Abstract
Rev3, the catalytic subunit of DNA polymerase ζ, is essential for translesion synthesis of cytotoxic DNA photolesions, whereas the Rev1 protein plays a noncatalytic role in translesion synthesis. Here, we reveal that mammalian Rev3−/− and Rev1−/− cell lines additionally display a nucleotide excision repair (NER) defect, specifically during S phase. This defect is correlated with the normal recruitment but protracted persistence at DNA damage sites of factors involved in an early stage of NER, while repair synthesis is affected. Remarkably, the NER defect becomes apparent only at 2 h post-irradiation indicating that Rev3 affects repair synthesis only indirectly, rather than performing an enzymatic role in NER. We provide evidence that the NER defect is caused by scarceness of Replication protein A (Rpa) available to NER, resulting from its sequestration at stalled replication forks. Also the induction of replicative stress using hydroxyurea precludes the accumulation of Rpa at photolesion sites, both in Rev3−/− and in wild-type cells. These data support a model in which the limited Rpa pool coordinates replicative stress and NER, resulting in increased cytotoxicity of ultraviolet light when replicative stress exceeds a threshold.
INTRODUCTION
Eukaryotic cells are equipped with a set of DNA repair pathways that each removes a different type of DNA damage. DNA damage that escapes repair will form an impediment for both replication and transcription, resulting in the arrest of replication forks and the perturbation of gene transcription, respectively.
The canonical nucleotide excision repair (NER) pathway removes helix-distorting nucleotide lesions, including ultraviolet light (UV)-induced (6-4)pyrimidine-pyrimidone photolesions [(6-4)PP]. In addition to global genome NER (GG-NER), that removes photolesions throughout the genome, the transcription-coupled NER [TC-NER; (1)] sub-pathway removes lesions that stall the transcription complex. Both NER sub-pathways differ in the initial recognition of the lesion but share downstream steps. After recognition of the lesion, a pre-incision complex is assembled, the damaged DNA strand is incised at the 5′ and 3′ sides of the lesion and a patch of ∼30 nt of single-stranded DNA (ssDNA) comprising the lesion is excised. Repair DNA synthesis and ligation ensure that the integrity of the DNA is restored (2). The coupling of NER-mediated incision to repair DNA synthesis is mediated by the heterotrimeric ssDNA binding replication protein A (Rpa), an essential factor in DNA replication and DNA damage repair (3,4).
NER has been studied best in quiescent cells, and little is known about NER during S phase (5–7). Timely removal of DNA lesions in replicating cells is important, as DNA lesions that escape repair will arrest the processive replication fork, leading to a patch of ssDNA surrounding the damage that will bind Rpa (8,9). When the stalled replicon is not rescued, these patches activate the S phase checkpoint. Prolonged stalling of replication forks can result in fork collapse. The ensuing double-strand DNA breaks ultimately may cause genomic instability or cell death. Alternatively, DNA translesion synthesis (TLS) can synthesize across and beyond the DNA lesion, thereby completing replication of the damaged template and quenching checkpoint responses. Eukaryotic TLS is characterized by insertion of a nucleotide opposite the lesion by one of a number of specialized DNA polymerases that are characterized by reduced stringency of the active site and a lack of proofreading activity. These characteristics imply that TLS is an inherently mutagenic process (10). A key player in TLS is the B-family polymerase ζ (Polζ). Polζ is a heterodimer, composed of catalytic Rev3 and accessory Rev7 subunits (11,12) and is essential for TLS of (6-4)PP (13). During TLS, Polζ acts most likely as an extender of nucleotides incorporated by one of the Y family TLS polymerases η, ι, κ and Rev1 opposite the damaged nucleotide (11,12).
It was found that in quiescent cells TLS polymerase κ can perform repair synthesis after NER-mediated excision. This result explains pleiotropic phenotypes of polymerase κ-deficient cells (14,15). To investigate whether also TLS polymerase ζ, in addition to TLS, is involved in NER, we have investigated NER in mouse embryonic fibroblasts (MEFs) with a targeted disruption in Rev3, the catalytic subunit of polymerase ζ. Surprisingly, our data show a significant defect in NER of (6-4)PP, but only in replicating Rev3-deficient MEFs, whereas quiescent Rev3-deficient cells display normal NER activity. We provide evidence that stalling of replication forks at photolesions reduces the amount of Rpa available for NER of photolesions. Moreover, also sequestration of Rpa at replication forks, arrested using hydroxyurea (HU), precludes its accumulation at sites of photolesions. These data reveal Rpa-mediated coordination in trans between replicative stress and NER.
MATERIALS AND METHODS
Generation and immortalization of MEF lines
Wild-type and Rev3−/− MEF lines were isolated and immortalized as described previously (13).
Immuno slot blot
Quantitative slot blotting for the determination of repair of (6-4)PP was performed as described (16). Cyclobutane pyrimidine dimer (CPD) lesions remain virtually unrepaired in mouse cells, and therefore staining for these lesions was used as a standard. The antibodies used were anti-(6-4)PP mouse monoclonal antibody 64M-2 and anti-CPD mouse monoclonal antibody TDM2 (both from MBL, Japan).
DNA repair replication
DNA repair replication in cells exposed to 10 J/m2 UV-C was measured by the radioisotope and density labeling technique as previously described (17) with minor modifications.
Transcription recovery
Recovery of transcription after treatment with 10 J/m2 UV-C was performed as described (18) with minor modifications. Briefly, cells were seeded at a density of 105 cells/well of a 24-wells plate in MEF medium [Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% Fetal calf serum (FCS) and antibiotics] containing [14C]thymidine (0.05 μCi/ml) and cultured at 37°C and 5% CO2 for 24 h to label genomic DNA. After removal of medium, cells were cultured for 2 days in DMEM containing 0.5% FCS and antibiotics. Cells were then washed once with phosphate-buffered saline, mock treated or exposed to 10 J/m2 UV-C and cultured in DMEM containing 0.5% FCS and antibiotics. At different times after treatment, cells were pulsed-labeled with [3H]uridine (10 μCi/ml) for 30 min and subsequently lysed in DNA lysis buffer (200 mM NaCl; 100 mM Tris–HCl pH8.0; 5 mM EDTA; 0.2% SDS). After incubation overnight at 37°C and 5% CO2, nucleic acids were precipitated in 10% trichloroacetic acid for 1 h at 4°C and collected on GF/C filters. Each filter was assayed for radioactivity using scintillation counting. Three wells per treatment per time point were analyzed.
Immuno-cytochemistry
To detect induction and persistence of (6-4)PP, MEFs were globally irradiated with 5 J/m2 UV-C light and immediately pulse labeled with 10 μM bromodeoxyuridine (BrdU) for 15 min. Cells were fixed 0 and 6 h after BrdU labeling. Detection by immunofluorescence of (6-4)PP using mouse monoclonal antibody 64 M-2 (MBL, Japan) and secondary antibody DαM AlexaFluor 488 (Molecular Probes), as described (19). BrdU staining was performed using rat monoclonal antibody BU1/75 (Abcam) and secondary DαR Cy3 (Jackson Immunoresearch).
Cells were locally irradiated with UV through a filter with pore size 8 μm as described previously (20). Detection of p89 using rabbit polyclonal α-p89 antibodies (Santa Cruz) by immunofluorescence was performed as described previously (19). Images were taken with equal exposure time, and 35–50 fluorescent images were analyzed for each time point. Signal intensities were determined as described (19).
To detect Rpa at locally induced UV damage, MEFs were either mock treated, globally exposed to 2 mM HU or to 1 J/m2 UV-C light and immediately pulse labeled with EdU for 1 h. After 1 h, cells were locally exposed to 100 J/m2 UV-C light through a filter with pore size 8 μm as described previously (20). Staining for Rpa was performed 1 h after local UV-C treatment using rat monoclonal antibody 4E4 (α-Rpa-32; cell signaling) and secondary DαR Cy3 (Jackson Immunoresearch). EdU-containing DNA was detected according to the manufacturer’s recommendations as described (21). Then, DNA was denatured and stained with mouse monoclonal antibody TDM2 (CPD; MBL, Japan). Images were taken with equal exposure time and 35–50 fluorescent images were analyzed for each time point. Signal intensities were determined as described (19).
Western blotting
Total cell lysates were prepared by adding Laemmli lysis buffer directly to 2 × 106 cells. Lysates containing chromatin-bound proteins were prepared as follows. IP lysis buffer (50 mM Tris–HCl pH7.5; 130 mM NaCl; 1 mM MgCl2; 0.5% Triton X-100; protease inhibitors) was added to 2 × 106 cells, 15 h after cells were mock treated or exposed to 5 J/m2 UV-C. After incubation on ice for 30 min and centrifugation, the pellet fraction was treated with Benzonase (Novagen) in IP lysis buffer on ice for 30 min. After centrifugation, the pellet fraction was resuspended in Laemmli lysis buffer. Proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and blotted onto a nitrocellulose membrane (Hybond-C extra, Amersham Biosciences). The membranes were incubated in 5% nonfat milk, 3% bovine serum albumin in phosphate-buffered saline and 0.1% Tween 20 for at least 1 h at room temperature. Thereafter, membranes were incubated overnight with primary antibodies at 4°C and with appropriate peroxidase-conjugated secondary antibodies (Bio-Rad) for 1 h at room temperature, subsequently. Proteins were visualized by enhanced chemiluminescence detection (ECL). The following antibodies were used: rat polyclonal α-Rpa-32 (Cell signaling), rabbit polyclonal α-histon H3 (Abcam) and rabbit polyclonal α-β-tubulin (Cell signaling).
RESULTS
Rev3−/− MEFs display a defect in GG-NER
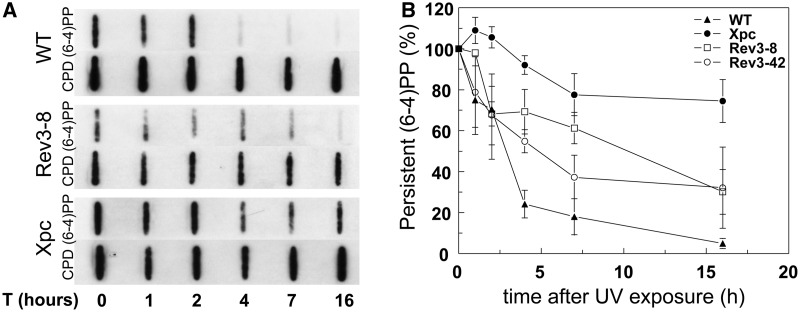
Recently, we have shown that Rev3−/− MEF lines are extremely sensitive to UV-C light (13,21). We investigated whether this hypersensitivity is caused by a NER defect, by measuring the removal of (6-4)PP in two independently derived Rev3−/− MEF lines, Rev3-8 and Rev3-42. The cells were cultured under conditions of low (0.5%) serum, which reduces, but does not abolish (Supplementary Figure S1), their proliferation. By using a quantitative slot blot assay, in GG-NER-deficient, Xpc−/− MEFs no removal of (6-4)PP was detected, emphasizing that this assay measures activity of the major NER sub-pathway (Figure 1). Up to 2 h after UV exposure, the kinetics of removal of (6-4)PP was normal in both Rev3−/− lines, indicating that Rev3 is not involved in the excision stage of NER itself, at least early after UV exposure. Remarkably, at later times after exposure, the removal of (6-4)PP was significantly affected, but not abolished, in the Rev3-8 and Rev3-42 MEF lines (Figure 1A and B).
Figure 1.
Defective repair of (6-4)PP in Rev3−/− MEFs. (A) Representative slot blot to measure the relative repair of (6-4)PP in wild-type, Rev3−/− and Xpc−/− MEF lines at different times following exposure to 5 J/m2 UV-C. CPD lesions are hardly repaired in mouse cells and therefore serve as internal control. (B) Average of four individual experiments. Error bars: Standard error of the mean (SEM).
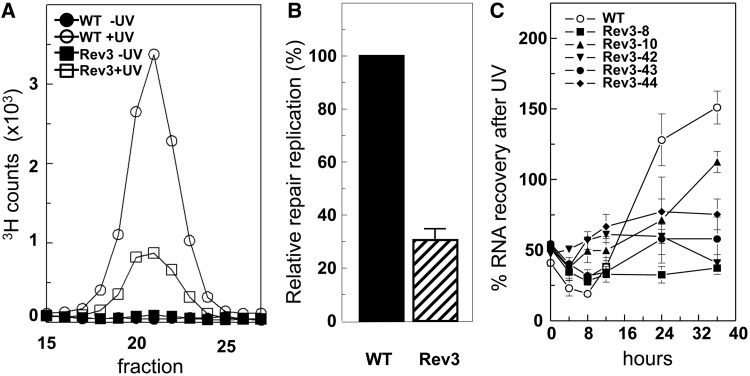
Because the GG-NER sub-pathway accounts for the large majority of all NER, loss of repair replication on UV exposure indicates a defect in GG-NER. To further establish this, we quantified NER-associated repair replication in DNA of wild-type and Rev3-8 MEF lines, exposed to 10 J/m2 of UV-C and cultured under low serum. Three hours after UV exposure, the Rev3-8 line displayed a reduction of repair replication of 70%, compared with wild-type MEFs (Figure 2A and B). This result confirms that GG-NER is affected in Rev3-deficient MEFs.
Figure 2.
Rev3 deficiency affects both TC-NER and GG-NER. All cell lines were cultured under low-serum conditions that reduce, rather than abolish (Supplementary Figure S1), proliferation of Rev3−/− MEFs. (A) Profiles of repair replication in wild-type and Rev3−/− MEF line 8, 3 h following exposure to 10 J/m2 UV-C. Repair replication was normalized to the amount of parental DNA in the gradient. (B) Relative repair replication in wild-type and Rev3−/− MEF line 8. Repair replication in wild-type cells was set at 100%. Repair replication was measured thrice. (C) Recovery of RNA synthesis in wild-type and five independent Rev3−/− MEF lines following exposure to 10 J/m2 UV-C. RNA synthesis in unexposed cells was set at 100%. Mean ± SEM.
Transcription recovery is perturbed in proliferating Rev3−/− MEFs
To test whether also TC-NER is affected in the absence of Rev3, we measured the extent of the recovery of transcription after UV treatment, a sensitive parameter of functional TC-NER (18). When cells were cultured under low serum, all tested independent Rev3−/− MEF lines displayed a defect in transcription recovery, suggestive of a role for Polζ in NER, at least under low-serum conditions (Figure 2C). Interestingly, when Rev3−/− MEF lines were cultured in high (10%) serum, the defect in transcription recovery was even more pronounced, indicating that the TC-NER defect conferred by Rev3 deficiency is correlated with cellular proliferation (Supplementary Figure S2A). To investigate TC-NER in the absence of any proliferation, we measured transcription recovery also in senescent primary Rev3−/− MEFs, cultured under low serum; in these cells proliferation was not measurable. These cells displayed virtually normal transcription recovery (Supplementary Figure S2B). Thus, Polζ does not appear to play a direct role in NER, but NER is perturbed in the absence of Polζ, though only in proliferating cells. We obtained comparable results with Rev1-deficient MEFs that also are defective in TLS of (6-4)PP [Supplementary Figure S2A; (22)].
The defect in the removal of (6-4)PP in Rev3−/− MEFs is restricted to replicating cells
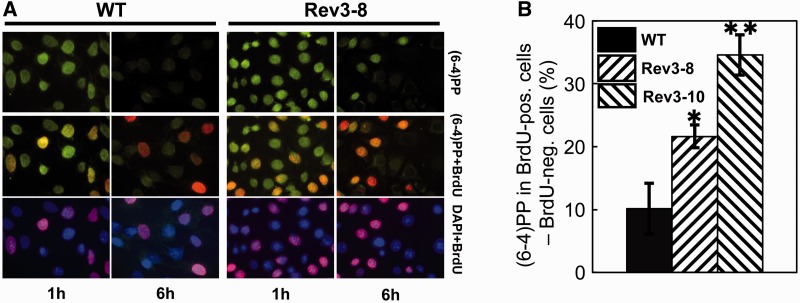
Based on these data, we hypothesized that Rev1 or Rev3 deficiency results in defective NER specifically during S phase. To investigate this, we measured the removal of (6-4)PP from the genomes of MEFs, both within and outside of S phase. This was done by pulse labeling S phase cells with BrdU, immediately after UV exposure, and immuno-staining of individual cells for BrdU and (6-4)PPs at 6 h after exposure (Figure 3A). The difference in (6-4)PP staining in BrdU-positive (replicating) cells versus BrdU-negative (nonreplicating) cells at 6 h after exposure was taken as a measure for differential NER within and outside of S phase. In wild-type cells, almost no difference in (6-4)PP repair was observed between S phase and non-S phase cells. Conversely, in two independent Rev3−/− MEF lines the repair of (6-4)PPs was affected specifically when the cells were exposed to UV during replication (Figure 3B). Thus, Rev3 deficiency results in a NER defect, selectively during S phase. Because NER is not affected by Rev3-deficiency at early time points on UV exposure (Figure 1), we infer that this represents an indirect defect of Rev3 in NER during S phase, rather than a direct involvement.
Figure 3.
Persistence of (6-4)PP in replicating Rev3−/− MEFs. (A) Immunostaining for (6-4)PP (green) and BrdU (replicating cells, red) in DAPI-stained nuclei (blue) of wild-type and Rev3-8 MEFs, 1 and 6 h after global exposure to 5 J/m2 UV-C. (B) Relative differences in (6-4)PP between BrdU positive (replicating) and BrdU negative (nonreplicating) wild-type and two independent Rev3−/− MEF lines, 6 h after UV treatment. Mean ± SEM. P = 0.03 (Rev3−/− line 8) or 0.004 (Rev3−/− line 10).
Persistence of early repair factors at photolesions in Rev3−/− MEFs
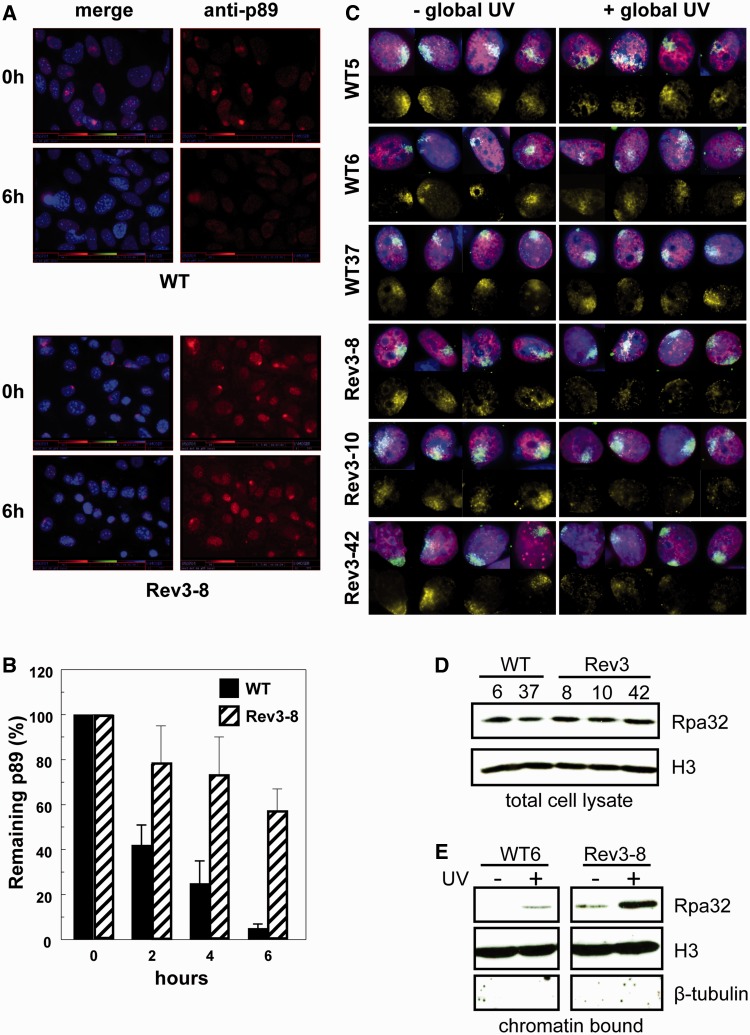
To investigate whether deficiency of Polζ affects an early or a late step of NER, we examined the accumulation and persistence of p89, a member of the pre-incision complex of NER (23), at locally induced photolesions in wild-type and Rev3-8 MEFs. Immediately after local UV exposure, wild-type and Rev3−/− MEFs displayed similar levels of p89 at local spots of (6-4)PP lesions (Figure 4A and B). This confirms that DNA damage recognition and assembly of the pre-incision complex do not depend on Rev3. In wild-type cells, p89-containing spots were virtually undetectable at 6 h after local irradiation (Figure 4A and B). In the Rev3−/− MEFs, however, the intensity of p89 spots was reduced by only 40% (Figure 4A and B). This phenotype mimics the phenotype of cells deficient in late steps of NER as in such cells pre-incision factors persist at the damage sites, making these factors unavailable for subsequent repair events (19,24). Therefore, this result suggests that Rev3−/− MEFs suffer from a defect in a late step of NER. Notwithstanding our efforts to investigate whether Rev3 or its heterodimeric partner Rev7 also localize at local UV damages, these attempts failed due to high nonspecific staining of the antibodies.
Figure 4.
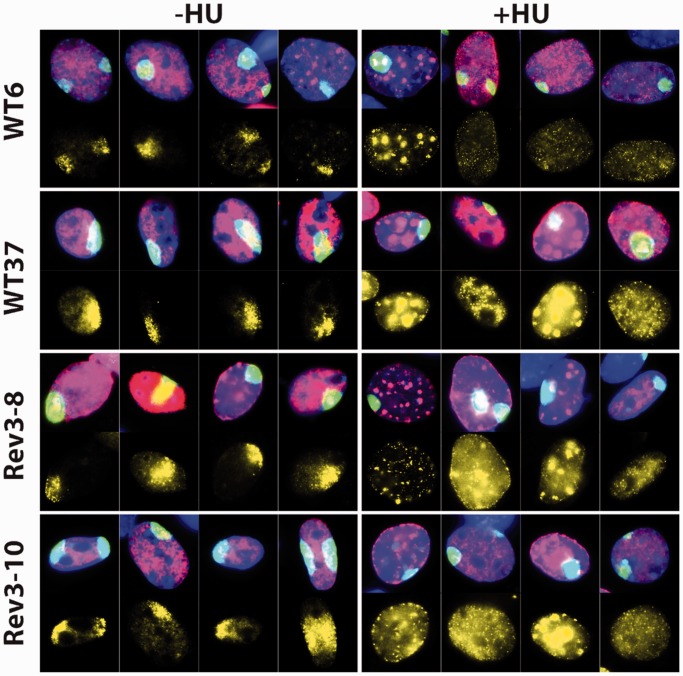
Recruitment of pre-incision factor p89 and Rpa to local UV damage. (A) Immunostaining for p89 in wild-type and Rev3-8 MEFs after local exposure to 30 J/m2 UV-C. (B) Prolonged persistence of p89 at locally induced UV damage in Rev3−/− MEFs. Mean ± SEM of 35–50 spots/time point. (C) Defective accumulation of Rpa in local damage spots in Rev3−/− MEFs, pretreated with low-dose UV-C. MEF lines of each genotype were mock treated (left panels) or globally exposed to 1 J/m2 UV-C (right panels) to induce the arrest of replication forks, followed by local UV-C treatment. Then, cells were stained for CPD (green), Rpa (yellow), EdU (red) and for nuclei (blue). Rpa is hardly recruited to local UV damage in the presence of globally arrested replication forks. (D) Total Rpa levels in wild-type and Rev3−/− MEF lines. (E) Levels of chromatin-associated Rpa upon UV exposure of wild-type and Rev3−/− MEF lines.
Defective recruitment of Rpa to local UV damage in Rev3−/− MEFs
Recently, it was shown that free Rpa is the rate-limiting factor in the initiation of new NER events (4). Based on this finding and our data, we hypothesized that in replicating Rev3−/− MEFs, stalled replication forks may sequester free Rpa, resulting in depletion of the pool of Rpa available for NER. The pool of free Rpa was investigated by monitoring its recruitment to local damage spots as readout. Three independent Rev3−/− and three independent wild-type MEF lines were pretreated globally with a dose of UV that is nontoxic to wild-type MEFs, to sequester Rpa at arrested replication forks only in Rev3−/− MEFs, followed by labeling of S phase cells with the nucleotide analog ethynyl-2′-deoxyuridine (EdU). Then, the cells were exposed locally to a high dose of UV and recruitment of Rpa to local damage spots was investigated. In the absence of low-dose pretreatment, Rpa was equally recruited to local spots of UV damage in the replicating (EdU-positive) Rev3−/− and wild-type control cells indicating that Rpa recruitment is not directly affected by Rev3 deficiency (Figure 4C, left panels). However, the recruitment of Rpa to locally induced photolesions was lost specifically in Rev3−/− cells that were pretreated globally with low-dose UV (Figure 4C, right panels). This loss of Rpa recruitment is not caused by reduced expression of Rpa in Rev3−/− cells, as all tested cell lines express similar levels of Rpa (Figure 4D). On the other hand, we observed strongly increased binding of Rpa to chromatin, specifically in Rev3−/− cells following global UV exposure (Figure 4E). Therefore, these data suggest that persistently stalled replication forks, presumably at (6-4)PP lesions, sequester Rpa. This limits the amount of free nuclear Rpa available for NER, causing the post-excision defect in NER in replicating Rev3−/− cells.
Replicative stress interferes with recruitment of Rpa to local photolesions also in wild-type MEFs
We then investigated whether the S phase-associated NER deficiency is confined to Rev3-deficient cells or represents a general phenotype of cells under replicative stress. To this aim, two independent Rev3−/− and two independent wild-type MEF lines were pretreated with the replication inhibitor HU. After 1 h cells were locally irradiated with UV, as above, and recruitment of Rpa to local damage spots was investigated.
In the absence of HU pretreatment, Rpa was recruited to the local damage spots, as expected (Figure 5). Treatment with HU resulted in the accumulation of chromatin-bound Rpa in replication foci, representing stressed replication forks. HU treatment before local UV exposure completely abolished Rpa recruitment to patches of photolesions, both in the Rev3−/− cells and in the wild-type cells (Figure 5). Thus, rather than being confined to Rev3 (and Rev1)-deficient cells, the sequestration of Rpa by ssDNA at stressed replication forks, resulting in its unavailability for NER, is a ubiquitous phenotype.
Figure 5.
Sequestration of Rpa by HU-induced replicative stress results in its unavailability for NER. MEF lines of each genotype were mock treated (left panels) or globally exposed to 2 mM HU (right panels) to induce the arrest of replication forks, followed by local UV-C treatment. Then, cells were stained for CPD (green), Rpa (yellow), EdU (red) and for nuclei (blue). Irrespective of the Rev3 status, Rpa is hardly recruited to local UV damage in the presence of replication forks, arrested by HU.
DISCUSSION
In the present study, we reveal a previously unrecognized function of Rpa in the coordination between replicative stress and NER. As a model, we have mainly used MEF lines deficient for the Rev3 subunit of Polζ, exposed to UV light. Our data show that Polζ is not directly involved in NER. Rather, the absence of Polζ results in the inhibition of repair of UV lesions in trans, specifically in S phase cells (Figure 6 for a model). We provide evidence that this in trans inhibition is caused by the depletion of free Rpa due to sequestering at persistently arrested replication forks (Figure 4C, right panels). It should be noted that subtle differences in the S phase-associated NER defect exist between the different Rev3-deficient MEFs lines. Thus, compared with the other Rev3-deficient lines tested, line Rev3-8 displays the most pronounced defect in transcription recovery (Figure 2C and Supplementary Figure S2) and the highest persistence of (6-4)PP lesions (Figure 1). This severe phenotype is correlated with the high proliferation rate of this line, both under low and high serum conditions (Supplementary Figure S1). The correlation between the proliferation rate of the cells, the extent of the TLS defect and the extent of the NER defect in replicating cells is in full agreement with squelching of Rpa at stalled replication forks during S phase in the Rev3-deficient cells. Our data also provide an explanation for the partial defect in NER during S phase, observed in xeroderma pigmentosum-variant cells, which lack TLS polymerase Polη, and in Seckel syndrome fibroblasts, defective for the checkpoint kinase Atr (25,26). The relevance of our findings is not confined to replicative stress in the absence of Rev3 (or Rev1), as HU treatment of wild-type cells results in a similar phenotype as low-dose global UV treatment of Rev3-deficient cells.
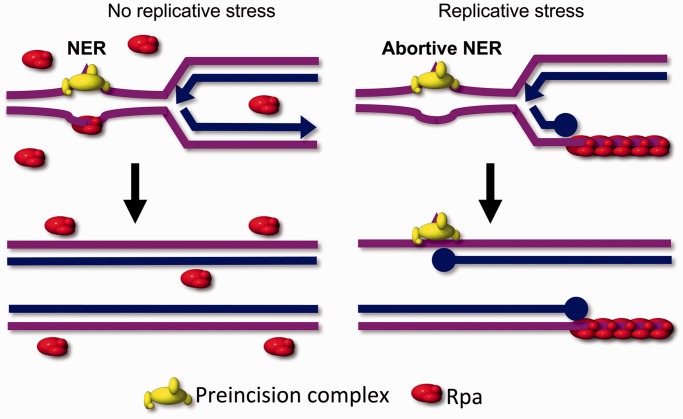
Figure 6.
Model for the interference between NER and replicative stress. Left: in the absence of replicative stress, free Rpa is abundant and available to perform coupling between pre-incision and post-incision complexes in NER. The removal of DNA lesions allows unperturbed progression of replication. Right: when excessive replicative stress is present, as in the treatment of Rev3−/− MEFs with low-dose UV, or of wild-type (or Rev3−/−) cells with HU, Rpa is sequestered at the resulting extended ssDNA tracts. Therefore, Rpa is unavailable to act during NER, resulting in the persistence of early NER factors at lesion sites and in an NER defect. This will further aggravate the consequences of replicative stress. This positive feedback loop explains the extreme UV sensitivity of proliferating Rev3-deficient cells (13,21).
The Rpa-dependent coordination in trans between stalled replication forks and NER events proposed here may have important physiological consequences for cytotoxic responses to genotoxic agents. Thus, an excessive load of arrested replication forks, or their persistence, entails a high risk for genomic instability in S phase cells. In such cells, NER is inhibited in trans, owing to the limited availability of free Rpa, which will result in further fork stalling, ultimately resulting in hypersensitivity to the insulting agent and cell death. We hypothesize that this positive feedback loop acts as a protective mechanism preventing detrimental genome instability in proliferating cells that suffer from excessive replicative stress. Recently, it was shown that an excessive amount of ssDNA in replicating cells leads to depletion of the nuclear pool of Rpa, particularly in the absence of the checkpoint kinase Atr. The consequent inability to protect ssDNA at replication forks results in exit from the cell cycle (27). Combined with our results this demonstrates that Rpa is instrumental in the induction of pleiotropic responses to replicative stress, which protects the cell against deleterious consequences of excessive genomic DNA damage.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for this work and for open access charge: EU [grant FP6-512113] and the Dutch Cancer Society [grant UL2010-4851].
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Fousteri MI, Mullenders LHF. Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res. 2008;18:73–84. doi: 10.1038/cr.2008.6. [DOI] [PubMed] [Google Scholar]
- 2.Shuck SC, Short EA, Turchi JJ. Eukaryotic nucleotide excision repair: from understanding mechanisms to influencing biology. Cell Res. 2008;18:64–72. doi: 10.1038/cr.2008.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fanning E, Klimovich V, Nager AR. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–4137. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Overmeer RM, Moser J, Volker M, Kool H, Tomkinson AE, van Zeeland AA, Mullenders LHF, Fousteri M. Replication protein A safeguards genome integrity by controlling NER incision events. J. Cell Biol. 2011;192:401–415. doi: 10.1083/jcb.201006011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilljam KM, Müller R, Liabakk NB, Otterlei M. Nucleotide excision repair is associated with the replisome and its efficiency depends on a direct interaction between XPA and PCNA. PloS One. 2012;7:e49199. doi: 10.1371/journal.pone.0049199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thyagarajan B, Anderson KE, Lessard CJ, Veltri G, Jacobs DR, Folsom AR, Gross MD. Alkaline unwinding flow cytometry assay to measure nucleotide excision repair. Mutagenesis. 2007;22:147–153. doi: 10.1093/mutage/gel071. [DOI] [PubMed] [Google Scholar]
- 7.Tommasi S, Oxyzoglou AB, Pfeifer GP. Cell cycle-independent removal of UV-induced pyrimidine dimers from the promoter and the transcription initiation domain of the human CDC2 gene. Nucleic Acids Res. 2000;28:3991–3998. doi: 10.1093/nar/28.20.3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- 9.Novarina D, Amara F, Lazzaro F, Plevani P, Muzi-Falconi M. Mind the gap: keeping UV lesions in check. DNA Repair. 2011;10:751–759. doi: 10.1016/j.dnarep.2011.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat. Rev. Mol. Cell Biol. 2012;13:141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gan GN, Wittschieben JP, Wittschieben BØ, Wood RD. DNA polymerase zeta (pol zeta) in higher eukaryotes. Cell Res. 2008;18:174–183. doi: 10.1038/cr.2007.117. [DOI] [PubMed] [Google Scholar]
- 12.Waters LS, Minesinger BK, Wiltrout ME, D'Souza S, Woodruff RV, Walker GC. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol. Mol. Biol. Rev. 2009;73:134–154. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jansen JG, Tsaalbi-Shtylik A, Hendriks G, Verspuy JWA, Gali H, Haracska L, de Wind N. Mammalian polymerase zeta is essential for post-replication repair of UV-induced DNA lesions. DNA Repair. 2009;8:1444–1451. doi: 10.1016/j.dnarep.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 14.Ogi T, Limsirichaikul S, Overmeer RM, Volker M, Takenaka K, Cloney R, Nakazawa Y, Niimi A, Miki Y, Jaspers NG, Mullenders LHF, Yamashita S, Fousteri MI, Lehmann AR. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol. Cell. 2010;37:714–727. doi: 10.1016/j.molcel.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 15.Ogi T, Lehmann AR. The Y-family DNA polymerase kappa (pol kappa) functions in mammalian nucleotide-excision repair. Nat. Cell Biol. 2006;8:640–642. doi: 10.1038/ncb1417. [DOI] [PubMed] [Google Scholar]
- 16.Ford JM, Hanawalt PC. Expression of wild-type p53 is required for efficient global genomic nucleotide excision repair in UV-irradiated human fibroblasts. J. Biol. Chem. 1997;272:28073–28080. doi: 10.1074/jbc.272.44.28073. [DOI] [PubMed] [Google Scholar]
- 17.van Zeeland AA, Bussmann CJ, Degrassi F, Filon AR, van Kesteren-van Leeuwen AC, Palitti F, Natarajan AT. Effects of aphidicolin on repair replication and induced chromosomal aberrations in mammalian cells. Mutat. Res. 1982;92:379–392. doi: 10.1016/0027-5107(82)90237-8. [DOI] [PubMed] [Google Scholar]
- 18.van Hoffen A, Natarajan AT, Mayne LV, van Zeeland AA, Mullenders LH, Venema J. Deficient repair of the transcribed strand of active genes in Cockayne's syndrome cells. Nucleic Acids Res. 1993;21:5890–5895. doi: 10.1093/nar/21.25.5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moser J, Kool H, Giakzidis I, Caldecott K, Mullenders LHF, Fousteri MI. Sealing of chromosomal DNA nicks during nucleotide excision repair requires XRCC1 and DNA ligase III alpha in a cell-cycle-specific manner. Mol. Cell. 2007;27:311–323. doi: 10.1016/j.molcel.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 20.Volker M, Moné MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JHJ, van Driel R, van Zeeland AA, Mullenders LH. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell. 2001;8:213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 21.Temviriyanukul P, van Hees-Stuivenberg S, Delbos F, Jacobs H, de Wind N, Jansen JG. Temporally distinct translesion synthesis pathways for ultraviolet light-induced photoproducts in the mammalian genome. DNA Repair. 2012;11:550–558. doi: 10.1016/j.dnarep.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 22.Jansen JG, Tsaalbi-Shtylik A, Hendriks G, Gali H, Hendel A, Johansson F, Erixon K, Livneh Z, Mullenders LHF, Haracska L, de Wind N. Separate domains of Rev1 mediate two modes of DNA damage bypass in mammalian cells. Mol. Cell. Biol. 2009;29:3113–3123. doi: 10.1128/MCB.00071-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaeffer L, Roy R, Humbert S, Moncollin V, Vermeulen W, Hoeijmakers JH, Chambon P, Egly JM. DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor. Science (New York, NY) 1993;260:58–63. doi: 10.1126/science.8465201. [DOI] [PubMed] [Google Scholar]
- 24.Mullenders LH, van Kesteren-van Leeuwen AC, van Zeeland AA, Natarajan AT. Analysis of the structure and spatial distribution of ultraviolet-induced DNA repair patches in human cells made in the presence of inhibitors of replicative synthesis. Biochim. Biophys. Acta. 1985;826:38–48. doi: 10.1016/s0167-4781(85)80006-3. [DOI] [PubMed] [Google Scholar]
- 25.Auclair Y, Rouget R, Belisle JM, Costantino S, Drobetsky EA. Requirement for functional DNA polymerase eta in genome-wide repair of UV-induced DNA damage during S phase. DNA Repair. 2010;9:754–764. doi: 10.1016/j.dnarep.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 26.Auclair Y, Rouget R, Affar EB, Drobetsky EA. ATR kinase is required for global genomic nucleotide excision repair exclusively during S phase in human cells. Proc. Natl Acad. Sci. USA. 2008;105:17896–17901. doi: 10.1073/pnas.0801585105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toledo LI, Altmeyer M, Rask M-B, Lukas C, Larsen DH, Povlsen LK, Bekker-Jensen S, Mailand N, Bartek J, Lukas J. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell. 2013;155:1088–1103. doi: 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.