Abstract
Folding messenger RNA into specific structures is a common regulatory mechanism involved in translation. In Escherichia coli, the operator of the rpsO gene transcript folds into a pseudoknot or double-hairpin conformation. S15, the gene product, binds only to the pseudoknot, thereby repressing its own synthesis when it is present in excess in the cell. The two RNA conformations have been proposed to exist in equilibrium. However, it remained unclear how structural changes can be achieved between these two topologically distinct conformations. We used optical tweezers to study the structural dynamics and rearrangements of the rpsO operator RNA at the single-molecule level. We discovered that the two RNA structures can be interchanged spontaneously and the pseudoknot can exist in conformations that exhibit various levels of stability. Conversion from the double hairpin to a pseudoknot through potential hairpin–hairpin interactions favoured the high-stability conformation. By contrast, mutations that blocked the formation of a hairpin typically resulted in alternative low-stability pseudoknots. These results demonstrate that specific tertiary interactions of RNA can be established and modulated based on the interactions and rearrangements between secondary structural components. Our findings provide new insight into the RNA folding pathway that leads to a regulatory conformation for target protein binding.
INTRODUCTION
Gene expression at the translation level is highly regulated in many systems. In prokaryotes, translation is initiated when a ribosome binds to the Shine–Dalgarno (SD) sequence and AUG start codon on messenger RNA (mRNA). Thus, modulating the accessibility of the ribosome binding site is a general strategy for controlling translation in the cell (1–3). For example, translation is turned off when the 5′-untranslated region (5′-UTR) of the mRNA folds into a structure that sequesters the ribosome-binding site. Alternatively, translation is turned on when a different structure forms to release the sequestered site. The switch of RNA structures from one to another is usually controlled by regulatory factors such as metabolites, non-coding RNA or proteins (4,5). These factors specifically bind to one of the structures and drive the conformational switch towards this end.
Expression of ribosomal proteins (r-proteins) provides examples of various modes for protein-controlled translation regulation. In addition to binding to the ribosomal RNA (rRNA) for ribosome assembly, an r-protein, when present in excess, may also bind to the operon of its cognate transcript to repress translation (1,6). The autogenous regulation controls the expression of more than half of the r-proteins in Escherichia coli and maintains cellular r-protein concentrations at the stoichiometric levels for ribosome biogenesis (6,7). The dual-binding nature of an r-protein to rRNA and mRNA suggests the existence of structural homology, or molecular mimicry, on the two targets (1,8,9). The feature of similar, but not identical, structural or sequence motifs on the RNAs enables the r-protein to distinguish between the targets. Because of its primary function for ribosome assembly, the r-protein usually binds with a higher affinity for the rRNA than for the mRNA (10–12). On r-protein binding, the ribosome-binding site of the mRNA may be occluded, and thus translation initiation is inhibited. This mode of action, referred to as the displacement mechanism (13), is based on competition between the r-proteins and ribosomes for their mutually exclusive binding sites on the mRNA. By contrast, in the entrapment mechanism, both the r-proteins and ribosomes can bind simultaneously to the mRNA, but conformational rearrangement of the mRNA into the translation-competent form is blocked in the presence of the r-proteins (13–15).
Interestingly, autogenous translational regulation of the S15 r-protein, encoded by the rpsO gene, may be controlled through different mechanisms in different species of bacteria. In Thermus thermophilus, binding of S15 triggers a structural rearrangement of the 5′-UTR of the rpsO mRNA into the three-way junction that mimics the conserved S15-binding site on rRNA (16). The structure sequesters the SD sequence to hinder ribosome binding. Similar patterns of S15 binding to its mRNA have also been observed in Bacillus stearothermophilus (17), but the mechanism of translational repression has not been experimentally determined. In E. coli, S15 binds to its mRNA only when the sequence of the regulatory region (including the 5′-UTR and a short coding region) folds into a pseudoknot structure (18,19). Although the pseudoknot shares little structural homology with the S15-binding site on the rRNA, similar binding motifs have been identified on both mRNA and rRNA (9,11). The ribosome binds to the SD sequence on an extensive loop of the pseudoknot (see Figure 1A). Unfolding of the RNA structure is inhibited by concomitant binding of S15 to the pseudoknot, resulting in entrapment of the pre-initiation complex (20,21).
Figure 1.
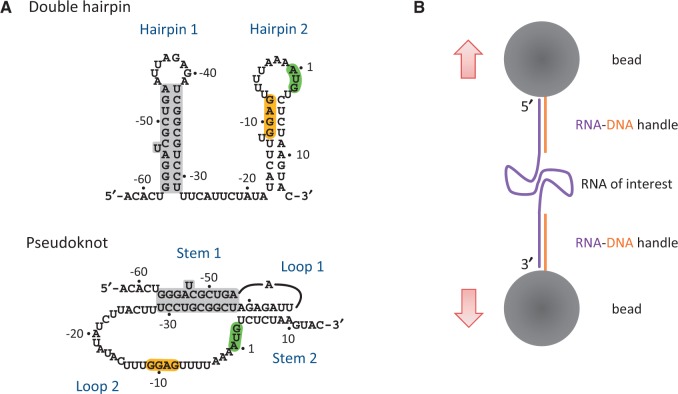
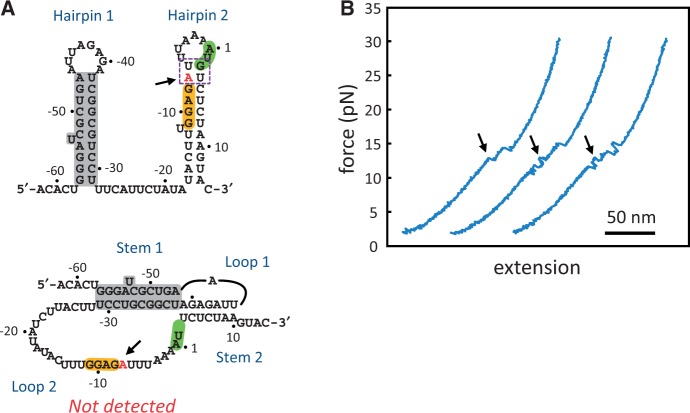
Schematic drawings of RNA structures and the experimental setup. (A) Two proposed RPSOutr structures, double hairpin (top) and pseudoknot (bottom). The common helices appearing on both conformations are highlighted in grey. The SD sequence (GGAG) and start codon (AUG) are also highlighted. (B) Design of optical tweezers for RNA pulling. RNA of interest was flanked by two handles of RNA–DNA duplexes, which were immobilized on the surface of two polystyrene beads through digoxigenin/anti-digoxigenin antibody (top) and biotin/streptavidin (bottom) interactions. The top bead was trapped by laser beams and the bottom bead was fixed on a micropipette.
The regulatory region of E. coli rpsO mRNA, called the ‘operator’, is extremely dynamic in structure. Experiments of chemical and enzymatic probing have shown that this region folds into either the pseudoknot (mentioned previously) or two hairpins (double hairpin) (22,23) (see Figure 1A). The population of the pseudoknot increases with an increase in magnesium concentrations (24). The SD sequence is accessible on the pseudoknot but is base paired in the second stem of the double hairpin. Thus, translation, as well as regulation through the binding of S15, can occur only on the pseudoknot conformation. The role of the other structure, the double hairpin, is vague in this respect and has not been thoroughly examined. In addition, it has been proposed that the two conformations are in a dynamic equilibrium (21–23), but direct experimental evidence of this is lacking. We used optical tweezers (25) to study the structural unfolding/refolding of the E. coli rpsO operator at the single-molecule level. Conformations of an RNA molecule may be inferred from the measured force and extension change, whereas the molecule undergoes a transition from folded to unfolded states (26,27). In addition to wild-type RNA, we constructed a series of mutants, each with one or more structural components strengthened or weakened, to determine how the components affect the overall folding of the pseudoknot and double hairpin.
MATERIALS AND METHODS
Sample preparation
DNA sequences for RPSOutr and the mutants were chemically synthesized and cloned into the pVE60hp plasmid (28) between the XmaI and BsrGI restriction sites. The resulting plasmids, containing a T7 promoter located ∼700 bp upstream from the insertion site, were cut at the BssSI site (∼900 bp downstream from the insertion site) and transcribed into RNA by using the MEGAscript T7 Kit (Invitrogen). Two DNA handles were prepared using PCR, tag labelled and then annealed to the RNA transcripts as described previously (29). In the finished RNA constructs, the 5′ handle was 686 bp long with a digoxigenin tag, and the 3′ handle was 921-bp long with a biotin tag.
Measurements using optical tweezers
The experimental setup for optical tweezers has been described previously (29,30). More detailed description for an instrument similar to the one we used can be found in a recent report (31). Briefly, the two ends of an RNA construct were attached to two polystyrene beads (2.1 µm in diameter; Spherotech) coated with streptavidin and anti-digoxigenin antibodies, respectively. One bead was fixed on a micropipette and the other was trapped by laser beams, the position of which can be tuned to control the relative distance between these two beads. For force-ramp experiments, the trap was moved at a rate of 100 nm/s. For constant-force experiments, the position of the trap was feed-backed (with a frequency of 2 KHz) to maintain a preset force. The experiments were performed in a buffer containing 10 mM Tris–HCl, pH 7.0, 200 mM KCl and 20 mM MgCl2, unless otherwise noted.
Data analysis
The data were recorded using optical tweezers at 1000 Hz and averaged to 100 Hz. Data analysis was performed using custom-written MATLAB programs. The extension change (Δx) for a structural transition was directly measured from the force-extension curve. The contour length (L) of the structure was calculated by the worm-like chain model (32):
where F is the unfolding force, kB is the Boltzmann constant, T is the absolute temperature and P is the persistent length [we used P = 1 nm for single-stranded RNA (33)]. In this calculation, the measured extension change has to be compensated for the finite distance between the termini of the folded structure. We used 2 nm for the diameter of a helix and 0.26 nm/bp (34) for the stacked helices of a pseudoknot. The number of nucleotides involved in folding the structure (i.e. released from the structure during transition) was then computed from the obtained contour length by a simple relationship of 0.59 nm/nt (35). The same worm-like chain model was also used to plot traces, as those shown in Figure 2C and D, to predict transitions when a given structure was unfolded.
Figure 2.
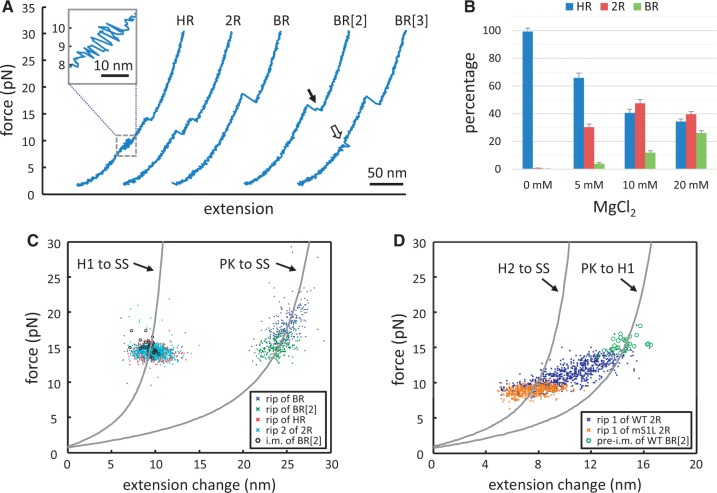
Characterization of the RPSOutr conformations. (A) Force-extension curves reveal distinctive unfolding patterns (HR, 2R and BR) measured by the force-ramp protocols. Some BR types exhibit an intermediate (BR[2], indicated by an arrow), and some reveal transient hopping at low forces (BR[3], indicated by an open arrow). Inset, detail of the hopping transition in HR. (B) Effects of magnesium concentrations on the relative occurrence of the patterns. Errors are estimated from N/√N, where N is the data number. (C and D) Distributions of the force and extension change for each type of unfolding pattern. The corresponding worm-like chain models for structural transitions are plotted for comparison (PK, pseudoknot; SS, single strand; H1, Hairpin 1; H2, Hairpin 2). (C) BR without intermediates (blue, N = 258), BR with an intermediate (BR[2], green, N = 234), the rip of HR (red, N = 670), the second rip of 2R (cyan, N = 832) and the intermediate (i.m.) of BR (i.m. of BR[2], black circles, N = 30). (D) The first rip of 2R (dark blue, N = 832) and the transition to the intermediate in BR (pre-i.m. of BR[2], green circles, N = 30). Transitions of the first rip of 2R from mS1L (brown, N = 507) are also shown for comparison.
RESULTS
The E. coli rpsO operator folds into double-hairpin or pseudoknot structures
To facilitate single-molecule measurements by using optical tweezers, the operator of E. coli rpsO mRNA, named RPSOutr (Figure 1A), was held through flanking ‘handles’ of RNA–DNA duplexes (Figure 1B). The force applied to pull the molecule was gradually increased (a process called force ramp), and the extension change between the two ends of the molecular construct was recorded. The result was plotted as a force-extension curve (Figure 2A). Unfolding of RNA structures during the process was reflected as characteristic transitions on the trace, from which the unfolding force (in pico-Newton, pN) and size (in nm) of the structural transition were measured. Structures were refolded by reversing the steps to relax the tension. From the force-extension curves, three distinctive transition patterns of structural unfolding were identified for RPSOutr (Figure 2A), denoted as HR (‘hopping’ followed by a ‘rip’), 2R (2 rips) and BR (one big rip). Small RNA structures are generally unfolded in a single step, called a rip, or through multiple unfolding-refolding cycles, called hopping (33). Table 1 shows a brief summary for the measured transitions. More details (including the results from mutants, see later in the text) can be found in Supplementary Table S1. The population of the patterns was dependent on magnesium concentrations; HR predominated in the absence of Mg2+, whereas 2R and BR gradually increased with Mg2+ (Figure 2B).
Table 1.
Quantification of unfolding transitions from RPSOutr and related mutants
| Construct | Wild-type | mS1L | mACd14 | |||||
|---|---|---|---|---|---|---|---|---|
| Transition | Rip 1 of 2R | Rip 2 of 2R | Rip of HR | Rip of BR | Rip of BR[2] | Rip 1 of 2R | Rip 1 of 2R | Rip 2 of 2R |
| Structural change | Mixed | H1 to SS | H1 to SS | PK to SS | PK to SS | H2 to SS | PK to H1 | H1 to SS |
| Number of nucleotides releaseda | Mixed | 27–29 nt | 27–29 nt | 67 nt | 67 nt | 26–30 nt | 24–26 nt | 27-29 nt |
| N (rips) | 810 | 826 | 649 | 258 | 232 | 220 | 273 | 273 |
| Unfolding force (pN) | 11.0 ± 1.7 | 14.3 ± 0.8 | 14.3 ± 0.7 | 18.0 ± 2.3 | 15.1 ± 1.4 | 9.0 ± 0.6 | 13.1 ± 1.2 | 14.3 ± 1.0 |
| Extension change (nm) | 10.0 ± 2.2 | 9.9 ± 1.4 | 9.8 ± 1.2 | 24.9 ± 1.4 | 23.9 ± 1.2 | 8.3 ± 0.9 | 7.1 ± 1.2 | 9.5 ± 1.0 |
| Number of nucleotide change, calibratedb | N/Ad | 28.3 ± 3.4 | 28.1 ± 2.8 | 66.8 ± 2.7 | 66.6 ± 2.4 | 27.8 ± 2.3 | 23.0 ± 2.6 | 27.4 ± 2.3 |
| ΔG (KJ/mol)c | N/Ad | 38.3 ± 2.2 | 40.6 ± 1.3 | 124.0 ± 5.2 | 110.5 ± 3.4 | 19.8 ± 1.3 | 22.7 ± 1.0 | 41.0 ± 2.9 |
A complete list can be found in Supplementary Table S1. Data are presented as mean ± SD.
Abbreviations: nt, nucleotide; H1, Hairpin 1; H2, Hairpin 2; SS, single strand; PK, pseudoknot.
aExpected numbers of nucleotides released from the designated structural change. A range is given for those involving Hairpin 1 or 2 because the weak G:U or A:U closing base pairs in the hairpins may or may not pair under force.
bCalibrated by the worm-like chain model for the designated structural change.
cFree energy change for the designated structural change. The RNA tethering energy has been subtracted. See Supplementary Table S1 for more details.
dNot applicable; including a variety of structural transitions.
Given the known structures of RPSOutr (see Figure 1A) and their magnesium dependence (24), the unfolding of the double hairpin was most accurately described by the HR pattern; the ∼10-pN hopping corresponded to fast unfolding/refolding of the A/U-rich Hairpin 2, and the ∼14-pN rip reflected the unwinding of the G/C-rich Hairpin 1. In addition, the size of the rip matched well to the model based on the Hairpin 1 structure (Table 1 and Figure 2C, red). The assignments were further supported by control experiments, in which the rip or hopping disappeared when the formation of one of the hairpins was selectively disabled (Supplementary Figure S1).
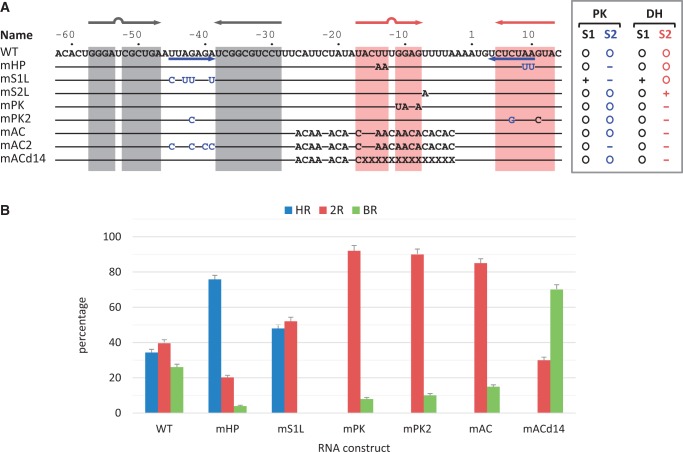
For the pseudoknot, the other reported RPSOutr structure, its unfolding process is most reliably depicted by the BR pattern, because the size of the measured transitions matched well to the model for the pseudoknot (Table 1 and Figure 2C, blue and green). To support the assignment, we made two mutants, mHP and mS1L (Figure 3A), in which the pseudoknot conformation is less likely to form due to partial (mHP) or extensive (mS1L) disruption of Stem 2 of the pseudoknot. As expected, the results show that the population of the BR transition was dramatically reduced in mHP and completely disappeared in mS1L (Figure 3B). A transient intermediate was found in some of the BR patterns from the wild-type sequence (Figure 2A, indicated by an arrow in BR[2]). Although the unfolding force was generally smaller in BR patterns with than in those without the intermediate, structural transitions from both matched equally well to the pseudoknot model (Table 1 and Figure 2C, green versus blue markers). Further analysis indicates that the intermediate was Hairpin 1 because the distribution of force and size of the intermediate (Figure 2C, black circles) was indistinguishable from that of the hairpin, and the transition to the intermediate was consistent with the model for the unfolding from the pseudoknot to Hairpin 1 (Figure 2D, green circles). These results reveal that the pseudoknot can be broken as a whole or through two-step unwinding of A/U-rich Stem 2 followed by G/C-rich Stem 1 (see Figure 1A, bottom).
Figure 3.
RPSOutr and the mutants. (A) List of sequences for the wild-type (WT) and mutants. The regions forming the stems of Hairpin 1 and Hairpin 2 of the double hairpin are highlighted in grey and red, respectively. Sequences forming Stem 2 of the pseudoknot are indicated by blue arrows. In the mutants, only the mutated nucleotides are shown; coloured in blue are those originally involved in pseudoknot base paring. X denotes deletion. The enclosed list to the right indicates stability changes relative to the wild-type in Stem 1 (S1) and Stem 2 (S2) of the pseudoknot (PK) and double-hairpin (DH) conformations. (B) Relative occurrence of unfolding patterns for each construct. Errors are estimated from N/√N, where N is the data number.
Identification of alternative pseudoknot conformations
Unlike the others, the third-pattern 2R observed during the unfolding of RPSOutr was apparently contributed by more than one unique structure. Whereas the second rip of 2R (at 14 pN) corresponded to Hairpin 1 as in the HR pattern (Table 1 and Figure 2C, cyan), transitions of the first rip were distributed in a wide range in sizes and forces (Figure 2D, dark blue), of which a fraction overlapped with those from mS1L (Figure 2D, brown). As mentioned, mutations in mS1L prevent it from forming the pseudoknot; the double hairpin becomes the dominant structure to fold. Approximately one-half of the unfolding traces from mS1L were the HR type (i.e. the double hairpin) and the other half were 2R (Figure 3B). Furthermore, the first and second rips of the 2R transitions matched well to Hairpin 2 (Table 1 and Figure 2D, ‘H2 to SS’) and Hairpin 1 (Supplementary Table S1 and Supplementary Figure S2), respectively. In other words, mS1L folded into only the double-hairpin conformation, and the unfolding of Hairpin 2 can appear as hopping (resulting in the HR pattern) or a rip (2R pattern). The clear rips in this case allowed us to calculate the unfolding free energy of Hairpin 2 (19.8 ± 1.3 KJ/mol, Table 1), which was otherwise difficult to measure unambiguously from fast hopping transitions. The value can serve as an estimate for the free energy of wild-type Hairpin 2, as its component sequence is identical to that of mS1L (see Figure 3A).
Based on the analysis for mS1L, some 2R transitions from the wild-type (overlapped with those of mS1L shown in Figure 2D) can be assigned to the double-hairpin conformation, whereas the majority may involve tertiary interactions between the loop of Hairpin 1 and a downstream sequence, because these transitions were missing in mS1L, of which Hairpin 1 loop was mutated. Such tertiary interactions can result in various pseudoknot conformations, including the ‘canonical’ conformation (as shown in Figure 1A) with the loop pairing to its cognate sequence UCUCUAA (positions 4–10, see Figure 3A) near the 3′ end. As shown in Figure 2D, only a minor fraction of the 2R transitions matched the two-step unfolding pattern of the canonical pseudoknot (‘PK to H1’), but most of them appeared to be smaller and less stable (compared with the green circles in Figure 2D). These ‘alternative’ pseudoknots could be the result of interactions between the loop of Hairpin 1 and a closer less-matched sequence (alternative base pairing), or between the loop and its cognate sequence with altered conformations (alternative folding). To suppress possible alternative base pairing, we constructed the mutant mAC (for A/C-rich; Figure 3A), in which most nucleotides between Hairpin 1 and the cognate sequence UCUCUAA were changed to A or C. The results reveal that the 2R patterns mainly persisted in mAC (Figure 3B), and their distribution was even wider than that of the wild-type (Supplementary Figure S3). By contrast, the 2R-type transitions were not detected when the UCUCUAA sequence of mAC was masked (Supplementary Figure S4A), indicating that base pairing between the cognate sequence and the loop of Hairpin 1 was crucial to result in the 2R-type transitions. Alternatively, we made another mutant mAC2 (based on mAC; Figure 3A) by mutating the loop of Hairpin 1 to prevent it from pairing to the downstream UCUCUAA sequence. Again, the results show that no 2R-type transitions were observed (Supplementary Figure S4B). Overall, all the data suggest that the loop of Hairpin 1 can interact with the downstream complementary sequence to form the high-stability pseudoknot (appearing as BR transitions) or alternatively folded less stable pseudoknots (as 2R transitions).
The low-stability, instead of high-stability, pseudoknots dominated in mutants with disabled Hairpin 2
In RPSOutr, the sequences involved in base pairing to form the pseudoknot or double hairpin mainly overlap. Thus, mutating the sequence uniquely to form one structure promotes the formation of the other. As demonstrated in the mS1L mutant, the double-hairpin conformation is predominant when mutations that could otherwise be uniquely base paired to form the pseudoknot are introduced to the Hairpin 1 loop. By contrast, the equilibrium was shifted to the pseudoknot conformations in mAC because the unique sequence to form Hairpin 2 of the double hairpin was mutated. However, the pseudoknots detected in mAC were predominantly the alternative conformers; the population of the high-stability pseudoknot (with BR transitions) was even lower than that of the wild-type (see Figure 3B). Similarly, another mutant mPK (Figure 3A) was constructed such that Hairpin 2 was destabilized, but the base pairs to form the pseudoknot were maintained. As in mAC, the data from mPK show a low population for the high-stability pseudoknot (Figure 3B).
Given the pseudoknot conformations, the Loop 2 sequences are the only difference between the wild-type and the Hairpin-2-disabled mutants, such as mAC and mPK. This extensive loop, containing 31 nt, may partly form a small internal stem-loop structure in the wild-type (Supplementary Figure S5), but it is essentially unstructured in mAC and mPK. The overall stability of the pseudoknots may be altered by structural perturbation of Loop 2, as has been demonstrated recently for a riboswitch (36). In this scenario, restoring the small stem-loop structure in Loop 2 of the Hairpin-2-disabled mutants would recover the structural stability. Accordingly, we constructed such a mutant mPK2 (Figure 3A), in which a G:C base pair was swapped to retain the pseudoknot but to disrupt Hairpin 2 of the double hairpin. The results reveal that the patterns of mPK2 structural transitions were comparable with those of mAC and mPK (Figure 3B). Thus, the capability to form Hairpin 2 in RPSOutr appears to play a key role in folding into the high-stability pseudoknot structure.
Formation of the high-stability pseudoknot was facilitated by the hairpin–hairpin interactions of the double-hairpin conformation
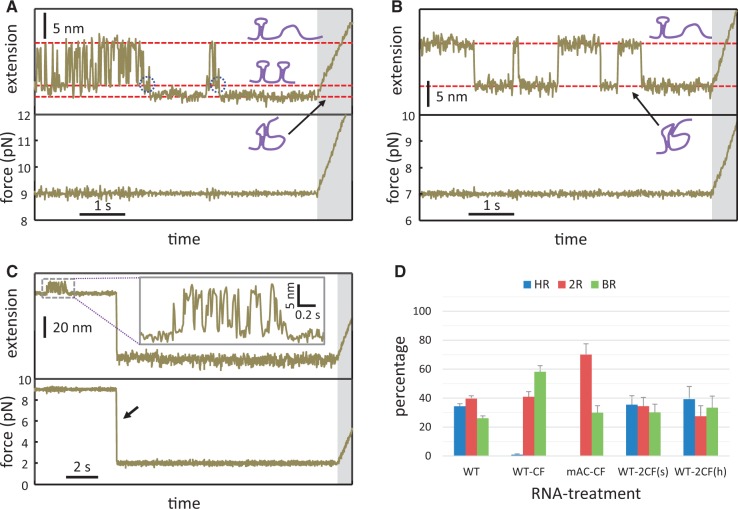
The unfolding patterns of wild-type RPSOutr occasionally appeared in a mixed mode. As shown in Figure 2A, a brief hopping transition at ∼9 pN (indicated by an open arrow in BR[3]) was followed by a big rip at ∼18 pN. The brief hopping transition corresponded to Hairpin 2 of the double hairpin (compared with the HR pattern in the same figure) and the big rip was a typical transition for the high-stability pseudoknot (BR pattern). Thus, this example suggests that the RNA can first fold into the double hairpin, from which structural rearrangement follows to form the pseudoknot. To test this hypothesis, we held the wild-type RNA at a constant force (e.g. 9 pN) such that Hairpin 1 was folded, whereas Hairpin 2 was bistable and underwent fast hopping reactions (i.e. changes between folded and unfolded structures; Figure 4A). In addition to fast hopping, the RNA occasionally experienced a smooth and stable state, which was much longer in lifetime and slightly shorter in extension than the state when Hairpin 2 was folded. To identify the structure, the RNA (while retained in the stable state) was pulled using the regular force-ramp protocol. The results show that almost 60% of these stable states appeared in BR transitions (Figure 4D, ‘WT-CF’). By contrast, mAC underwent slow transitions between only two smooth states (Figure 4B); fast hopping was absent because the mutant did not form any bistable structures as did Hairpin 2 of the wild-type. Structural determination by the force-ramp protocol demonstrated that only 30% of the folded state (with a smaller extension in Figure 4B) appeared in BR transitions (Figure 4D, ‘mAC-CF’). Thus, the probability of forming the high-stability pseudoknots was considerably decreased in the Hairpin-2-disabled mutant.
Figure 4.
Constant-force experiments. Time-evolved extension change of the wild-type (A) and mAC (B) under a preset force (9 and 7 pN, respectively). RNA conformations corresponding to each extension state are schematically illustrated. Possible folding intermediates (in panel A) to the pseudoknot are indicated by dotted circles. The force-ramp protocol (shaded regions) was followed to determine the folded structure. (C) Two-step constant-force experiments for the wild-type. The same procedure was conducted as in panel A, except that the preset force was quickly dropped to 2 pN (indicated by an arrow) and maintained for 15 s before proceeding to the force-ramp protocol. (D) Relative occurrence of unfolding patterns after constant-force experiments for the wild-type or mAC. Errors are estimated from N/√N, where N is the data number. CF, constant force with a smooth-extension state (as in panel A); 2CF(s), two-step constant force with a smooth-extension state (as in panel C); 2CF(h), two-step constant force with a hopping-extension state (see the text for details).
The rationale for these observations is provided as follows. A pseudoknot forms when the loop of Hairpin 1 base pairs to its far downstream complementary sequence. This process is entropically facilitated when the sequence is brought closer in space through the folding of Hairpin 2, the bistable nature of which makes this sequence accessible. In other words, interactions between Hairpin 1 and Hairpin 2 can result in the formation of pseudoknots, preferentially with a high-stability conformation. Specifically, the transition to the pseudoknot directly from the state when Hairpin 2 was folded can be detected from the time-evolved extension trajectories in the constant-force experiments (Figure 4A, indicated by dotted circles; see Supplementary Figure S6 for more examples). Consistent with the model, pseudoknot formation is suppressed when Hairpin 2 is further stabilized. This was confirmed by the mutant mS2L (Figure 3A), in which the stability of Hairpin 2 was increased by a point mutation that resulted in the formation of two extra base pairs in the stem (Figure 5A), a structure predicted by mfold (37). We discovered that all the unfolding patterns of mS2L were from the two individual hairpins, and pseudoknot transitions were not detected (Figure 5B).
Figure 5.
Characterization of the mS2L mutant. (A) The predicted double-hairpin structure from mfold (top). The single mutated nucleotide is indicated by arrows. Compared with the wild-type, Hairpin 2 in mS2L was further stabilized by two extra base pairs (boxed). The corresponding pseudoknot conformation (bottom) was not detected in our measurements, though all the involved base pairs were retained. (B) Representative force-extension curves. Because of its increased stability, Hairpin 2 was unfolded at 11–13 pN (indicated by arrows) and appeared as a rip (first trace) or hopping (second and third traces). The measured unfolding free energy was 28.1 ± 0.7 KJ/mol (Supplementary Table S1). As in the wild-type, the unfolding of Hairpin 1 still occurred at 14–15 pN, with an unfolding free energy of 38.8 ± 1.9 KJ/mol (Supplementary Table S1).
In addition, if shortening the effective distance between the Hairpin-1 loop and its complementary sequence through forming an intermediate hairpin is necessary to facilitate the folding of stable pseudoknots, then similar results may be obtained by partially deleting the connecting sequence. This was tested by using another mutant mACd14 (Figure 3A), derived from mAC by removing 14 nt (approximately half the size of Hairpin 2) from the A/C-rich region. The results show that the population of the high-stability pseudoknot jumped to 70% (BR, Figure 3B), indicating that the length of the connecting sequence did play a role in the pseudoknot folding process. As in mAC, the remaining 2R transitions were mostly contributed by low-stability pseudoknots.
Conformations of the pseudoknot and double hairpin are mutually convertible
We showed that wild-type RPSOutr underwent repeated structural rearrangements between the pseudoknot and double hairpin at a moderate force (8.5–11 pN), causing Hairpin 2 to become bistable. To test whether such rearrangements can occur at a lower tension in which each structural component of the RNA is not destabilized by the external mechanical force, we programmed a two-step constant-force procedure. As demonstrated in Figure 4C, while the RNA was held at 9 pN and retained in the stable (pseudoknot) state (referred to as the initial state), the force was quickly dropped to and held at a low value (2 pN) for 15 s to allow spontaneous structural rearrangements, if any. The finally folded structures were determined by the regular force-ramp protocol. This procedure was also applied to the initial state exhibiting hopping (with double hairpin). As shown in Figure 4D [‘WT-2CF(s)’ and ‘WT-2CF(h)’], regardless of the initial conformational state of the RNA, populations of the HR (double hairpin), BR (pseudoknot) and 2R (mixed) transitions were similar and comparable with those measured from only the regular force-ramp experiments. The results strongly suggest that the RPSOutr RNA continuously and reversibly samples the pseudoknot and double-hairpin conformations in the low- or even zero-force regimes.
DISCUSSION
In this study, we used optical tweezers to investigate the structural dynamics of RPSOutr, which is the operator of E. coli rpsO mRNA involved in binding the ribosome and S15 protein for translational regulation. We discovered that the RNA underwent spontaneous rearrangements between two structures, the pseudoknot and double hairpin. The pseudoknot is the binding target for regulation. Such rearrangements appear to be essential for the RNA to fold into a stable pseudoknot conformation because the Hairpin-2-disabled mutants (mAC, mPK and mPK2) tended to form alternative low-stability pseudoknots. Interestingly, alternatively or partially folded pseudoknots have also been detected in a variety of RNA molecules (38–43).
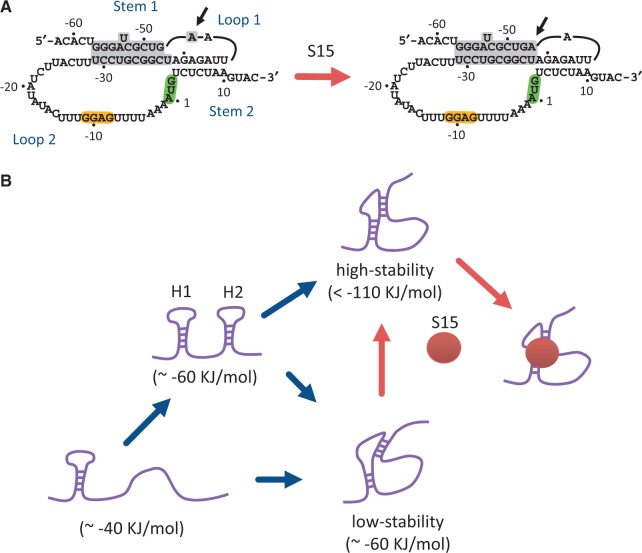
Although detailed X-ray or nuclear magnetic resonance structures of RPSOutr are not currently available, previous studies using chemical and enzymatic probing have provided great insight into the base pairing states, from which possible folded conformations can be inferred. Whereas wild-type RNA exists in both double-hairpin and pseudoknot structures, a mutant having a destabilized Hairpin 2 (possessing a point mutation in the stem) appears primarily as a pseudoknot (44). However, Loop 1 of this pseudoknot contains an additional nucleotide A(−47), which is base paired to U(−38) when the pseudoknot is bound by the regulatory r-protein S15 (Figure 6A) (19,44). This mutant is analogous to our Hairpin-2-disabled mutants (mAC, mPK and mPK2). In the absence of S15, the broken A(−47):U(−38) base pair on the helix junction is likely to disrupt the integrity of the pseudoknot, resulting in reduced mechanical stability, as we observed for these mutants. By contrast, a minimal RPSOutr pseudoknot that does not lose S15-binding affinity was identified (24); the mutant is truncated by 22 nt in Loop 2 (only 9 nt remaining). The reactivity of the corresponding A(−47) to the probes appeared to be decreased in the absence of S15 (24), indicating that the A(−47):U(−38) base pair tends to be retained in this truncated pseudoknot to increase its stability. These results can account for the increased population of high-stability pseudoknots from the measurements of mACd14, in which 14 nt were deleted from Loop 2 (see Figure 3). When combined with the probing data, our results suggest that the Hairpin-2-disabled mutants prefer to fold into an alternative pseudoknot with a distorted stacking helix and reduced mechanical stability, but that the stacking state can be effectively reshaped when the linker connecting the two stems is shortened, when the RNA is bound by S15 or when the stacking is established through the rearrangement of two preformed hairpins.
Figure 6.
A model for RPSOutr folding and S15 binding. (A) A possible conformational change from a low-stability (left) to a high-stability (right) pseudoknot induced by the binding of r-protein S15. The nucleotide A (−47), indicated by arrows, is moved from Loop 1 to Stem 1 and paired with U(−38). (B) A schematic of RNA folding and S15-binding pathways. G/C-rich Hairpin 1 usually forms first, followed by A/U-rich Hairpin 2 when its formation is allowed. Structural rearrangement through hairpin–hairpin interactions preferentially results in the high-stability pseudoknot, the conformation of which can be readily bound by S15. RNA folding free energies are shown in parentheses. H1, Hairpin 1; H2, Hairpin 2.
By conducting constant-force experiments, we demonstrated that wild-type RPSOutr is subject to spontaneous rearrangement between the double-hairpin and pseudoknot conformations when Hairpin 2 is bistable. Because Hairpin 1 is a shared and stable component in both conformations (see Figure 1A), it is straightforward to assume that the rearrangement from the double hairpin is through the unwinding of Hairpin 2 into a single strand, followed by the base pairing of the Hairpin-1 loop to its complementary sequence on the unstructured strand. However, based on the following evidence, we suggest that the transition to the pseudoknot is directly caused by hairpin–hairpin interactions on the double-hairpin structure. First, a transient intermediate in the transition to the pseudoknot was captured in some of the constant-force trajectories (Figure 4A and Supplementary Figure S6); the end-to-end distance of the intermediate was apparently shorter than the Hairpin-1-only structure. Second, most of the wild-type pseudoknots formed through the rearrangement under a constant force fell in the high-stability group (∼60% with BR transitions, see Figure 4D), but less (30%) were from the Hairpin-2-disabled mutant mAC (thus without possible hairpin–hairpin interactions). Finally, analysis of the folding process reveals that RPSOutr usually folded first into the double-hairpin conformation, but the high-stability BR transition may appear in the following unfolding process (Supplementary Figure S7), indicating that structural rearrangements must have occurred in these cases. Such structural rearrangement is most likely to occur through hairpin–hairpin interactions because both hairpins remain adequately stable under low forces.
The mechanical unfolding experiments using optical tweezers allowed us to measure the unfolding free energy (ΔG) corresponding to most of the detectable structural transitions, as summarized in Table 1 and Supplementary Table S1. For the wild-type RPSOutr, ΔG of Hairpin 1 (G/C-rich) was around 40 KJ/mol, as reflected in the HR and 2R transitions. For Hairpin 2 (A/U-rich), its fast hopping nature prevented us from correctly measuring the extension change. Instead, we estimate ΔG of this hairpin from the corresponding transition in mS1L (see ‘Results’ section and Table 1), and it is 19.8 ± 1.3 KJ/mol. Thus, ΔG for the double-hairpin structure is ∼60 KJ/mol, assuming no interactions between these two component hairpins. For the high-stability pseudoknot, the measured ΔG were 124.0 ± 5.2 (unfolded in one step; BR) or 110.5 ± 3.4 KJ/mol (unfolded with an intermediate; BR[2]). The ∼10% energy difference may account for a subtle conformational variation on the pseudoknot. Finally, the diverse distribution of structural transitions from the low-stability pseudoknot (see Figure 2D and Supplementary Figure S3A and B) suggests that this type of structures exists in various energy states. To get an approximation, we estimate ΔG of the low-stability pseudoknot from the truncated mutant mACd14, of which the corresponding transition was rather consistent (Supplementary Figure S3C); the value is 63.7 ± 3.9 KJ/mol (summation of Rip 1 and Rip 2 of 2R, Table 1). In summary, both the double-hairpin structure and low-stability pseudoknot are similar in the energy state, which is about half that of the high-stability pseudoknot.
The folding of RNA is generally considered hierarchical; a nucleotide sequence folds into secondary structures, followed by interactions among them to form a tertiary conformation (45). Because of their higher stability, the preformed secondary structures can be minimally distorted in the tertiary interactions (46,47). However, global rearrangements involving secondary and tertiary structural elements have also been observed in a variety of RNA (48–50). These structural transitions are usually slow or unfavourable unless induced by specific triggers, such as divalent cations, metabolites and proteins. By contrast, we demonstrated that two secondary structures (hairpins) can interact and spontaneously refold into a globally different tertiary structure (pseudoknot) in the absence of external triggers. In this case, long-range tertiary interactions can be quickly established through forming an intermediate hairpin (Hairpin 2) that brings the complementary sequence to the proximity of Loop 1 to facilitate base pairing. The detailed molecular mechanism for this process remains elusive, but the presence of an unstable hairpin seems to be a prerequisite, as demonstrated by the Hairpin-2-stabilized mutant mS2L. From this point of view, Hairpin 2 acts in cis to assist in folding the pseudoknot, preferentially into a stable conformation; an alternative and less stable pseudoknot is predominantly formed when the intermediate hairpin is absent (Figure 6B). Thus, our data suggest a biological function for the double-hairpin conformation, which directs the RPSOutr RNA through structural rearrangement to form a binding site specific for the regulatory protein.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Council [98-2311-B-002-017-MY2]; National Health Research Institutes [EX100-10016BC]; National Taiwan University [startup fund to J.-D.W.]. Funding for open access charge: National Science Council.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Ignacio Tinoco, Jr, Ms Yi-Lan Chen and Mr Kai-Chun Chang for critically reading the manuscript.
REFERENCES
- 1.Babitzke P, Baker CS, Romeo T. Regulation of translation initiation by RNA binding proteins. Annu. Rev. Microbiol. 2009;63:27–44. doi: 10.1146/annurev.micro.091208.073514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Unoson C, Wagner EGH. Dealing with stable structures at ribosome binding sites: bacterial translation and ribosome standby. RNA Biol. 2007;4:113–117. doi: 10.4161/rna.4.3.5350. [DOI] [PubMed] [Google Scholar]
- 3.Geissmann T, Marzi S, Romby P. The role of mRNA structure in translational control in bacteria. RNA Biol. 2009;6:153–160. doi: 10.4161/rna.6.2.8047. [DOI] [PubMed] [Google Scholar]
- 4.Serganov A, Nudler E. A decade of riboswitches. Cell. 2013;152:17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 6.Fu Y, Deiorio-Haggar K, Anthony J, Meyer MM. Most RNAs regulating ribosomal protein biosynthesis in Escherichia coli are narrowly distributed to Gammaproteobacteria. Nucleic Acids Res. 2013;41:3491–3503. doi: 10.1093/nar/gkt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zengel JM, Lindahl L. Diverse mechanisms for regulating ribosomal protein synthesis in Escherichia coli. Prog. Nucleic Acid Res. Mol. Biol. 1994;47:331–370. doi: 10.1016/s0079-6603(08)60256-1. [DOI] [PubMed] [Google Scholar]
- 8.Nomura M, Yates JL, Dean D, Post LE. Feedback regulation of ribosomal protein gene expression in Escherichia coli: structural homology of ribosomal RNA and ribosomal protein mRNA. Proc. Natl Acad. Sci. USA. 1980;77:7084–7088. doi: 10.1073/pnas.77.12.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehresmann C, Ehresmann B, Ennifar E, Dumas P, Garber M, Mathy N, Nikulin A, Portier C, Patel D, Serganov A. Molecular mimicry in translational regulation: the case of ribosomal protein S15. RNA Biol. 2004;1:66–73. [PubMed] [Google Scholar]
- 10.Gregory RJ, Cahill PB, Thurlow DL, Zimmermann RA. Interaction of Escherichia coli ribosomal protein S8 with its binding sites in ribosomal RNA and messenger RNA. J. Mol. Biol. 1988;204:295–307. doi: 10.1016/0022-2836(88)90577-3. [DOI] [PubMed] [Google Scholar]
- 11.Mathy N, Pellegrini O, Serganov A, Patel DJ, Ehresmann C, Portier C. Specific recognition of rpsO mRNA and 16S rRNA by Escherichia coli ribosomal protein S15 relies on both mimicry and site differentiation. Mol. Microbiol. 2004;52:661–675. doi: 10.1111/j.1365-2958.2004.04005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nomura M, Gourse R, Baughman G. Regulation of the synthesis of ribosomes and ribosomal components. Annu. Rev. Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- 13.Draper DE. In: Translational Regulation of Gene Expression. Ilan J, editor. New York: Plenum Press; 1987. pp. 1–26. [Google Scholar]
- 14.Schlax PJ, Xavier KA, Gluick TC, Draper DE. Translational repression of the Escherichia coli alpha operon mRNA: importance of an mRNA conformational switch and a ternary entrapment complex. J. Biol. Chem. 2001;276:38494–38501. doi: 10.1074/jbc.M106934200. [DOI] [PubMed] [Google Scholar]
- 15.Spedding G, Draper DE. Allosteric mechanism for translational repression in the Escherichia coli α operon. Proc. Natl Acad. Sci. USA. 1993;90:4399–4403. doi: 10.1073/pnas.90.10.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Serganov A, Polonskaia A, Ehresmann B, Ehresmann C, Patel DJ. Ribosomal protein S15 represses its own translation via adaptation of an rRNA-like fold within its mRNA. EMBO J. 2003;22:1898–1908. doi: 10.1093/emboj/cdg170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scott LG, Williamson JR. The binding interface between Bacillus stearothermophilus ribosomal protein S15 and its 5′-translational operator mRNA. J. Mol. Biol. 2005;351:280–290. doi: 10.1016/j.jmb.2005.06.030. [DOI] [PubMed] [Google Scholar]
- 18.Ehresmann C, Philippe C, Westhof E, Benard L, Portier C, Ehresmann B. A pseudoknot is required for efficient translational initiation and regulation of the Escherichia coli rpsO gene coding for ribosomal protein S15. Biochem. Cell Biol. 1995;73:1131–1140. doi: 10.1139/o95-122. [DOI] [PubMed] [Google Scholar]
- 19.Benard L, Philippe C, Dondon L, Grunberg-Manago M, Ehresmann B, Ehresmann C, Portier C. Mutational analysis of the pseudoknot structure of the S15 translational operator from Escherichia coli. Mol. Microbiol. 1994;14:31–40. doi: 10.1111/j.1365-2958.1994.tb01264.x. [DOI] [PubMed] [Google Scholar]
- 20.Marzi S, Myasnikov AG, Serganov A, Ehresmann C, Romby P, Yusupov M, Klaholz BP. Structured mRNAs regulate translation initiation by binding to the platform of the ribosome. Cell. 2007;130:1019–1031. doi: 10.1016/j.cell.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 21.Philippe C, Eyermann F, Benard L, Portier C, Ehresmann B, Ehresmann C. Ribosomal protein S15 from Escherichia coli modulates its own translation by trapping the ribosome on the mRNA initiation loading site. Proc. Natl Acad. Sci. USA. 1993;90:4394–4398. doi: 10.1073/pnas.90.10.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Philippe C, Portier C, Mougel M, Grunberg-Manago M, Ebel JP, Ehresmann B, Ehresmann C. Target site of Escherichia coli ribosomal protein S15 on its messenger RNA. Conformation and interaction with the protein. J. Mol. Biol. 1990;211:415–426. doi: 10.1016/0022-2836(90)90362-P. [DOI] [PubMed] [Google Scholar]
- 23.Portier C, Philippe C, Dondon L, Grunberg-Manago M, Ebel JP, Ehresmann B, Ehresmann C. Translational control of ribosomal protein S15. Biochim Biophys. Acta. 1990;1050:328–336. doi: 10.1016/0167-4781(90)90190-d. [DOI] [PubMed] [Google Scholar]
- 24.Serganov A, Ennifar E, Portier C, Ehresmann B, Ehresmann C. Do mRNA and rRNA binding sites of E. coli ribosomal protein S15 share common structural determinants? J. Mol. Biol. 2002;320:963–978. doi: 10.1016/s0022-2836(02)00553-3. [DOI] [PubMed] [Google Scholar]
- 25.Moffitt JR, Chemla YR, Smith SB, Bustamante C. Recent advances in optical tweezers. Annu. Rev. Biochem. 2008;77:205–228. doi: 10.1146/annurev.biochem.77.043007.090225. [DOI] [PubMed] [Google Scholar]
- 26.Greenleaf WJ, Frieda KL, Foster DA, Woodside MT, Block SM. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 2008;319:630–633. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Onoa B, Dumont S, Liphardt J, Smith SB, Tinoco I, Jr, Bustamante C. Identifying kinetic barriers to mechanical unfolding of the T. thermophila ribozyme. Science. 2003;299:1892–1895. doi: 10.1126/science.1081338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wen JD, Lancaster L, Hodges C, Zeri AC, Yoshimura SH, Noller HF, Bustamante C, Tinoco I., Jr Following translation by single ribosomes one codon at a time. Nature. 2008;452:598–603. doi: 10.1038/nature06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wen JD, Manosas M, Li PT, Smith SB, Bustamante C, Ritort F, Tinoco I., Jr Force unfolding kinetics of RNA using optical tweezers. I. Effects of experimental variables on measured results. Biophys. J. 2007;92:2996–3009. doi: 10.1529/biophysj.106.094052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qu X, Wen JD, Lancaster L, Noller HF, Bustamante C, Tinoco I., Jr The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature. 2011;475:118–121. doi: 10.1038/nature10126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elms PJ, Chodera JD, Bustamante C, Marqusee S. Limitations of constant-force-feedback experiments. Biophys. J. 2012;103:1490–1499. doi: 10.1016/j.bpj.2012.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bustamante C, Marko JF, Siggia ED, Smith S. Entropic elasticity of lambda-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 33.Liphardt J, Onoa B, Smith SB, Tinoco I, Jr, Bustamante C. Reversible unfolding of single RNA molecules by mechanical force. Science. 2001;292:733–737. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- 34.Holbrook SR, Kim SH. RNA crystallography. Biopolymers. 1997;44:3–21. doi: 10.1002/(SICI)1097-0282(1997)44:1<3::AID-BIP2>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 35.Saenger W. Principles of Nucleic Acid Structure. New York: Springer-Verlag; 1984. [Google Scholar]
- 36.Souliere MF, Altman RB, Schwarz V, Haller A, Blanchard SC, Micura R. Tuning a riboswitch response through structural extension of a pseudoknot. Proc. Natl Acad. Sci. USA. 2013;110:E3256–E3264. doi: 10.1073/pnas.1304585110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen G, Chang KY, Chou MY, Bustamante C, Tinoco I., Jr Triplex structures in an RNA pseudoknot enhance mechanical stability and increase efficiency of -1 ribosomal frameshifting. Proc. Natl Acad. Sci. USA. 2009;106:12706–12711. doi: 10.1073/pnas.0905046106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen G, Wen JD, Tinoco I., Jr Single-molecule mechanical unfolding and folding of a pseudoknot in human telomerase RNA. RNA. 2007;13:2175–2188. doi: 10.1261/rna.676707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Green L, Kim CH, Bustamante C, Tinoco I., Jr Characterization of the mechanical unfolding of RNA pseudoknots. J. Mol. Biol. 2008;375:511–528. doi: 10.1016/j.jmb.2007.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hansen TM, Reihani SN, Oddershede LB, Sorensen MA. Correlation between mechanical strength of messenger RNA pseudoknots and ribosomal frameshifting. Proc. Natl Acad. Sci. USA. 2007;104:5830–5835. doi: 10.1073/pnas.0608668104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ritchie DB, Foster DA, Woodside MT. Programmed -1 frameshifting efficiency correlates with RNA pseudoknot conformational plasticity, not resistance to mechanical unfolding. Proc. Natl Acad. Sci. USA. 2012;109:16167–16172. doi: 10.1073/pnas.1204114109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.White KH, Orzechowski M, Fourmy D, Visscher K. Mechanical unfolding of the beet western yellow virus -1 frameshift signal. J. Am. Chem. Soc. 2011;133:9775–9782. doi: 10.1021/ja111281f. [DOI] [PubMed] [Google Scholar]
- 44.Philippe C, Benard L, Portier C, Westhof E, Ehresmann B, Ehresmann C. Molecular dissection of the pseudoknot governing the translational regulation of Escherichia coli ribosomal protein S15. Nucleic Acids Res. 1995;23:18–28. doi: 10.1093/nar/23.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brion P, Westhof E. Hierarchy and dynamics of RNA folding. Annu. Rev. Biophys. Biomol. Struct. 1997;26:113–137. doi: 10.1146/annurev.biophys.26.1.113. [DOI] [PubMed] [Google Scholar]
- 46.Tinoco I, Jr, Bustamante C. How RNA folds. J. Mol. Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 47.Wu M, Tinoco I., Jr RNA folding causes secondary structure rearrangement. Proc. Natl Acad. Sci. USA. 1998;95:11555–11560. doi: 10.1073/pnas.95.20.11555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cruz JA, Westhof E. The dynamic landscapes of RNA architecture. Cell. 2009;136:604–609. doi: 10.1016/j.cell.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 49.Dethoff EA, Chugh J, Mustoe AM, Al-Hashimi HM. Functional complexity and regulation through RNA dynamics. Nature. 2012;482:322–330. doi: 10.1038/nature10885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Micura R, Hobartner C. On secondary structure rearrangements and equilibria of small RNAs. ChemBioChem. 2003;4:984–990. doi: 10.1002/cbic.200300664. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.