Abstract
The eubacterial RNA polymerase core, a transcription machinery performing DNA-dependent RNA polymerization, consists of two α subunits and β, β′ and ω subunits. An additional σ subunit is recruited for promoter recognition and transcription initiation. Cyanobacteria, a group of eubacteria characterized by oxygenic photosynthesis, have a unique composition of the RNA polymerase (RNAP) core due to splitting of the β′ subunit to N-terminal γ and C-terminal β′ subunits. The physiological roles of the small ω subunit of RNAP, encoded by the rpoZ gene, are not yet completely understood in any bacteria. We found that although ω is non-essential in cyanobacteria, it has a major impact on the overall gene expression pattern. In ΔrpoZ strain, recruitment of the primary σ factor into the RNAP holoenzyme is inefficient, which causes downregulation of highly expressed genes and upregulation of many low-expression genes. Especially, genes encoding proteins of photosynthetic carbon concentrating and carbon fixing complexes were down, and the ΔrpoZ mutant showed low light-saturated photosynthetic activity and accumulated photoprotective carotenoids and α-tocopherol. The results indicate that the ω subunit facilitates the association of the primary σ factor with the RNAP core, thereby allowing efficient transcription of highly expressed genes.
INTRODUCTION
The eubacterial RNA polymerase (RNAP) core, a transcription machinery performing DNA-dependent RNA polymerization, consists of two α subunits and β, β′ and ω subunits. For the RNAP holoenzyme, capable of transcription initiation, an additional σ subunit is recruited. The function of the small ω subunit of RNAP, encoded by the rpoZ gene, is not yet completely understood. A structural model of the Thermus thermophilus RNAP holoenzyme shows that the ω subunit contacts with the N and C-terminal parts of the β′ subunit (1). Studies with Escherichia coli have shown that the ω subunit binds to full length β′ and assists the final step of RNAP core assembly when β′ associates with an α2β sub-complex (2). However, completely segregated rpoZ deletion strains of E. coli (3), Streptomyces kasugaensis (4), Streptomyces coelicolor (5) and Mycobacterium smegmatis (6) have revealed that ω is not an essential subunit. On the contrary, the RPB6 subunit of eukaryotic RNAP I, II and III, and the archaean RpoK subunit, which show sequence, structure and function homology with the ω subunit, are essential proteins (7).
The rpoZ deletion strains in M. smegmatis and S. coelicolor show altered cell surface properties and colony morphology, and the deletion strain of S. coelicolor lost antibiotic production capacity (5,6). In E. coli, the slow-growth phenotype of an rpoZ inactivation strain was shown to be caused by a polarity effect on the adjacent spoT gene encoding a pyrophosphatase that regulates the amounts of the stringent response alarmone molecules ppGpp and pppGpp (3). Recently, lack of the ω subunit was suggested to increase DNA relaxation in E. coli (8).
Cyanobacteria are eubacteria characterized by oxygenic photosynthesis. The cyanobacterial RNAP has unique features. First, the β′ subunit is split, the N-terminal part is called γ and the C-terminal part retains the name β′ (9). Second, the β′ subunits harbors a large and >600 amino acid long lineage-specific insertion (10). A similar assembly function of the ω subunit as suggested in the other eubacteria would require simultaneous connection of the ω subunit to both γ and β′ subunits.
In this work, we constructed a ΔrpoZ strain of the cyanobacterium Synechocystis sp. PCC 6803 (hereafter Synechocystis). The ΔrpoZ strain grew as well as the control strain (CS) in standard conditions and contained a normal amount of RNA, but showed major physiological differences to CS, including a low light-saturated photosynthetic activity and a high carotenoid content. Our results show that the RNAP holoenzyme contains less of the primary σ factor SigA in ΔrpoZ than in CS. We suggest that this explains downregulation of many highly expressed genes and simultaneous upregulation of low-expression genes in ΔrpoZ.
MATERIALS AND METHODS
Strains and growth conditions
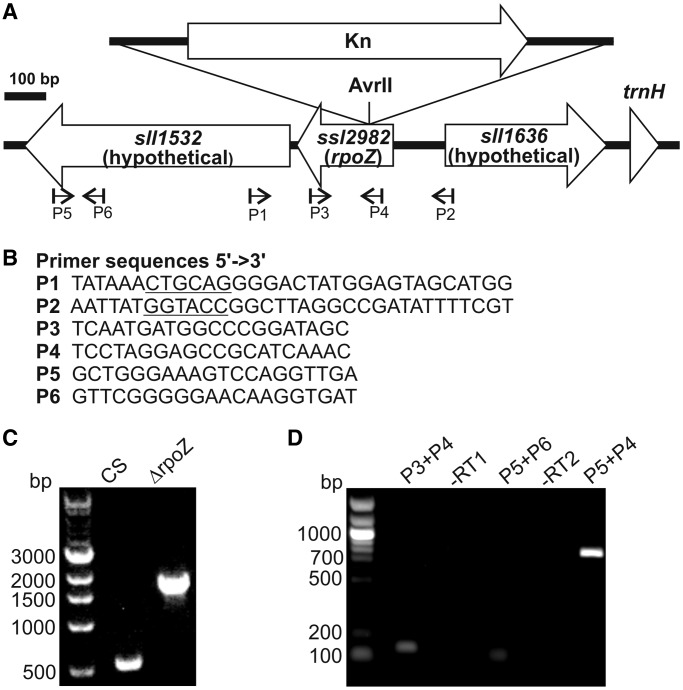
The glucose-tolerant strain of Synechocystis sp. PCC 6803 (11) was used as a CS. To construct the ΔrpoZ inactivation strain (Figure 1A), the rpoZ gene with flanking sequences was amplified by polymerase chain reaction (PCR) using primers P1 and P2 (Figure 1B) and ligated into KpnI- and PstI-digested pUC19. A fragment containing the kanamycin resistance cassette was amplified from pUC4K and inserted into the AvrII site in the pUC19-rpoZ. CS was transformed with the pUC19-rpoZ:Kn. Transformants were selected on BG-11 agar plates supplemented with kanamycin (50 µg/ml). Complete segregation was confirmed with PCR analysis (Figure 1C).
Figure 1.
The rpoZ gene region of the genome and construction of the ΔrpoZ strain (A) A schematic drawing of the rpoZ region of the genome and a ΔrpoZ strain. Primers P1 and P2 were used for construction and PCR verification of the mutant strain, the kanamycin cassette (Kn) was inserted in the middle of the rpoZ gene in antisense orientation. Primers P3, P4, P5 and P6 were used in the reverse transcription and subsequent PCR analysis. (B) Sequences of primers P1-P6. (C) PCR analysis of the ΔrpoZ strain. Genomic DNA was isolated from the control (CS) and ΔrpoZ strains, and the rpoZ gene was amplified. Expected fragment sizes are 533 and 1745 bp for the control and ΔrpoZ strains, respectively. (D) RNA was isolated from CS, and reverse transcription was performed using the primer P3 or P5, and then complementary DNA was amplified by PCR. The –RT1 and –RT2 denotes control reactions for primers P3 and P5, respectively, where no reverse transcriptase was added. The predicted PCR product sizes are 130 bp (P3 + P4), 78 bp (P5 + P6) and 795 bp (P5 + P4).
For the complementation strain, rpoZ was amplified with 5′-AATTATGGTACCGGGACTATGGAGTAGCATGG-3′ and 5′-ATATTACATATGATGACCAAGCGTAGTAATTTGG-3′ and ligated into KpnI- and NdeI-digested pAII:Sm (a generous gift from Dr Marion Eisenhut), a plasmid containing the up- and downstream regions of the psbA2 gene, and this construct was used to transform ΔrpoZ cells to replace the coding region of psbA2 with the coding region of rpoZ. Transformants were selected on BG-11 agar plates supplemented with kanamycin (50 µg/ml), spectinomycin (20 µg/ml) and streptomycin (10 µg/ml).
Cells were grown in standard conditions, in BG-11 medium supplemented with 20 mM Hepes-NaOH, pH 7.5, under continuous illumination at the photosynthetic photon flux density (PPFD) of 40 µmol/(m2 s), at 32°C in ambient CO2. The BG-11 agar plates for ΔrpoZ were supplemented with kanamycin (50 µg/ml) and those for complementation strain with kanamycin (50 µg/ml), spectinomycin (20 µg/ml) and streptomycin (10 µg/ml). For the experiments, all strains were grown without antibiotics in liquid BG-11 medium for 1–5 days.
The rpoZ-sll1532 operon
Reverse transcription-PCR was performed to judge whether of the rpoZ and sll1532 genes belong to the same operon. DNA-free RNA was extracted from CS cells as described for the DNA microarray experiments. Reverse transcription (RT) was performed for 900 ng RNA using SuperScript III kit (Invitrogen) according to the manufacturer’s instructions using the primer P3 for rpoZ or the primer P5 for sll1532 (Figure 1B). For control reactions, no reverse transcriptase was added. Then 2 µl of each RT reaction was used as a template to amplify rpoZ (primers P3 and P4), sll1532 (primers P5 and P6) and possible dicistronic rpoZ-sll1532 (primers P5 and P4) using Phusion HeatShock II (Thermo Scientific) enzyme (Figure 1B). The following program was used: initial denaturation (30 s at 98°C); 25 cycles of denaturation (10 s at 98°C), annealing (30 s at 51°C) and polymerization (30 s at 72°C); final polymerization (5 min at 72°C). Five microliters of the PCR reaction was loaded on 1% agarose gel and stained with ethidium bromide (Figure 1D).
Isolation of total RNA and DNA microarray
Total RNA was isolated from 1 ml of cell culture (OD730 = 1) as described previously (12) from three independent biological replicates, and RNA concentration was measured; for visualization, RNAs were separated in 1.2% agarose gel and stained with ethidium bromide.
For transcription profiling, CS and ΔrpoZ were grown in standard conditions for 72 h (OD730 = 1; 3.5 µg of chlorophyll (Chl) a/ml; 20 ml). Total RNA was isolated from three (ΔrpoZ) or four (CS) independent biological replicates with the hot-phenol method and then purified with RNeasy kit (Qiagen). Microarray experiments were performed using an Agilent 8 × 15 K custom cyanobacterium Synechocystis sp. PCC 6803 array (13). RNA labeling, data collection and normalization were done as described previously (14). A gene was considered upregulated if log2 of the fold change (FC) was ≥1 and downregulated if FC was less than and equal to −1, and the results were statistically significant (P < 0.05).
For estimation of gene expression levels in CS, both raw microarray data and data after normalization with the quantile method were analyzed. The signal intensity for each gene was calculated as the mean intensity obtained from all probes representing the gene in the DNA microarray, and the mean of the log2 of the signal value in four independent biological replicates was used as the final value. The data shown are after normalization but both methods gave similar results, as the lists of 100 most highly expressed genes were 96% similar.
Sequence logos of promoter regions
The 60-nt long 5′ upstream sequences of Synechocystis genes for which a transcription start site has been experimentally determined (15) and for which FC less than and equal to −1 or FC more than and equal to +1 in ΔrpoZ compared with CS were aligned. To calculate the −10 region, the sequences were aligned so that the information content (16) of a 6-nt long subsequence between nucleotides −14 and −5 was maximized. The −35 region was then obtained by maximizing the information content of a 6-nt long subsequence placed 16–18 nt upstream from the −10 region. The logos were drawn with Weblogo (http://weblogo.berkeley.edu).
Western blotting
Cells (25 ml; OD730 = 1; 3.5 µg of Chl a/ml) were harvested from standard growth conditions. Total proteins and membrane proteins were isolated as described previously (17). Protein samples containing 1.6 µg (allophycocyanin, phycocyanin), 5 µg (CP43, rubisco), 10 ug (PsaB, NdhJ, NdhK), 20 µg (ω), 40 µg (Flv3) or 50 µg (α and β subunits of RNAP, SigA, HspA, NdhD3, Flv2) total protein were solubilized for 10 min at 75°C and separated by 10% NEXT GEL™ sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) (Amresco) according to the manufacturer's instructions. For AtpE, 60 µg of membrane proteins was used instead of total proteins. Proteins were transferred to Immobilon-P membrane (Millipore). Antibodies against allophycocyanin (AS08 277), phycocyanin (AS08 278), PsaB (AS10 695), CP43 (AS11 1787) and HspA (AS08 286) were purchased from Agrisera; antibodies against Flv2, Flv3, NdhJ, NdhK and NdhD3 were gifts from Prof. E-M. Aro. Custom polyclonal antibodies against peptides CKSYTDQPQIGRLTA, CIRVQPHSPDNPAEK, CVAATEGKEKKVRKI and CMSDELTRPEIISDN recognizing α, β, SigA and ω subunits of RNAP, respectively, were purchased from Agrisera. The goat anti-rabbit IgG (H+L) alkaline phosphatase conjugate (Zymed) and the CDP star chemiluminescence kit (New England Biolabs) were used for detection in western blotting. At least three independent biological replicates were analyzed, and immunoblots were quantified with a FluorChem image analyzer (Alpha Innotech Corp.).
Free and RNAP bound SigA
Cells (30 ml, OD730 = 1) were harvested from the standard growth conditions, and soluble proteins were isolated as described previously (18) without a freezing step, and using buffer 50 mM Tris–HCl, pH 7.6, 0.15 M NaCl, 10 mM EDTA supplemented with protease inhibitor cocktail tablet (Roche). Protein samples containing 250 μg of soluble proteins were filtered through an Amicon Ultra-0.5 ml 100K column (Millipore), and the flow through was further filtered with 30 K column, to obtain fractions containing the RNAP complex and free SigA and α subunits, respectively. Equal amounts of fractions were loaded on gels. Samples were analyzed as in western blotting using antibodies against α and SigA subunits of RNAP.
Growth, photosynthetic activity and 77K fluorescence measurements
Growth was measured as described previously (19). Light-saturated photosynthetic activity in vivo was measured (1 ml samples, 3.5 µg of Chl a/ml) with a Clark-type oxygen electrode (Hansatech Ltd) at 32°C in BG-11 medium supplemented with 10 mM NaHCO3. Fluorescence emission spectra were measured at 77 K with an Ocean Optics S2000 spectrometer by exciting the sample (OD730 = 5, 50 µl) with light from a slide projector through a 440-nm line filter (Corion, Dunedin FL, USA). The spectra were corrected by subtracting a background at 615 nm, smoothened by a moving median with a 2-nm window and normalized by dividing by the peak value of PSI emission at 723 nm.
Pigment, α-tocopherol and lipid contents
In vivo absorption spectra were measured with a UV-3000 spectrophotometer (Shimadzu) from 400 to 800 nm. The carotenoid peak at 678 nm, the phycobilin peak at 625 nm and the Chl a peak at 678 nm were used for pigment analysis. The Chl a content of intact cells was measured as described previously (19). Carotenoids and α-tocopherol were extracted with methanol and detected by high pressure (or high performance) liquid chromatography (HPLC) as described earlier, except that a washing step of cell pellets was omitted (19). Lipids were extracted from intact cells by the method of Blight and Dyer (20). Lipid classes were separated with thin-layer chromatography and quantified by gas chromatography as described previously (21).
RESULTS AND DISCUSSION
The ω subunit is non-essential in cyanobacteria
Cyanobacterial ω subunits form a monophyletic clade in a phylogenetic tree containing representatives of the main eubacterial groups (Supplementary Figure S1). The cyanobacterial clade includes chloroplast-encoded ω subunits of green and red algae and cyanelle-encoded ones of glaucocystophytes, but the ω subunit appears to be lost from plastomes of higher plants (Supplementary Figure S1).
Like many other Synechocystis sp. PCC 6803 laboratory strains, our glucose tolerant CS does not contain the slr1635 transposase next to the rpoZ (ssl2982) gene, unlike the originally sequenced Kazusa strain (Figure 1 and Supplementary Figure S2). The coding regions of rpoZ and adjacent sll1532 are separated only by 31 bp. Our analysis showed that the rpoZ gene forms an operon together with sll1532 that encodes a hypothetical protein (Figure 1D).
The ΔrpoZ mutant was constructed by inserting a kanamycin resistance cassette in antisense orientation to the middle of the rpoZ gene (Figure 1A). Complete segregation of the ΔrpoZ mutant strain was verified by PCR (Figure 1C). A complementation strain was constructed to verify that the phenotype of ΔrpoZ was due to inactivation of the rpoZ gene. In the complementation strain, the coding region of the psbA2 gene was replaced by the coding region of the rpoZ gene in the ΔrpoZ strain. The phenotype of the complementation strain was similar to that of CS, except that the ω subunit was overexpressed, as it was under the strong psbA2 promoter (Supplementary Figure S3).
The ΔrpoZ strain grew as well as CS in standard growth conditions (Figure 2A). Western blot analysis confirmed the absence of the ω subunit in ΔrpoZ (Figure 2B). Similar amounts of α and β subunits of RNAP were detected in ΔrpoZ and CS, but the amount of the primary σ factor, SigA, was slightly lower in ΔrpoZ than in CS (Figure 2B). The RNA contents of ΔrpoZ and CS were similar, 1 ml of cells culture (OD730 = 1) contained 1.28 ± 0.05 and 1.18 ± 0.14 µg RNA in CS and ΔrpoZ, respectively (Figure 2C). This indicates that at least the abundant ribosomal RNA is similarly produced in both strains. Our data also suggest that the RNAP core content remains normal in cyanobacteria lacking the ω subunit, but the lowered SigA content of ΔrpoZ may cause changes in the transcriptional pattern.
Figure 2.
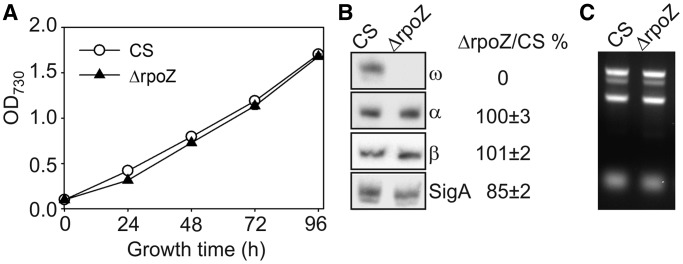
Growth, RNAP and RNA contents of the control (CS) and ΔrpoZ strains. (A) Cells were grown in standard conditions under continuous illumination at the PPFD of 40 µmol/(m2s), 32°C, air level CO2. Each data point represents an average of six independent cultures, and the error bars denote SE (shown if larger than the symbols). (B) Total proteins were isolated from cells grown in standard conditions, and α, β, ω and SigA subunits of RNAP were detected by western blotting. Subunit content in ΔrpoZ is expressed as percentage of that measured in CS. (C) Total RNA was isolated from 1 ml of cell culture with OD730 = 1, and the RNAs were separated with 1.2% agarose gel and stained with ethidium bromide.
Inactivation of rpoZ reorganizes transcription pattern
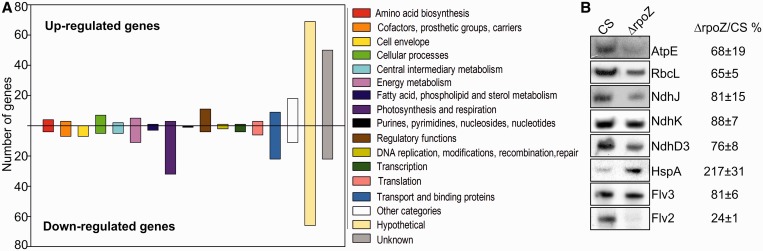
A DNA microarray analysis was performed in standard conditions. In ΔrpoZ, 187 genes were at least 2-fold upregulated and 212 genes were downregulated to one half or less. The distribution of upregulated and downregulated genes to different gene categories according to Cyanobase (http://genome.microbedb.jp/cyanobase) is shown in Figure 3A and, full lists of upregulated and downregulated genes are shown in Supplementary Tables S1 and S2, respectively. All microarray data are available in GEO (accession GSE51647). Genes for energy metabolism, photosynthesis and transport were largely downregulated in ΔrpoZ, whereas many regulatory genes and genes with unknown functions were upregulated (Figure 3A). The downregulated genes included both the rpoZ gene and the adjacent sll1532.
Figure 3.
Upregulated and downregulated genes in ΔrpoZ. (A) The 187 genes upregulated at least 2-fold, and the 212 genes downregulated to one half or less were arranged to gene categories according to Cyanobase. (B) Total proteins were isolated and the amounts of AtpE, RbcL, NdhJ, NdhK, NdhD3, HspA, Flv3 and Flv2 proteins were detected by western blotting.
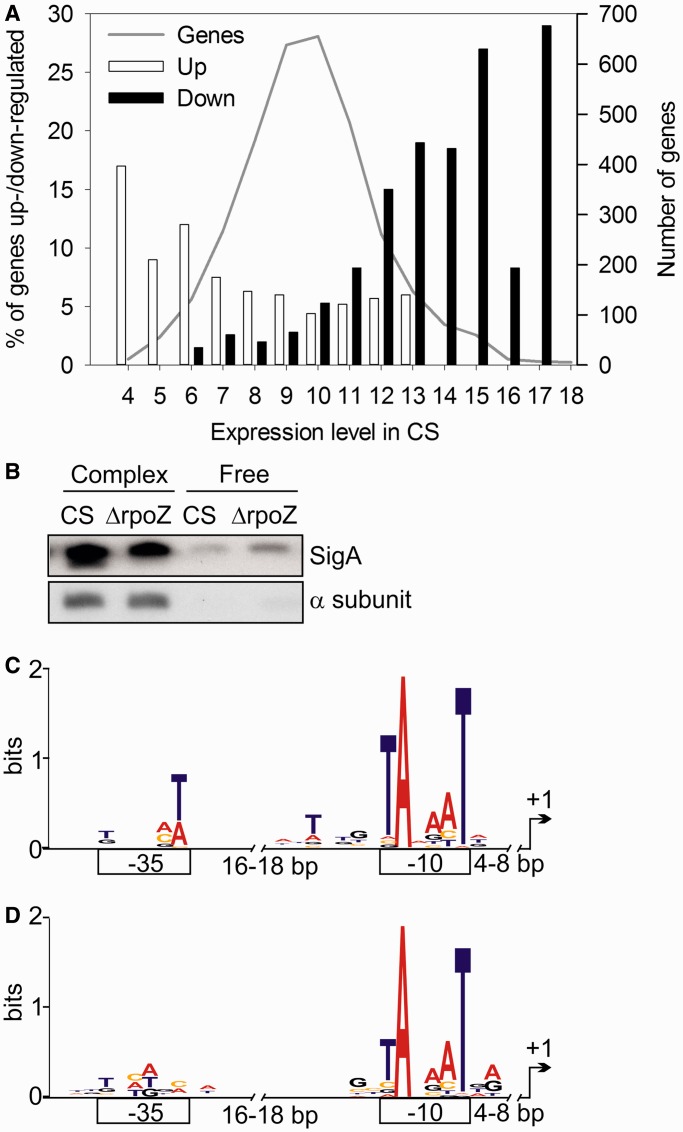
The findings that many downregulated genes encoded abundant proteins like photosynthetic proteins and that many upregulated genes belong to categories hypothetical or unknown (Figure 3) raised the question if deletion of the ω subunit affects differentially genes with high- and low-expression levels. Estimation of the gene expression level from the DNA microarray data showed that the most highly expressed genes of CS-encoded protein subunits of the light harvesting phycobilisome antenna, photosynthetic light reactions, ATP synthase, rubisco, carbon concentrating mechanisms, ribosomes, the nicotinamide adenine dinucleotide dehydrogenase (NDH) complex, the groES/EL chaperone and few others (see Supplementary Table S3 for a complete list of genes with log2 of the raw signal value >14, representing 3.8% of the genes). Interestingly, genes that were downregulated in ΔrpoZ mainly belonged to the highly expressed genes, whereas genes upregulated in ΔrpoZ showed only low or moderate expression in CS (Figure 4A).
Figure 4.
Characteristics of upregulated and downregulated genes in ΔrpoZ. (A) Relationship between gene expression level and upregulation or downregulation of the gene in ΔrpoZ. The log2 value of the signal intensity in DNA microarray of CS (gray line, y-axis on the right side) was used as an estimate of the expression level for each gene. The percentage of upregulated (white bars) and downregulated (black bars) genes in ΔrpoZ in each expression category is shown. (B) Soluble proteins were isolated and fractionated to >100 kDa (contains RNAP core and holoenzyme) and 30–100 kDa fractions (free α and σ subunits). Equal amounts of fraction were loaded on SDS-PAGE gel, proteins were separated and the amounts of the α and SigA subunits were detected by western blotting. (C) A sequence logo of the promoter regions of genes downregulated in ΔrpoZ. (D) A sequence logo for genes upregulated in ΔrpoZ.
Genes showing high expression in standard growth conditions can be assumed to be mainly transcribed by the primary σ factor SigA. To estimate the amount of SigA functioning in transcription, the distribution of SigA protein in the RNAP holoenzyme and in the free protein pool was measured. For this, soluble proteins were isolated and fractionated to >100 kDa fraction that contains RNAP complexes (>400 kDa) and to a 30–100 kDa fraction that contains free α subunits (41 kDa) and free sigA (60 kDa). The α subunit of RNAP was only found from the >100 kDa fraction, and similar quantities of α subunits in both strains confirmed that the same amount of RNAP complexes were present in ΔrpoZ and CS (Figure 4B). The SigA protein was found both in the >100 kDa (RNAP holoenzyme) and the 30–100 kDa (free SigA protein) fractions (Figure 4B). The SigA content of the RNAP holoenzyme in ΔrpoZ was only 79 ± 7% of that measured in CS, whereas free SigA was more abundant (171 ± 16%) in ΔrpoZ than in CS. The results suggest that the presence of the ω subunit favors the binding of the primary σ factor to the RNAP core, and downregulation of many highly expressed genes in ΔrpoZ might be due to the lowered SigA content of the RNAP holoenzyme. Further in vivo and in vitro experiments would be required to reveal why the absence of the ω subunit leads to a lower SigA content in RNAP holoenzyme.
To compare the promoter regions of upregulated and downregulated genes in ΔrpoZ, the −10 and −35 promoter regions were predicted for genes whose transcription initiation sites have been determined (15). The alignments and information contents of the promoter elements of upregulated and downregulated genes are shown in Supplementary Table S4. The sequence logos of promoter regions of genes that were downregulated in ΔrpoZ (Figure 4C) had a higher information content than those of upregulated genes (Figure 4D and Supplementary Table S4), although both had a highly conserved −10 region that resembles the −10 element of E. coli promoters (22). An extended −10 element, typical for highly expressed genes in E. coli (23), was found in the sequence logo of the downregulated genes (Figure 4C), suggesting that an extended −10 element is typical for highly expressed genes also in cyanobacteria. The −35 elements of upregulated and downregulated genes differed from each other (Figure 4C and D), and neither of them was similar to the E. coli −35 consensus sequence (22). The information content of the −35 region of downregulated genes was higher than that of the upregulated genes. An AT-rich sequence between the −10 and −35 elements was only detected in the sequence logo of genes that were downregulated in ΔrpoZ (Figure 4). The differences in the promoter elements of upregulated and downregulated genes further support the idea that transcriptional differences between CS and ΔrpoZ are caused by differences in σ factor recruitment.
Most genes showing upregulation in ΔrpoZ were weakly expressed in CS in standard conditions; expression of those genes may depend on the primary-like group 2 σ factors that closely resemble SigA but are non-essential in optimal conditions or on alternative group 3 σ factors that show considerable variation in amino acid sequence when compared with SigA. The hspA gene, recognized by SigB (24), and pil genes, recognized by SigF (25), provide examples of genes that are upregulated in ΔrpoZ and depend on a group 2 and a group 3 σ factor, respectively. Results from ΔrpoZ strains in different eubacteria support the role of the ω subunit as a factor influencing σ factor recruitment. In the rpoZ deletion mutant of E. coli, the primary σ factor σ70 is present in almost normal quantity but the RNAP largely recruits the group 2 σ factor σS instead of σ70, and DNA relaxation of ΔrpoZ strain can be suppressed by overexpressing σ70 in the ΔrpoZ strain (8). However, the interpretation of E. coli results is complicated because the inactivation of rpoZ decreases the expression of the downstream spoT gene affecting accumulation of (p)ppGpp (3). The (p)ppGpp, in turn, binds to RNAP core at the interface of β′ and ω (26) and facilitates recruitment of σS and enhances expression of stress responsive genes (27). Although ω-less E. coli strains are fully viable, some ω mutants are lethal, and in vitro studies showed that in one of the mutants transcription initiation was defective (28). The finding that cell surface properties change due to inactivation of the ω subunit in M. smegmatis and S. coelicolor (5,6) points to interplay between ω and σ subunits, as cell surface properties are known to be affected by alternative σ factors.
Genes encoding proteins of ATP synthase, carbon concentrating mechanisms, CO2 fixation, NDH-1 and ATP synthase are downregulated in ΔrpoZ
The DNA microarray analysis revealed that the expression of several central photosynthetic protein complexes and biochemical pathways were downregulated in ΔrpoZ (Supplementary Table S2). All subunits of the ATP synthase were downregulated at the transcript level in ΔrpoZ, and downregulation of AtpE was verified at protein level as well (Figure 3B). The messenger RNA (mRNA) levels of rbcS and rbcL genes encoding the small and large subunit of rubisco, respectively, and the rbcX gene encoding rubisco activase were downregulated. Downregulation of rubisco large subunit was verified at protein level (Figure 3B). In addition to rubisco, many other genes encoding proteins of the carbon fixation pathway were downregulated. The central carboxysome operon ccmK2K1LMN (29) was downregulated but the carbonic anhydrase gene ccaA and the ccmA, ccmK3, ccmK4 and ccmO genes were expressed similarly as in CS.
Different forms of the NDH-1 complex function either in CO2 uptake or in respiration and cyclic electron flow (30). All genes (ndhA-C, E, G-K, M-N, S), encoding NDH-1 core subunits (30,31), except ndhO, were downregulated in ΔrpoZ (Supplementary Table S2), and downregulation at protein level was verified for the NdhJ and NdhK proteins (Figure 3B). The ndhF4 and ndhD4 genes (but not cupB) for the CO2 uptake form of NDH-1 were downregulated, and also the other constitutively expressed low-affinity inorganic carbon transporter system, encoded by the bicA gene, was down. In addition, the sbtAB operon and ndhF-ndhD3-cupA-cupS operon, encoding low-CO2-inducible transporters of inorganic carbon, showed low expression in ΔrpoZ (Supplementary Table S2). Downregulation of ndhD3 was verified at the protein level (Figure 3B). Furthermore, some subunits of the respiration and cyclic electron flow-specific form of NDH-1 and an operon including ndhD6 and ndhD5 genes were downregulated in ΔrpoZ (Supplementary Table S2).
Interestingly, downregulated genes included the genes ccmR (32) and sll0822 (33), which encode repressor proteins for sbt, ndhF3-ndhD3-cupA-cupS and mnh (slr2006-2010/ssr3409-3410/slr2011-2013) operons, and ccmR and ubiX genes. Because both repressor protein genes and their target genes (except ubiX) were downregulated, absence of the ω subunit reveals a new common regulation system for a variety of genes involved in carbon concentrating and fixation.
Genes upregulated in ΔrpoZ include many pili genes and the hspA heat shock gene
Genes upregulated in ΔrpoZ included mainly genes belonging to categories unknown or hypothetical in Cyanobase (Figure 3A). Many genes encoding pili proteins were upregulated in ΔrpoZ (Supplementary Table S1), and several genes encoding envelope components were downregulated (Supplementary Table S2). Despite the upregulation of many pili proteins, ΔrpoZ remained non-motile like our glucose tolerant host strain, CS. The visual appearance of ΔrpoZ colonies was similar to that of CS colonies but ΔrpoZ cells adhered more firmly to BG-11 plates and to walls of centrifuge tubes, suggesting that inactivation of the rpoZ gene causes changes in cell surface properties also in cyanobacteria.
In E. coli, inactivation of rpoZ increases the activity of the GroEL heat shock protein, and GroEL has been suggested to replace the chaperon activity of the ω subunit in the ΔrpoZ strain (34). In our ΔrpoZ strain, expression of groEL was not changed but instead the hspA heat shock gene was upregulated both at mRNA (Supplementary Table S1) and protein levels (Figure 3B).
Physiological properties of ΔrpoZ
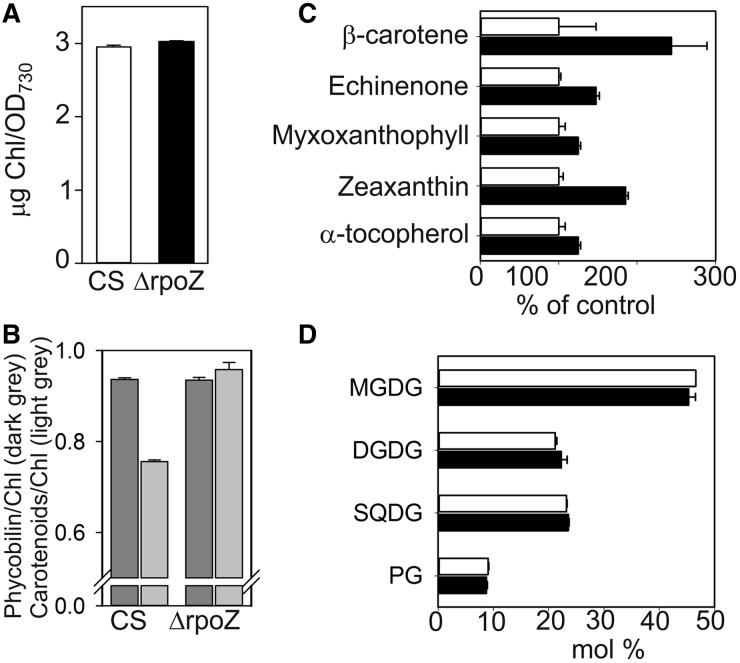
Cell cultures of the ΔrpoZ strain appeared yellowish, prompting us to analyze pigments. Chl a contents were similar in CS and ΔrpoZ (Figure 5A). In vivo absorption spectra revealed that the ratio of carotenoids to Chl a was high in the ΔrpoZ strain, whereas the ratio of phycobilisomes to Chl a was similar in ΔrpoZ and CS (Figure 5B), indicating that the high carotenoid content caused the yellowish appearance of ΔrpoZ cultures. HPLC analysis of carotenoids (Figure 5C) showed that the amounts of β-carotene and zeaxanthin were ∼2-fold higher in ΔrpoZ than in CS. The ΔrpoZ also contained high concentrations of the two other main carotenoids echinenone (1.5-fold) and myxoxanthopyll (1.25-fold). Membrane-bound carotenoids function as non-enzymatic antioxidants in cyanobacteria, mainly quenching and scavenging 1O2 (35). Another important 1O2 scavenger, α-tocopherol, was upregulated in ΔrpoZ as well (Figure 5C). The lipid composition was analyzed to find out whether the high carotenoid content of membranes is accompanied by changes in membrane lipids. Similar amounts of the main lipids, monogalactosyldiacylglycerol, digalactosyldiacylglycerol, sulfoquinovosyldiacylglycerol and phosphatidylglycerol were detected in ΔrpoZ and CS (Figure 5D). The fatty acid composition of lipids showed only some minor modifications in ΔrpoZ (Supplementary Figure S4).
Figure 5.
Pigment and lipid compositions of CS and ΔrpoZ in standard conditions. (A) Chl a content in 1 ml of cell culture (OD730 = 1). (B) The ratios of phycobilin (625 nm) to Chl a (678 nm) (dark gray bars) and carotenoids (485 nm) to Chl a (light gray bars) were calculated from in vivo absorption spectra. (C) Carotenoids were extracted with methanol and analyzed with HPLC, and the amount of each carotenoid in ΔrpoZ (black bars) was compared with that in CS (white bars). (D) Composition of membrane lipids in CS and ΔrpoZ. Each bar (A–D) represents an average of three independent experiments, and the error bars denote SE.
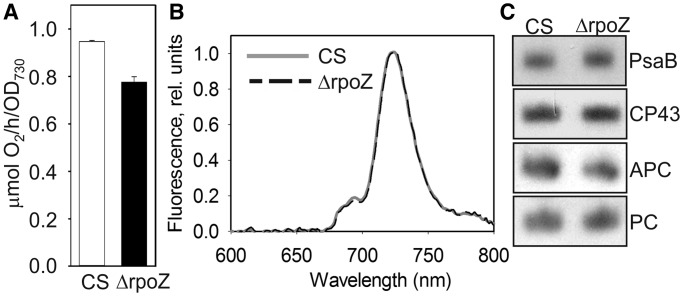
Although similar growth rates were measured for ΔrpoZ and CS, the light-saturated photosynthetic activity of ΔrpoZ was 20% lower than that of CS (Figure 6A), in accordance with a decreased rubisco content of ΔrpoZ (Figure 3B). Fluorescence emission spectra, measured at 77 K using Chl a excitation, were similar in ΔrpoZ and CS (Figure 6B), indicating a similar PSII:PSI ratio in both strains, and equal amounts of the PSI reaction center protein PsaB and the PSII core protein CP43 were detected in both strains (Figure 6C). The amounts of phycobilisome proteins, phycocyannin and allophycocyanin, were similar in both strains (Figure 6C). The mRNA levels encoding these light reaction proteins were also equal in ΔrpoZ and CS. Normal amounts of PSI and PSII and a reduced amount of rubisco might lead to imbalance of photosynthetic light reactions and carbon fixation, which in turn may increase the risk of production of reactive oxygen species (ROS). Lowered expression of the flavodiiron protein Flv3 [involved in a Mehler-like reaction that transfers electron from PSI to O2 without production of ROS (36)] in ΔrpoZ both at mRNA (Supplementary Table S3) and protein levels (Figure 3B) further increases the risk of ROS production. The flv4-sll0218-flv2 operon, providing photoprotection for PSII (14,37), showed only low expression at the mRNA (Supplementary Table S2) and protein levels in ΔrpoZ (Figure 3B). Apparently, upregulation of carotenoids and α-tocopherol in ΔrpoZ provides protection against photoinhibition and adverse effects of ROS (14).
Figure 6.
Photosynthetic properties of ΔrpoZ. (A) Light-saturated photosynthetic activity of a 1 ml of culture (OD730 = 1) of CS (white bar) and ΔrpoZ (black bar) in standard conditions. Each bar represents an average of three independent biological replicates, and the error bars denote SE. (B) Fluorescence at 77 K was measured using 440-nm light that excites Chl. The data were normalized by dividing with the height of the PSI emission peak at 723 nm. (C) Total proteins were isolated, separated with SDS-PAGE and the amounts of PSI reaction center protein PsaB, PSII core protein CP43 and the phycobilisome proteins allophycocyanin (APC) and phycocyanin (PC) were measured by western blotting. The protein contents of PsaB, CP43, ACP and PC were 99 ± 6%, 97 ± 7, 103 ± 7% and 99 ± 4, respectively, in ΔrpoZ of that measured in CS.
Our results show that the ω subunit is not an essential protein in cyanobacteria, in accordance with the results from other eubacteria. In cyanobacteria, the absence of the ω subunit has a negative effect on recruitment of the primary σ factor, which causes downregulation of highly expressed genes, and leads to a distinguished phenotype including low light-saturated photosynthetic activity and high carotenoid content of the mutant cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Academy of Finland. Funding for open access charge: Academy of Finland.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The Ndh and Flv antibodies were provided by Prof. Eva-Mari Aro and the pAII-Sm plasmid by Dr Marion Eisenhut. The Finnish Microarray and Sequencing Centre is thanked for microarray analyses services and Dr Hans CP Matthijs for allowing the authors to use his custom Synechocystis sp. PCC 6803 array. Dr Mika Keränen is acknowledged for help with HPLC measurements and Prof. Jussi Meriluoto for use of UV-3000 spectrophotometer. Dr Paula Mulo is thanked for useful comments.
REFERENCES
- 1.Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 2.Ghosh P, Ishihama A, Chatterji D. Escherichia coli RNA polymerase subunit ω and its N-terminal domain bind full-length β' to facilitate incorporation into the α2β subassembly. Eur. J. Biochem. 2001;268:4621–4627. doi: 10.1046/j.1432-1327.2001.02381.x. [DOI] [PubMed] [Google Scholar]
- 3.Gentry DR, Burgess RR. rpoZ, encoding the omega subunit of Escherichia coli RNA polymerase, is in the same operon as spoT. J. Bacteriol. 1989;171:1271–1277. doi: 10.1128/jb.171.3.1271-1277.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kojima I, Kasuga K, Kobayashi M, Fukasawa A, Mizuno S, Arisawa A, Akagawa H. The rpoZ gene, encoding the RNA polymerase omega subunit, is required for antibiotic production and morphological differentiation in Streptomyces kasugaensis. J. Bacteriol. 2002;184:6417–6423. doi: 10.1128/JB.184.23.6417-6423.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santos-Beneit F, Barriuso-Iglesias M, Fernández-Martínez LT, Martínez-Castro M, Sola-Landa A, Rodríguez-García A, Martín JF. The RNA polymerase omega factor RpoZ is regulated by PhoP and has an important role in antibiotic biosynthesis and morphological differentiation in Streptomyces coelicolor. Appl. Environ. Microbiol. 2011;77:7586–7594. doi: 10.1128/AEM.00465-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mathew R, Mukherjee R, Balachandar R, Chatterji D. Deletion of the rpoZ gene, encoding the ω subunit of RNA polymerase, results in pleiotropic surface-related phenotypes in Mycobacterium smegmatis. Microbiology. 2006;152:1741–1750. doi: 10.1099/mic.0.28879-0. [DOI] [PubMed] [Google Scholar]
- 7.Minakhin L, Bhagat S, Brunning A, Campbell EA, Darst SA, Ebright RH, Severinov K. Bacterial RNA polymerase subunit ω and eukaryotic RNA polymerase subunit RPB6 are sequence, structural, and functional homologs and promote RNA polymerase assembly. Proc. Natl Acad. Sci. USA. 2001;98:892–897. doi: 10.1073/pnas.98.3.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geertz M, Travers A, Mehandziska S, Sobetzko P, Janga SC, Shimamoto N, Muskhelishvili G. Structural coupling between RNA polymerase composition and DNA supercoiling in coordinating transcription: a global role for the omega subunit? MBio. 2011;2:e00034–11. doi: 10.1128/mBio.00034-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schneider GJ, Haselkorn R. RNA polymerase subunit homology among cyanobacteria, other eubacteria and archaebacteria. J. Bacteriol. 1988;170:4136–4140. doi: 10.1128/jb.170.9.4136-4140.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iyer LM, Koonin EV, Aravind L. Evolution of bacterial RNA polymerase: implications for large-scale bacterial phylogeny, domain accretion, and horizontal gene transfer. Gene. 2004;335:73–88. doi: 10.1016/j.gene.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 11.Williams JGK. Construction of specific mutations in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. Methods Enzymol. 1988;167:766–778. [Google Scholar]
- 12.Tyystjärvi T, Herranen M, Aro EM. Regulation of translation elongation in cyanobacteria: membrane targeting of the ribosome nascent-chain complexes controls the synthesis of D1 protein. Mol. Microbiol. 2001;40:476–484. doi: 10.1046/j.1365-2958.2001.02402.x. [DOI] [PubMed] [Google Scholar]
- 13.Eisenhut M, von Wobeser EA, Jonas L, Schubert H, Ibelings BW, Bauwe H, Matthijs HCP, Hagemann M. Long-term response toward inorganic carbon limitation in wild type and glycolate turnover mutants of the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Physiol. 2007;144:1946–1959. doi: 10.1104/pp.107.103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hakkila K, Antal T, Gunnelius L, Kurkela J, Matthijs HCP, Tyystjärvi E, Tyystjärvi T. Group 2 sigma factor mutant ΔsigCDE of the cyanobacterium Synechocystis sp. PCC 6803 reveals functionality of both carotenoids and flavodiiron proteins in photoprotection of photosystem II. Plant Cell Physiol. 2013;54:1780–1790. doi: 10.1093/pcp/pct123. [DOI] [PubMed] [Google Scholar]
- 15.Mitschke J, Georg J, Scholz I, Sharma CM, Dienst D, Bantscheff J, Voss B, Steglich C, Wilde A, Vogel J, et al. An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc. Natl Acad. Sci. USA. 2011;108:2124–2129. doi: 10.1073/pnas.1015154108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pollari M, Rantamäki S, Huokko T, Kårlund-Marttila A, Virjamo V, Tyystjärvi E, Tyystjärvi T. Effects of deficiency and overdose of group 2 sigma factors in triple inactivation strains of Synechocystis sp. strain PCC 6803. J. Bacteriol. 2011;193:265–273. doi: 10.1128/JB.01045-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gunnelius L, Tuominen I, Rantamäki S, Pollari M, Ruotsalainen V, Tyystjärvi E, Tyystjärvi T. SigC sigma factor is involved in acclimation to low inorganic carbon at high temperature in Synechocystis sp. PCC 6803. Microbiology. 2010;156:220–229. doi: 10.1099/mic.0.032565-0. [DOI] [PubMed] [Google Scholar]
- 19.Nikkinen HL, Hakkila K, Gunnelius L, Huokko T, Pollari M, Tyystjärvi T. The SigB σ factor regulates multiple salt acclimation responses of the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 2012;158:514–523. doi: 10.1104/pp.111.190058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 21.Wada H, Murata N. Synechocystis PCC6803 mutants defective in desaturation of fatty acids. Plant Cell Physiol. 1989;30:971–978. [Google Scholar]
- 22.Lisser S, Margalit H. Compilation of E. coli mRNA promoter sequences. Nucleic Acids Res. 1993;21:1507–1516. doi: 10.1093/nar/21.7.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barne KA, Bown JA, Busby SJW, Minchin SD. Region 2.5 of the Escherichia coli RNA polymerase σ70 subunit is responsible for the recognition of the ‘extended 10′ motif at promoters. EMBO J. 1997;16:4034–4040. doi: 10.1093/emboj/16.13.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tuominen I, Pollari M, Tyystjärvi E, Tyystjärvi T. The SigB σ factor mediates high-temperature responses in the cyanobacterium Synechocystis sp. PCC6803. FEBS Lett. 2006;580:319–323. doi: 10.1016/j.febslet.2005.11.082. [DOI] [PubMed] [Google Scholar]
- 25.Asayama M, Imamura S. Stringent promoter recognition and autoregulation by the group 3 σ-factor SigF in the cyanobacterium Synechocystis sp. strain PCC 6803. Nucleic Acids Res. 2008;36:5297–5305. doi: 10.1093/nar/gkn453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ross W, Vrentas CE, Sanchez-Vazquez P, Gaal T, Gourse RL. The magic spot: A ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol. Cell. 2013;50:420–429. doi: 10.1016/j.molcel.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dalebroux ZD, Swanson MS. ppGpp: magic beyond RNA polymerase. Nat. Rev. Microbiol. 2012;10:203–212. doi: 10.1038/nrmicro2720. [DOI] [PubMed] [Google Scholar]
- 28.Sarkar P, Sardesai AA, Murakami KS, Chatterji D. Inactivation of the bacterial RNA polymerase due to acquisition of secondary structure by the ω subunit. J. Biol. Chem. 2013;288:25076–25087. doi: 10.1074/jbc.M113.468520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanaka S, Sawaya MR, Phillips M, Yeates TO. Insights from multiple structures of the shell proteins from the β-carboxysome. Protein Sci. 2009;18:108–120. doi: 10.1002/pro.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Battchikova N, Eisenhut M, Aro E-M. Cyanobacterial NDH-1 complexes: Novel insights and remaining puzzles. Biochim. Biophys. Acta. 2011;1807:935–944. doi: 10.1016/j.bbabio.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 31.Battchikova N, Wei L, Du L, Bersanini L, Aro E-M, Ma W. Identification of novel Ssl0352 protein (NdhS), essential for efficient operation of cyclic electron transport around photosystem I, in NADPH:plastoquinone oxidoreductase (NDH-1) complexes of Synechocystis sp. PCC 6803. J. Biol. Chem. 2011;286:36992–37001. doi: 10.1074/jbc.M111.263780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang HL, Postier BL, Burnap RL. Alterations in global patterns of gene expression in Synechocystis sp. PCC 6803 in response to inorganic carbon limitation and the inactivation of ndhR, a LysR family regulator. J. Biol. Chem. 2004;279:5739–5751. doi: 10.1074/jbc.M311336200. [DOI] [PubMed] [Google Scholar]
- 33.Lieman-Hurwitz J, Haimovich M, Shalev-Malul G, Ishii A, Hihara Y, Gaathon A, Lebendiker M, Kaplan A. A cyanobacterial AbrB-like protein affects the apparent photosynthetic affinity for CO2 by modulating low-CO2-induced gene expression. Environ. Microbiol. 2009;11:927–936. doi: 10.1111/j.1462-2920.2008.01818.x. [DOI] [PubMed] [Google Scholar]
- 34.Mukherjee K, Nagai H, Shimamoto N, Chatterji D. GroEL is involved in activation of Escherichia coli RNA polymerase devoid of the ω subunit in vivo. Eur. J. Biochem. 1999;266:228–235. doi: 10.1046/j.1432-1327.1999.00848.x. [DOI] [PubMed] [Google Scholar]
- 35.Latifi A, Ruiz M, Zhang C-C. Oxidative stress in cyanobacteria. FEMS Microbiol. Rev. 2009;33:258–278. doi: 10.1111/j.1574-6976.2008.00134.x. [DOI] [PubMed] [Google Scholar]
- 36.Helman Y, Tchernov D, Reinhold L, Shibata M, Ogawa T, Schwarz R, Ohad I, Kaplan A. Genes encoding A-type flavoproteins are essential for photoreduction of O2 in cyanobacteria. Curr. Biol. 2003;13:230–235. doi: 10.1016/s0960-9822(03)00046-0. [DOI] [PubMed] [Google Scholar]
- 37.Zhang P, Eisenhut M, Brandt A, Carmel D, Silén HM, Vass I, Allahverdiyeva Y, Salminen TA, Aro E-M. Operon flv4-flv2 provides cyanobacterial photosystem II with flexibility of electron transfer. Plant Cell. 2012;24:1952–1971. doi: 10.1105/tpc.111.094417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.