (41). To directly compare the pause-inducing properties of wild-type and mutant σs in the absence of the promoter escape step, we analyzed σ-dependent pausing in a model elongation complex assembled on a synthetic nucleic acid scaffold containing a −10-like pause-inducing signal optimally positioned for σ binding in respect to the RNA 3′-end (Supplementary Figure S3A) (23). The σ subunits were added at varying concentrations to pre-assembled elongation complexes, followed by nucleotide addition. We observed comparable pausing with wild-type, 513–516A and Δ513–519 σ subunits (Supplementary Figure S3B). In particular, wild-type and mutant σs had similar affinities to the elongation complex (apparent Kds of about 310 ± 65, 290 ± 30 and 240 ± 50 nM for wild-type, 513–516A and Δ513–519 σ subunits, respectively) and did not significantly differ in the maximal pause efficiencies measured at saturating concentrations of σ (65 ± 4, 74 ± 9 and 73 ± 6%, respectively). Thus, the wild-type and mutant RNAPs do not differ significantly in the pause-recognition or backtracking properties. The observed small differences apparently cannot explain the greatly enhanced promoter-proximal pausing by the mutant RNAPs, suggesting that it indeed results from more efficient σ retention in the transcription complexes during the initiation-to-elongation transition.
DISCUSSION
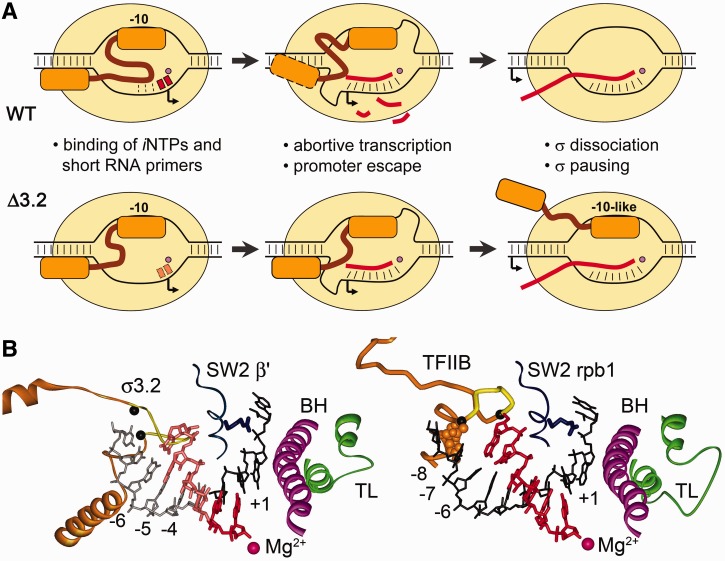
The results of the work demonstrate that region σ3.2 plays important and distinct functions in transcription initiation by bacterial RNAP. The series of events that occur during transcription initiation by RNAPs containing either wild-type or mutant σ subunits is schematically shown in Figure 6A.
Figure 6.
Functions of region σ3.2 at different steps of transcription initiation. (A) Scheme illustrating the roles of region σ3.2 during initiation by wild-type RNAP (upper raw) and the effects of σ3.2 mutations on different steps of initiation (bottom row). See comments in the text. (B) Structural parallels between the initiation complexes of bacterial RNAP [left, 4G7O, (7)] and Saccharomyces cerevisiae RNAPII [right, 4BBS, (20)]. The trigger loop (TL) and bridge helix (BH) elements of the active centre are shown in green and violet, respectively; region σ3.2 and the B-reader element of factor TFIIB are orange; the colour code for other elements corresponds to Figure 1B. Positions of the DNA template relative to the active site are indicated. In the bacterial structure, the upstream part of the RNA–DNA hybrid (shown in lighter colours) is superimposed from the elongation complex structure [2O5J, (44)]. Region σ3.2 clashes with the RNA 5′-end 5 nt upstream of the active site, in a perfect agreement with the range of abortive products stabilized by the σ3.2 deletions (see Figure 4). TFIIB residues that contact the −7/−8 DNA bases are shown in CPK mode. The borders of the Δ507–519 deletion in region σ3.2 and a B-reader loop deletion that was shown to impair initiation (20) are shown with black Cα-atoms.
In the open promoter complex, region σ3.2 participates in the iNTP binding in the RNAP active centre and in the first RNA bond formation (Figure 6A, upper row, left). Deletions in region σ3.2 and substitutions of amino acids that directly contact the template DNA strand impaired iNTP binding and made transcription dependent on short RNA primers (Figure 6A, lower row, left). At the same time, as we previously showed, deletion of the σ3.2 loop did not impair promoter melting by RNAP and did not change promoter complex stability (11). Amino acid substitutions in region σ3.2 had a cumulative impact on transcription: while individual substitutions only marginally affected transcription initiation, the quadruple substitution at the tip of the σ3.2 loop had the strongest effect on the first phosphodiester bond formation, comparable to the σ3.2 deletions (Figure 1 and Table 1). The binding of the 3′-NTP was affected to a greater extent, suggesting that the 5′-NTP binding is less dependent on the σ subunit, in agreement with previous observations (13,14,27). In addition to increasing apparent KMs for iNTPs, substitutions and deletions in region σ3.2 increased the amounts of trinucleotide RNA products synthesized during initiation (Figure 4), likely due to destabilization of their binding in the RNAP active centre. Similar effects of various changes in region σ3.2 on the first steps of RNA synthesis are most likely explained by changes in the positioning of the template DNA strand in the active centre, which requires specific contacts of individual σ3.2 residues with the DNA bases [Figure 1B; (7)]. In addition, region σ3.2 may affect the template DNA positioning through β′ region switch2 that contacts both σ and DNA [Figure 1B; (21)].
The proposed role of region σ3.2 in the template positioning likely explains the observed inhibitory effects of Rif antibiotics on the iNTP binding and the first phosphodiester bond formation [Table 1 and (29)]. As was recently proposed, Rif may affect the conformation of the template DNA strand through region 3.2 (10); our demonstration that the effects of Rif on the KMs for iNTPs are much weaker in the case of σ3.2 mutations provide an experimental support to this hypothesis. Importantly, region σ3.2 was proposed to be targeted by another antibiotic, lipiarmycin, which was proposed to inhibit transcription by blocking the DNA template fitting in the RNAP active centre (45). This identifies region σ3.2 as a potential target for development of new antibacterial compounds.
After starting the RNA synthesis, region σ3.2 directly participates in abortive transcription and promoter escape (Figure 6A, middle). Deletions in region σ3.2 led to pronounced defects in promoter clearance and significantly affected the pattern of abortive products synthesized by RNAP on the T7A1cons promoter, which is characterized by hampered promoter escape even in the case of wild-type RNAP, and, similarly, on the galP1 promoter. All three deletions promoted more efficient extension of middle-size (∼5–10 nt) abortive products, apparently by stabilizing their binding in the initiating complex, but dramatically decreased productive RNA synthesis. Thus, region σ3.2 facilitates efficient promoter clearance by RNAP, likely through physical competition with the growing RNA in the RNA exit channel, further leading to σ dissociation and disruption of specific RNAP-promoter contacts (see below). In support of this hypothesis, the largest deletion in region σ3.2 (Δ507–519) affected the synthesis of longer abortive RNA products than the two smaller deletions, likely because it removed a larger part of the σ3.2 linker located within the RNA exit channel. Notably, amino acid substitutions in region σ3.2 had much weaker effects on the pattern of abortive products and the efficiency of promoter clearance. Thus, the function of region σ3.2 in abortive synthesis and promoter escape is less dependent on the amino acid–specific contacts with the template DNA strand and likely requires its physical presence in the RNA exit channel.
Finally, region σ3.2 likely plays a role in σ dissociation during initiation and, as a consequence, modulates the efficiency of σ-dependent promoter-proximal pausing (Figure 6A, right). Based on structural data, σ dissociation was proposed to be initiated by extrusion of regions σ3.2 and σ4 from the RNA exit channel coupled to RNA extension, ultimately leading to disruption of σ-dependent RNAP-promoter interactions (8,9,33,39). However, structural modelling suggests that σ may retain contacts with the elongation complex through interactions of region σ2 with a conserved coiled-coil element of the RNAP clamp domain, explaining the ability of σ to stimulate transcription pausing (39). We now demonstrate that mutations in region σ3.2 strongly stimulate σ-dependent promoter-proximal pausing, directly suggesting that this region promotes complete σ dissociation during initiation. The pausing was stimulated by both deletions and substitutions in region σ3.2, suggesting that both the physical presence and the intact structure of region σ3.2 are required for the proper extrusion of σ from the transcription complex. Deletion of the largest size (Δ507–519) led to the most pronounced stimulation of σ-dependent pausing, suggesting that removal of a larger part of the σ3.2 linker has a stronger effect on σ dissociation. This correlates with the strengths of the effects of various deletions on the pattern of abortive RNA products and promoter escape by RNAP (Figure 4). Point substitution F522A that changed a residue located upstream from the active site in the RNA exit path (see Figure 1B) also stimulated σ-dependent pausing, but did not affect RNA priming. Thus, this conserved residue may have a specific role in σ dissociation, by promoting the displacement of region σ3.2 by the growing RNA. This substitution decreased the amounts of short 3–6 nt RNAs synthesized during initiation (Figure 2), suggesting that the conserved phenylalanine residue may clash with the growing RNA, resulting in either RNA dissociation or σ displacement. Interestingly, the σ3.2 changes had stronger effects on the pausing than on promoter escape by RNAP, which was not significantly affected on the T7A1 templates (Figures 2A and 5B). Thus, proper extrusion of region σ3.2 from the RNA exit channel likely has a specific role in σ dissociation during promoter escape.
The reported effects of changes in region σ3.2 on the iNTP binding, RNA priming, promoter escape and pausing raise the possibility that this region might serve as a target for transcription regulation. This may be particularly important for highly regulated rRNA promoters that have low open complex stability (24,25,46) and, probably, differ in the positioning of the template DNA strand contacted by σ3.2. We observed that mutations in σ3.2 significantly affected transcription initiation on the rrnB P1 promoter. Previously, two point substitutions near the σ3.2 loop, P504L and S506F, were shown to partially suppress growth defects of E. coli strain lacking ppGpp, apparently due to a decreased activity of rRNA promoters (47). Interestingly, in contrast to the σ3.2 loop mutations studied in our work, the P504L and S506F substitutions enhanced promoter escape by RNAP, probably by changing the conformation of region σ3.2 and facilitating σ dissociation (48). An interesting goal of the future studies will be to reveal a possible interplay between region σ3.2 and known regulators of stringent response promoters, such as ppGpp and DksA, during transcription initiation. Another important goal will be to determine the functions of regions σ3.2 in alternative σ factors in transcription initiation from their respective promoters. Recently, reduced promoter melting activity of alternative σs has been proposed to serve as a regulatory mechanism that increases the stringency of promoter recognition and enables a focused response of the target regulons to altered conditions (49). Variations in the RNA priming functions of region σ3.2 in alternative σ factors (or σ2–σ4 linkers in ECF σs lacking σ3.2) might also serve for transcription regulation, for example, by linking σ activities to intracellular NTP concentrations, similar to rRNA promoters (25).
In eukaryotic RNAPII, the general transcription factor TFIIB likely plays similar functions in transcription initiation and promoter escape as region σ3.2 in the bacterial system. The B-reader region of TFIIB occupies a similar position in the RNA exit channel and directly contacts −7/−8 bases of the template DNA strand, playing a role in stabilization of the template DNA and short RNA–DNA hybrids in the active centre (Figure 6B) (20). Similar to region σ3.2, the B-reader loop blocks extension of the RNA–DNA hybrid past 6 nt (Figure 6B) and was proposed to play a role in separation of the RNA product and destabilization of the TFIIB binding to RNAPII (20). Interestingly, both region σ3.2 and TFIIB B-reader contain conserved negatively charged residues at the tips of their loops [Figure 1 and (20)], which may be involved in charge repulsion with the nascent RNA. Substitutions in the B-reader were shown to affect start site selection, and deletions in the B-reader loop dramatically impaired transcription initiation by yeast and human RNAPs [Figure 6B; see (20) and references therein]. Structural parallels between the σ subunit and TFIIB likely reflect the basic similarities of the underlying process of transcription initiation that involves specific DNA recognition, primer-independent RNA initiation and promoter complex dissociation, and may suggest an ancient orthology of these factors (50).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Russian Academy of Sciences Presidium Program in Molecular and Cellular Biology; Russian Foundation for Basic Research [12-04-33187 and 12-04-32042]; Federal Targeted Program ‘Scientific and scientific-pedagogical personnel of innovative Russia 2009-2013’ [8129 and 8475]. Funding for open access charge: Russian Academy of Sciences Presidium Program in Molecular and Cellular Biology.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank I. Artsimovitch for critical reading of the manuscript, helpful discussions and plasmids, L. Minakhin for plasmids.
REFERENCES
- 1.Murakami KS, Darst SA. Bacterial RNA polymerases: the wholo story. Curr. Opin. Struct. Biol. 2003;13:31–39. doi: 10.1016/s0959-440x(02)00005-2. [DOI] [PubMed] [Google Scholar]
- 2.Haugen SP, Ross W, Gourse RL. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat. Rev. Microbiol. 2008;6:507–519. doi: 10.1038/nrmicro1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saecker RM, Record MT, Jr, Dehaseth PL. Mechanism of bacterial transcription initiation: RNA polymerase - promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J. Mol. Biol. 2011;412:754–771. doi: 10.1016/j.jmb.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feklistov A, Barinova N, Sevostyanova A, Heyduk E, Bass I, Vvedenskaya I, Kuznedelov K, Merkiene E, Stavrovskaya E, Klimasauskas S, et al. A basal promoter element recognized by free RNA polymerase sigma subunit determines promoter recognition by RNA polymerase holoenzyme. Mol. Cell. 2006;23:97–107. doi: 10.1016/j.molcel.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 5.Haugen SP, Berkmen MB, Ross W, Gaal T, Ward C, Gourse RL. rRNA promoter regulation by nonoptimal binding of sigma region 1.2: an additional recognition element for RNA polymerase. Cell. 2006;125:1069–1082. doi: 10.1016/j.cell.2006.04.034. [DOI] [PubMed] [Google Scholar]
- 6.Feklistov A, Darst SA. Structural basis for promoter-10 element recognition by the bacterial RNA polymerase sigma subunit. Cell. 2011;147:1257–1269. doi: 10.1016/j.cell.2011.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, Feng Y, Chatterjee S, Tuske S, Ho MX, Arnold E, Ebright RH. Structural basis of transcription initiation. Science. 2012;338:1076–1080. doi: 10.1126/science.1227786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murakami KS, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution. Science. 2002;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- 9.Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 10.Molodtsov V, Nawarathne IN, Scharf NT, Kirchhoff PD, Showalter HD, Garcia GA, Murakami KS. X-ray crystal structures of the Escherichia coli RNA polymerase in complex with Benzoxazinorifamycins. J. Med. Chem. 2013;56:4758–4763. doi: 10.1021/jm4004889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kulbachinskiy A, Mustaev A. Region 3.2 of the sigma subunit contributes to the binding of the 3′-initiating nucleotide in the RNA polymerase active center and facilitates promoter clearance during initiation. J. Biol. Chem. 2006;281:18273–18276. doi: 10.1074/jbc.C600060200. [DOI] [PubMed] [Google Scholar]
- 12.Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science. 2002;296:1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- 13.Hansen UM, McClure WR. A noncycling activity assay for the sigma subunit of Escherichia coli RNA polymerase. J. Biol. Chem. 1979;254:5713–5717. [PubMed] [Google Scholar]
- 14.Hansen UM, McClure WR. Role of the sigma subunit of Escherichia coli RNA polymerase in initiation. I. Characterization of core enzyme open complexes. J. Biol. Chem. 1980;255:9556–9563. [PubMed] [Google Scholar]
- 15.Zenkin N, Severinov K. The role of RNA polymerase sigma subunit in promoter-independent initiation of transcription. Proc. Natl Acad. Sci. USA. 2004;101:4396–4400. doi: 10.1073/pnas.0400886101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- 17.Campbell EA, Tupy JL, Gruber TM, Wang S, Sharp MM, Gross CA, Darst SA. Crystal structure of Escherichia coli sigmaE with the cytoplasmic domain of its anti-sigma RseA. Mol. Cell. 2003;11:1067–1078. doi: 10.1016/s1097-2765(03)00148-5. [DOI] [PubMed] [Google Scholar]
- 18.Zhou YN, Walter WA, Gross CA. A mutant sigma 32 with a small deletion in conserved region 3 of sigma has reduced affinity for core RNA polymerase. J. Bacteriol. 1992;174:5005–5012. doi: 10.1128/jb.174.15.5005-5012.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kostrewa D, Zeller ME, Armache KJ, Seizl M, Leike K, Thomm M, Cramer P. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature. 2009;462:323–330. doi: 10.1038/nature08548. [DOI] [PubMed] [Google Scholar]
- 20.Sainsbury S, Niesser J, Cramer P. Structure and function of the initially transcribing RNA polymerase II-TFIIB complex. Nature. 2013;493:437–440. doi: 10.1038/nature11715. [DOI] [PubMed] [Google Scholar]
- 21.Pupov D, Miropolskaya N, Sevostyanova A, Bass I, Artsimovitch I, Kulbachinskiy A. Multiple roles of the RNA polymerase β' SW2 region in transcription initiation, promoter escape, and RNA elongation. Nucleic Acids Res. 2010;38:5784–5796. doi: 10.1093/nar/gkq355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kulbachinskiy A, Bass I, Bogdanova E, Goldfarb A, Nikiforov V. Cold sensitivity of thermophilic and mesophilic RNA polymerases. J. Bacteriol. 2004;186:7818–7820. doi: 10.1128/JB.186.22.7818-7820.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhilina E, Esyunina D, Brodolin K, Kulbachinskiy A. Structural transitions in the transcription elongation complexes of bacterial RNA polymerase during sigma-dependent pausing. Nucleic Acids Res. 2012;40:3078–3091. doi: 10.1093/nar/gkr1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gourse RL. Visualization and quantitative analysis of complex formation between E. coli RNA polymerase and an rRNA promoter in vitro. Nucleic Acids Res. 1988;16:9789–9809. doi: 10.1093/nar/16.20.9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaal T, Bartlett MS, Ross W, Turnbough CL, Jr, Gourse RL. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science. 1997;278:2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- 26.McClure WR, Cech CL, Johnston DE. A steady state assay for the RNA polymerase initiation reaction. J. Biol. Chem. 1978;253:8941–8948. [PubMed] [Google Scholar]
- 27.Naryshkina T, Mustaev A, Darst SA, Severinov K. The beta ‘subunit of Escherichia coli RNA polymerase is not required for interaction with initiating nucleotide but is necessary for interaction with rifampicin. J. Biol. Chem. 2001;276:13308–13313. doi: 10.1074/jbc.M011041200. [DOI] [PubMed] [Google Scholar]
- 28.Artsimovitch I, Vassylyeva MN, Svetlov D, Svetlov V, Perederina A, Igarashi N, Matsugaki N, Wakatsuki S, Tahirov TH, Vassylyev DG. Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins. Cell. 2005;122:351–363. doi: 10.1016/j.cell.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 29.McClure WR, Cech CL. On the mechanism of rifampicin inhibition of RNA synthesis. J. Biol. Chem. 1978;253:8949–8956. [PubMed] [Google Scholar]
- 30.Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, Darst SA. Structural mechanism for rifampicin inhibition of bacterial rna polymerase. Cell. 2001;104:901–912. doi: 10.1016/s0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- 31.Feklistov A, Mekler V, Jiang Q, Westblade LF, Irschik H, Jansen R, Mustaev A, Darst SA, Ebright RH. Rifamycins do not function by allosteric modulation of binding of Mg2+ to the RNA polymerase active center. Proc. Natl Acad. Sci. USA. 2008;105:14820–14825. doi: 10.1073/pnas.0802822105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campbell EA, Pavlova O, Zenkin N, Leon F, Irschik H, Jansen R, Severinov K, Darst SA. Structural, functional, and genetic analysis of sorangicin inhibition of bacterial RNA polymerase. EMBO J. 2005;24:674–682. doi: 10.1038/sj.emboj.7600499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nickels BE, Garrity SJ, Mekler V, Minakhin L, Severinov K, Ebright RH, Hochschild A. The interaction between sigma70 and the beta-flap of Escherichia coli RNA polymerase inhibits extension of nascent RNA during early elongation. Proc. Natl Acad. Sci. USA. 2005;102:4488–4493. doi: 10.1073/pnas.0409850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burns HD, Belyaeva TA, Busby SJ, Minchin SD. Temperature-dependence of open-complex formation at two Escherichia coli promoters with extended -10 sequences. Biochem J. 1996;317(Pt. 1):305–311. doi: 10.1042/bj3170305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minakhin L, Severinov K. On the role of the Escherichia coli RNA polymerase sigma 70 region 4.2 and alpha-subunit C-terminal domains in promoter complex formation on the extended -10 galP1 promoter. J. Biol. Chem. 2003;278:29710–29718. doi: 10.1074/jbc.M304906200. [DOI] [PubMed] [Google Scholar]
- 36.Ring BZ, Yarnell WS, Roberts JW. Function of E. coli RNA polymerase sigma factor sigma 70 in promoter-proximal pausing. Cell. 1996;86:485–493. doi: 10.1016/s0092-8674(00)80121-x. [DOI] [PubMed] [Google Scholar]
- 37.Brodolin K, Zenkin N, Mustaev A, Mamaeva D, Heumann H. The sigma 70 subunit of RNA polymerase induces lacUV5 promoter-proximal pausing of transcription. Nat. Struct. Mol. Biol. 2004;11:551–557. doi: 10.1038/nsmb768. [DOI] [PubMed] [Google Scholar]
- 38.Nickels BE, Mukhopadhyay J, Garrity SJ, Ebright RH, Hochschild A. The sigma 70 subunit of RNA polymerase mediates a promoter-proximal pause at the lac promoter. Nat. Struct. Mol. Biol. 2004;11:544–550. doi: 10.1038/nsmb757. [DOI] [PubMed] [Google Scholar]
- 39.Mooney RA, Darst SA, Landick R. Sigma and RNA polymerase: an on-again, off-again relationship? Mol. Cell. 2005;20:335–345. doi: 10.1016/j.molcel.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 40.Zenkin N, Kulbachinskiy A, Yuzenkova Y, Mustaev A, Bass I, Severinov K, Brodolin K. Region 1.2 of the RNA polymerase sigma subunit controls recognition of the -10 promoter element. EMBO J. 2007;26:955–964. doi: 10.1038/sj.emboj.7601555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perdue SA, Roberts JW. sigma(70)-dependent transcription pausing in Escherichia coli. J. Mol. Biol. 2011;412:782–792. doi: 10.1016/j.jmb.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 42.Deighan P, Pukhrambam C, Nickels BE, Hochschild A. Initial transcribed region sequences influence the composition and functional properties of the bacterial elongation complex. Genes Dev. 2011;25:77–88. doi: 10.1101/gad.1991811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perdue SA, Roberts JW. A backtrack-inducing sequence is an essential component of Escherichia coli sigma(70)-dependent promoter-proximal pausing. Mol. Microbiol. 2010;78:636–650. doi: 10.1111/j.1365-2958.2010.07347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vassylyev DG, Vassylyeva MN, Zhang J, Palangat M, Artsimovitch I, Landick R. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448:163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- 45.Tupin A, Gualtieri M, Leonetti JP, Brodolin K. The transcription inhibitor lipiarmycin blocks DNA fitting into the RNA polymerase catalytic site. EMBO J. 2010;29:2527–2537. doi: 10.1038/emboj.2010.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rutherford ST, Villers CL, Lee JH, Ross W, Gourse RL. Allosteric control of Escherichia coli rRNA promoter complexes by DksA. Genes Dev. 2009;23:236–248. doi: 10.1101/gad.1745409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hernandez VJ, Cashel M. Changes in conserved region 3 of Escherichia coli sigma 70 mediate ppGpp-dependent functions in vivo. J. Mol. Biol. 1995;252:536–549. doi: 10.1006/jmbi.1995.0518. [DOI] [PubMed] [Google Scholar]
- 48.Cashel M, Hsu LM, Hernandez VJ. Changes in conserved region 3 of Escherichia coli sigma 70 reduce abortive transcription and enhance promoter escape. J. Biol. Chem. 2003;278:5539–5547. doi: 10.1074/jbc.M211430200. [DOI] [PubMed] [Google Scholar]
- 49.Koo BM, Rhodius VA, Nonaka G, deHaseth PL, Gross CA. Reduced capacity of alternative sigmas to melt promoters ensures stringent promoter recognition. Genes Dev. 2009;23:2426–2436. doi: 10.1101/gad.1843709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iyer LM, Aravind L. Insights from the architecture of the bacterial transcription apparatus. J. Struct. Biol. 2012;179:299–319. doi: 10.1016/j.jsb.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.