Abstract
MicroRNAs (miRNAs) are central regulators of gene expression, and a large fraction of them are encoded in introns of RNA polymerase II transcripts. Thus, the biogenesis of intronic miRNAs by the microprocessor and the splicing of their host introns by the spliceosome require coordination between these processing events. This cross-talk is addressed here. We show that key microprocessor proteins Drosha and DGCR8 as well as pre-miRNAs cosediment with supraspliceosomes, where nuclear posttranscriptional processing is executed. We further show that inhibition of splicing increases miRNAs expression, whereas knock-down of Drosha increases splicing. We identified a novel splicing event in intron 13 of MCM7, where the miR-106b-25 cluster is located. The unique splice isoform includes a hosted pre-miRNA in the extended exon and excludes its processing. This indicates a possible mechanism of altering the levels of different miRNAs originating from the same transcript. Altogether, our study indicates interplay between the splicing and microprocessor machineries within a supraspliceosome context.
INTRODUCTION
MicroRNAs (miRNAs) are small ∼22-nt long molecules involved in the negative control of gene expression by binding mainly to the 3′UTR of target messenger RNA (mRNA) transcripts (1–3). A large fraction of miRNA genes are located in introns (4–6). The canonical biogenesis of intronic miRNAs from RNA polymerase II (Pol II) transcripts involves two main steps. The first takes place in the nucleus and is performed by the microprocessor. Key proteins of the microprocessor are DGCR8, which binds the RNA molecule, and Drosha, an RNase III type enzyme, which cleaves the primary (pri) miRNA transcript into a precursor (pre) miRNA stem-loop molecule of ∼70–80 bases (7–11). In the second step, which occurs after its export by exportin-5 to the cytoplasm (12,13), the pre-miRNA is cleaved by the RNase III Dicer yielding mature miRNA and its complementary miRNA* (14–18). The miRNA is then loaded on the RNA-induced silencing complex (RISC) (19–21), which directs its binding to its target gene.
Another cleavage pathway that takes place on introns is the pre-mRNA splicing process, where the introns are excised out of the pre-mRNA transcript and the exons are ligated. Splicing as well as other processing events of Pol II transcripts occur in the cell nucleus within a huge and highly dynamic ribonucleoprotein (RNP) machine—the supraspliceosome. The supraspliceosome is a 21 (±1.6)-MDa complex of RNA and proteins composed of four native spliceosomes connected by the pre-mRNA (22,23). The entire repertoire of nuclear pre-mRNAs, independent of their length and number of introns, is individually found assembled in supraspliceosomes [reviewed in (24)]. Components of the supraspliceosome include the spliceosomal U small nuclear RNPs (U snRNPs) and splicing factors, among which are Sm proteins; alternative splicing proteins such as SR proteins; the splicing regulatory factor heterogeneous RNP G (hnRNP G) hnRNP G (25); the alternative splicing factors RBM4 and WT1, which cointeract to influence alternative splicing (26); the alternative splicing regulator ZRANB2 (27); and other proteins that process the pre-mRNA, among which are the editing enzymes ADAR1 and ADAR2 (24). The supraspliceosome was shown to have both splicing and editing activities (28,29). Alternative splicing events were also shown to occur within the supraspliceosome (25,30,31). Splicing is a major event in the processing of Pol II transcripts. Therefore, the interplay between the processing of intronic pri-miRNAs and the processing of pre-mRNA is intriguing (32,33).
One way of coordination between intronic miRNAs processing and splicing occurs in short introns. In this case, the entire intron is a pre-miRNA, and the first step of miRNA biogenesis is the splicing of the intron (34,35). The biogenesis pathway of these miRNAs, called ‘mirtrons’, does not involve the microprocessor. There are also mirtron-like splicing-independent miRNAs that require Drosha, but neither DGCR8 nor Dicer, for their processing and are called ‘simtrons’ (36).
However, most intronic miRNAs are processed by the microprocessor and, it seems, from the same pre-mRNA molecule as the mRNA (5,37,38). Several reports, with different conclusions, were published in recent years about the processing of the transcripts into mRNAs and miRNAs. Comparison of the level of pri-miRNA transcription expressed from either an intronic sequence or an intronic sequence flanked by exons, showed that the presence of the flanking exons increased the level of transcription, possibly due to prolonged time at the site of transcription and splicing (39). Microprocessing was shown to take place cotranscriptionally before splicing, and it was suggested that this processing enhanced splicing (40). Another study showed that pre-miRNA processing could occur in an intron before its splicing (5). Knock-down of Drosha did not reveal a strong effect on splicing, but introns without a pre-miRNA were spliced more rapidly than those with pre-miRNA. Supporting this, an in vitro system demonstrated that cleavage by the microprocessor can occur faster than splicing and that anti-Drosha antibodies can precipitate Y-shaped splicing intermediates (41). In contrast, an artificial intron harboring an miRNA was shown to be spliced before the miRNA processing (42), similarly to what occurs with small nucleolar RNA (snoRNA) processing (43).
A recent study demonstrated a more complex interaction between splicing and microprocessing—it was shown that processing of miR-211, located in intron 6 of melastatin, promoted the splicing of that intron. An importance for the 5′ splice site (SS) in miRNA biogenesis was also shown (44). In addition, a link between splicing and miRNA processing was found when splicing components were reported to associate or cosediment with the microprocessor and with pre-miRNA (41,45).
Because a large proportion of miRNA genes are located in introns, we searched for a cross-talk between their processing and pre-mRNA splicing. Here we show interplay between pre-mRNA splicing and intronic miRNA biogenesis. We demonstrate that inhibition of splicing increases miRNAs expression, whereas knock-down of Drosha increases splicing. We further show that the microprocessor components, Drosha and DGCR8, and the processed pri-miRNAs of the miR-106b-25 cluster of intron 13 of MCM7 (mini chromosome maintenance 7) are found in supraspliceosomes, together with pri-miRNAs. Importantly, we identified two novel alternative 3′ splicing events, each between a miRNA pair of the miR-106 b-25 cluster, which extend exon 14 of MCM7 and change its coding capacity. In the novel splice isoform, miRNAs are included in the extended exon and are thus excluded from being processed into miRNA. Thus, changes in these splicing events were shown to differentially affect the levels of miRNAs originating from the same cluster. Overall, we introduce an extended interplay between the miRNA biogenesis and the pre-mRNA splicing within the supraspliceosome.
MATERIALS AND METHODS
RNA extraction and real-time polymerase chain reaction
Total RNA from cell lines was prepared and treated with RNase-free DNase I as previously described (46). cDNA was synthesized by using 0.5–5 μg of RNA with dT15 primer and Moloney murine leukemia virus reverse transcriptase (Promega). Polymerase chain reactions (PCRs) (20 μl) contained 10 pmol of each of the indicated primer pairs (see later in the text) and 1× taq master mix, purple (λ biotech). The amplified products were run on a 2% agarose gel. All experiments were performed with at least 2 or 3 biological repeats.
Drosha knock-down by small interfering RNA
Small interfering RNA (siRNA) targeted to Drosha 5′ AACGAGUAGGCUUCGUGACUU 3′ (7) and siGENOME Nontargeting siRNA (Dharmacon) were transfected into HeLa cells with DharmaFECT 1 (Dharmacon) according to the manufacturer’s instructions with some modifications. Cells grown in 6-cm plates were transfected with 75-nM siRNA by using 0.176% DharmaFECT 1. After 72 h, total proteins and RNA were extracted and analyzed.
Western blot
Western blot (WB) analyses were performed as previously described (29), using Drosha (D28B1, Cell Signaling Technology) monoclonal antibodies and polyclonal antibodies raised against the Drosha peptide corresponding to amino acids 9–28 (RMSFHPGRGCPRGRGGHGAR, as described in the anti-Drosha 07-717 Millipore datasheet); polyclonal anti-DGCR8 (10996-1-AP, proteintech); anti-hnRNP G provided by Dr Stefan Stamm (University of Kentucky, Lexington); Mab 104 for SRSF5 (SRp40) protein; and anti-CBP antibodies (38A1) provided by Dr Mutsuhito Ohno (Laboratory of Biochemistry Institute for Virus Research, Kyoto University). All the above were visualized with horseradish peroxidase conjugated to affinity-pure goat anti-rabbit IgG (H + L) diluted 1:5000.
Preparation of supraspliceosomes, and analyses of protein and RNA
Supraspliceosomes were prepared from HeLa cells (CILBIOTECH) as previously described (47,48). Briefly, nuclear supernatant was prepared from clean cell nuclei by microsonication of the nuclei and precipitation of the chromatin in the presence of transfer RNA. The nuclear supernatant was fractionated on 10–45% (vol/vol) glycerol gradients. Centrifugations were carried out at 4°C in an SW41 rotor run at 41 krpm for 90 min (or an equivalent w2t). The gradients were calibrated with tobacco mosaic virus as a 200S sedimentation marker. Supraspliceosome peak fractions were confirmed by electron microscopy visualization. For a second fractionation, the 200S peak fractions were pooled, and the glycerol was removed by dialysis. The sample was then concentrated using Vivaspin 15R hydrosart 5 KDa (Sartorius, according to the instructions) and subsequently loaded on a second 10–45% glycerol gradient and centrifuged using the same parameters. For protein analysis, fractions were acetone-precipitated. For RNA extraction, fractions of the glycerol gradients (520 μl) were mixed with 150 μl of extraction buffer [50 mM Tris–HCl (pH 7.5), 300 mM NaCl] and 50 μl of 10% (w/v) sodium dodecyl sulphate, and the RNA was recovered by extraction with phenol and precipitation in ethanol. Supraspliceosome fractions (sedimenting at 200S), were pooled, concentrated using vivaspin 15R 5000 MWCO HY (Sartorius) and analyzed.
Preparation and analysis of nuclear and cytoplasmic fractions
The purification was performed according to Qiagen’s cytoplasmic RNA extraction using RNeasy protocol. Six-well plates were washed with phosphate-buffered saline followed by the addition of 175 μl of cold RLN buffer (50 mM Tris, pH 8, 140 mM NaCl, 1.5 mM MgCl2 and 0.5% NP40). The cells were then scraped and moved to an Eppendorf tube on ice for 5 min. Centrifugation for 2 min at 300g at 4°C was then performed. The supernatant was transferred to a new tube, and both tubes (supernatant–cytoplasm and pellet–nucleus) were centrifuged again for better purification. RNA was then extracted from both fractions using the TRI-reagent method.
Quantitative RT-PCR for analysis of miRNA molecules
miRNA levels were measured using TaqMan miRNA Assay [Applied Biosystem (49)] according to the manufacturer’s instructions. Amplification was carried out using an ABI PRISM 7700 sequence detector (Applied Biosystem). Analysis was performed using the delta–delta CT, 2−ΔΔCT method (50). Assays were performed on hsa-miR-25 (Assay ID 403), mmu-miR-93 (Assay ID 1090), hsa-miR-106b (Assay ID 442), hsa-let-7g (Assay ID 2282) and U6 snRNA (Assay ID 1973) for normalization. All experiments were performed in triplicate of at least two or three biological repeats.
Morpholino transfections
Antisense morpholino (Gene Tools) against the 3′SS between miR-93 and miR-25, 5′ TCCTGTGAGGGAGACCAGACCCTTT 3′, was transfected to HeLa cells with endo-porter (Gene Tools). One milliliter of medium without serum was added to 3.5-cm plates. Morpholino was added to a concentration of 10 µM followed by the addition of endo-porter, final concentration 0.6%. RNA was extracted 24 h later. The results represent two biological repeats and at least three repeats of each.
Spliceostatin A treatment
Spliceostatin A (SSA) (51), provided by Dr Minoru Yoshida (Chemical Genomics Research Group and Chemical Genetics Laboratory, RIKEN, The Institute of Physical and Chemical Research, Wako City, Saitama, Japan), was diluted in methanol and was transfected into HeLa cells. One microliter of 100 ng/µl SSA was added to a 3.5-cm plate with 1 ml of medium (final concentration 100 µM). As a control, 1 µl of methanol was added to 1 ml of medium. RNA was extracted 5 h later. The results represent two biological repeats and at least three repeats from each.
Primers
To amplify exon 13 of MCM7, we used primer a, 5′ ATCACAGCAGCATACGTGGA 3′ (sense), and primer b, 5′ AGTGGAAAGGCGCAGGATAG 3′ (antisense).
To amplify exon 13–exon 14 of MCM7, we used primer a and primer c, 5′ TAGCTGTCTGCCCCTTGTCT 3′ (antisense).
To amplify exon 14–exon 15, we used primer g, 5′ AGACAAGGGGCAGACAGCTA 3′ (sense) and primer h, 5′ GGAACTGGGCGGGTGTGAAG 3′ (antisense).
To amplify exon 14–intron 14, we used primer g and primer i, 5′ AGTGGGTGTGTAAGGTCAGGA 3′ (antisense).
To amplify pri-miR-25, we used primer d, 5′ ACAGCTGAACTCCGGGACTG 3′ (sense) and primer e, 5′ CCCAGCATCCGCAGTGTTGG 3′ (antisense); both, d (miR-25-fw) and e (miR-25-rev), from (40).
To amplify exon 13 through miR-25 (spliced and nonspliced), we used primer a and primer e.
To amplify exon 13 through miR-93 (spliced and nonspliced), we used primer a and primer f, 5′ CAGAGAGAACGTGTCCCG 3′ (antisense).
To amplify exon 13 through the intron 13 exon 14 splice junction, we used primer a (sense) and primer j, 5′ CATTCTCAGACGTGCCTAAGGG 3′ (antisense).
To amplify pri-miR-330, we used miR-330-fw, 5′ CCTTCTTCCAGGATCGCGTC 3′ (sense) and miR-330-rev, 5′ GAGGTCTCCGATGAAAACGG 3′ (antisense), both from (40).
To amplify GAPDH, we used GAPDH fwd, 5′ TGCACCACCAACTGCTTAGC 3′ (sense) and GAPDH rev, 5′ GGCATGGACTGTGGTCATGAG 3′ (antisense).
To amplify exon 12 through miR-25 (spliced and nonspliced) in rat, we used rMCM7-12-s, 5′ CTACATCACTGCAGCGTATG 3′ and rmiR25-as, 5′ CCACATCTGCAGTGTTGG 3′.
To amplify exon 12 through miR93 in rat, we used rMCM7-12-s and rmiR93 as, 5′ ACTGTCAGAGGCTGTGTCCT 3′.
RESULTS
The microprocessor and pri-miRNAs are found within the supraspliceosome
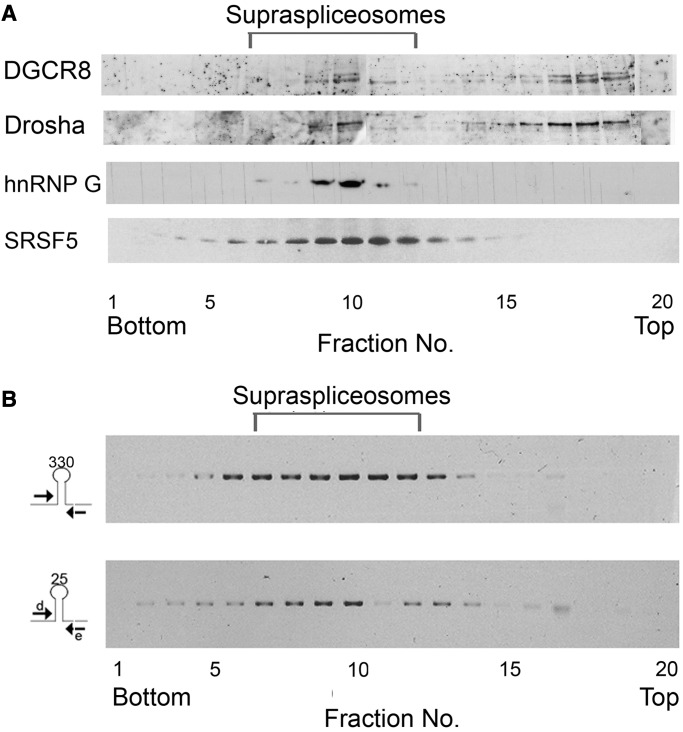
Given that a large proportion of miRNA genes are located within introns of genes transcribed by Pol II, and that RNA processing activities are performed within the supraspliceosome, we asked whether key microprocessor components are found in supraspliceosomes. To answer this question, we fractionated, on glycerol gradients, nuclear supernatants enriched with supraspliceosomes prepared from HeLa cells (24) and then re-fractionated the 200S fractions, where supraspliceosomes sediment, on a second glycerol gradient. WB analyses across the second gradient, with antibodies against the two microprocessor core proteins Drosha and DGCR8, show a similar distribution of the two proteins (Figure 1A). Both peak at the 200S—supraspliceosome—region of the gradient, where Pol II transcripts as well as spliceosomal proteins and splicing regulatory factors (e.g. SR proteins, hnRNP G, previously shown to be associated with supraspliceosomes) sediment (Figure 1A, bottom) (24,25,28,30,47,52–54). In addition, both proteins appear as free proteins (or part of small complexes) at the top of the gradient, indicating dissociation from the large complexes during fractionation.
Figure 1.
Microprocessor components and pri-miRNA sequences are found in supraspliceosomes. (A) Nuclear supernatants enriched for supraspliceosomes were prepared from HeLa cells and were fractionated in 10–45% glycerol gradients (28). Supraspliceosome peak fractions (8–10) were pooled and refractionated on a second glycerol gradient. Aliquots from gradient fractions were analyzed by WB using anti-Drosha and anti-DGCR8 antibodies. For comparison, distribution across glycerol gradients of regulatory splicing factors associated with supraspliceosomes, hnRNP G and SRSF5 SR proteins is shown (B) Nuclear supernatants enriched for supraspliceosomes were prepared from HeLa cells and were fractionated in 10–45% glycerol gradients (28). RT-PCR analyses of the distribution of pri-miR-330 and pri-miR-25 across the gradient using the indicated primer pairs that flank the respective pri-miRs.
The presence of microprocessor key components in supraspliceosomes fractions prompted us to further analyze the possible cross-talk between the microprocessor and the supraspliceosome by studying specific pri-miRNAs within the supraspliceosome. For this aim, we chose to study miR-25, a member of the miR-106b-25 cluster in intron 13 of MCM7, which harbors miRNAs 106 b, 93 and 25, and was already investigated with regard to splicing (5,40). This cluster is conserved in mammals, whereas the miR-25 and miR-93 are also conserved in Xenopous tropicalis and zebrafish (Supplementary Figure S1). The miR 106b-25 cluster has two paralogs—miR 17-92 and miR 106a-363. It is over-expressed in several cancers and was found to be pro-oncogenic, like miR 17-92 (55). To analyze how pri-miR-25 of this cluster is distributed across a glycerol gradient of fractionated nuclear supernatants enriched with supraspliceosomes prepared from HeLa cells nuclei, RNA was extracted from each gradient fraction, and RT-PCR was performed with primers flanking the pre-miRNA. It can be seen that pri-miRNA of miR-25 peaks at the 200S region where supraspliceosomes sediment (Figure 1B). For comparison, we also analyzed the distribution across a glycerol gradient of a pri-miRNA of a single intronic miRNA. We chose pri-miRNA of miR-330 located in intron 4 of the echinoderm microtubule-associated protein like 2 (EML2) gene. RT-PCR analysis revealed that pri-miRNA of this miRNA also peaks in supraspliceosome fractions (Figure 1B) indicating that more than miR-25 is present in these complexes.
Processed pri-miRNAs of the miR-106b-25 cluster are found in supraspliceosomes
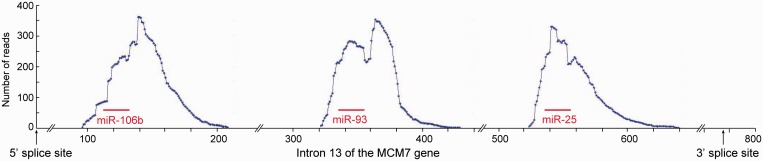
The finding of key microprocessor components and pri-miRNA sequences in supraspliceosomes fractions suggests interaction between the microprocessor and the splicing machine. To further substantiate this interaction, focusing on the miR-106b-25 cluster in intron 13 of MCM7, we searched for sequences of pre-miRNAs of the 106b-25 cluster within the deep sequencing data of small RNAs (<200 nt) present in supraspliceosome (24) fractions (8–10) isolated from HeLa cells nuclei (analysis of full sequencing data will be published elsewhere). Figure 2 shows that the supraspliceosome fractions harbor processed pri-miRNA-106b, pri-miRNA-93 and pri-miRNA-25. The data are in support of the previous results. Importantly, the analysis revealed that the sequences found in supraspliceosomes are not only of pri-miRNA (which might be part of an unprocessed intron) but of processed pri-miRNA-106b, pri-miRNA-93 and pri-miRNA-25, as manifested by the decline in the number of reads flanking each pre-miRNA of the 106 b-25 cluster (see also Supplementary Figure S2). Some are full-length pre-miRNAs and some are smaller that might be further cleavage products from the pre-miRNAs. The finding of both pri-miRNAs and processed pri-miRNAs of the 106b-25 cluster in supraspliceosome fractions indicates that the processing of these pri-miRNAs to pre-miRNAs likely occurs within the supraspliceosome.
Figure 2.
Processing of pri-miRNA in supraspliceosomes. RNA was extracted from supraspliceosomes prepared from frozen HeLa cells, as previously described (28). Searching for sequences of pre-miRNAs of the 106b-25 cluster within the deep sequencing data of small RNAs (<200 nt), from this supraspliceosomal RNA (fractions 8–10), revealed that sequences of the pre-miRNA 106 b, 93 and 25 were found in supraspliceosomes.
Novel splicing events in the miR-106b-miR-25 cluster
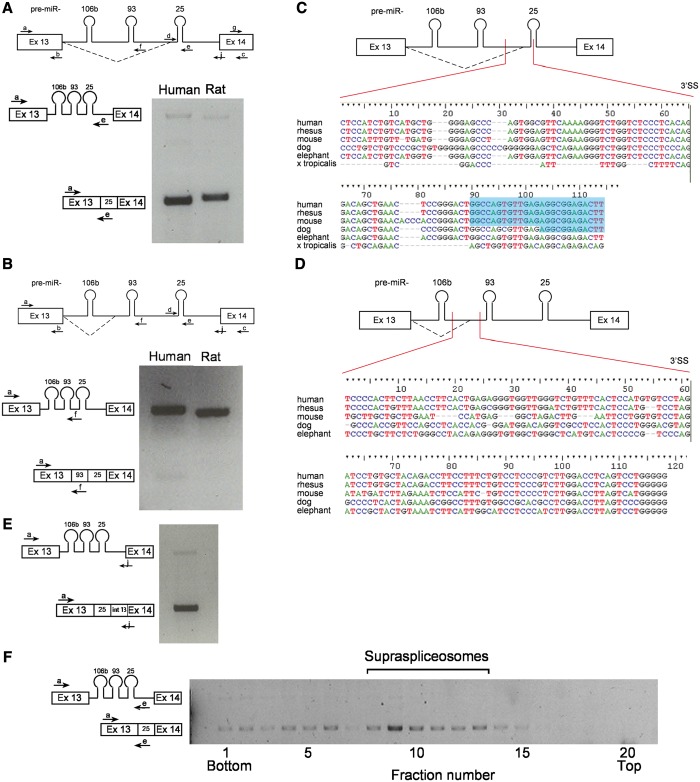
To further explore the relationship between splicing and miRNA processing, we focused on the miR-106b-25 cluster in intron 13 of MCM7 (5,40), encouraged by the finding of both pri-miR 25 and pre-miR 25 sequences in supraspliceosome fractions. We next analyzed the expression of sequences of intron 13 of MCM7 using RT-PCR and different combinations of primer pairs (Figure 3A). The analyses revealed two previously unknown alternative splicing events, alternative to the constitutive 3′SS of intron 13, that use two alternative 3′SSs, which are located within the cluster (Figure 3A and B). One of the alternative 3′SSs is located between pre-miR-93 and pre-miR-25 (hence termed 93-25 3′SS, Figure 3A). The other is located between pre-miR-106b and pre-miR-93 (hence 106b-93 3′SS, Figure 3B). The identity of the bands corresponding to the two new splice variants was confirmed by DNA sequencing. The new splice isoform, the product of the 93-25 3′SS, seems to incorporate the sequence of pre-miR-25 into exon 14 of MCM7. We validated this result (Figure 3E) by performing RT-PCR using primer a, (a sense primer from exon 13) and primer j (antisense primer, that spans the 3′ splice junction of intron 13 and exon 14). The identity of this band was confirmed by DNA sequencing.
Figure 3.
Conserved novel 3′SS in intron 13 of MCM7. (A) A novel alternative 3′SS between pre-miR-93 and 25. A diagram of MCM7 exon 13 through exon 14 with splicing at the novel 93-25 3′SS (broken line) and the primers used for RT-PCR (upper panel) indicated. RT-PCR on RNA extracted from HeLa cells using primer pair a/e (lower panel). The amplified RT-PCR products are depicted on the left. (B) A second, yet minor, novel 3′SS between pre-miR-106 b and 93. A diagram of MCM7 exon 13 through exon 14 with splicing at the novel 106 b-93 3′SS (broken line) and the primers used for RT-PCR (upper panel) are indicated. RT-PCR using primer pair a/f (lower panel). The amplified RT-PCR products are depicted on the left. (C) Conservation of the sequences flanking the novel 93-25 3′SS. Sequence alignment of the region surrounding the novel 3′SS between pre-miRNAs 93 and 25. The different organisms are marked on the left. The splice point is marked by a vertical line. Above is a diagram of exon 13 through exon 14 of MCM7 with the aligned region marked by two vertical lines. The 5′-end of sequences of pre-miR-25 are highlighted in blue. (D) Conservation of the sequences flanking the novel 106b-93 3′SS. The same as in (C), with sequence alignment of the region surrounding the novel 3′SS between pre-miRNAs 106 b and 93. (E) The novel splice isoform, the product of splicing at the 93-25 3′SS, incorporates the sequence of pre-miR-25 into exon 14 of MCM7. RT-PCR analysis using primer pair a/j. (F) The novel splice isoform at the 93-25 3'SS is found in supraspliceosomes. RT-PCR analysis of aliquots of RNA is extracted from gradient fractions, using the indicated primer pairs.
Both splicing events seem much less dominant than the constitutive splicing of intron 13 as RT-PCR with primers from exons 13 and 14 (primer pair a/c) gives only the constitutive product (see later in the text). The splicing from the 93-25 3′SS seems stronger than the 106b-93 3′SS one, again because the use of primer pair a/e gives this product predominantly (Figure 3B). Also, RT-PCR with primer pair a/f gives more of the unspliced form than of the spliced one (Figure 3B). The finding that the splicing from the 106b-93 3′SS occurs at a lower frequency than that from the 93-25 3′SS, might be due to the fact that this SS is newer in evolution (Figure 3D). The finding of the novel 3′SS events introduces an additional aspect to the interplay between splicing and miRNA biogenesis. This is because transcripts that undergo splicing at the novel 3′SS include the relevant miRNA sequences within the extended exon, an uncommon feature of miRNAs (4). At the same time, the inclusion of the miRNA sequences in the extended exon changes the coding capacity of the MCM7 3′-end.
Conservation of the novel alternative splicing event
Because the miR 106b-25 cluster is conserved in mammals, we asked whether the sequences of the novel 3′SS, which might have implications on alterations in the levels of miR-25, miR-93 and of MCM7, are conserved in evolution. Comparison of the sequences of the miR-93-25 3′SS and its upstream poly-pyrimidine tract in different organisms shows a high level of conservation (Figure 3C). The 93-25 3′SS is also found in other vertebrates besides mammals. Similar comparison of the sequences of the 106b-93 3′SS and its upstream poly-pyrimidine tract shows that they are less conserved (Figure 3D). To test whether the 93-25 3′SS and 106b-93 3′SS are functioning in other organisms besides human, we tested the splicing pattern in rats. RT-PCR analyses of RNA prepared from the rat PC12 cell line showed that the 93-25 3′SS was also used in rat cells (Figure 3A), but the splicing from the 106b-93 3′SS did not take place (Figure 3B). The conservation of the novel 3′SS sequences indicates their biological relevance, and the use of the 93-25 3′SS suggests that this splicing isoform plays a functional role.
Because splicing at the 106b-93 3′SS is found in low levels, in the following sections, we focus our analyses on the more abundant 93-25 3′SS. We next asked whether the novel spliced isoform is found in supraspliceosomes. Interestingly, RT-PCR analysis of the distribution of the novel splice isoform (93-25 3′SS) across the glycerol gradients reveals that it cosediments with supraspliceosmes (Figure 3F). This suggests that the constitutive, the novel alternative splicing events as well as the pri-miRNA processing events that MCM7 intron 13 undergoes, occur in supraspliceosomes.
The novel splice isoform is found in the nucleus and cytoplasm
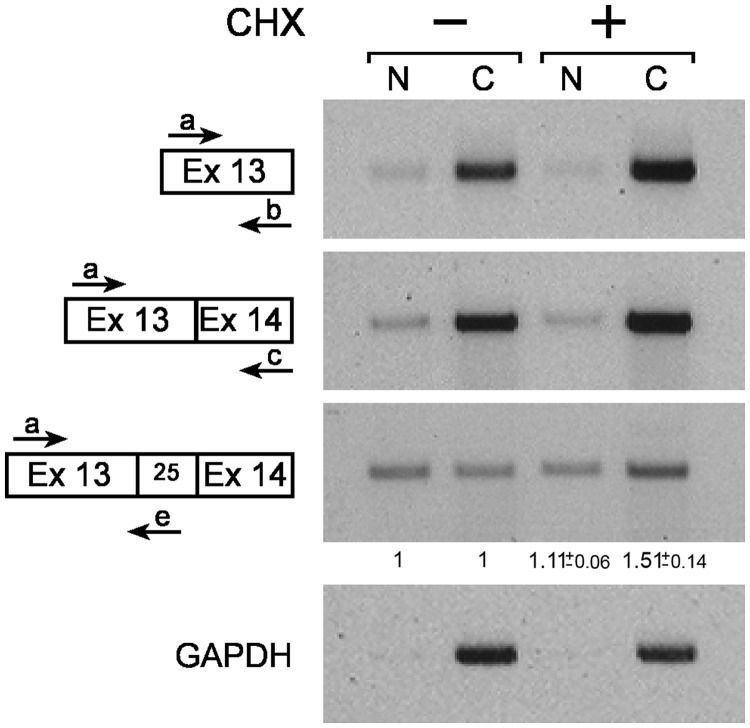
The new splice isoform, the product of the 93-25 3′SS, incorporates the sequence of pre-miR-25 into exon 14 of MCM7. To investigate whether the new alternative splice isoform that changes the coding capacity of the MCM7 mRNA is exported to the cytoplasm, we analyzed the distribution of the spliced product between the nucleus and the cytoplasm (Figure 4). HeLa cells were fractionated into nuclear and cytoplasmic fractions, and the splicing of the MCM7 products were analyzed by RT-PCR (positive and negative controls for the fractionation are given in Supplementary Figure S3). Because the new spliced product harbors premature termination codons in all three reading frames, it is likely to be a substrate for the nonsense-mediated mRNA decay pathway (56,57). Thus, we performed the same experiment also in the presence of cycloheximide (CHX), which is known to inhibit nonsense-mediated mRNA decay by translational block (58). We observed that GAPDH and MCM7 exon 13–exon 14 mRNAs are located mostly in the cytoplasm, and RNA from exon 13 of MCM7, which is expected to be in all the MCM7 RNA molecules, is similarly enriched in the cytoplasm. However, the product of splicing at the 93-25 3′SS is distributed evenly between the nucleus and the cytoplasm in the untreated cells. The addition of CHX elevated the level of this isoform in the cytoplasm (1.51 ± 0.14-fold compared with changes in mRNA level, measured using primers from exons 13 and 14). Yet, the distribution between nuclear and cytoplasmic fractions of this isoform remains relatively even compared with the distribution of the constitutive mRNA molecules, which is mainly cytoplasmic. First, these results show that the new 93-25 3′SS isoform is transported to the cytoplasm. Second, regardless of the CHX treatment, the 93-25 3′ splice isoform is dispersed more evenly between the cytoplasm and the nucleus than the constitutive MCM7 mRNA, and the part of it that is found in the nucleus indicates that it might also have some function in the nucleus, presumably reflecting the competition between the novel splicing event and the production of the miRNA.
Figure 4.
The novel alternative 93-25 3′ splice isoform is found in the cytoplasm. RNA was extracted from nuclear (N) and cytoplasmic (C) fractions of HeLa cells, either treated (+) or untreated (−) with 50 µg/ml CHX for 2 h, and analyzed by RT-PCR. Fold change of the novel alternative 93-25 3′ splice isoform in nucleus and cytoplasm after CHX treatment, compared with fold change in the MCM7 constitutive mRNA, is indicated below the specific lanes with standard error (n = 2).
Inhibition of the splicing at the 93-25 3′SS results in elevated miRNA levels
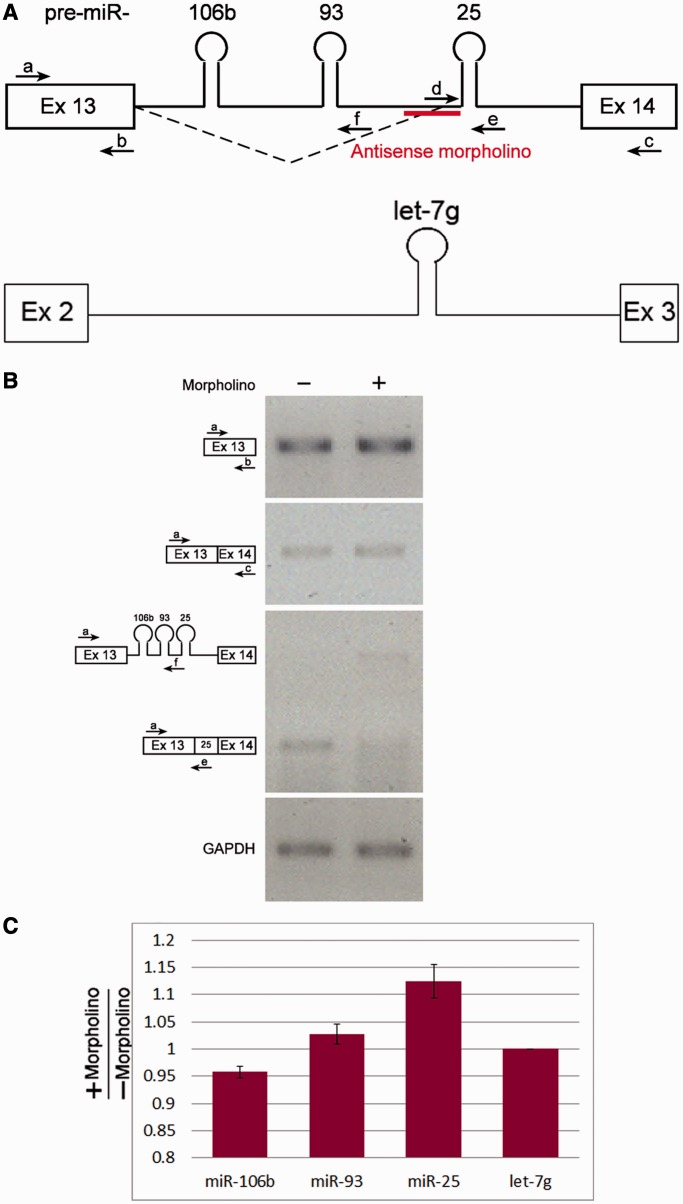
Given that the 93-25 3′SS is located between the sequences of pre-miRNA 93 and 25, it is likely that changes in splicing at that site affect the level of the miRNAs of the cluster. To test this, we decided to inhibit this splicing event in a specific manner, and then test the effect on miRNA levels. For this purpose, we used antisense molecules in the form of morpholinos, antisense to the 93-25 3′SS and the polypyrimidine tract, 25 nt in length (Figure 5A). Following incubation, RNA was extracted and analyzed by RT-PCR for mRNA and miRNA levels. The analyses confirmed the inhibition of splicing of the novel isoform and not of the canonical isoform (Figure 5B).
Figure 5.
An antisense morpholino oligonucleotide abolishes splicing at the alternative 3'SS. Morpholino molecules antisense to the 93-25 3'SS were transfected into HeLa cells. RNA was then extracted and analyzed. (A) Schematic representation of exon 13– exon 14 of the MCM7 gene and the antisense morpholino is indicated; and schematic representation of exon 2–exon 3 of the WDR82 gene. (B) RT-PCR analyses of RNA extracted from the cells treated with antisense morpholino and from control untreated cells were performed using the indicated primers. (C) Graph showing the fold change of the indicated miRNAs after morpholino treatment measured using the miRNA TaqMan assay, normalized to the change in let-7 g miRNA. Standard errors are shown. P < 0.05 for miR-106 b and miR-25 (t-test). The results represent triplicates of each of two independent experiments.
To test the effect of the inhibition of splicing at the 93-25 3′SS on the level of miRNAs processed from this cluster, we performed quantitative RT-PCR using Taq-Man microRNA assay (Figure 5C). For comparison, we tested the effect of the morpholino on the level of let-7g (an intronic miRNA expressed in HeLa cells). Normalizing the effect of morpholino on the 106b-25 cluster to that on let-7g, it seems that the levels of miRs-106b and 93 were not so affected. However, the level of miR-25 was upregulated (although to a small extent, possibly due to the relatively small effect of the morpholino on the splicing reaction). Although other explanations cannot be ruled out, the results indicate that for the efficient processing of pre-miRNA-25, its inclusion in the intron, rather than in the exon, is favored. This is because inhibition of the splicing event that excludes it from the intron and leads to its inclusion in the alternatively spliced mRNA resulted in the increase of the level of this miRNA.
Inhibition of splicing by spliceostatin A upregulates the levels of the intronic miRNAs
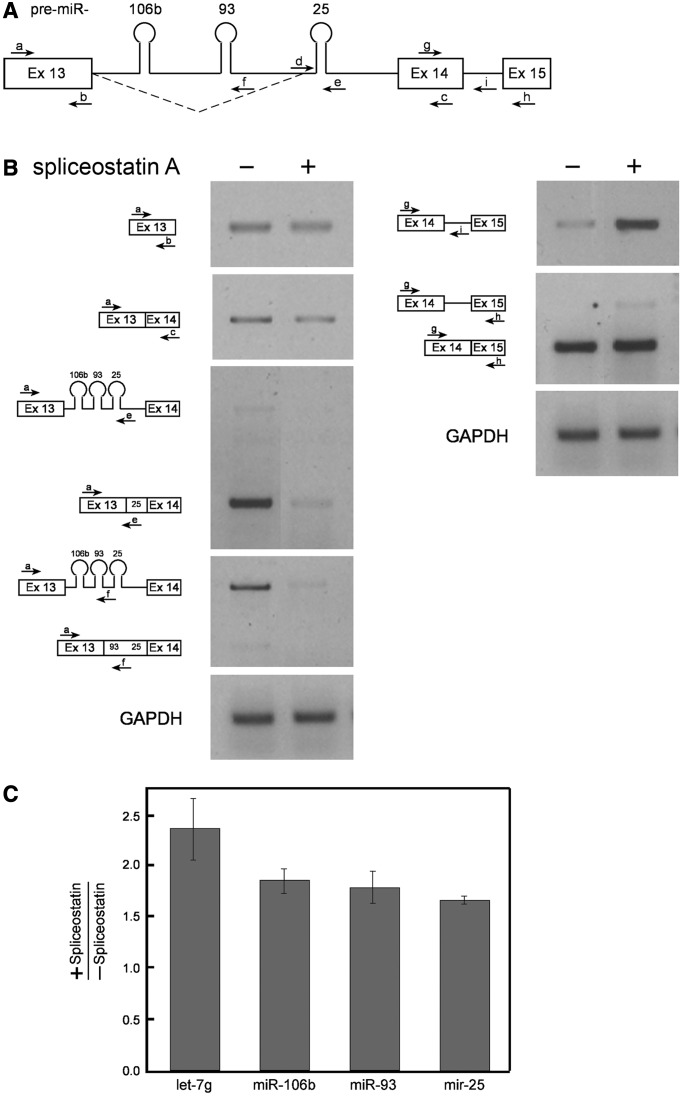
After observing the effect of the specific inhibition of the novel splicing event on the interplay between splicing and miRNA biogenesis, we wanted to test the effect of general inhibition of splicing on this interplay. The inhibition was achieved using spliceostatin A (SSA), which inhibits splicing by binding to SF3b, a subcomplex of U2 snRNP, and keeping the splicing reaction in its primary state (51,59). As can be seen in Figure 6B, SSA inhibited both splicing reactions; namely, the constitutive splicing and the use of the 93-25 3′SS. The constitutive splicing seems less affected, perhaps because of its initial high level (Figure 4). The splicing of exon 14–exon 15 is also inhibited (Figure 6B, right panel). When we tested the effect of SSA on the level of the miRNAs, using quantitative RT-PCR (Taq-Man microRNA assay), we found that all three miRNAs of the cluster were upregulated (Figure 6C). The level of the intronic let-7g was also upregulated (Figure 6C). A number of possible interpretations comes to mind; for example, pri-miRNA processing from an intact intron is more efficient; retaining the intron by inhibiting splicing allows an extended period for processing of pri-miRNA. After splicing inhibition, RT-PCR with primer pair a/e did not amplify more of the unspliced intron 13 as expected (Figure 6B), possibly because the pre-miRNAs were cleaved from the intron, thereby precluding it from being amplified. The amplification of the adjacent intron 14 (RT-PCR with primer pairs g/h and g/i), which lacks any miRNAs, was higher after splicing inhibition (Figure 6B). Independent of the interpretation, this experiment shows interplay between splicing and pri-miRNA processing.
Figure 6.
SSA upregulates the levels of intronic miRNAs. SSA was added to HeLa cells (100 ng/ml for 5 h), and RNA was then extracted. (A) Schematic representation of exon 13– exon 15 of the MCM7 gene. The novel 93-25 3′ splicing pattern is indicated (broken line), and so are the primers used. (B) RT-PCR analyses of RNA extracted from cells treated (+) or untreated (−) with SSA, using the indicated primer pairs from MCM7 and with primers from GAPDH. (C) Graph showing the fold change of the indicated miRNAs after SSA treatment measured using the miRNA TaqMan assay. Standard errors are shown. P < 0.05 for let-7 g and miR-93; <0.01 for miR-106 b; and <0.005 for miR-25 (t test). The results represent triplicates of each of two independent experiments.
Drosha knock-down upregulates the novel splice isoform of MCM7
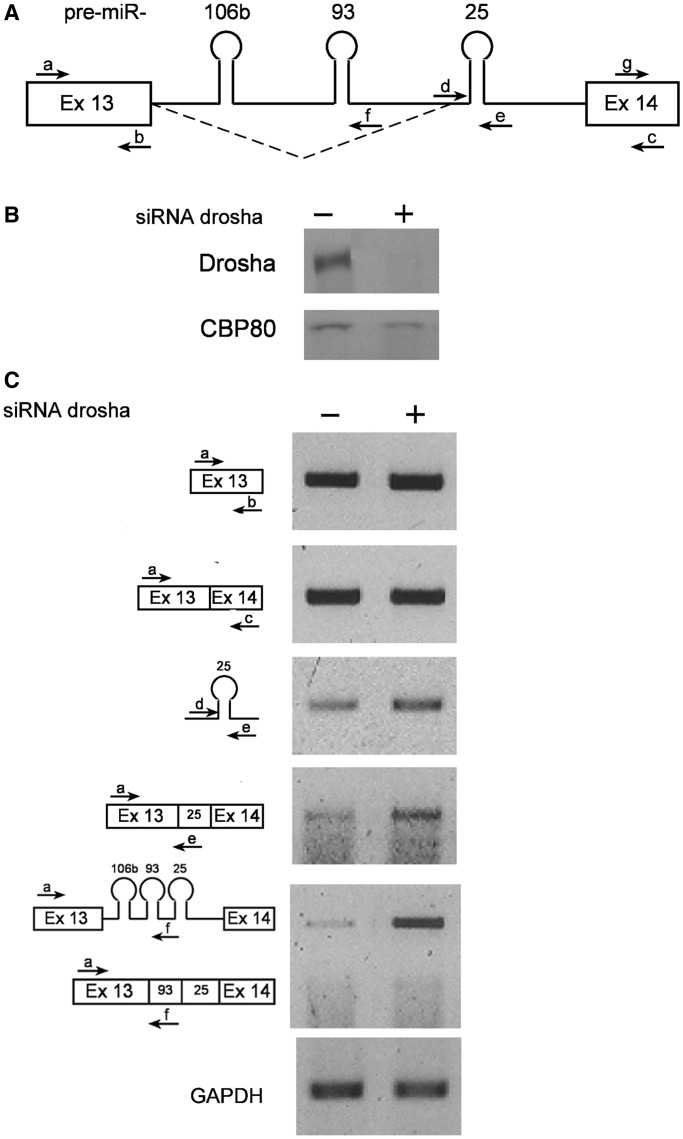
To test whether the microprocessing affects the splicing reaction—Drosha was knocked-down from cells by siRNA. Figure 7B shows WB with anti-Drosha antibodies verifying that the level of Drosha was reduced significantly following the addition of the siRNA. As cleavage of the pre-miRNAs out of the intron is downregulated after Drosha knock-down, it is expected that there would be more of the intact intron. This seems to be the case as on knock-down of Drosha, primer pair a/f gives significantly higher levels of the pri-miRNA 106b and 93 (5.25-fold increase, Figure 7C). The level of the pri-miRNA 25 (primer pair d/e, amplifying a part of intron 13 surrounding pri-miRNA 25), another substrate of Drosha, was also upregulated, but to a lesser extent when Drosha's levels decreased (1.62-fold increase, Figure 7C). This is not surprising, as knock-down of Drosha increased splicing from the novel alternative 93-25 3′SS, including pre-miRNA 25 in the exon of the novel splice isoform (Figure 7C). These results demonstrate an effect of the microprocessor on splicing and further substantiate the cross-talk between these two pathways.
Figure 7.
Knock-down of Drosha is accompanied by increase in the level of the novel 3′ splice isoform of MCM7 mRNA. HeLa cells were treated with siRNA molecules against Drosha. Proteins and RNAs were extracted from siRNA treated (+), and untreated (−) cells, and were analyzed. (A) Schematic representation of exon 13–exon 14 of the MCM7 gene. The novel 93-25 3′ splicing pattern is indicated (broken line), and so are the primers used. (B) WB with anti-Drosha and anti-CBP80 antibodies as a reference. Drosha was knocked-down by siRNA to 18%. (C) RT-PCR analyses with the indicated primers.
DISCUSSION
The finding that a large proportion of miRNAs are processed from pre-mRNA introns raises the question of how the two processing reactions, that of splicing and pri-miRNA processing, are coordinated. Our results show interplay between splicing and pri-miRNA processing. Coordination between splicing and pri-miRNA processing is likely facilitated by the presence of Drosha and DGCR8, the core of the microprocessor, in supraspliceosome fractions (Figure 1A). Our finding of processed pri-miRNA in supraspliceosomes (Figure 2) brings further support to the cross-talk between the splicing and miRNA processing machines and to the hypothesis that the processing of the tested intronic miRNAs occurs in supraspliceosomes. Earlier findings of association between certain splicing components and the microprocessor and pre-miRNAs (41,45) are in support of our findings.
To analyze the effect of splicing inhibition on miRNA biogenesis, we chose the splicing inhibitor SSA, which was shown to inhibit splicing by binding to SF3b and preventing the pre-spliced spliceosome from moving to an active state (51,59,60). A number of previous studies showed that intronic pre-miRNAs are processed by the microprocessor before splicing, but when assembled in spliceosomes (5,40,44). In the presence of SSA, introns are inhibited from being spliced out, as exemplified by intron 14 of MCM7, which does not harbor a pre-miRNA (Figure 6). Importantly, the likelihood of cleavage of the pre-miRNAs from the looped-out intron increased upon SSA treatment, resulting in elevated levels of the miRNAs (miRs 106b, 93, 25 and let-7g, Figure 6). Accordingly, intron 13, harboring the 106b, 93 and 25 miRNAs is cleaved by the microprocessor and therefore is not detected by RT-PCR. These results show that splicing inhibition increases the levels of miRNAs, indicating competition between microprocessing and splicing.
Focusing on the miR-106b-25 cluster, we found two new 3′ alternative splicing events in intron 13 of MCM7. One between pre-miRNAs 93 and 25 (93-25 3′SS) and the other, much less abundant, between pre-miRNAs 106 b and 93 (106 b 93 3′SS) (Figure 3). The calculated 3′SS scores (http://ibis.tau.ac.il/ssat/SpliceSiteFrame.htm) of the novel 3′SSs are 84.79 and 73.25 for the 93-25 and 106b-93 3′SSs, respectively, compared with a score of 79.46 for the normal 3′SS. Therefore, we focused our analyses here on the 93-25 3′SS.
Interestingly, the new splice isoform, the product of splicing at the 93-25 3′SS, incorporates the sequence of pre-miR-25 into exon 14 of MCM7 (Figure 3B and E). miRNAs are usually not found in exons, and if so, they are predominantly in noncoding RNAs (4). We found that the inclusion of miR-25 in a coding mRNA affected its processing. The inhibition of splicing at the 93-25 3′SS by antisense morpholino (Figure 5) led to an increase in the level of miR-25, in accordance with the model that the miRNAs are cleaved out of the intron and that the use of the 93-25 3′SS excludes pre-miR-25 from the intron. In addition, the processing of miR-25 was the least upregulated by the general inhibition of splicing by SSA that keeps the pre-spliced state (Figure 6).
When we tested the effect of inhibition of the microprocessor on splicing, by knock-down of Drosha, we found that the level of the novel spliced isoform increased, likely because the SS was more available as less cleavage occurred by the microprocessor within the intron (Figure 7). These results add further support to the cross-talk between splicing and miRNA biogenesis.
Although both miRNA 25 and MCM7 mRNA can be generated from an MCM7 pre-mRNA spliced at the normal SSs, this is not the case when splicing occurs at the novel 3′SS. As shown in Figure 4, a detectable level of the new splice isoform (the product of splicing at the 93-25 3′SS), is found in the cytoplasm that might be translated into proteins. This alternative isoform encodes for a shortened protein that lacks the 3′ carboxy terminus because the alternative exon harbors premature termination codons in all three reading frames. In view of our results, we suggest that expression of the novel splice isoform can lead to a modulation in the levels of the different miRNAs of the cluster and of the hosting mRNA isoforms originating from the same transcript.
A recent study of the effect of alternative splicing on miRNA levels in plants (61) showed that an alternative splicing event occurring on heat shock included intronic miR-400 in the alternative exon and excluded it out of the intron, leading to a reduction in miR-400 level. These results are consistent with our findings of the effect of the novel 3′SS on the level of miR-25. Both studies demonstrate the importance of alternative splicing in controlling miRNA levels.
The evolutionary conservation of the sequences of the pre-miR-106 b-25 cluster as well as the sequences of the novel 93-25 3′SS indicate the importance of these miRNAs and the biological relevance of the interplay between their processing and the splicing at the novel 3′SS. The 5′-end of intron 13 of MCM7 including pre-miRNA 106b up to pre-miRNA 93 is newer in evolution. Compatible with that, splicing from the 93-25 3′SS is more abundant than that from the 106b-93 3′SS and is also found in rat (Figure 3). One can speculate that early on, when there were only two miRNAs in that intron, an SS evolved to separate them. Now, a new SS is emerging separating the new miRNA from the older ones.
MCM7 plays a critical role in the G1/S phase transition, ensuring that the entire genome is replicated once and not more than once at each cell cycle (55). Because MCM7 pre-mRNA and pri-miR-106b-25 cluster are cotranscribed, they are both regulated by the transcription factor E2F1. Members of the miR-106b-25 cluster are key modulators of the TGFß tumor suppressor signaling pathway in tumors. While miR-106b and miR-93 inhibit cell cycle arrest through inhibition of p21, miR-25 inhibits apoptosis through affecting Bim (55). Being members of the same cluster, they are likely transcribed to a similar level along with MCM7 mRNA, and posttranscriptional processing events are likely responsible for the final levels of these three miRNAs and mRNA. The finding that both miR-106b and miR-93 inhibit cell cycle arrest, while miR-25 inhibits apoptosis, calls for different processing of miR-25 compared with the other two. The novel 3′ splice isoform that we identified enables a differential control of the levels of miR-25 compared with miR-106b and miR-93. Although the miR-106b-25 cluster is coexpressed, in some cell lines such as human cholangiocarcinoma (H69, Mz-ChA-1 and KMCH), the miRNAs are expressed differentially, with miR-25 expressed at lower levels than miRs 106b and 93 (62,63). Furthermore, the novel 3′ splice isoform, not only eliminates production of miR-25 from these transcripts, but also at the same time encodes for a defective MCM7 protein isoform with a shortened carboxy-terminal end. In view of the important role of MCM7 in replication, and in view of the fact that both MCM7 and miR-106b-25 cluster are pro-oncogenic, understanding the regulation of their processing is important for human health and disease. Importantly, it is likely that the level of the novel 3′ splice isoform can be influenced by a number of cellular or environmental conditions that affect alternative splicing, thus changing the level of miR-25 compared with that of miR-106b and miR-93.
In conclusion, here we shed light on the interplay between intronic miRNA processing and splicing. We show an association between the two machineries—the splicing and the microprocessor, Drosha and DGCR8, within the supraspliceosome. We also suggest how miRNA processing can use alternative splicing to manipulate the levels of different clustered intronic miRNAs. Specifically, we show how miRNAs of the miR-106b-25 cluster, involved in the inhibition of several apoptotic pathways can be differentially expressed.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The US NIH [RO1 GM079549 to R. S. and J.S.]; the Helen and Milton Kimmelman Center for Biomolecular Structure and Assembly at the Weizmann Institute of Science (to J.S.) for financial support. Funding for open access charge: Waived by Oxford University Press.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Aviva Pecho for excellent technical assistance and Dr Nitzan Kol for bioinformatics assistance. They are grateful to Dr Minoru Yoshida of The Chemical Genomics Research Group and Chemical Genetics Laboratory, RIKEN, The Institute of Physical and Chemical Research, Wako City, Saitama, Japan, for spliceostatin A, and to Drs Mutsuhito Ohno and Stefan Stamm for antibodies.
REFERENCES
- 1.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat. Struct. Mol. Biol. 2012;19:586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shomron N, Golan D, Hornstein E. An evolutionary perspective of animal microRNAs and their targets. J. Biomed. Biotechnol. 2009 doi: 10.1155/2009/594738. 2009, 594738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 8.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 10.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 11.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 15.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 16.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 17.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 20.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 21.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 22.Azubel M, Wolf SG, Sperling J, Sperling R. Three-dimensional structure of the native spliceosome by cryo-electron microscopy. Mol. Cell. 2004;15:833–839. doi: 10.1016/j.molcel.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 23.Cohen-Krausz S, Sperling R, Sperling J. Exploring the architecture of the intact supraspliceosome using electron microscopy. J. Mol. Biol. 2007;368:319–327. doi: 10.1016/j.jmb.2007.01.090. [DOI] [PubMed] [Google Scholar]
- 24.Sperling J, Azubel M, Sperling R. Structure and function of the Pre-mRNA splicing machine. Structure. 2008;16:1605–1615. doi: 10.1016/j.str.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 25.Heinrich B, Zhang Z, Raitskin O, Hiller M, Benderska N, Hartmann AM, Bracco L, Elliott D, Ben-Ari S, Soreq H, et al. Heterogeneous nuclear ribonucleoprotein G regulates splice site selection by binding to CC(A/C)-rich regions in Pre-mRNA. J. Biol. Chem. 2009;284:14303–14315. doi: 10.1074/jbc.M901026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Markus MA, Heinrich B, Raitskin O, Adams DJ, Mangs H, Goy C, Ladomery M, Sperling R, Stamm S, Morris BJ. WT1 interacts with the splicing protein RBM4 and regulates its ability to modulate alternative splicing in vivo. Exp. Cell Res. 2006;312:3379–3388. doi: 10.1016/j.yexcr.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 27.Yang Y-HJ, Markus AM, Mangs HA, Raitskin O, Sperling R, Morris BJ. ZRANB2 localizes to supraspliceosomes and influences the alternative splicing of multiple genes in the transcriptome. Mol. Biol. Rep. 2013;40:5381–5395. doi: 10.1007/s11033-013-2637-9. [DOI] [PubMed] [Google Scholar]
- 28.Azubel M, Habib N, Sperling J, Sperling R. Native spliceosomes assemble with pre-mRNA to form supraspliceosomes. J. Mol. Biol. 2006;356:955–966. doi: 10.1016/j.jmb.2005.11.078. [DOI] [PubMed] [Google Scholar]
- 29.Raitskin O, Cho DS, Sperling J, Nishikura K, Sperling R. RNA editing activity is associated with splicing factors in lnRNP particles: The nuclear pre-mRNA processing machinery. Proc. Natl Acad. Sci. USA. 2001;98:6571–6576. doi: 10.1073/pnas.111153798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sebbag-Sznajder N, Raitskin O, Angenitzki M, Sato TA, Sperling J, Sperling R. Regulation of alternative splicing within the supraspliceosome. J. Struct. Biol. 2012;177:152–159. doi: 10.1016/j.jsb.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen M, Bellaousov S, Hiller M, de La Grange P, Creame TP, Malina O, Sperling R, Mathews D, Stoilov P, Stamm S. Pyrvinium Pamoate changes alternative splicing of the serotonin receptor 2C by influencing its RNA structure. Nucleic Acids Res. 2013;41:3819–3832. doi: 10.1093/nar/gkt063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shomron N, Levy C. MicroRNA-biogenesis and Pre-mRNA splicing crosstalk. J. Biomed. Biotechnol. 2009 doi: 10.1155/2009/594678. 2009, 594678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pawlicki JM, Steitz JA. Nuclear networking fashions pre-messenger RNA and primary microRNA transcripts for function. Trends Cell Biol. 2010;20:52–61. doi: 10.1016/j.tcb.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Havens MA, Reich AA, Duelli DM, Hastings ML. Biogenesis of mammalian microRNAs by a non-canonical processing pathway. Nucleic Acids Res. 2012;40:4626–4640. doi: 10.1093/nar/gks026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smalheiser NR. EST analyses predict the existence of a population of chimeric microRNA precursor-mRNA transcripts expressed in normal human and mouse tissues. Genome Biol. 2003;4:403. doi: 10.1186/gb-2003-4-7-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pawlicki JM, Steitz JA. Primary microRNA transcript retention at sites of transcription leads to enhanced microRNA production. J. Cell Biol. 2008;182:61–76. doi: 10.1083/jcb.200803111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morlando M, Ballarino M, Gromak N, Pagano F, Bozzoni I, Proudfoot NJ. Primary microRNA transcripts are processed co-transcriptionally. Nat. Struct. Mol. Biol. 2008;15:902–909. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kataoka N, Fujita M, Ohno M. Functional association of the Microprocessor complex with the spliceosome. Mol. Cell. Biol. 2009;29:3243–3254. doi: 10.1128/MCB.00360-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ying SY, Lin SL. Current perspectives in intronic micro RNAs (miRNAs) J. Biomed. Sci. 2006;13:5–15. doi: 10.1007/s11373-005-9036-8. [DOI] [PubMed] [Google Scholar]
- 43.Filipowicz W, Pogacic V. Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol. 2002;14:319–327. doi: 10.1016/s0955-0674(02)00334-4. [DOI] [PubMed] [Google Scholar]
- 44.Janas MM, Khaled M, Schubert S, Bernstein JG, Golan D, Veguilla RA, Fisher DE, Shomron N, Levy C, Novina CD. Feed-forward microprocessing and splicing activities at a microRNA-containing intron. PLoS Genet. 2011;7:e1002330. doi: 10.1371/journal.pgen.1002330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shiohama A, Sasaki T, Noda S, Minoshima S, Shimizu N. Nucleolar localization of DGCR8 and identification of eleven DGCR8-associated proteins. Exp. Cell Res. 2007;313:4196–4207. doi: 10.1016/j.yexcr.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 46.Agranat L, Raitskin O, Sperling J, Sperling R. The editing enzyme ADAR1 and the mRNA surveillance protein hUpf1 interact in the cell nucleus. Proc. Natl Acad. Sci. USA. 2008;105:5028–5033. doi: 10.1073/pnas.0710576105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spann P, Feinerman M, Sperling J, Sperling R. Isolation and visualization of large compact ribonucleoprotein particles of specific nuclear RNAs. Proc. Natl Acad. Sci. USA. 1989;86:466–470. doi: 10.1073/pnas.86.2.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sperling R, Sperling J. In: RNP Particles, Splicing and Autoimmune Diseases. Schenkel J, editor. Heidelberg: Springer; 1998. pp. 29–47. [Google Scholar]
- 49.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:1–9. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 51.Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, Watanabe H, Kitahara T, Yoshida T, Nakajima H, et al. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat. Chem. Biol. 2007;3:576–583. doi: 10.1038/nchembio.2007.18. [DOI] [PubMed] [Google Scholar]
- 52.Sperling R, Spann P, Offen D, Sperling J. U1, U2 and U6 small nuclear ribonucleoprtoeins (snRNPs) are associated with large nuclear RNP particles containing transcripts of an amplified gene in vivo. Proc. Natl Acad. Sci. USA. 1986;83:6721–6725. doi: 10.1073/pnas.83.18.6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yitzhaki S, Miriami E, Sperling J, Sperling R. Phosphorylated Ser/Arg-rich proteins: Limiting factors in the assembly of 200S large nuclear ribonucleoprotein particles. Proc. Natl Acad. Sci. USA. 1996;93:8830–8835. doi: 10.1073/pnas.93.17.8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miriami E, Angenitzki M, Sperling R, Sperling J. Magnesium cations are required for the association of U small nuclear ribonucleoproteins and SR proteins with pre-mRNA in 200 S large nuclear ribonucleoprotein particles. J. Mol. Biol. 1995;246:254–263. doi: 10.1006/jmbi.1994.0081. [DOI] [PubMed] [Google Scholar]
- 55.Petrocca F, Vecchione A, Croce CM. Emerging role of miR-106b-25/miR-17-92 clusters in the control of transforming growth factor beta signaling. Cancer Res. 2008;68:8191–8194. doi: 10.1158/0008-5472.CAN-08-1768. [DOI] [PubMed] [Google Scholar]
- 56.Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Muhlemann O. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell Mol. Life Sci. 2010;67:677–700. doi: 10.1007/s00018-009-0177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- 58.Rajavel KS, Neufeld EF. Nonsense-mediated decay of human HEXA mRNA. Mol. Cell. Biol. 2001;21:5512–5519. doi: 10.1128/MCB.21.16.5512-5519.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roybal GA, Jurica MS. Spliceostatin A inhibits spliceosome assembly subsequent to prespliceosome formation. Nucleic Acids Res. 2010;38:6664–6672. doi: 10.1093/nar/gkq494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martins SB, Rino J, Carvalho T, Carvalho C, Yoshida M, Klose JM, de Almeida SF, Carmo-Fonseca M. Spliceosome assembly is coupled to RNA polymerase II dynamics at the 3' end of human genes. Nat. Struct. Mol. Biol. 2011;18:1115–1123. doi: 10.1038/nsmb.2124. [DOI] [PubMed] [Google Scholar]
- 61.Yan K, Liu P, Wu CA, Yang GD, Xu R, Guo QH, Huang JG, Zheng CC. Stress-induced alternative splicing provides a mechanism for the regulation of microRNA processing in Arabidopsis thaliana. Mol. Cell. 2012;48:521–531. doi: 10.1016/j.molcel.2012.08.032. [DOI] [PubMed] [Google Scholar]
- 62.Razumilava N, Bronk SF, Smoot RL, Fingas CD, Werneburg NW, Roberts LR, Mott JL. miR-25 targets TNF-related apoptosis inducing ligand (TRAIL) death receptor-4 and promotes apoptosis resistance in cholangiocarcinoma. Hepatology. 2012;55:465–475. doi: 10.1002/hep.24698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gupta S, Read DE, Deepti A, Cawley K, Gupta A, Oommen D, Verfaillie T, Matus S, Smith MA, Mott JL, et al. Perk-dependent repression of miR-106b-25 cluster is required for ER stress-induced apoptosis. Cell Death Dis. 2012;3:e333. doi: 10.1038/cddis.2012.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.