Abstract
The trypanosome zinc finger protein ZC3H11 binds to AU-rich elements in mRNAs. It is essential for survival of the mammalian-infective bloodstream form, where it stabilizes several mRNAs including some encoding chaperones, and is also required for stabilization of chaperone mRNAs during the heat-shock response in the vector-infective procyclic form. When ZC3H11 was artificially ‘tethered’ to a reporter mRNA in bloodstream forms it increased reporter expression. We here show that ZC3H11 interacts with trypanosome MKT1 and PBP1, and that domains required for both interactions are necessary for function in the bloodstream-form tethering assay. PBP1 interacts with MKT1, LSM12 and poly(A) binding protein, and localizes to granules during parasite starvation. All of these proteins are essential for bloodstream-form trypanosome survival and increase gene expression in the tethering assay. MKT1 is cytosolic and polysome associated. Using a yeast two-hybrid screen and tandem affinity purification we found that trypanosome MKT1 interacts with multiple RNA-binding proteins and other potential RNA regulators, placing it at the centre of a post-transcriptional regulatory network. A consensus interaction sequence, H(E/D/N/Q)PY, was identified. Recruitment of MKT1-containing regulatory complexes to mRNAs via sequence-specific mRNA-binding proteins could thus control several different post-transcriptional regulons.
INTRODUCTION
The degradation of mRNAs is a vital component of eukaryotic homeostasis. This is especially true in Kinetoplastid protists, such as trypanosomes and leishmanias, where protein-coding genes are arranged in polycistronic units. Kinetoplastids thus lack individual control of transcription initiation, and as a consequence rely almost exclusively on post-transcriptional mechanisms to control gene expression. As in other eukaryotes so far studied, degradation of normal (non-defective) mRNAs in trypanosomes is mainly initiated by removal of the poly(A) tail, followed by decapping and degradation by 5′-3′ exoribonucleases and/or the exosome (1–6). It is expected that, as in other eukaryotes, trypanosome polysomal mRNAs are initially shielded from the degradation machinery. At the 5′ end, the cap is bound by a complex of eIF4E, eIF4G and eIF4A (see, e.g. (7,8)), while the 3′ poly(A) tail is protected by poly(A) binding protein (PABP) (e.g. (9)). PABP can interact with the cap-binding complex (10); not only can this promote translation, but may also further stabilize mRNAs through adding cooperativity to protection of the 5′ end (11). In both yeast and trypanosomes, artificial ‘tethering’ of PABP to the 3′-untranslated region of an mRNA can also stabilize that mRNA (12,13).
In the African trypanosome Trypanosoma brucei, most mRNAs are relatively unstable (6). Sequences that regulate degradation have been found in 3′-UTRs, and several regulatory RNA-binding proteins have been characterized (14). Strikingly, all of the regulatory proteins identified so far in T. brucei have been shown to stabilize, rather than to destabilize, their target RNAs (e.g. (14,15)). Only a few examples of stabilization have been investigated in mammalian cells, and so far as we know, none in other eukaryotic species. For example, many mammalian mRNAs are rendered unstable by the presence of an AU-rich element in the 3′ untranslated region (3′-UTR). HuR family proteins stabilize these mRNAs (16,17) by competing with destabilizing proteins for binding to the AU-rich element (18,19). Similarly, the transferrin and alpha globin mRNAs are destroyed by endonuclease cleavage in the 3′-UTR, but this is inhibited by binding of the iron response protein (20) and alpha-CP (21,22). The globin mRNA also binds to nucleolin (23), AUF-1 and YB-1 (24,25).
Trypanosoma brucei has two life-cycle stages that are routinely cultured in the laboratory: the bloodstream form, which is the form that multiplies in mammals at 37°C, and the procyclic form, which multiplies in the midgut of the tsetse fly and is usually cultured at 27°C. Recently, we showed that in T. brucei, the zinc finger protein ZC3H11 specifically binds to an AU-rich element that is found in mRNAs encoding chaperones that are required for the heat-shock response (26). Procyclic forms were unaffected by ZC3H11 depletion at their normal growth temperature, but they were severely impaired at 37°C and exquisitely sensitive to a 41°C heat shock. This correlated with a failure to retain ZC3H11 target chaperone mRNAs. In contrast, depletion of ZC3H11 via RNAi rapidly killed bloodstream-form trypanosomes, which grow at 37°C. Using microarrays, northern blotting and a single RNASeq sample, we showed that in bloodstream forms, ZC3H11 depletion caused ∼2-fold decreases in the abundance of a few mRNAs—including that encoding HSP70. We also showed that the regulation of the HSP70 mRNA in bloodstream forms was mediated by an AU-rich element in the 3′-UTR. Artificial tethering of ZC3H11 to a chloramphenicol acetyltransferase (CAT) reporter in bloodstream forms increased the amounts of CAT activity and mRNA (26). This indicated that in bloodstream forms, growing at their normal temperature, ZC3H11 is not just protecting target mRNAs from a destabilizing factor, but instead has an active ability to stabilize a bound transcript.
The work described in this article, we started by investigating the mechanisms by which ZC3H11 can increase mRNA levels and protein expression in bloodstream forms. We present evidence that the activity of trypanosome ZC3H11 in the tethering assay requires interactions with trypanosome MKT1 and PBP1. MKT1 and PBP1are homologues of Saccharomyces cerevisiae Mkt1p and Pbp1p. S. cerevisiae MKT1 is not an essential gene (27,28). Mkt1p is required for maintenance of dsRNA killer virus above 30°C (29,30), possibly through a link to RNA degradation by the exosome (31). It is also involved in translation of the HO mRNA (27), which may be linked to its role in sporulation (32). The presence of wild-type Mkt1p promotes mitochondrial genome maintenance (33) and survival at high temperature (34), in low glucose (28), or after exposure to DNA-damaging agents (34) or high ethanol levels (35). All of these results suggest a role of Mkt1p in stress resistance, but the precise function of Mkt1p is unknown.
A two-hybrid screen with Mkt1p revealed only one interaction partner in yeast, Pbp1p (27). It too is not essential. Pbp1p can multimerize, is associated with polysomes (27), and interacts with Lsm12p (36,37) and PABP (38). Pbp1p and Lsm12p co-sediment with ribosomes (37); both were also found in stress granules and Pbp1p can promote granule formation (39). The interaction of Mkt1p with Pbp1p was shown to be necessary for positive regulation of HO mRNA and for association of Mkt1p with polysomes (27). Other evidence, however, suggests that Pbp1p’s main role is in regulating poly(A) tail length during polyadenylation, rather than in mRNA decay or translation (38,40). Polyglutamine expansions in the human homologue of Pbp1p, Ataxin 2, cause a neurodegenerative disease; like its yeast equivalent, Ataxin 2 interacts with PABP (36).
Beyond the evidence for the role of MKT1 in ZC3H11 action, we show below that MKT1 can interact with several other RNA-binding proteins, suggesting that MKT1 has a pivotal role in trypanosome mRNA metabolism.
MATERIALS AND METHODS
Cells and plasmids
Details of all plasmids and oligonucleotides are provided in Supplementary Table S1. All experiments were done with Lister 427 monomorphic procyclic or bloodstream-form parasites expressing the Tet-repressor (41). Procyclic forms were grown in MEM-Pros medium at 27°C (unless stated otherwise) at a densities lower than 8 × 106 cells/ml. The bloodstream stage parasites were cultivated in HMI-9 medium in an incubator at 37°C with 5% CO2 at densities lower than 1.5 × 106 cells/ml.
Stable cell lines were made with in situ V5-tagged ORFs (42) or tetracycline-inducible expression of various tagged proteins, or dsRNA (41,43,44). For the tethering assays, cell lines constitutively expressing CAT reporter with actin 3′-UTR or boxB actin 3′-UTR were co-transfected with plasmids for inducible expression of inducible λN-ZC3H11-myc fusion proteins (13,45). RNAi was done either with an MKT1 stem-loop plasmid or with either MKT1 or PBP1 plasmids with opposing T7 promoters. Transfection of normal amounts (10 µg/cuvette) of tetracycline-inducible MKT1 RNAi plasmid killed the parasites even in the absence of tetracycline; survival was possible only if the amount of DNA was decreased 5-fold. Cultivation of the RNAi cell lines resulted in a rapid decrease in RNAi effectiveness, and freezing had the same effect.
Protein detection and manipulation
Sub-cellular fractionation (46), dephosphorylation assays (47), tandem affinity purification (TAP) (43,48) and co-immunoprecipitation assays (49) were done as previously described. Phosphatase inhibitors used were Sodium Orthovanadate (2 mM) and Sodium Fluoride (8 mM). For co-immunoprecipitation with V5-ZC3H11, 4 × 107 procyclic trypanosomes or 5 × 107 bloodstream forms were pre-treated for 1 h with the proteasome inhibitor MG-132 (Calbiochem) at a concentration of 10 µg/ml. Cells were lysed in hypotonic buffer (10 mM NaCl, 10 mM Tris-Cl pH7.5, 0.1%NP40 with complete protease inhibitor (Roche), and precipitation was with anti-myc (Biomol) or anti-V5 (Biomol) -coupled beads after the salt was adjusted to 150 mM NaCl.
Proteins were detected by western blotting. Antibodies used were to the V5 tag (AbD seroTec, 1:1000), the Myc tag (Santa Cruz Laboratories, 1:1000), aldolase (rabbit, 1:50000 (50)) ribosomal protein S9 (rabbit antibody raised against recombinant protein prepared by C. Helbig, ZMBH) and trypanothione reductase (kind gift of Prof. L Krauth-Siegel, BZH, Heidelberg). Detection was done using ECL solutions (GE Healthcare). CAT was measured in a kinetic assay involving partition of 14C-buturyl chloramphenicol from the aqueous to the organic phase of scintillation fluid (51).
Targeted yeast two-hybrid analysis
The Matchmaker Yeast Two-Hybrid System (Clontech) was used for protein–protein interaction analysis following the manufacturer's protocol. The DNAs of the protein ORFs were PCR-amplified and cloned into both pGBKT7 and pGADT7. The bait has an N-terminally fused GAL4 DNA binding domain and myc tag, while the prey has N-terminally fused GAL4 activation domain and HA tag. The plasmids were co-transformed pairwise into AH109 yeast strains (Matchmaker 3 System, Clontech), and selected on quadruple drop-out medium (minimal SD media lacking Trp, Leu, His and Ade). Positive interactions were indicated by growth on QDO and also by a blue colour-change due to X-α-gal present in the medium. The interaction between murine p53 and SV40 large T-antigen served as positive control, with Lamin and SV40 large T-antigen as negative bait and prey controls. Expression of the proteins was checked by western blotting, using the myc and HA tags for detection.
High-throughput yeast two-hybrid screen
A library was made with genomic DNA since only two introns are known (52). Fifteen micrograms of T. brucei strain 427 DNA were sheared using a Covaris S2 acoustic sonicator. End repair of the resultant fragments was performed with 15 U of T4 DNA polymerase, 15 U of Klenow polymerase and dNTPs in FastDigest® buffer (Fermentas) for 15 min at 20°C. The repaired DNA was ligated to a 50-fold molar excess of synthetic semi-Xho adapters. The semi-Xho adapters were formed by annealing two synthetic oligonucleotides having the following sequences: CGAGCTACGTCAACG and 5′ phosphorylated CGTTGACGTAGC. After ligation, the semi-Xho-adapted DNA was size fractionated by agarose gel electrophoresis and DNA fragments in the size range from 0.7 to 2.5 Kbp were gel purified. The yeast expression vector pGADT7 (Clontech) was first linearized at the unique XhoI site, filled in with the large fragment of DNA polymerase I in the presence of dTTP, and ligated to the semi-Xho-adapted DNA (53). The ligation reaction was used to transform Escherichia coli NEB 5-alpha cells by electroporation. Of the 3 × 106 independent clones obtained, 1-2 × 105 should encode in-frame fusions. To assess the quality of the library, E. coli cells were re-transformed with purified pGADT7 library, plated and plasmids purified from individual colonies. Inserts were found in 97% of plasmids and the average size was ∼1.2 Kbp. Excluding pseudogenes, the T. brucei genome has ∼7000 ORFs, so on average each ORF should be represented by ∼10 fusions.
As bait for the library screen, the MKT1 ORF was cloned into the pGBKT7 plasmid. The pGADT7 T. brucei genomic library was screened by transformation of AH109 yeast expressing pGBKT7-MKT1. Transformants were selected on QDO medium after 5 days of incubation at 30°C. Approximately 60 individual colonies were picked and checked by plasmid retransformation. For the high-throughput screen, after selection on plates, yeast cells were harvested en masse and plasmid DNA was isolated by cell wall digestion with lyticase, followed by the NucleoSpin Plasmid alkaline lysis spin kit (Macherey Nagel). To sequence the putative interactors, PCR amplification was carried out with pGADT7 vector-specific primers (Supplementary Table S3). PCR reaction conditions were as follows: initial denaturation cycle at 95°C for 2 min, followed by 22 cycles of 95°C for 1 min, 57.5°C for 30 s, 72°C for 3 min and a final extension of 5 min at 72°C. Agarose gel electrophoresis of a small sample of the PCR products showed a DNA smear of expected size distribution. The PCR products were purified, sheared and prepared for Illumina sequencing using standard Illumina kits, then sequenced for 50 cycles on an Illumina HiSeq.
RNASeq and Bioinformatics
Poly(A)+ mRNA was sequenced using Illumina HiSeq with multiplexing and the data were analysed as described previously (5,26). For replicate independent samples of ‘wild-type’ bloodstream forms expressing the Tet-repressor (pHD1313), ∼24 million reads were mapped to the unique gene dataset; while 30 million reads were mapped for a single sample from cells 24 h after induction of XRNA RNAi.
The yeast two-hybrid data sequences were analysed using custom scripts designed to identify only mRNA sequences that were fused in-frame to the pGADT7 activation domain. Briefly, sequences containing a terminal insert-vector junction sequence were identified, the vector sequence was removed, then the remainder was mapped to the T. brucei 927 reference genome (http://tritrypdb.org/tritrypdb/, http://www.genedb.org/Homepage/Tbruceibrucei927) using Bowtie, allowing one base mis-match. SAMtools (54) and custom-made PERL scripts were then used to select only sequences that were in frame with an annotated ATG start codon. In-frame sequences in annotated 5′ untranslated regions (5′-UTR) were also included. This generated Supplementary Table S4.
Tandem affinity purification
Approximately 1.5 × 1010 trypanosomes expressing TAP-tagged protein were harvested and used for TAP as previously described (43). Bloodstream forms were used for MKT1, and procyclic forms for ZC3H11. The eluate was run 2 cm into a 10% SDS-polyacrylamide gel electrophoresis (PAGE) resolving gel, stained with colloidal Coomassie and then, the protein-containing gel area was analysed by mass spectrometry.
Polysomes
5 × 108 bloodstream-form cells (1 × 106 cells/ml) were taken into centrifuge flasks and cycloheximide was added to a concentration of 25 µg/ml. The cells were incubated for 1 min at room temperature and then chilled in a dry ice ethanol bath. The cells were then spun down (4°C, 2300g, 20 min), transferred to Eppendorf tubes and spun down again at 2300 g for 2 min. The pellet was washed twice in polysome buffer (20 mM Tris pH:7.4–7.5, 20 mM MgCl2, 600 mM KCl) containing 200 mM sucrose. The pellet was then forced five times through a 27 gauge needle in ice cold lysis buffer containing 10 mM Tris-HCl, pH 7.6, 300 mM KCl, 10 mM MgCl2 and protease inhibitors (Protease Inhibitor Mixture [EDTA free]; Roche Applied Science), 0.4 mg/ml Heparin, 1 mM DTT and 10 μg/ml leupeptin. IGEPAL CA-630 (Sigma) was added to a final concentration of 0.1% and the cells were again forced through the needle five times. For the RNase A control, RNase A (Sigma) was directly added in the cell lysate to a final concentration of 1 mg/ml and the mixture incubated for 20 min at room temperature. The lysates were layered onto 15–50% sucrose gradients prepared in polysome buffer and centrifuged at 4°C for 2 h at 40 000 rpm in a Beckman SW60i rotor. Fractions were eluted from the top of the gradient using a Teledyne Isco (Lincoln, NE) gradient elution system; polysome profiles were obtained by measuring absorbance at 254 nm.
Immunofluorescence
For the intracellular detection of Myc-tagged MKT1 and PBP1, 106 procyclic trypanosomes were sedimented, re-suspended in PBS, fixed in 4% paraformaldehyde (w/v) in 1 × PBS for 18 min, sedimented again for 2 min, re-suspended in PBS and allowed to settle down on a poly-lysine-coated glass slides overnight. The cells were then permeabilized with pre-chilled methanol (–20°C) for 10 min. Before staining, slides were blocked with 0.5% (w/v) gelatin in PBS for 20 min. Slides were then incubated with 1:500 diluted anti c-Myc primary antibody (Santa Cruz Biotechnology, Germany) and 1:1000 SCD6 antibody (55) (kind gift from Susanne Krämer and Mark Carrington, University of Cambridge) for 1 h and with a 1:1000 dilution of the second antibody Alexa Fluor 488 goat anti-mouse IgG and CY5 goat anti-rabbit for 40 min (Molecular probes). Kinetoplast and the nuclear DNA were then stained with 100 ng/ml DAPI/1 x PBS for 10 min.
For heat-shock experiments, the procyclic cells were incubated in a water bath at 41°C for 1 h before fixation. For starvation, cells were sedimented, washed twice in 1 × PBS and then re-suspended in 1 x PBS for 3 h at 27°C before fixation.
Z-stacks of the cells were taken and the images were deconvoluted (Wiener algorithm) using the Olympus Cell-R microscope.
RESULTS
The effects of ZC3H11 RNAi in bloodstream forms
We had previously investigated the effect of ZC3H11 on bloodstream-form transcriptomes using microarrays, supported by a single biological replicate analysed by RNASeq with previously published wild-type data as a control. To enable more quantitative analysis, we generated two new RNASeq datasets for wild-type (repressor expressing) bloodstream forms, and an additional set for cells after 24 h of ZC3H11 RNAi (Supplementary Table S2 and Supplementary Figure S1). Decreases in ZC3H11 target chaperone mRNAs were confirmed. There were, however, clear differences from the results in heat-shocked procyclic forms. Of the 12 chaperone mRNAs that bind to ZC3H11 in procyclic forms, all of which depend on ZC3H11 after heat shock, 6 showed 1.5–2.5-fold decreases after ZC3H11 RNAi in the bloodstream-form RNASeq datasets (Supplementary Table S2, sheet 2), but the other 6 were unaffected. Decreases in Tb927.10.16100 (FK506-binding protein (FKBP)-type peptidyl-prolyl isomerase), Tb927.2.5160 (DNAj-like), Tb927.11.11330 (HSP70) and Tb927.10.12710 (HSP110) were the most robust. Superimposed on this were decreases in other mRNAs, encoding (for example) glycolytic enzymes, cytoskeletal proteins and translation factors. These latter changes may reflect the onset of growth inhibition. A few mRNAs that can bind to ZC3H11 reproducibly increased: GPEET procyclin mRNA and mRNAs encoding hypothetical proteins (Tb927.11.2160, Tb927.10.780 and Tb927.9.15080). Other mRNA increases may be a normalization artifact since the mRNAs that decrease upon ZC3H11 RNAi are usually rather abundant and account for a large number of reads in the wild-type dataset.
ZC3H11 co-purifies MKT1, PBP1 and LSM12
To analyse the mechanism by which ZC3H11 stabilizes bound RNAs, we looked for interaction partners by TAP of ZC3H11. These experiments were done with procyclic forms expressing ZC3H11 with a TAP tag. 205 proteins co-purified with TAP-ZC3H11 (Table 1 and Supplementary Table S3). The most interesting ones with high coverage were homologues of yeast Mkt1p, Pbp1p and Lsm12p. Mkt1 homologues from fungi, trypanosomes, an amoeba and Apicomplexa have a PIN-like domain at the N-terminus, and limited matches further towards the C-terminus (Supplementary Figure S2). The PIN domains resemble those of human Fen1 and XPD endonucleases (27), but Mkt1s lack acidic residues that are essential for endonuclease activity (Supplementary Figure S2) (56). The laboratory yeast strain S288C has a G->D mutation that results in reduced fitness after stress; all other homologues examined, including that from trypanosomes, retained a D and also a neighbouring acidic residue which is essential for both Fen1 endonuclease (56) activity and Mkt1p effects on HO translation (27) (Supplementary Figure S2).
Table 1.
Selected proteins associated with ZC3H11
| GeneDB no. | Function | MWt kDa | Unique peptides | Percentage of coverage (%) |
|---|---|---|---|---|
| Verified complex | ||||
| Tb927.5.810 | ZC3H11 | 40 | 19 | 61 |
| Tb927.6.4770 | MKT1 | 83 | 33 | 48 |
| Tb927.9.9060 | LSM12 | 21 | 10 | 47 |
| Tb927.8.4540 | PBP1 | 60 | 9 | 20 |
| Unknown function | ||||
| Tb927.4.1300 | unknown | 42 | 6 | 24 |
| Tb927.7.2780 | unknown | 13 | 2 | 21 |
| Tb927.1.3070 | unknown | 45 | 5 | 15 |
| Tb927.9.11840 | unknown | 22 | 2 | 13 |
| RNA related | ||||
| Tb927.6.4440 | RBP42 | 38 | 8 | 36 |
| Tb927.8.6440 | RBP20 | 20 | 4 | 20 |
| Tb927.9.8740 | DRBD3 (PTB1) | 37 | 5 | 17 |
| Tb927.10.3990 | DHH1 | 46 | 5 | 16 |
| Tb927.11.510 | UBP2 | 20 | 2 | 15 |
| Tb927.11.1980 | ZC3H41 | 58 | 4 | 10 |
The listed proteins had at least 10% representation in the ZC3H11-TAP purification. For the complete list see Supplementary Table S3.
Pbp1 proteins and Ataxin-2 have an LSm AD domain towards the N-terminus. The C-terminus is less well conserved (Supplementary Figure S3): for yeast and trypanosomes, 17% identity was found, usually with matching of isolated residues. Lsm12 was the most conserved and ubiquitous protein, with cross-kingdom sequence identities ranging between 21% and 32% (Supplementary Figure S4).
The ZC3H11 affinity purification contained a large number of ribosomal proteins, abundant translation factors, various helicases and RNA-binding proteins and the poly(A) binding protein PABP2 (Table 1 and Supplementary Table S3). Some of these interactions are probably specific, but many of the proteins concerned have been found in numerous other purifications (57). Although the purification did not contain RNase inhibitors, some RNA-mediated associations could have persisted. DHH1, the trypanosome homologue of Dhh1p, was present; yeast Dhh1p was identified as a partner of Pbp1p (36).
MKT1 is associated with ZC3H11 and PBP1 in trypanosomes
We verified the interaction of MKT1 with ZC3H11 and PBP1 by co-immunoprecipitation using tagged proteins. DNA encoding the V5 tag was inserted into the genome by homologous recombination, in frame with the open reading frames of interest. This should give expression levels that are similar to those of the endogenous protein unless protein degradation is affected (42). C-terminally myc-tagged proteins were expressed from a tetracycline-inducible RNA polymerase I promoter, which is likely to result in over-expression.
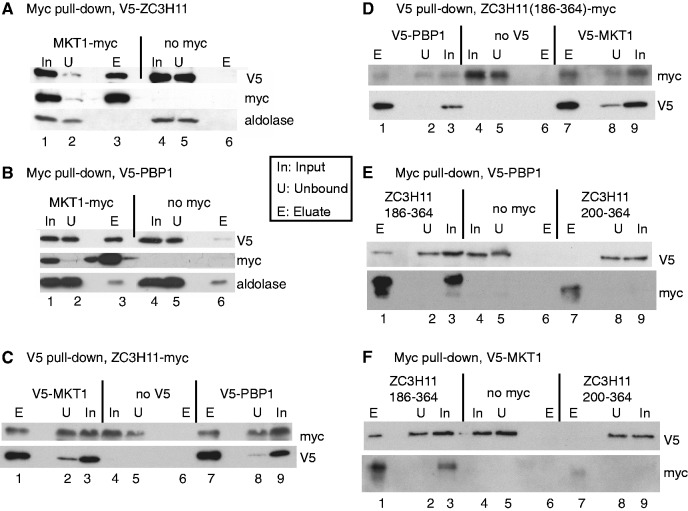
All experiments involving V5-ZC3H11 were done after a 1-h treatment with MG132 to allow detection; this may act by inhibiting proteolysis and/or by inducing a stress response. Using extracts from procyclic-form trypanosomes co-expressing MKT1-myc and V5-ZC3H11, V5-ZC3H11 was quantitatively precipitated by anti-myc antibody (Figure 1A). No precipitation of V5-ZC3H11 was seen for cells lacking MKT1-myc. (Figure 1A, lane 6). ZC3H11 is phosphorylated (26); since phosphatase inhibitors were in the lysis buffer, the results indicate that phosphorylated ZC3H11 binds to MKT1. Since we were principally interested in the function of ZC3H11 in bloodstream-form trypanosomes, the subsequent co-immunoprecipitations were done in that form. With bloodstream-forms co-expressing MKT1-myc and V5-PBP1, V5-PBP1 was seen after precipitation by an anti-myc antibody (Figure 1B, lane 3) and very little V5-PBP1 was seen in the control without MKT1-myc (Figure 1B, lane 6). Similarly, ZC3H11-myc was pulled down by either V5-MKT1 (Figure 1C, lane 1 and Supplementary Figure S5A) or V5-PBP1 (Figure 1C, lane 7 and Supplementary Figure S5A), showing that the interaction between MKT1 and ZC3H11 was present in the bloodstream form. The interactions were unaffected by addition of RNase (Supplementary Figure S5A). In procyclic forms, a small degree of homodimerization of ZC3H11 was also observed (Supplementary Figure S5B and C); this was unaffected by addition of RNase but could have been via other proteins.
Figure 1.
Interactions of ZC3H11, MKT1 and PBP1 in trypanosomes. (A) ZC3H11 interacts with MKT1. Extracts from cells expressing V5-ZC3H11, with (lanes 1–3) or without (lanes 4–6) additional expression of MKT1-myc, were subjected to immunoprecipitation with anti-myc. The precipitated proteins were analysed by western blotting, using anti-myc, anti-V5 and, as control, anti-aldolase. In: input, U: unbound (2 × 106 cell-equivalents), E: eluate (3.8 × 107 cell-equivalents). In all cases cells were pre-incubated with MG132 for an hour prior to lysis to facilitate detection of the ZC3H11. This panel is for procyclic forms but all others are for bloodstream forms. (B) MKT1 interacts with PBP1. As in (A), but here the cells express V5-PBP1 with or without MKT1-myc. (C). ZC3H11 interacts with PBP1 and MKT1. Cells expressed ZC3H11-myc without (lanes 4–6) or with either V5-PBP1 (lanes 7–9) or V5-MKT1 (lanes 1–3). The pull-down was with anti-V5. (D) The C-terminal half of ZC3H11, ZC3H11(186-364), is pulled down by immunoprecipitation of PBP1 and MKT1. Cells expressed ZC3H11(186-364)-myc. Other details are as in (C). (E) PBP1 is pulled down by immunoprecipitation of the C-terminal half of ZC3H11, ZC3H11(186-364). Cells expressed V5-PBP1 either without myc-tagged protein (lanes 4–6), or with ZC3H11(186-364)-myc (lanes 1–3) or ZC3H11(200-364)-myc (lanes 7–9). Pull-down was with anti-myc. ZC3H11(200-364)-myc was too poorly expressed to draw conclusions. (F) MKT1 is pulled down by immunoprecipitation of the C-terminal half of ZC3H11, ZC3H11(186-364). Experiment is as for (F), except that the V5-tagged protein was MKT1.
The levels of in situ V5-PBP1 and V5-MKT1 in bloodstream-form trypanosome lysates appeared similar (Figure 1C, lanes 3 and 9), and both were very easily detected by western blotting, whereas V5-tagged ZC3H11 is not detectable at all under normal conditions (26). MKT1 and PBP1 also showed high peptide coverage after mass spectrometry of total cell lysates, whereas ZC3H11 was not detected at all (58). This suggests that both MKT1 and PBP1 are present in considerable excess relative to ZC3H11. Indeed, even in procyclic forms grown at 37°C, V5 in situ-tagged MKT1 was way more abundant than equivalently tagged ZC3H11 (Supplementary Figure S5D). Even when ZC3H11-myc was inducibly expressed using a polymerase I promoter, about half of the protein could be pulled down by endogenously tagged V5-PBP1 or V5-MKT1 (Figure 1C). This suggests that in normal cells, most ZC3H11 is likely to be associated with MKT1 and PBP1. Over-expression of the tagged PBP1 or MKT1 in bloodstream forms sometimes caused mild growth inhibition (not shown) as previously reported for Pbp1p in yeast (39).
ZC3H11, MKT1 and PBP1 interact in yeast
To further investigate the interactions between ZC3H11, MKT1 and PBP1, and also with LSM12, we used the yeast two-hybrid system (Table 2, Figure 2 and Supplementary Figure S6). PBP1 fused to the DNA-binding domain (‘bait’ configuration) was able to activate gene expression when expressed alone. We therefore could not test whether PBP1 interacted with itself, as in yeast (59). As prey, PBP1 interacted with ZC3H11 and MKT1 (Figure 2), LSM12 and both poly(A) binding proteins (PABP1 and PABP2) (Supplementary Figure S6A). ZC3H11 and MKT1 interacted with each other in both configurations; and ZC3H11 interacted with itself (Figure 2); MKT1 did not interact with the PABPs (Supplementary Figure S6B). We were worried that some of the interactions that we had seen could have been caused by the trypanosome proteins interacting with yeast Mkt1p, so we tested this directly. Importantly, none of the trypanosome proteins showed any interaction with S. cerevisiae Mkt1p expressed as bait (Supplementary Figure S6B), and ZC3H11, MKT1, PBP1 and LSM12 all did not interact with yeast Pbp1p (tested in both directions, not shown). These controls ruled out interactions via yeast Mkt1p and Pbp1p, but as always with the yeast two-hybrid system, interactions via other unknown yeast proteins cannot be ruled out. Overall the results so far indicated that the complex of MKT1, PBP1 and LSM12 is conserved from yeast to trypanosomes.
Table 2.
Summary of two-hybrid interactions
| MKT1 bait | MKT1 prey | PBP1 prey | ||
|---|---|---|---|---|
| Prey | ZC3H11 | + | ||
| Prey | ZC3H11 C->S | + | ||
| Prey | ZC3H11 PY->AA | – | ||
| Prey | ZC3H11 1–128 | – | ||
| Prey | ZC3H11 101–364 | + | ||
| Prey | ZC3H11 101–301 | + | ||
| Prey | ZC3H11 101–265 | + | ||
| Prey | ZC3H11 101–202 | + | ||
| Prey | ZC3H11 101–195 | – | ||
| Bait | ZC3H11 | + | + | |
| Bait | ZC3H11 186–364 | + | + | |
| Bait | ZC3H11 200–364 | – | + | |
| Bait | LSM12 | – | + | |
| Bait | MKT1 | – | + | |
| Bait | MKT1 1–494 | – | ||
| Bait | MKT1 232–735 | – | ||
| Bait | PABP1 | – | + | |
| Bait | PABP2 | – | + | |
| Bait | CFB1 | – | ||
| Prey | CFB1 | + | ||
| Prey | CFB1 500–527 | + | ||
| Prey | CFB1 PRRFRHDPY | + | ||
| Prey | EF2a | + | ||
| Prey | eIF-5Aa | + | ||
| Prey | ERF1a | + | ||
| Prey | EF1alpha | – | ||
| Prey | ERF3 | – | ||
| Prey | eIF-2B | – |
Data are provided in Supplementary Figure S5.
aThese interactions were not found with above-threshold reads in the high-throughput screen although plasmids expressing the protein were present (Supplementary Table S2).
Figure 2.
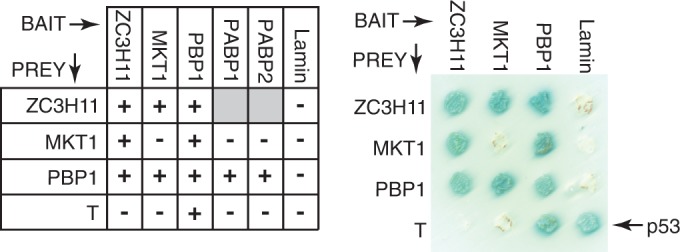
Two-hybrid interactions of ZC3H11, PBP1 and MKT1. Table: Interactions marked + were positive both in quadruple drop-out plates and via alpha-galactosidase assay. Grey boxes: not done. The plate on the right illustrates some of the interactions.
We next investigated which parts of ZC3H11 were capable of interacting with MKT1. ZC3H11 has a predicted molecular weight of 40 KDa but the recombinant protein migrates at ∼50 kDa; the phosphorylated version migrates at ∼60 KDa. The activation and DNA-binding domains are ∼20 kDa, and in yeast, the full-length ZC3H11 fusion proteins migrated ∼70 KDa, suggesting little or no phosphorylation. Analysis of deletion mutants (Table 2 and Supplementary Figure S6C) implicated the region between residues 186 and 202 in the MKT1 interaction. The sequence H(N/D)PY, at residues 196–199, is conserved in all Kinetoplastid ZC3H11s (26), and ZC3H11 with a PY->AA mutation failed to interact with MKT1 (Table 2 and Supplementary Figure S6C). The C-terminal 164 residues of ZC3H11 (200–364), lacking HNPY, were in contrast sufficient to interact with PBP1 (Table 2 and Supplementary Figure S6C).
Deletion analyses of Mkt1p indicated that a portion lacking the C-terminal 230 residues was unable to interact with Pbp1p; the smallest fragment giving a positive interaction contained the C-terminal 247 amino acids (475–722) (27). In contrast, neither an N-terminal (1–494) nor a C-terminal (233–735) fragment of trypanosome MKT1 was active in the yeast-two-hybrid assay with either PBP1 or ZC3H11 (Table 2 and Supplementary Figure S6D).
To verify the two-hybrid results in bloodstream-form trypanosomes, we repeated the co-immunoprecipitations using C-terminally myc-tagged versions of ZC3H11. As expected, the 186-364 fragment of ZC3H11 co-immunoprecipitated both V5-PBP1 (Figure 1D, lane 3) and V5-MKT1 (Figure 1D, lane 7) and the reciprocal pull-downs were also positive (Figure 1E, lane 1 and Figure 1F, lane 1). Both myc-tagged C-terminal ZC3H11 fragments were rather poorly expressed (Figure 1E and F); for the 202–364 fragment expression was too poor to assess interactions.
We concluded that the central portion of ZC3H11 containing the HNPY motif is necessary and sufficient for binding of MKT1, while PBP1 interacts further towards the ZC3H11 C-terminus.
MKT1 and LSM12 can actively stabilize a bound mRNA
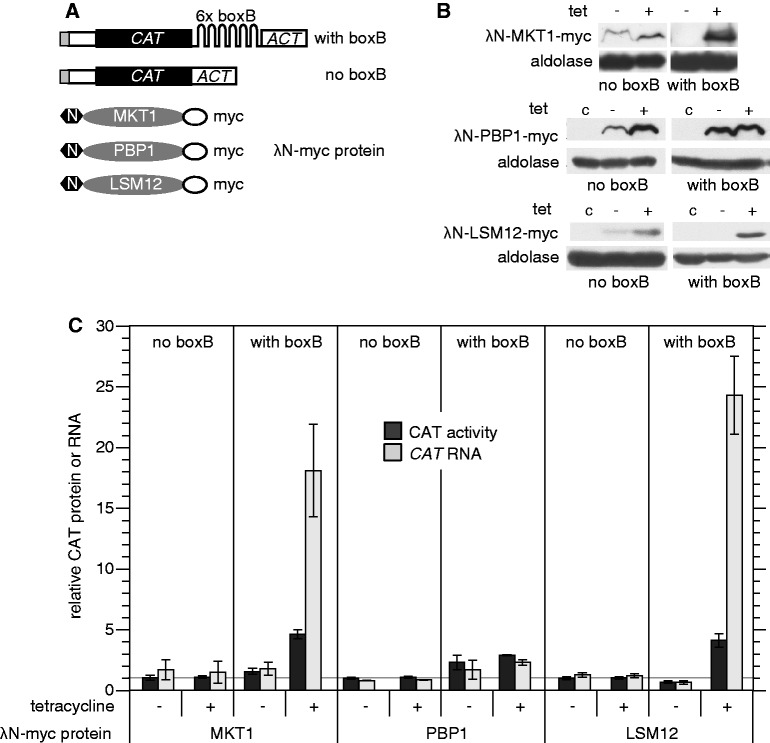
We previously found that in bloodstream forms, tethering of an λN-ZC3H11 to a CAT reporter mRNA with the actin 3′-UTR and five copies of boxB (Figure 3A) resulted in a 4-fold increase in reporter RNA, and a 2-fold increase in CAT protein (26). Tethering of λN-GFP, in contrast, caused a moderate decrease in expression (45). If ZC3H11 stabilizes mRNAs via the interactions with MKT1 and/or PBP1 and LSM12, then these proteins should also be active in the tethering assay. To explore this, we expressed MKT1, PBP1 and LSM12 as λN fusions in the reporter lines (Figure 3B). Tethering of each caused significant increases in CAT mRNA and CAT activity (Figure 3C). The increases in RNA for MKT1 and LSM12 were particularly large. Expression of CAT protein the protein increased less, but this could simply be because we induced expression for only 1 day and changes in the expression of stable proteins changes lags behind changes in mRNAs. PBP1 was less active in the assay than LSM12 and MKT1.
Figure 3.
MKT1, PBP1 and LSM12 can all increase mRNA abundance. (A) Diagram (not to scale) of the CAT reporter mRNA, with or without 6 ‘boxB’ recognition sites in the 3′-UTRs, and fusion proteins with an N-terminal λ-N peptide and a C-terminal myc tag. (B) Expression of MKT1, PBP1 and LSM12 fusion proteins in both cell lines, with and without 1 day of tetracycline induction. Western blot with anti-myc, 5 × 106 cells per lane. C: control cells with no myc tag. Aldolase serves as a loading control. (C) Expression of CAT activity, and CAT RNA, in cells with or without inducible expression of λ-N-myc fusion proteins. RNA and protein extracts were prepared from cells with or without tetracycline treatment to induce fusion protein expression (100 ng/ml, 24 h). RNA was measured by northern blotting and CAT enzyme activity was assayed. Results are arithmetic mean ± standard deviation of at least three measurements. The increase for PBP1 without tetracycline addition is probably due to leaky expression (see (B)).
We also tested which parts of MKT1 were required for activation in the tethering assay. The N-terminus (positions 1–494) and C-terminus (233–735, lacking the PIN domain) were both inactive (Table 3). This might be because neither fragment is able to interact with PBP1, or for other reasons. Mutation of selected residues conserved in kinetoplastid MKT1s had no effect on tethering activity (Table 3).
Table 3.
Activities of ZC3H11 and MKT1 fragments in the tethering assay
| Box B | Tet | CAT activity | CAT RNA | |
|---|---|---|---|---|
| ZC3H11 101–195 | + | – | 0.9 ± 0.1 | 0.8 ± 0.2 |
| + | + | 1.0 ± 0.1 | 0.8 ± 0.2 | |
| ZC3H11 101–202 | + | – | 1.0 ± 0.1 | 1.1 ± 0.3 |
| + | + | 1.0 ± 0.1 | 1.2 ± 0.2 | |
| ZC3H11 200–364 | + | – | 0.9 ± 0.1 | 1.6 ± 0.3 |
| + | + | 1.0 ± 0.0 | 1.1 ± 0.6 | |
| ZC3H11 186–364 | + | – | 1.0 ± 0.1 | 0.7 ± 0.2 |
| + | + | a3.5 ± 0.6 | a5.0 ± 0.6 | |
| MKT1 233–735 | + | – | 1.0 ± 0.1 | 1.3 ± 0.1 |
| + | + | 1.1 ± 0.1 | 1.9 ± 0.2 | |
| – | – | 1.0 ± 0.1 | 1.0 ± 0.1 | |
| – | + | 1.0 ± 0.1 | 1.1 ± 0.1 | |
| MKT1 1–494 | + | – | 0.6 ± 0.1 | 1.0 ± 0.2 |
| + | + | 0.5 ± 0.1 | 1.0 ± 0.2 | |
| – | – | 0.9 ± 0.1 | 1.1 ± 0.1 | |
| – | + | 0.9 ± 0.1 | 1.2 ± 0.1 | |
| MKT1 SRV571->AAA | + | – | 1.0 ± 0.2 | 1.0 ± 0.3 |
| + | + | a3.9 ± 0.6 | a3.8 ± 0.8 | |
| – | – | 1.0 ± 0.1 | 1.0 ± 0.2 | |
| – | + | 1.0 ± 0.0 | 1.1 ± 0.3 | |
| MKT1 D590A | + | – | 1.0 ± 0.0 | 0.9 ± 0.1 |
| + | + | a4.0 ± 0.6 | a4.4 ± 0.5 | |
| – | – | 1.0 ± 0.0 | 1.2 ± 0.2 | |
| – | + | 1.0 ± 0.1 | 1.2 ± 0.2 |
Results are arithmetic mean and standard deviation for at least three independent experiments. aIncreased CAT protein and RNA.
These results confirm that MKT1 and PBP1 can increase mRNA and protein in the tethering assay, although the activity of PBP1 was weak.
ZC3H11 activity in the tethering assay requires the regions that interact with MKT1 and PBP1
We now investigated whether the activity of ZC3H11 in the tethering assay required the domains that interacted with MKT1 and PBP1 (Table 3, Supplementary Figure S7 and Figure 4). The minimal fragment that was active in the tethering assay was ZC3H11(186–364). A further N-terminal deletion (200–364) abolished the tethered activity, just as it had prevented the interaction with MKT1 in the yeast two-hybrid assay. Interaction with MKT1 was, however, not sufficient to activate CAT expression, since ZC3H11(101-202), which lacks the PBP1-interacting region, was inactive. This suggested that the activity of ZC3H11 in the tethering assay requires interaction with both MKT1 and PBP1. Two ZC3H11 proteins with mutations in the central HNPY sequence (AAPY or HNAA) were unfortunately extremely unstable in trypanosomes so their activities in the tethering assay could not be measured.
Figure 4.
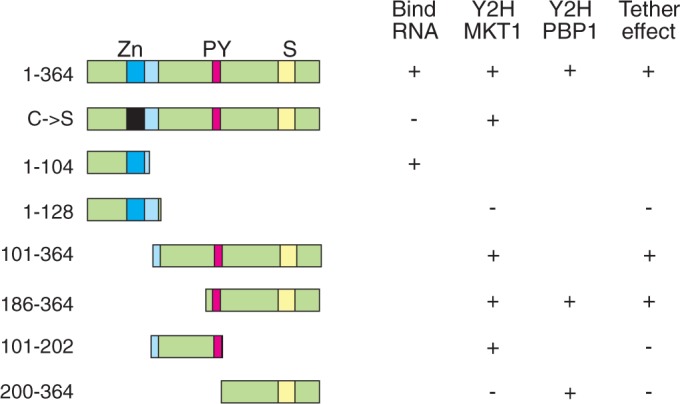
ZC3H11 activity requires the interactions with MKT1 and PBP1. The diagrams on the left illustrate selected mutant versions of ZC3H11. Zn: zinc finger; PY: HNPY motif; S: serine-rich domain. Properties of the mutants are shown in the columns. Only selected, critical, mutants are shown. Bind RNA: ability to bind (UAU) repeats (26). Y2H MKT1: binding to MKT1 in the yeast two-hybrid assay. Y2H PBP1: binding to PBP1 in the yeast two-hybrid assay; Tether effect: ability to increase CAT activity and CAT mRNA in the tethering assay. + , positive result; –, negative result; gap, not done.
Tethering of trypanosome PABP1 caused large increases in CAT RNA and CAT activity (13,45), as has also been seen in other systems (12,60). Our results so far supported the idea that the stabilizing activity of tethered ZC3H11 in bloodstream forms might be due to recruitment of MKT1 and PBP1, which in turn recruits PABP.
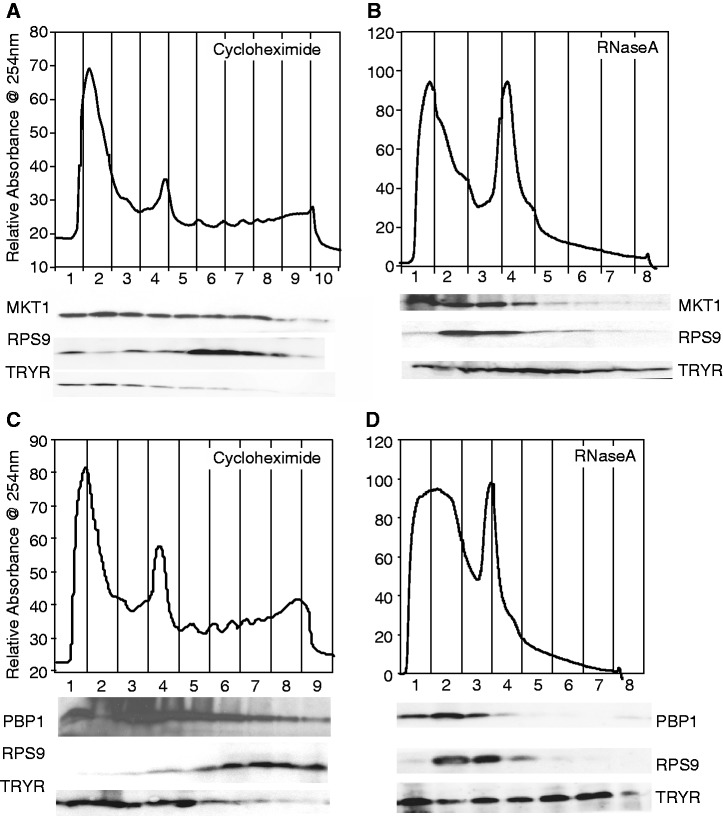
MKT1 is essential in bloodstream-form trypanosomes and is associated with polysomes
MKT1, LSM12 and PBP1 were previously shown to be required for normal growth of bloodstream- and procyclic-form trypanosomes in a high-throughput screen (61). Indeed, RNAi targeting both MKT1 and PBP1 caused death of bloodstream trypanosomes within 24 h. This precluded using RNAi to further investigate the role of either in the ZC3H11 tethering assay in bloodstream forms. In procyclic trypanosomes, depletion of either MKT1 or PBP1 decreased the growth rate (Supplementary Figure S8A). Since some protein always remains after the RNAi, we cannot yet tell whether MKT1 and PBP1 are essential in procyclics at 27°C or not, but the growth inhibition in this form suggests that both proteins have broader roles than ZC3H11.
As reported for yeast Mkt1p, trypanosome MKT1 was partially associated with polysomes (Figure 5A and B). We therefore speculated that the rapid lethality of MKT1 depletion might be caused by defects in translation. Indeed, trypanosomes with MKT1 RNAi showed decreased incorporation of [35S]-methionine into protein (Supplementary Figure S8B). However, cell morphology alterations, growth cessation and translation inhibition occurred roughly simultaneously. We were therefore unable to work out whether translation inhibition caused, or was a consequence of, growth inhibition. We looked for interactions of MKT1 with various translation factors. We found positive yeast two-hybrid interactions with EF2, eIF5A and ERF1, and no interaction with EF1-alpha, ERF3 or eIF-2B (Supplementary Figure S6E).
Figure 5.
Tagged MKT1 and PBP1 partially co-migrate with polysomes on sucrose gradients. Extracts from ∼5 × 108 bloodstream-form trypanosomes were separated on sucrose gradients. Proteins were extracted and analysed by western blotting using antibodies to myc, RPS9 (small ribosomal subunit protein S9) and trypanothione reductase (TRYR). TRYR is a soluble cytosolic protein but seems to run throughout the gradients. (A and B): Cells expressing MKT1-myc; (C and D): Cells expressing PBP1-myc. (A and C): Cycloheximide (100 µg/ml) was added immediately prior to parasite lysis; (B and D): RNase A (1 g/ml) added immediately after lysis.
Upon heat shock of procyclics, ZC3H11 levels are increased, and this is required for the retention of chaperone mRNAs. We had so far shown that the ZC3H11 sequences that are required for interaction with MKT1 are also required for the activity of ZC3H11 in the tethering assay in bloodstream forms. We therefore decided to investigate whether MKT1 is also required for the procyclic-form heat-shock response. In procyclics expressing V5-MKT1, the level of V5-MKT1 was decreased to ∼3% of normal 2 days after RNAi induction (Supplementary Figure S9). To assay the heat-shock response, cells were incubated at 41°C for an hour, then protein synthesis was measured. In heat-shocked cells without RNAi, as expected, most protein synthesis was inhibited but two bands that are probably HSP70 and HSP83 proteins were spared. MKT1 RNAi had no effect on this pattern (Supplementary Figure S9), and also did not affect cell survival at either 37°C or 41°C (not shown). This suggests either that MKT1 is not required for the procyclic-form heat-shock response. Unfortunately, given the vast excess of MKT1 relative to ZC3H11 (Supplementary Figure S5D) it is also possible that MKT1 is required for the heat-shock response, but the amount that remains after RNAi is sufficient.
MKT1 can interact with many proteins with RNA-binding domains
We performed a two-hybrid screen to find further interactions of MKT1. Randomly sheared trypanosome genomic DNA was used to make a library of ‘prey’ trypanosome proteins bearing the activation domain. About 3 × 106 independent clones were obtained with insert sizes of 0.6–2.3 kbp; 1-2 × 105 of these should encode in-frame fusions. The T. brucei genome has ∼7000 different ORFs, so on average each ORF should be represented by ∼10 fusions.
The library was transfected into yeast expressing MKT1 fused to the DNA-binding domain. From 60 independent colonies, 55 interactions were verified; these represented 21 different protein sequences, plus 2 outside annotated open reading frames. Later, we took a larger-scale approach, as in (62,63). Seven hundred colonies which grew on highly stringent plates were pooled, plasmid DNA was isolated and the trypanosome DNA inserts were PCR-amplified and sequenced. The inserts in the input yeast library (before high-stringency selection) served as controls: 9302 annotated open reading frames were represented (Supplementary Table S4, sheet 4). Selection for MKT1 interaction reduced this complexity (Supplementary Table S4, sheet 1), with 177 ORFs showing at least 20 sequence reads (Supplementary Table S4, sheets 2 and 3). Reassuringly, 5 different (>20 read) locations were found for ZC3H11, and 8 for PBP1 (Supplementary Table S4, sheet 1) and 20 of the 21 manually identified protein sequences were also found. The exception was a fragment of the SMC1 gene (Tb927.9.11850), for which only a single sequence had been found with 15 reads. The translation factors EF1-alpha, ERF3 and eIF-2B were not found. One possible explanation is that full-length proteins were required for these interactions, and the complete ORFs were not present in the two-hybrid library.
The potential MKT1 partners in Supplementary Table S4 represent 2% of all predicted proteins. RNA-binding proteins were highly enriched in the partner list (P = 0.0006); cytoskeletal components (P = 0.03), RNA helicases (P = 0.06) and RNA degradation (P = 0.07) were also over-represented. Of the 16 predicted RNA-binding proteins, 12 were represented by more than one cloned fragment (Table 4). The 16 include 3 of the 10 trypanosome pumilio domain proteins, and 10 of the ∼40 zinc finger proteins. Other RNA-related two-hybrid interactors included three RNA helicases, the deadenylase complex component NOT2, the exosome subunit EAP2, the possible translation factor eIF4G1, tRNA modification enzymes and two nucleoporins. The results suggest that MKT1 mediates the action not just of ZC3H11, but also that of RNA-binding proteins with other specificities.
Table 4.
Selected interaction partners of MKT1
| GeneDB no. | Annotation | MKT 2H | 2H coverage | Motif? | Manual | MKT1- TAP (%) | ZC3H11- TAP (%) |
|---|---|---|---|---|---|---|---|
| Verified complex | |||||||
| Tb927.6.4770 | MKT1 | 0 | 45 | No | ND | 69 | 48 |
| Tb927.5.810 | ZC3H11 | 5 | 13 | HNPY | Yes | 12 | 61 |
| Tb927.8.4540 | PBP1 | 8 | 11 | No | Yes | 45 | 20 |
| Tb927.9.9060 | LSM12 | 0 | 10 | No | ND | 60 | 47 |
| Y2H and TAP | |||||||
| Tb927.10.5250 | ZC3H32 | 16 | 21 | NNPY | Yes | 5 | 0 |
| Tb927.1.4650 | CFB2, CFB1 | 9 | 33 | HDPY | Yes | 40 | 0 |
| Tb927.7.2670 | ZC3H21 | 9 | 13 | HNPY | ND | 4 | 0 |
| Tb927.11.980 | Nucleoporin (TbNup158) | 4 | 64 | No | ND | 13 | 0 |
| Tb927.6.850 | NOT2 | 2 | 14 | (Q)n | ND | 19 | 0 |
| Tb927.5.1990 | Unknown | 2 | 25 | (Q)n | Yes | 26 | 0 |
| Tb927.11.3440 | Unknown | 2 | 20 | (Q)n | Yes | 9 | 0 |
| Tb927.10.8330 | PSP1 homologue? | 2 | 5 | HDPY | ND | 6 | 0 |
| Tb927.2.5810 | Unknown | 1 | 55 | No | ND | 41 | 0 |
| Tb927.9.4080 | Interacts with DRBD3 | 1 | 37 | No | ND | 17 | 2 |
| Tb927.3.5010 | Unknown | 1 | 11 | No | ND | 16 | 0 |
| Tb927.3.2600 | RNA helicase | 1 | 77 | No | ND | 15 | 1 |
| Tb927.5.2820 | Protein kinase | 1 | 22 | No | Yes | 4 | 0 |
| Tb927.8.8050 | Nucleoporin NUP75 | 1 | 14 | No | ND | 4 | 0 |
| Y2H only | |||||||
| Tb927.11.510 | UBP2 | 1 | 5 | (Q)n | ND | 0 | 15 |
| Tb927.11.5850 | RBP38 | 1 | 15 | No | ND | 0 | 0 |
| Tb927.3.4800 | Unknown | 1 | 30 | (Q)n | Yes | 0 | 0 |
| Tb927.4.3470 | Unknown | 1 | 24 | No | Yes | 0 | 0 |
| Tb927.7.4730 | PUF5 | 1 | 14 | HNPY | ND | 0 | 0 |
| Tb927.8.4020/70/120 | ZC3H24, 25, 26 | 1 | 11 | No | ND | 0 | 0 |
| Tb927.10.5150 | ZC3H31 | 2 | 11 | NNPY | ND | 0 | 0 |
| Tb927.5.1490 | eIF4G1 | 2 | 26 | HNPY | ND | 0 | 0 |
| Tb927.7.1500 | Thiomodification of cytosolic tRNAs (NBP 2) | 2 | 10 | No | ND | 0 | 0 |
| Tb927.10.12780 | ZC3H37 | 3 | 5 | HNPY | Yes | 0 | 0 |
| Tb927.11.2830 | Unknown | 3 | 33 | No | Yes | 0 | 0 |
| Tb927.8.3850 | PSP1 domain | 3 | 10 | HNPY | ND | 0 | 0 |
| Tb927.10.12800 | ZC3H38 | 4 | 3 | HNPY | Yes | 0 | 0 |
| Tb927.10.4010 | Unknown | 4 | 13 | No | Yes | 0 | 0 |
| Tb927.6.740 | ATP-dependent DEAD/H RNA helicase | 2 | 63 | No | ND | 0 | 0 |
| Tb927.6.3090 | ARM repeat, microtubule associated? | 4 | 88 | No | Yes | 0 | 0 |
| Tb927.7.2140 | ZC3H18 | 4 | 18 | HNPY | ND | 0 | 0 |
| Tb927.8.1590 | ubiquitin-protein ligase | 4 | 153 | No | Yes | 0 | 0 |
| Tb927.10.840 | ERGIC53 like | 5 | 61 | No | Yes | 0 | 0 |
| Tb927.10.11760 | PUF6 | 6 | 23 | No | Yes | 0 | 0 |
| Tb927.1.2600 | PUF9 | 7 | 22 | HQPY, (Q)n | Yes | 0 | 0 |
| Tb927.11.10930 | Delta tubulin | 8 | 21 | No | Yes | 0 | 0 |
| Tb927.3.3960 | DRBD6A | 8 | 17 | HEPY | Yes | 0 | 0 |
| Tb927.8.1290 | Unknown | 8 | 52 | HNPY | Yes | 0 | 0 |
| Tb927.3.3930 | DRBD6B | 9 | 18 | HDPY | Yes | 0 | 0 |
| Tb927.7.2660 | ZC3H20 | 11 | 9 | HYNPY | ND | 0 | 0 |
| TAP only | |||||||
| Tb927.7.2780 | Unknown | 0 | 2 | No | ND | 71 | 21 |
| Tb927.11.6130 | Skp1 family protein | 0 | 7 | No | ND | 56 | 0 |
| Tb927.4.2040 | ALBA3 | 0 | 8 | No | ND | 51 | 0 |
| Tb927.10.8660 | Unknown | 0 | 11 | No | ND | 42 | 0 |
| Tb927.11.3340 | RBP34 | 0 | 5 | No | ND | 24 | 0 |
| Tb927.8.6440 | RBP20 | 0 | 3 | 2 | ND | 23 | 20 |
| Tb927.3.740 | ZC3H5 | 0 | 13 | No | ND | 17 | 0 |
| Tb927.1.3070 | Unknown | 0 | 14 | (Q)n | ND | 14 | 15 |
| Tb927.10.12330 | ZC3H34 | 0 | 12 | No | ND | 13 | 0 |
| Tb927.11.1980 | ZC3H41 | 0 | 17 | No | ND | 11 | 10 |
| Tb927.10.15360 | P-loop hydrolase? | 0 | 0 | No | ND | 3 | 0 |
The table lists a selection of ORFs identified by the yeast high-throughput screen and/or MKT1-TAP purification. Supplementary Tables S3 and S4 are full lists. All proteins of unknown function listed are conserved in kinetoplastids but have no evident homologues in other eukaryotes. ZC3H11-TAP: percentage of coverage in ZC3H11-TAP purification. MKT1 Y2H: ‘yes’ denotes at least 20 reads in the MKT1 high-throughput two-hybrid screen. Annotation: TritrypDB annotation, manually modified; ‘MKT 2H’: number of different sequences represented by 20 reads in the selected population after yeast two-hybrid screening; 2H coverage: number of different in-frame sequences in the original yeast two-hybrid library; ‘Motif’: presence of (H/N)(D/N/E/Q)PY or poly(Q) motif; Manual: found as an interaction partner either by colony picking and re-transformation, or targeted verification. MKT1-TAP: peptide coverage in MKT1 TAP. ZC3H11-TAP: peptide coverage in ZC3H11 TAP.
Deducing interaction domains from the two-hybrid screen
A scan using MEME (minimum motif length set to 6) gave three motifs that were enriched in the MKT1 partner set (Figure 6A and Supplementary Table S4, sheet 3). A kinesin motif (not shown) was found because five kinesins are in the list of potential partners. A highly glutamine-rich 10mer was found at 14 sites (P = 1.9 e-11) in 14 different proteins; in 3 cases, however, two-hybrid interactions were detected from protein fragments in which the poly(Q) was absent.
Figure 6.
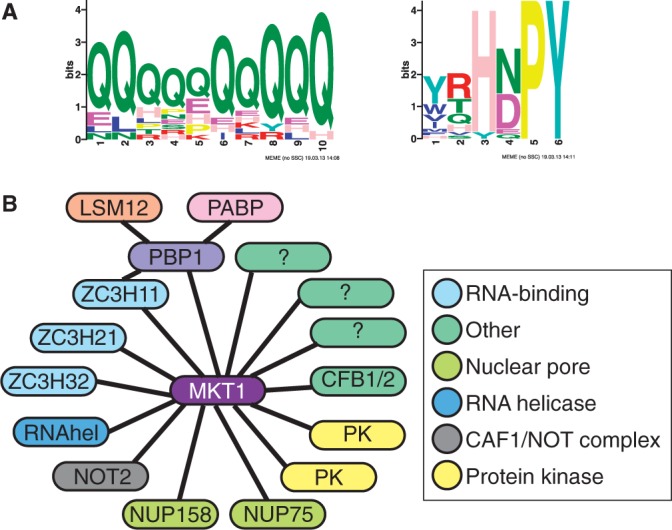
Interactions of MKT1. (A) Two motifs enriched in MKT1 yeast two-hybrid interaction partners. The output is directly from MEME. (B) Interaction partners of MKT1 and connections via PBP1. Proteins are shown as oval labels and interactions as solid lines.
The third motif was (Y/W/T)(R/T/Q)H(N/D)PY at 18 sites (P = 1 e-7); this is the ZC3H11 motif. Concentrating on confirmed partners, we derived the consensus, (H/N)(D/E/N/Q)PY; this is present in 75 trypanosome ORFs, of which 22 were positive the two-hybrid screen (Supplementary Table S4, sheet 3 and Table 4). For all 22 of these open reading frames all sequence tags with more than 20 reads mapped N-terminal to the start of the HNPY motif (Supplementary Table S4, sheet 3). Some examples are illustrated in Supplementary Figure S10. PBP1 lacks both poly(Q) and HNPY motifs, and its interaction domain must be downstream of residue 165 (Supplementary Table S3).
The cyclin F box protein genes CFB1 and CFB2 (64,65) were found multiple times. CFB1 as prey interacted with MKT1 bait, but not in the opposite configuration (Table 2 and Supplementary Figure S6F). The shortest interacting CFB2 clone encoded the C-terminal 27 amino acids, starting 4 residues before the potential interaction motif, FRHDPY. Another interactor, ZC3H37, has HNPY as the last four residues (Table 4). Correspondingly, a two-hybrid interaction was obtained with a nonapeptide from CFB2, PRRFRHDPY (Table 2 and Supplementary Figure S6F).
Confirmation of MKT1 interactions in trypanosomes
We tested the interactions of V5-tagged ZC3H38 (HNPY motif), Tb927.5.1990 (poly(Q) motif) and Tb927.6.3090 (no evident motif) with MKT1-myc in bloodstream forms. Low levels of interaction were seen for V5-ZC3H38 (Supplementary Figure S11A and C) and Tb927.5.1990 (Supplementary Figure S11B and C), but none was seen for Tb927.6.3090 (Supplementary Figure S11C and D).
Next we examined interactions of MKT-TAP in bloodstream forms by TAP. Four hundred and twenty different proteins were identified from at least two peptides (Supplementary Table S5). Reassuringly, LSM12 and PBP1 were strongly represented, and despite its low abundance in these un-stressed cells, ZC3H11 was also found (Supplementary Table S5 and Table 4). CFB2 was associated but no CFB1-specific peptides were found. After removal of possible contaminants (Supplementary Table S5, sheet 2), 144 possible partners remained, of which 15 had been positive in the two-hybrid screen (Table 4 and Figure 6B). Seven putative RNA-binding proteins were found by TAP but not two-hybrid; 2 components of the NOT complex, NOT1 and CAF40, might have been associated via NOT2; and components of ubiquitin pathways might have been associated via CFB2. There was also a large number of polysome-associated proteins; these could have associated indirectly, via PBP1 or LSM12, or via mRNA, although no RNase inhibitor was included during the purification. Nuclear proteins were also found; intriguingly, they included not only splicing factors and RNA polymerase subunits, but also five subunits of a DNA replication factor complex (Supplementary Table S5, sheet 2).
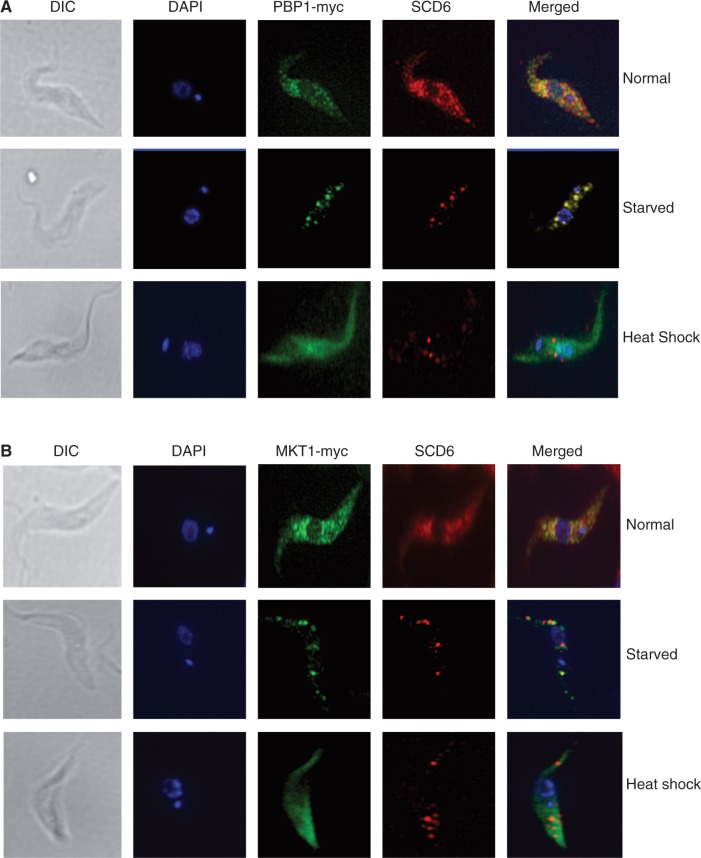
MKT1 and PBP1 relocate to granules in response to starvation, but not heat shock
Both MKT1 and PBP1 are in the cytosol (Figure 7). Pbp1p and Ataxin-2 were previously shown to associate with stress granules in yeast and mammalian cells, respectively (36,39). The formation of cytosolic granules in response to heat shock and starvation has been extensively investigated in procyclic-form trypanosomes, where starvation has been shown to induce the formation of granules containing mRNA, DHH1 and SCD6 (66,67). We therefore used the same protocols to find out whether MKT1 and PBP1 associate with stress granules. PBP1 indeed associated with starvation-stress granules (Figure 7A), whereas MKT1 showed only partial association (Figure 7B and Supplementary Figure S8C). As previously described (55) heat shock also caused the formation of SCD6-containing granules, but contained neither MKT1 nor PBP1, which instead remained dispersed in the cytosol (Figure 7A and B), as does ZC3H11 (26).
Figure 7.
MKT1 and PBP1 are associated with starvation-stress granules, but not heat-shock granules. Procyclic trypanosomes expressing either PBP1-myc (A) or MKT1-myc (B) were stained for both RFP and myc. Starved cells were incubated in PBS for 3 h at 27°C, and heat-shocked cells were incubated at 41°C for an hour prior to fixation. Further images for MKT1 after starvation are in Supplementary Figure S8.
DISCUSSION
We previously showed that ZC3H11 is required for preservation of chaperone mRNAs during heat shock in procyclic trypanosomes (26). In bloodstream forms, ZC3H11 was required for survival. Its depletion causes decreases in some target mRNAs, including those encoding four chaperones, while others remain unaffected and four increased. There was also dysregulation of other mRNAs that are not known to bind ZC3H11; this was presumably linked to growth inhibition. We now suggest a mechanism for ZC3H11 stabilization of mRNA in bloodstream forms: while the N-terminal zinc finger binds to RNA (26), the C-terminus interacts with MKT1 and PBP1, and the latter recruits LSM12 and PABP1 or 2 (Figure 8), which in turn interacts with the poly(A) tail and/or eIF4G. Since interaction of ZC3H11 with PBP1 was not sufficient for activity in the tethering assay, MKT1 might be needed to stabilize recruitment of PBP1 to the complex, or for activities independent of PABP, such as recruitment of translation factors.
Figure 8.
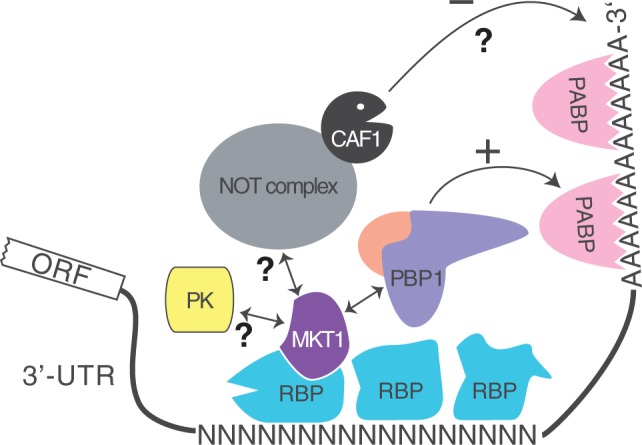
A possible mechanism for mRNA stabilization by ZC3H11 in bloodstream forms. For example ZC3H11 is bound to UAUU elements on the mRNA, and recruits PABP via MKT1 and PBP1.
Binding of a single UAUU by ZC3H11 would not give RNA specificity, and most chaperone mRNAs have multiple contiguous copies of UAU. This suggests binding of multiple copies. Self-interaction of ZC3H11 was seen in the two-hybrid assay (Figure 2), but was extremely weak by co-immunoprecipitation (Supplementary Figure S4). Since yeast PBP1 interacts with itself (38), we suggest that the specificity of ZC3H11 for its poly(UAU) targets relies on the assembly of ZC3H11–MKT1–PBP1 complexes, held together mainly via PBP1–PBP1 interactions (Figure 8).
Yeast Pbp1p and Lsm12p accumulate in stress granules induced by glucose deprivation (39) and in COS-1 cells, Ataxin-2 accumulated in granules induced by heat shock (36). Trypanosomes also form stress granules after both starvation and heat shock (55,68). It is thought that no translation occurs in stress granules, so association of PBP1 or MKT1 with heat-shock granules would probably be incompatible with roles in promoting mRNA translation. Although PBP1 was associated with starvation granules, both MKT1 and PBP1 remained distributed throughout the cytosol after heat shock.
Trypanosomes have two poly(A)-binding proteins, PABP1 and PABP2, both of which partially localize to heat-shock granules. After starvation, PABP1 shows little granule formation whereas PABP2 and eIF4E1 are together in granules (9). The other three eIF4E orthologues, including the likely major translation factor eIF4E3, are not in starvation granules (9). Intriguingly, PABP2 was much more strongly represented than PABP1 in the ZC3H11 and MKT1 TAP purifications (32%:0% for ZC3H11, and 41%:6% for MKT1; Supplementary Tables S1 and S3, sheet 2). It is tempting to speculate that binding of MKT1 to mRNAs may select them for binding of PABP2 and translation using eIF4E1. Intriguingly, in Leishmania, eIF4E1 seems to be specifically involved in translation at higher temperatures (10).
The procyclic form heat-shock response was unaffected by MKT1 RNAi. It is quite possible that MKT1 is required but enough MKT1 remained after RNAi. Alternatively, the mechanism by which ZC3H11 stabilizes mRNAs after heat shock is not the same as the one that operates constitutively in bloodstream forms. The RNASeq results also showed differences between procyclic heat shock and bloodstream forms in the regulation of mRNAs with ZC3H11 binding sites. Although the central ZC3H11–MKT1–PBP1 interaction was found in both stages, differences in regulation could easily arise, for example through binding of stage-specific proteins to other motifs in the mRNA (69,70).
In Drosophila, Ataxin-2 is involved in regulation of circadian rhythms. In addition to binding PABP, it also interacts with ‘TWENTY-FOUR’, which in turn activates translation of the PERIOD locus by recruiting eIF4F (71,72). As for trypanosome PBP1, tethering of Ataxin-2 caused an increase in reporter expression (71). Although the LSm domain might interact with RNA, the attachment of Drosophila Ataxin-2 to clock mRNA depended on the domain that interacts with PABP (71). TWENTY-FOUR is specifically associated with two circadian transcripts (73), but it lacks canonical RNA-binding domains and there is so far no evidence for direct RNA binding. PBP1 and Ataxin-2 have been found associated with yeast and mammalian mRNA as well (69,74,75), but again there is no information so far concerning specificity. Human Ataxin 2 can interact with A2BP1, a protein containing an RNA-recognition motif (76), but the implications of this interaction are unknown. We suggest that, as in trypanosomes, animal Ataxin-2 may be recruited to specific mRNAs via sequence-specific mRNA-binding proteins.
A two-hybrid screen and TAP yielded very intriguing lists of possible interaction partners of MKT1. The enrichment of proteins with RNA-binding domains, and especially of Puf and zinc finger proteins, among the MKT1 two-hybrid interaction partners was remarkable. Several of them-PUF5, DRBD6A/B, PUF9, ZC3H18, ZC3H21, ZC3H31, ZC3H32, ZC3H37 and ZC3H38, have the HNPY motif. Although not all were found after TAP of MKT1-TAP from bloodstream forms, their absence could have been caused by because of low abundance, or because interaction with MKT1 is transient or regulated, occurring only at a particular life-cycle stage or under specific conditions. The affinity purification also yielded various RNA-binding proteins that were not positive in the two-hybrid assay and could have associated indirectly. Recruitment of MKT1 to an RNA may not always lead to stabilization: at least in the case of ZC3H11, an independent interaction of the RNA-binding protein with PBP1 appeared to be necessary. Notably, the two-hybrid screen revealed an interaction of MKT1 with NOT2, a component of the CAF1-NOT deadenylation complex (Supplementary Table S4), and TAP yielded additional components, NOT1 and CAF40 (Supplementary Table S5). Thus under some circumstances, recruitment of MKT1 might lead to deadenylation. The three associated RNA helicases could also either promote or repress expression, and protein kinases might have regulatory functions.
Our model for the function of MKT1 in mRNA metabolism is not without precedent, although the idea has not previously been developed in detail. Promotion of ethanol tolerance by Mkt1p (35) might be related to the function of Puf3p in controlling expression of mitochondrial proteins (28,77–80), although there is no evidence for a direct interaction. The regulation of HO mRNA also involves a pumilio domain protein as well as Mkt1p (81). Tethering of the globin mRNA-stabilizer alpha-CP to a reporter mRNA caused very mild (1.3-fold) stabilization (82). Alpha-CP Interacts with both PABP (83,84), and AUF1 (85). It is therefore possible that alpha-CP could act via PABP recruitment, although so far there is no evidence for this and the tethering effect is not nearly as strong as that obtained with PABP alone. In an example of possible opposing actions, the mammalian TOB proteins, which promote deadenylation, bind to both PABP and to CAF1 (86).
MKT1 interacts with CFB2, an F-box protein which is important for trypanosome proliferation (64). F-box proteins bind to substrate adaptors, such as Skp1, and to Cullins to recruit E2 and E3 ubiquitin ligases (87). Since TAP of MKT1 pulled down a trypanosome homologue of Skp1 (87), we speculate that the interaction of MKT1 with CFB2 could result in ubiquitination and degradation of other proteins associated with MKT1, or even of MKT1 itself. CFB2 is, however, unlikely to be responsible for proteasomal degradation of ZC3H11: since both proteins have an HNPY motif, they probably cannot interact with MKT1 simultaneously. Correspondingly, CFB2 was not found in the ZC3H11 TAP.
RNAi targeting MKT1 rapidly killed in bloodstream forms, and was also—unlike RNAi targeting ZC3H11—deleterious in procyclics. We suggest that MKT1 has broader functions than ZC3H11 because it is recruited to various mRNA classes via different RNA-binding partners (Figure 7). As a consequence, it could be involved in control of multiple mRNA regulons.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft (DFG) [Cl112/17-1]; Bundesministerium für Bildung und Forschung, through the Systems Microbiology of Microorganisms (Sysmo) project ‘The silicon trypanosome’; Core funding from the DFG excellence initiative and the state of Baden-Württemberg. Funding for open access charge: Deutsche Forschungsgemeinschaft and core funds.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Mass spectrometry was done at the ZMBH facility led by Thomas Ruppert. RNA sequencing libraries were built by David Ibberson at the Bioquant facility and sequenced either at that facility or at EMBL, Heidelberg. We thank Claudia Hartmann and Ute Leibfried for technical assistance, João Paulo Linhares Velloso for some of the co-immunoprecipitations, Luise Krauth—Siegel (BZH, Heidelberg) for antibody to trypanothione reductase, Mark Carrington and Susanne Kramer for the antibody to SCD6 and Elisha Mugo for the two-hybrid plasmids of translation factors.
REFERENCES
- 1.Fernández-Moya S, Estévez A. Posttranscriptional control and the role of RNA-binding proteins in gene regulation in trypanosomatid protozoan parasites. WIREs RNA. 2010;1:34–46. doi: 10.1002/wrna.6. [DOI] [PubMed] [Google Scholar]
- 2.Utter C, Garcia S, Milone J, Bellofatto V. Poly(A)-specific Ribonuclease (PARN-1) function in stage-specific mRNA turnover in Trypanosoma brucei. Eukaryot Cell. 2011;10:1230–1240. doi: 10.1128/EC.05097-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee H, Palenchar J, Lukaszewicz M, Bojarska E, Stepinski J, Jemielity J, Guranowski A, Ng S, Wah D, Darzynkiewicz E, et al. Identification of the HIT-45 protein from Trypanosoma brucei as an FHIT protein/dinucleoside triphosphatase: substrate specificity studies on the recombinant and endogenous proteins. RNA. 2009;15:1554–1564. doi: 10.1261/rna.1426609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwede A, Manful T, Jha B, Helbig C, Bercovich N, Stewart M, Clayton C. The role of deadenylation in the degradation of unstable mRNAs in trypanosomes. Nucleic Acids Res. 2009;37:5511–5528. doi: 10.1093/nar/gkp571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fadda A, Färber V, Droll D, Clayton C. The roles of 3′-exoribonucleases and the exosome in trypanosome mRNA degradation. RNA. 2013;19:937–947. doi: 10.1261/rna.038430.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manful T, Fadda A, Clayton C. The role of the 5′-3′ exoribonuclease XRNA in transcriptome-wide mRNA degradation. RNA. 2011;17:2039–2047. doi: 10.1261/rna.2837311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoffe Y, Leger M, Zinoviev A, Zuberek J, Darzynkiewicz E, Wagner G, Shapira M. Evolutionary changes in the Leishmania eIF4F complex involve variations in the eIF4E-eIF4G interactions. Nucleic Acids Res. 2009;37:3243–3253. doi: 10.1093/nar/gkp190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freire E, Dhalia R, Mouraa D, da Costa Lima T, Lima R, Reisa C, Hughes K, Figueiredo R, Standart N, Carrington M, et al. The four trypanosomatid eIF4E homologues fall into two separate groups, with distinct features in primary sequence and biological properties. Mol. Biochem. Parasitol. 2011;176:25–36. doi: 10.1016/j.molbiopara.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kramer S, Bannerman-Chukualim B, Ellis L, Boulde nE, Kelly S, Field M, Carrington M. Differential localization of the two T. brucei poly(A) binding proteins to the nucleus and RNP granules suggests binding to distinct mRNA pools. PLoS ONE. 2013;8:e54004. doi: 10.1371/journal.pone.0054004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zinoviev A, Leger M, Wagner G, Shapira M. A novel 4E-interacting protein in Leishmania is involved in stage-specific translation pathways. Nucleic Acids Res. 2011;39:8404–8415. doi: 10.1093/nar/gkr555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Preiss T, Hentze MW. From factors to mechanisms: translation and translational control in eukaryotes. Curr. Opin. Genet. Dev. 1999;9:515–521. doi: 10.1016/s0959-437x(99)00005-2. [DOI] [PubMed] [Google Scholar]
- 12.Tsuboi T, Inada T. Tethering of poly(A)-binding protein interferes with non-translated mRNA decay from 5′ end in yeast. J. Biol. Chem. 2010;285:33589–33601. doi: 10.1074/jbc.M110.117150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delhi P, Queiroz R, Inchaustegui D, Carrington M, Clayton C. Is there a classical nonsense-mediated decay pathway in trypanosomes? PLoS ONE. 2011;6:e25112. doi: 10.1371/journal.pone.0025112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kramer S, Carrington M. Trans-acting proteins regulating mRNA maturation, stability and translation in trypanosomatids. Trends Parasitol. 2011;27:23–30. doi: 10.1016/j.pt.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das A, Morales R, Banday M, Garcia S, Hao L, Cross GA, Estevez AM, Bellofatto V. The essential polysome-associated RNA-binding protein RBP42 targets mRNAs involved in Trypanosoma brucei energy metabolism. RNA. 2012;18:1968–1983. doi: 10.1261/rna.033829.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raineri I, Wegmueller D, Gross B, Certa U, Moroni C. Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Res. 2004;32:1279–1288. doi: 10.1093/nar/gkh282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brennan CM, Steitz JA. HuR and mRNA stability. Cell. Mol. Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Tanaka Hall T. Structural basis for recognition of AU-rich element RNA by the HuD protein. Nat. Struct. Biol. 2001;8:141–145. doi: 10.1038/84131. [DOI] [PubMed] [Google Scholar]
- 19.Chen CY, Xu N, Shyu AB. Highly selective actions of HuR in antagonizing AU-rich element-mediated mRNA destabilization. Mol. Cell. Biol. 2002;22:7268–7278. doi: 10.1128/MCB.22.20.7268-7278.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pantopoulos K. Iron metabolism and the IRE/IRP regulatory system: an update. Ann. N. Y. Acad. Sci. 2004;1012:1–13. doi: 10.1196/annals.1306.001. [DOI] [PubMed] [Google Scholar]
- 21.Chkheidze AN, Lyakhov DL, Makeyev AV, Morales J, Kong J, Liebhaber SA. Assembly of the alpha-globin mRNA stability complex reflects binary interaction between the pyrimidine-rich 3′ untranslated region determinant and poly(C) binding protein alphaCP. Mol. Cell. Biol. 1999;19:4572–4581. doi: 10.1128/mcb.19.7.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji X, Kong J, Liebhaber SA. In vivo association of the stability control protein alphaCP with actively translating mRNAs. Mol. Cell. Biol. 2003;23:899–907. doi: 10.1128/MCB.23.3.899-907.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang Y, Xu XS, Russell JE. A nucleolin-binding 3′ untranslated region element stabilizes beta-globin mRNA in vivo. Mol. Cell. Biol. 2006;26:2419–2429. doi: 10.1128/MCB.26.6.2419-2429.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Zalen S, Jeschke GR, Hexner EO, Russell JE. AUF-1 and YB-1 are critical determinants of beta-globin mRNA expression in erythroid cells. Blood. 2012;119:1045–1053. doi: 10.1182/blood-2011-10-387316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gratacós F, Brewer G. The role of AUF1 in regulated mRNA decay. Wiley Interdiscip. Rev. RNA. 2010;1:457–473. doi: 10.1002/wrna.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Droll D, Minia I, Fadda A, Singh A, Stewart M, Queiroz R, Clayton C. Post-transcriptional regulation of the trypanosome heat shock response by a zinc finger protein. PLoS Pathog. 2013;9:e1003286. doi: 10.1371/journal.ppat.1003286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tadauchi T, Inada T, Matsumoto K, Irie K. Posttranscriptional regulation of HO expression by the Mkt1-Pbp1 complex. Mol. Cell. Biol. 2004;24:3670–3681. doi: 10.1128/MCB.24.9.3670-3681.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parreiras LS, Kohn LM, Anderson JB. Cellular effects and epistasis among three determinants of adaptation in experimental populations of Saccharomyces cerevisiae. Eukaryot. Cell. 2011;10:1348–1356. doi: 10.1128/EC.05083-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wickner R. Plasmids controlling exclusion of the K2 killer double-stranded RNA plasmid of yeast. Cell. 1980;21:217–226. doi: 10.1016/0092-8674(80)90129-4. [DOI] [PubMed] [Google Scholar]
- 30.Wickner R. MKT1, a nonessential Saccharomyces cerevisiae gene with a temperature-dependent effect on replication of M2 double-stranded RNA. J. Bacteriol. 1987;169:4941–4945. doi: 10.1128/jb.169.11.4941-4945.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wickner RB. Prions and RNA viruses of Saccharomyces cerevisiae. Annu. Rev. Genet. 1996;30:109–139. doi: 10.1146/annurev.genet.30.1.109. [DOI] [PubMed] [Google Scholar]
- 32.Deutschbauer A, Davis R. Quantitative trait loci mapped to single-nucleotide resolution in yeast. Nat. Genet. 2005;37:1333–1340. doi: 10.1038/ng1674. [DOI] [PubMed] [Google Scholar]
- 33.Dimitrov L, Brem R, Kruglyak L, Gottschling D. Polymorphisms in multiple genes contribute to the spontaneous mitochondrial genome instability of Saccharomyces cerevisiae S288C strains. Genetics. 2009;183:365–383. doi: 10.1534/genetics.109.104497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Demogines A, Smith E, Kruglyak L, Alani E. Identification and dissection of a complex DNA repair sensitivity phenotype in Baker's yeast. PLoS Genet. 2008;4:e1000123. doi: 10.1371/journal.pgen.1000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swinnen S, Schaerlaekens K, Pais T, Claesen J, Hubmann G, Yang Y, Demeke M, Foulquie-Moreno MR, Goovaerts A, Souvereyns K, et al. Identification of novel causative genes determining the complex trait of high ethanol tolerance in yeast using pooled-segregant whole-genome sequence analysis. Genome Res. 2012;22:975–984. doi: 10.1101/gr.131698.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ralser M, Albrecht M, Nonhoff U, Lengauer T, Lehrach H, Krobitsch S. An integrative approach to gain insights into the cellular function of human ataxin-2. J. Mol. Biol. 2005;346:203–214. doi: 10.1016/j.jmb.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 37.Fleischer T, Weaver C, McAfee K, Jennings J, Link A. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes Dev. 2006;20:1294–1307. doi: 10.1101/gad.1422006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mangus D, Amrani N, Jacobson A. Pbp1p, a factor interacting with Saccharomyces cerevisiae poly(A)-binding protein, regulates polyadenylation. Mol. Cell. Biol. 1998;18:7383–7396. doi: 10.1128/mcb.18.12.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swisher K, Parker R. Localization to, and effects of Pbp1, Pbp4, Lsm12, Dhh1, and Pab1 on stress granules in Saccharomyces cerevisiae. PLoS ONE. 2010;5:e10006. doi: 10.1371/journal.pone.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mangus DA, Smith MM, McSweeney JM, Jacobson A. Identification of factors regulating poly(A) tail synthesis and maturation. Mol. Cell. Biol. 2004;24:4196–4206. doi: 10.1128/MCB.24.10.4196-4206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alibu VP, Storm L, Haile S, Clayton C, Horn D. A doubly inducible system for RNA interference and rapid RNAi plasmid construction in Trypanosoma brucei. Mol. Biochem. Parasitol. 2004;139:75–82. doi: 10.1016/j.molbiopara.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 42.Shen S, Arhin GK, Ullu E, Tschudi C. In vivo epitope tagging of Trypanosoma brucei genes using a one step PCR-based strategy. Mol. Biochem. Parasitol. 2001;113:171–173. doi: 10.1016/s0166-6851(00)00383-2. [DOI] [PubMed] [Google Scholar]
- 43.Estévez A, Kempf T, Clayton CE. The exosome of Trypanosoma brucei. EMBO J. 2001;20:3831–3839. doi: 10.1093/emboj/20.14.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Colasante C, Alibu VP, Kirchberger S, Tjaden J, Clayton C, Voncken F. Characterisation and developmentally regulated localisation of the mitochondrial carrier protein homologue MCP6 from Trypanosoma brucei. Eukaryot. Cell. 2006;5:1194–1205. doi: 10.1128/EC.00096-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wurst M, Selinger B, Jha B, Klein C, Queiroz R, Clayton C. Expression of the RNA Recognition Motif protein RBP10 promotes a bloodstream-form transcript pattern in Trypanosoma brucei. Mol. Microbiol. 2012;83:1048–1063. doi: 10.1111/j.1365-2958.2012.07988.x. [DOI] [PubMed] [Google Scholar]
- 46.Zeiner GM, Sturm NR, Campbell DA. Exportin 1 mediates nuclear export of the Kinetoplastid spliced leader RNA. Eucaryot. Cell. 2003;2:222–230. doi: 10.1128/EC.2.2.222-230.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benz C, Mulindwa J, Ouna B, Clayton C. The Trypanosoma brucei zinc finger protein ZC3H18 is involved in differentiation. Mol. Biochem. Parasitol. 2011;177:148–151. doi: 10.1016/j.molbiopara.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 48.Estévez AM, Lehner B, Sanderson CM, Ruppert T, Clayton C. The roles of inter-subunit interactions in exosome stability. J. Biol. Chem. 2003;278:34943–34951. doi: 10.1074/jbc.M305333200. [DOI] [PubMed] [Google Scholar]
- 49.Ouna B, Stewart M, Helbig C, Clayton C. The Trypanosoma brucei CCCH zinc finger proteins ZC3H12 and ZC3H13. Mol. Biochem. Parasitol. 2012;183:184–188. doi: 10.1016/j.molbiopara.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 50.Clayton CE. Import of fructose bisphosphate aldolase into the glycosomes of Trypanosoma brucei. J. Cell. Biol. 1987;105:2649–2653. doi: 10.1083/jcb.105.6.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clayton CE. Genetic manipulation of Kinetoplastida. Parasitol. Today. 1999;15:372–378. doi: 10.1016/s0169-4758(99)01498-2. [DOI] [PubMed] [Google Scholar]
- 52.Gunzl A. The pre-mRNA splicing machinery of trypanosomes: complex or simplified? Eukaryot. Cell. 2010;9:1159–1170. doi: 10.1128/EC.00113-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang YC, Ciarletta AB, Temple PA, Chung MP, Kovacic S, Witek-Giannotti JS, Leary AC, Kriz R, Donahue RE, Wong GG, et al. Human IL-3 (multi-CSF): identification by expression cloning of a novel hematopoietic growth factor related to murine IL-3. Cell. 1986;47:3–10. doi: 10.1016/0092-8674(86)90360-0. [DOI] [PubMed] [Google Scholar]
- 54.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kramer S, Queiroz R, Ellis L, Webb H, Hoheisel J, Clayton C, Carrington M. Stress granules and the heat shock response in Trypanosoma brucei. J. Cell Sci. 2008;121:3002–3014. doi: 10.1242/jcs.031823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen B, Nolan J, Sklar L, Park M. Functional analysis of point mutations in human flap endonuclease-1 active site. Nucelic Acids Res. 1997;25:3332–3338. doi: 10.1093/nar/25.16.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ouna B, Nyambega B, Manful T, Helbig C, Males M, Fadda A, Clayton C. Depletion of trypanosome CTR9 leads to gene expression defects. PLoS ONE. 2012;7:e34256. doi: 10.1371/journal.pone.0034256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Urbaniak MD, Guther ML, Ferguson MA. Comparative SILAC proteomic analysis of Trypanosoma brucei bloodstream and procyclic lifecycle stages. PLoS ONE. 2012;7:e36619. doi: 10.1371/journal.pone.0036619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mangus DA, Evans MC, Agrin NS, Smith M, Gongidi P, Jacobson A. Positive and negative regulation of poly(A) nuclease. Mol. Cell. Biol. 2004;24:5521–5533. doi: 10.1128/MCB.24.12.5521-5533.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rebbapragada I, Lykke-Andersen J. Execution of nonsense-mediated mRNA decay: what defines a substrate? Curr. Opin. Cell Biol. 2009;21:394–402. doi: 10.1016/j.ceb.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 61.Alsford S, Turner D, Obado S, Sanchez-Flores A, Glover L, Berriman M, Hertz-Fowler C, Horn D. High throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res. 2011;21:915–924. doi: 10.1101/gr.115089.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lewis JD, Wan J, Ford R, Gong Y, Fung P, Nahal H, Wang PW, Desveaux D, Guttman DS. Quantitative interactor screening with next-generation sequencing (QIS-Seq) identifies Arabidopsis thaliana MLO2 as a target of the Pseudomonas syringae type III effector HopZ2. BMC Genomics. 2012;13:8. doi: 10.1186/1471-2164-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weimann M, Grossmann A, Woodsmith J, Oüzkan Z, Birth P, Meierhofer D, Benlasfer N, Valovka T, Timmermann B, Wanker E, et al. A Y2H-seq approach defines the human protein methyltransferase interactome. Nature Meth. 2013;10:339–342. doi: 10.1038/nmeth.2397. [DOI] [PubMed] [Google Scholar]
- 64.Benz C, Clayton C. The cyclin F box protein CFB2 is required for cytokinesis of bloodstream-form Trypanosoma brucei. Mol. Biochem. Parasitol. 2007;156:217–224. doi: 10.1016/j.molbiopara.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 65.Hartmann C, Benz C, Brems S, Ellis L, Luu V-D, Stewart M, D'Orso I, Busold C, Fellenberg K, Frasch ACC, et al. The small trypanosome RNA-binding proteins TbUBP1 and TbUBP2 influence expression of F box protein mRNAs in bloodstream trypanosomes. Eukaryot. Cell. 2007;6:1964–1978. doi: 10.1128/EC.00279-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cassola A, De Gaudenzi J, Frasch A. Recruitment of mRNAs to cytoplasmic ribonucleoprotein granules in trypanosomes. Mol. Microbiol. 2007;65:655–670. doi: 10.1111/j.1365-2958.2007.05833.x. [DOI] [PubMed] [Google Scholar]
- 67.Holetz F, Correa A, Avila A, Nakamura C, Krieger M, Goldenberg S. Evidence of P-body-like structures in Trypanosoma cruzi. Biochem. Biophys. Res. Commun. 2007;356:1062–1067. doi: 10.1016/j.bbrc.2007.03.104. [DOI] [PubMed] [Google Scholar]
- 68.Walrad PB, Capewell P, Fenn K, Matthews KR. The post-transcriptional trans-acting regulator, TbZFP3, co-ordinates transmission-stage enriched mRNAs in Trypanosoma brucei. Nucleic Acids Res. 2012;40:2869–2883. doi: 10.1093/nar/gkr1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baltz AG, Munschauer M, Schwanhausser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell. 2012;46:674–690. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 70.Oldenbroek M, Robertson SM, Guven-Ozkan T, Gore S, Nishi Y, Lin R. Multiple RNA-binding proteins function combinatorially to control the soma-restricted expression pattern of the E3 ligase subunit ZIF-1. Dev. Biol. 2012;363:388–398. doi: 10.1016/j.ydbio.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lim C, Allada R. ATAXIN-2 activates PERIOD translation to sustain circadian rhythms in Drosophila. Science. 2013;340:875–879. doi: 10.1126/science.1234785. [DOI] [PubMed] [Google Scholar]
- 72.Zhang Y, Ling J, Yuan C, Dubruille R, Emery P. A role for Drosophila ATX2 in activation of PER translation and circadian behavior. Science. 2013;340:879–882. doi: 10.1126/science.1234746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lim C, Lee J, Choi C, Kilman VL, Kim J, Park SM, Jang SK, Allada R, Choe J. The novel gene twenty-four defines a critical translational step in the Drosophila clock. Nature. 2011;470:399–403. doi: 10.1038/nature09728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Castello,A., Fischer,B., Eichelbaum,K., Horos,R., MORE AUTHORS, Preiss,T., Steinmetz,L., Krijgsveld,J. and Hentze,M. (2012) Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell, 149, 1393–1406. [DOI] [PubMed]
- 75.Tsvetanova NG, Klass DM, Salzman J, Brown PO. Proteome-wide search reveals unexpected RNA-binding proteins in Saccharomyces cerevisiae. PLoS One. 2010;5:e12671. doi: 10.1371/journal.pone.0012671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shibata H, Huynh D, Pulst SM. A novel protein with RNA-binding motifs interacts with ataxin-2. Hum. Mol. Genet. 2000;9:1303–1313. doi: 10.1093/hmg/9.9.1303. [DOI] [PubMed] [Google Scholar]
- 77.Saint-Georges Y, Garcia M, Delaveau T, Jourdren L, Le Crom S, Lemoine S, Tanty V, Devaux F, Jacq C. Yeast mitochondrial biogenesis: a role for the PUF RNA-binding protein Puf3p in mRNA localization. PLoS ONE. 2008;3:e2293. doi: 10.1371/journal.pone.0002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Garcia-Rodriguez L, Gay A, Pon L. Puf3p, a Pumilio family RNA binding protein, localizes to mitochondria and regulates mitochondrial biogenesis and motility in budding yeast. J. Cell Biol. 2007;176:197–207. doi: 10.1083/jcb.200606054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chatenay-Lapointe M, Shadel GS. Repression of mitochondrial translation, respiration and a metabolic cycle-regulated gene, SLF1, by the yeast Pumilio-family protein Puf3p. PLoS ONE. 2011;6:e20441. doi: 10.1371/journal.pone.0020441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee SI, Dudley AM, Drubin D, Silver PA, Krogan NJ, Pe'er D, Koller D. Learning a priori on regulatory potential from eQTL data. PLoS Genet. 2009;5:e1000358. doi: 10.1371/journal.pgen.1000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chritton J, Wickens M. A role for the Poly(A)-binding protein Pab1p in PUF protein-mediated repression. J. Biol. Chem. 2011;286:33268–33278. doi: 10.1074/jbc.M111.264572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kong J, Ji X, Liebhaber SA. The KH-domain protein alpha CP has a direct role in mRNA stabilization independent of its cognate binding site. Mol. Cell. Biol. 2003;23:1125–1134. doi: 10.1128/MCB.23.4.1125-1134.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang Z, Day N, Trifillis P, Kiledjian M. An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA In vitro. Mol. Cell. Biol. 1999;19:4552–4560. doi: 10.1128/mcb.19.7.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang Z, Kiledjian M. The poly(A)-binding protein and an mRNA stability protein jointly regulate an endoribonuclease activity. Mol. Cell. Biol. 2000;20:6334–6341. doi: 10.1128/mcb.20.17.6334-6341.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kiledjian M, DeMaria C, Brewer G, Novick K. Identification of AUF1 (heterogeneous nuclear ribonucleoprotein D) as a component of the alpha-globin mRNA stability complex. Mol. Cell. Biol. 1997;17:4870–4876. doi: 10.1128/mcb.17.8.4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ezzeddine N, Chen CY, Shyu AB. Evidence providing new insights into TOB-promoted deadenylation and supporting a link between TOB's deadenylation-enhancing and antiproliferative activities. Mol. Cell. Biol. 2012;32:1089–1098. doi: 10.1128/MCB.06370-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Skaar JR, Pagan JK, Pagano M. Mechanisms and function of substrate recruitment by F-box proteins. Nat. Rev. Mol. Cell Biol. 2013;14:369–381. doi: 10.1038/nrm3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.