Abstract
RydC pseudoknot aided by Hfq is a dynamic regulatory module. We report that RydC reduces expression of curli-specific gene D transcription factor required for adhesion and biofilm production in enterobacteria. During curli formation, csgD messenger RNA (mRNA) synthesis increases when endogenous levels of RydC are lacking. In Escherichia coli and Salmonella enterica, stimulation of RydC expression also reduces biofilm formation by impairing curli synthesis. Inducing RydC early on in growth lowers CsgA, -B and -D protein and mRNA levels. RydC’s 5′-domain interacts with csgD mRNA translation initiation signals to prevent initiation. Translation inhibition occurs by an antisense mechanism, blocking the translation initiation signals through pairing, and that mechanism is facilitated by Hfq. Although Hfq represses csgD mRNA translation without a small RNA (sRNA), it forms a ternary complex with RydC and facilitates pseudoknot unfolding to interact with the csgD mRNA translation initiation signals. RydC action implies Hfq-assisted unfolding and mRNA rearrangements, but once the pseudoknot is disrupted, Hfq is unnecessary for regulation. RydC is the sixth sRNA that negatively controls CsgD synthesis. Hfq induces structural changes in the mRNA domains targeted by these six sRNAs. What we describe is an ingenious process whereby pseudoknot opening is orchestrated by a chaperone to allow RNA control of gene expression.
INTRODUCTION
Many bacterial small RNAs (sRNAs) modulate gene expression by base pairing with target messenger RNAs (mRNAs) (1). Trans-encoded sRNAs regulate mRNA expression through small discontinuous ‘seed-pairings’, which are usually at or near the translation initiation signals (TIS) of their targets, whereas cis-antisense sRNAs are encoded on the DNA strand opposite to that of their targets (2). In the cellular transcript overflow, each of these base-pairing sRNAs has to efficiently locate and bind to its mRNA target, recognizing these through high-affinity contacts made by a few accessible nucleotides. These are usually situated in single strands (i.e. C-rich stretches), in loops of the regulator, in the targets or in both places. After this primary interaction, the structure of the two RNAs is generally rearranged and additional base pairs are formed. In gram-negative bacteria, the Hfq RNA-binding protein is usually required for trans-encoded sRNA stability and operation (2). Hfq facilitates sRNA–mRNA base pairing by binding both RNAs simultaneously and/or by changing one or both of the RNA structures (3), but its exact contribution at a molecular level remains, for the most part, unresolved.
RydC is a trans-encoded sRNA expressed by enteric bacteria that folds as a pseudoknot and interacts with Hfq, a protein that positively influences sRNA stability in vivo (4). In Escherichia coli, RydC controls yejABEF mRNA expression producing an inner membrane ATP Binding Cassette (ABC) transporter (4). The yejABEF allows the uptake of translation inhibitor microcin C, a peptide-nucleotide antibiotic targeting aspartyl-tRNA (transfer RNA) synthetase (5). In Salmonella, the yej operon is involved in virulence, interferes with Major Histocompatibility Complex (MHC) I presentation, counteracts antimicrobial peptides and provides a nutritious peptide source for survival and proliferation inside the host (6). In intracellular Salmonella typhimurium, RydC expression is repressed (7), and perhaps, as is the case for E. coli, this is to reduce nutrient uptake by lowering yej mRNA levels (4). In Salmonella, RydC selectively activates the longer isoform of the cyclopropane fatty acid (CFA) synthase mRNA to regulate membrane stability (8).
Here, we report that RydC negatively controls curli and biofilm production in both E. coli and Salmonella enterica. Many bacteria switch between a single-cell motile lifestyle and multicellular sessile adhesive states forming biofilm, resulting in a protected growth mode that allows cells to survive and thrive in hostile environments (9). Biofilm formation is a complex process involving numerous sensory signals linked to elaborate gene regulations via a transcription factor array. When enteric bacteria construct biofilms, they involve curli-specific genes (csg) organized in the csgDEFG and csgBAC bicistronic operon. csgEFG is required for export, and CsgD is a member of the LuxR family of transcriptional regulators that activate csgBA to synthesize the structural components of curli fimbriae. CsgD governs the synthesis of the extracellular matrix components cellulose and curli fimbriae in enteric bacteria responsible for the ‘rdar’ morphotype (10). A collection of environmental alerts adjust CsgD expression, causing it to swap from a mobile to an attached mode (11). The csgD promoter is positively regulated by several transcription factors (11) and by small signalling molecules (12), whereas its expression is negatively controlled at the post-transcriptional level by five sRNAs acting in collaboration with Hfq. In response to various environmental signals, OmrA/B (13), McaS (14), RprA (15) and GcvB (16) all downregulate CsgD translation by binding at specific locations onto the csgD mRNA 5′-untranslated region (UTR), which is a signal perception platform (17).
Experimental evidence provided in this report shows that RydC, with the help of Hfq, negatively controls csgD mRNA and protein levels. It diminishes csgA and csgBA mRNA and protein levels as well, thus attenuating curli synthesis and biofilm production. CsgD regulation by RydC occurs by direct pairing at the csgD mRNA TIS, preventing translation initiation. On complex formation with the csgD mRNA, probing and mutational data indicate that RydC induces a structural rearrangement of the csgD mRNA TIS, and the sRNA pseudoknot partially unfolds its 5′-domain to pair with its mRNA target. In the absence of sRNA, Hfq acts as a repressor of csgD mRNA translation, but it promotes complex formation between the two RNAs, presumably by facilitating pseudoknot opening to increase accessibility to the RydC 5′-domain. This makes RydC the sixth sRNA to negatively influence the expression of the csgD transcription factor that regulates collective behaviour in enteric bacteria, determining progression from a planktonic to a sessile condition.
MATERIALS AND METHODS
Bacterial strains, media and growth conditions
E scherichia coli K-12 MG1655Z1, Shigella sonnei and S. enterica strains and their derivatives were used (Supplementary Table S1). RydC gene disruption and overexpression in E. coli cells were done as previously described (4). The biofilm assays were performed in 96-well polystyrene plates, as previously described (18). E. coli, S. enterica and S. sonnei cells were grown aerobically under static conditions at 28°C in half-diluted M9 media supplemented with a 0.4% glucose carbon source. After 48-h growth, planktonic cells were discarded and kept for growth evaluation at OD600 nm. Each well was washed twice with phosphate-buffered saline and put into a ‘swimming pool’, pooled with the initial supernatant. Biofilm was developed in plates then dyed with crystal violet for 15 min at room temperature. The biofilm was recovered through application of an 80% absolute ethanol and 20% acetone solution and by pipetting up and down. After two further washes in ‘ethanol/acetone’, the number of surface-attached bacteria was estimated from the optical density at 590 nm and divided by the evaluation of growth at 600 nm. Curli expression was monitored for 48 h at 28°C on Congo red plates (1% casamino acids, 0.1% yeast extract, 20 µg/ml Congo red and 10 µg/ml Coomassie brilliant blue G). Expression of csg proteins and csg genes was accomplished by growing cells on YESCA agar (1% casamino acids, 0.1% yeast extract and 2% agar) at 28°C and for various time frames. When required, the growth media were supplemented with spectinomycin (10 µg/ml) or ampicillin (50 µg/ml).
Northern blots and quantitative RT-PCR experiments
After 8-, 10-, 15-, 24- and 48-h incubation at 28°C on YESCA plates, cells were scraped with fresh ethanol containing 5% phenol and immediately centrifuged for 10 min at 4500 rpm at 4°C. Total RNA extraction was performed on the cell pellet by the hot acid phenol method as described previously (4). For csgD and csgA mRNA analysis, 20 µg total RNA was fractionated by 1% agarose gel containing 2.2 M formaldehyde, then transferred onto nylon membranes (Zeta-Probe GT, Bio-Rad) using a Vacuum Blotter (Bio-Rad) as per the manufacturer’s protocol. For RydC analysis, northern blot analysis was carried out by loading 10 µg total RNA/lane onto a 5% PAGE containing 8 M urea. The gel was then electroblotted in 0.5× Tris-HCl, Borate, EDTA (TBE) onto nylon membrane (Zeta-Probe GT) at 30 V for 1 h 30 min. Prehybridization and hybridization were performed in ExpressHyb (Clontech). CsgD mRNA, csgA mRNA, RydC, transfer-messenger RNA (tmRNA) and 5S ribosomal RNA (rRNA) were analysed using 5′-end-labelled DNA oligonucleotides (Supplementary Table S2). Signals were detected using a PhosphorImager and quantified using ImageQuant NT 5.2 (both from Molecular Dynamics). CsgD mRNA and RydC expression levels in the E. coli strains were monitored by quantitative PCR. After an overnight culture in YESCA broth and then incubations for 2, 4 and 8 h on YESCA plates at 28°C, total RNA were extracted as described for the northern blots. The complementary DNAs (cDNAs) were produced using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). RT-PCR was performed using RealMasterMIX SYBR kit (5′PRIME) on a StepOnePlus Real-Time PCR (Applied Biosytems). Using the comparative ΔΔCt method, the amount of csgD mRNA was normalized against the tmrna reference gene.
Western blots
After 8-, 10-, 15-, 24- and 48-h incubation at 28°C on YESCA plates, cells were scraped with phosphate-buffered saline and immediately centrifuged for 10 min at 4500 rpm at 4°C. Cell pellets were then treated with formic acid in ice during 5 min. After evaporation in Speedvac, each pellet was dissolved in sample loading buffer (Laemmli 1X with 10% ß-mercaptoethanol) and heated at 90°C for 5 min. Samples were separated onto 15% SDS–PAGE gels and transferred to PolyVinyliDene Fluoride (PVDF) membranes (GE Healthcare) at 100 V for 1 h. Membranes were blocked in TBS containing 5% milk. Incubation with primary antibodies was performed for 2 h at room temperature at a 1:1000 dilution for anti-CsgA and at 1:5000 for anti-CsgD. After the incubation with the secondary antibody for 2 h at room temperature, the blots were washed in Tris-Buffered Saline (TBS) containing 0.05% Tween and then developed in ECL Western Blotting Detection Reagent (GE Healthcare). Results were obtained by exposing the blots with an ImageQuant LAS 4000 (GE Healthcare) for incremental incubation times. The signals were quantified using Image-Quant NT 5.2.
In vitro transcription, purification and end labelling
To generate the various csgD mRNA fragments as well as the sRNA’s RydC, DNA templates containing a T7 promoter sequence were generated by PCR using the appropriate primers (Supplementary Table S2) followed by in vitro transcription using a MEGAscript kit (Ambion) as per the manufacturer’s protocol. Transcription products were then electrophoresed onto a 6% PAGE containing 8 M urea, excised from the gel, then precipitated and after elution from the gel and ethanol. When necessary, purified RNA was dephosphorylated using CIP (New England Biolabs), 5′-end labelled with ATP γ-32P (PerkinElmer) and T4 polynucleotide kinase (NE Biolabs), then treated with gel purification, passive elution and ethanol precipitation.
Structural analysis of RNAs
Structural analysis of end-labelled and gel-purified csgD mRNA or RydC was performed as described previously (4). Two pmol of 5′-end-labelled csgD mRNA was mixed with 100 pmol of cold RydC or 40 pmol of Hfq and incubated 30 or 10 min at 37°C, respectively. After the incubation, V1 (5.10−5 or 15.10−5 U), S1 (0.5, 1 or 2 U) or lead acetate (0.5 or 1 mM final) were added, and the mixes were incubated for 10 min more at 37°C. The reactions were precipitated and the pellets dissolved in loading buffer (Ambion). Samples were loaded onto an 8% PAGE containing 8 M urea. Gels were dried and visualized (Phosphor-Imager).
Hfq purification
Hfq was purified as previously described (4). Escherichia coli BL21(DE3) harbouring the pTE607 plasmid and grown at 37°C to an OD600 of 0.4. After induction with 1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) during 3 h, cells were pelleted, dissolved in a buffer solution (20 mM Tris–HCL, 500 mM NaCl, 10% glycerol and 0.1% Triton X-100), sonicated, heated at 80°C for 10 min and finally centrifuged. Supernatant was then charged onto an ‘AKTA purifier’ (GE Healthcare) equipped with a Ni2+ column. Washes were performed with buffer containing 10 mM imidazole; the Hfq protein was eluted with the same buffer but with 300 mM imidazole. The purity of the protein was visualized on a 12% SDS–PAGE and concentration estimated by Bradford assay.
Toeprint and gel shift assays
After denaturation followed by renaturation at room temperature, annealing mixes containing 0.2 pmol csgD mRNA and 1 pmol of labelled primer were incubated for 15 min with or without various concentrations of RydC or Hfq. The fMet-tRNAfMet was then added for 5 min. Reverse transcription was started by adding 2 µl of AMV RT (NE Biolabs) and dNTPs for 15 min and then stopped by adding 10 µl of Buffer II (Ambion). The cDNAs and sequencing reactions were run on polyacrylamide gels, and signals detected using a PhosphorImager. Gel retardation assays are performed as previously described (4). In all, 0.5 pmol of labelled RydC were incubated for 10 min at 37°C with various concentrations (0–500 pmols) of unlabelled csgD mRNA215, csgD mRNAΔ5′UTR or csgD mRNA100 in 1× Tris-MgCl2-NaCl (TMN) buffer (20 mM Tris-acetate pH 7.6, 100 mM sodium acetate and 5 mM magnesium acetate). Samples were loaded onto a native 5% acrylamide gel and separated with 0.5× TBE at 4°C. Gels are dried and visualized (PhosphorImager).
In vitro translation assays
In vitro translation of CsgD mRNA503 (2.5 pmols) using [35S]-methionine was carried out with a PURESYSTEM (Cosmo Bio) following the manufacturer's instructions and as described previously (13). After RNA denaturation for 2 min at 85°C, chilling 2 min on ice and then renaturation for 5 min at 37°C in TMN 1× buffer, a pre-complex between RydC and 10 pmol of Hfq was performed during 10 min at 37°C. To form the complex with the csgD mRNA, we again incubated for 10 min at 37°C and then translation assays were initiated by adding [35S]-methionine and the PURESYSTEM classic II. Each reaction was denatured in a 1× Laemmli buffer at 95°C for 5 min, loaded onto a 16% Tris-glycine gel and visualized on a PhosphorImager.
RESULTS
RydC induction reduces biofilm formation in two enteric bacteria
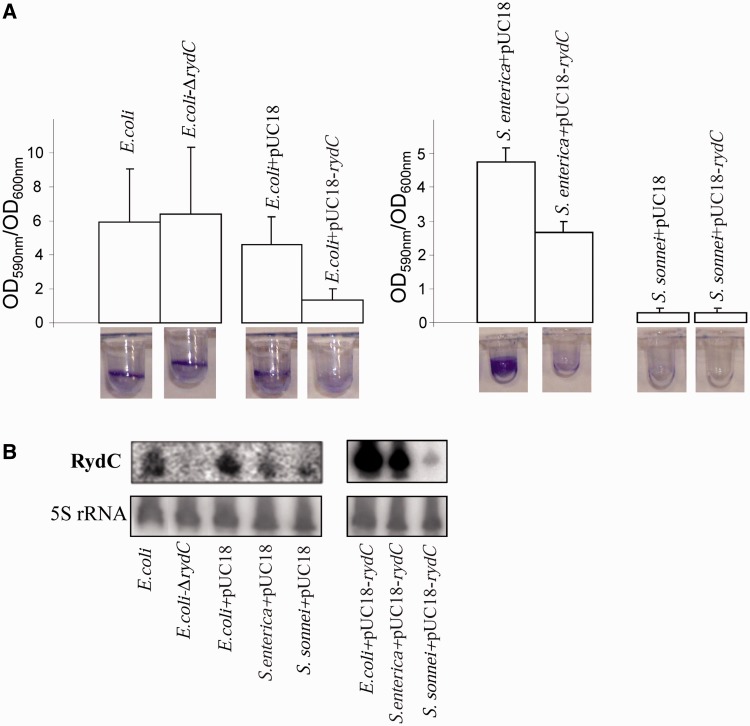
Bacterial sRNAs often regulate the expression of several targets (19). To search for a phenotype associated with the expression of RydC, E. coli strains either deficient in RydC expression (ΔRydC) or harbouring a multicopy plasmid stimulating RydC expression was used. Interestingly, a ‘RydC-dependent’ biofilm phenotype was detected. After 48 h incubation at 28°C, in both E. coli and S. enterica, increasing RydC expression reduces biofilm formation by about one-third and one-half, respectively, when compared with isogenic strains (Figure 1A). The ΔRydC E. coli strain essentially forms biofilms in the same way the wild-type (wt) strain (Figure 1A). In the closely related Shigella genus, biofilm formation is impaired by mutations in the curli gene locus (20). Accordingly and irrespective of RydC, S. sonnei bacteria neither synthesize curli nor produce biofilms. Total RNAs were extracted from the biofilms and RydC levels were monitored by northern blots (Figure 1B). In the E. coli and S. enterica cells, RydC expression is low, probably after reduction by unknown factors to facilitate biofilm synthesis. This could explain the absence of phenotypic differences between the wt and ΔrydC E. coli strains. In the three enteric bacteria transformed with pUC-rydC, RydC induction was verified during biofilm formation (Figure 1B). It can be concluded that stimulation of RydC expression reduces biofilm formation in E. coli and S. enterica.
Figure 1.
RydC regulates biofilm synthesis in E. coli and S. enterica. (A) Microtiter dish biofilm mass measured by crystal violet staining in E. coli, S. enterica (bongori) and S. sonnei after 48-h incubation. (left) The E. coli strains are wild-type MG1655Z1, its isogenic ΔRydC derivative (ΔrydC), strain containing a multicopy plasmid pUC18 and one containing pUC18 encoding RydC expressed from its endogenous promoter sequence (pUC18-rydC). (right) The same pUC18 constructs were added to Salmonella and Shigella. The data represent the means and standard deviations of at least 10 replicates. (B) RydC expression levels in the recombinant strains from the three bacteria monitored by northern blots on total RNAs directly extracted from the biofilms. The 5S rRNAs are internal loading controls.
RydC lowers curli synthesis by reducing CsgA and CsgB protein and mRNA levels
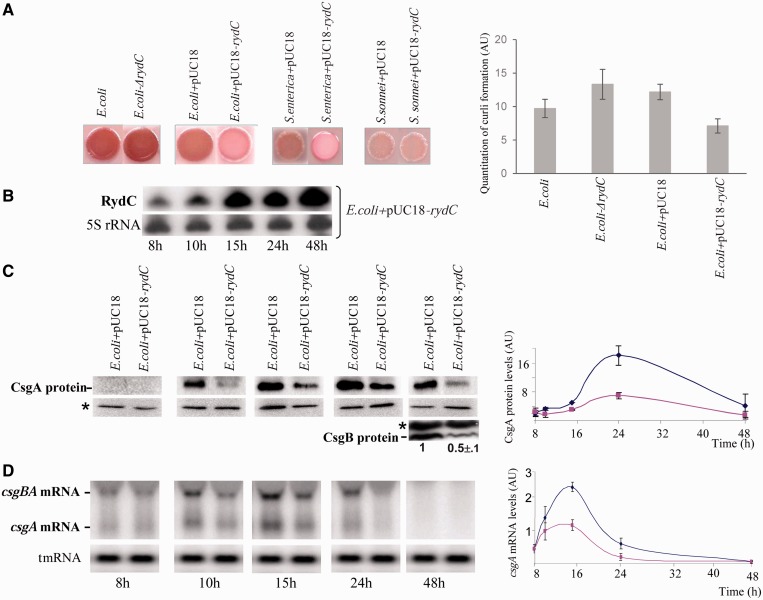
In enteric bacteria, curli fibres are involved in surface adhesion, cell aggregation and biofilm formation (21). One possible explanation for the involvement of RydC in E. coli and S. enterica biofilm formation might be linked to curli biogenesis. Curliated bacteria stain red when grown on YESCA plates supplemented with Congo red diazo dye (22). After 48-h incubation at 28°C, stimulating RydC expression results in lowered curli formation in E. coli and S. enterica cells (Figure 2A). The ΔrydC E. coli strain forms consistently slightly more curli than isogenic cells (Figure 2A, right panel). During curli synthesis on YESCA plates with a RydC-overproducing strain, RydC gradually accumulates up to 15 h and remains high afterwards (Figure 2B). In E. coli, at least six proteins encoded by the csgBA and csgDEFG operons are dedicated to curli formation (22). Homologous agfBA and agfDEFG operons were also identified in Salmonella (23). In E. coli, csgBA encodes the two curli structural subunits (24): CsgA is the major structural subunit, whereas CsgB is a nucleator. To determine whether RydC influences csgBA expression in E. coli, the effect of RydC accumulation on steady-state levels of the CsgA protein was monitored by western blots using anti-CsgA antibodies at several time points (0–48 h) during curli formation on YESCA plates. CsgA had a similar overall profile in a wt strain transformed with an empty vector as that of cells overexpressing RydC. CsgA is detected after 10-h incubation, increases up to 24 h and then decreases (Figure 2C). However, western blots show that stimulating RydC expression reduces the quantity of the CsgA structural protein by up to 2.5 times as compared with the wt (Figure 2C). After 24-h incubation, RydC induction strongly reduces CsgA levels. CsgB expression was also monitored after 48 h by western blots using anti-CsgB antibodies. Compared with an isogenic strain, promoting RydC expression also reduces the CsgB nucleator protein by about 2-fold (Figure 2C).
Figure 2.
RydC induction lowers curli synthesis by reducing CsgA and CsgB protein and mRNA levels in enteric bacteria. (A) (left) Congo red (diazo dye) YESCA agar plates grown at 28°C for 48 h added to E. coli, S. enterica (bongori) and S. sonnei. The experiments were repeated at least three times. The Shigella strain does not form curli because its csg locus is disrupted by insertions and deletions (20), an action considered to be an internal negative control. (right) The graph shows quantitation of curli formation in the four isogenic strains using the GelQuant.NET software (Arbitrary Units, AU). The data are derived from three independent experiments. (B) Northern blots monitoring of 8–48 h of RydC expression in ‘pUC18-rydC’ isogenic strains, resulting in curli formations. As loading controls, the blots were also probed for 5S rRNA. (C) Immunoblots with anti-CsgA and anti-CsgB antibodies showing CsgA and CsgB protein expression in an E. coli strain harbouring pUC18-rydC versus an isogenic strain containing the empty plasmid (E. coli +pUC18). Curli formation was a result of 8–48-h of incubation on YESCA agar plates at 28°C for CsgA and 48 h for CsgB protein. The asterisks mark two aspecific protein bands, each revealed by one antibody. The graph shows CsgA protein quantification in the two isogenic strains (E. coli+pUC18 is blue; E. coli+pUC18-rydC is pink, Arbitrary Units, AU) relative to the levels of the aspecific protein. (D) Northern blot analysis of the csgA and csgBA mRNAs in the two strains during curli formation at identical time points, as in panel A. The blots were also probed for tmRNA as loading internal controls. The graph shows csgA mRNA quantification in the strains relative to tmRNA (using a similar colour code as panel C).
CsgA and CsgB proteins are produced from a single operon, and their RydC-induced reduction could originate from an mRNA-level regulation. Using a DNA probe targeting the csgA mRNA, two ∼0.65- and ∼1.15-kb-long transcripts were detected in the wt and RydC-overproducing strains by northern blots (Figure 2D), and these correspond, respectively, to the csgA and csgBA mRNAs (25). In the wt cells, the two mRNAs are detected early, and as expected their highest expression is around 15 h before optimal expression of the CsgA protein (Figure 2C), decreases thereafter, and is undetectable after 48 h (Figure 2D). During curli synthesis, after 10-h incubation high levels of RydC decrease the steady-state levels of csgBA and csgA mRNA transcripts by half. The stronger reduction of csgBA mRNA expression in the RydC-overproducing strain occurs after 15-h incubation. Thus, the maximum reduction of csgBA mRNA expression in the RydC-overexpressing strain occurs when RydC expression is highest (Figure 2B). In both strains, there is a 9 h interval between the peaks of csgA transcription and translation, which could be ascribed to unknown CsgA regulators acting at the post-transcriptional level. Additional time points between 15 and 24 h would be required to investigate this further. In summary, RydC induction impairs curli synthesis by lowering CsgA and B, mRNA and protein levels, in turn reducing biofilm formation.
RydC controls CsgD protein and mRNA expression levels
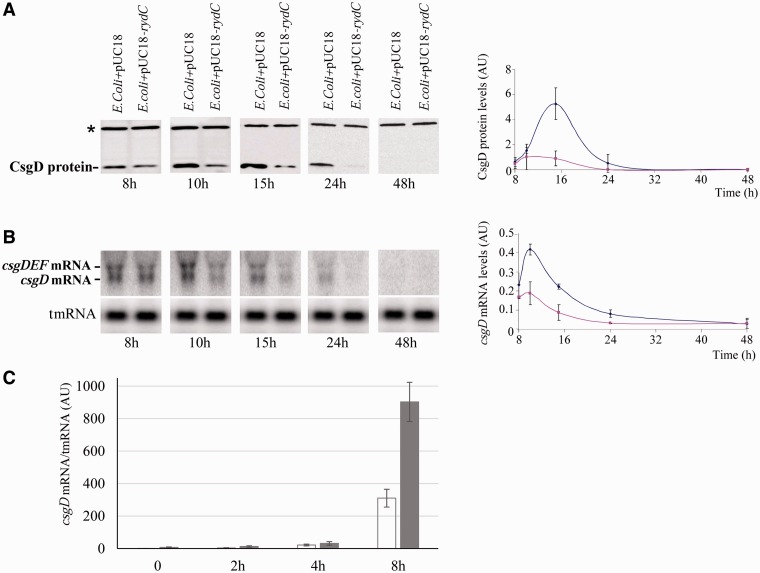
CsgD is a transcriptional activator of the csgBA operon required for curli and biofilm synthesis in E. coli (22). To assess whether RydC influences CsgD expression in E. coli, the effect of RydC expression on steady-state levels of the CsgD protein was monitored by western blots using anti-CsgD antibodies at several times during curli synthesis on YESCA plates (Figure 3A). CsgD protein expression was detected after 8-h incubation and increased to a maximum at 15 h, which as expected for a csgBA transcriptional activator corresponds to the peak of csgBA mRNA expression (Figure 2D), then slowly decreased down to zero after 48 h. This indicates that curli formation is substantially induced after 15-h incubation in E coli. Compared with an isogenic strain, at all times during curli synthesis, induction of RydC expression reduced the CsgD protein up to five-fold (Figure 3A). Thus, RydC impairs curli and biofilm synthesis by lowering CsgD protein levels. RydC may regulate CsgD expression at the mRNA level. During curli formation, RydC involvement in csgD mRNA levels was monitored by northern blots using a DNA probe specific for csgD mRNA. Hybridization of total RNAs extracted from curli-producing cells identified two ∼0.9- and ∼1.6-kb-long transcripts (Figure 3B), compatible, respectively, with the csgD and csgDEF mRNAs (22). The highest expression of the two mRNAs is at ∼10 h and then it decreases to nothing after 48 h (Figure 3B). During curli formation, the csgD mRNA and protein expressions peak before that of csgBA mRNA and proteins, which is as expected for a transcriptional regulator when compared with its target genes. Throughout curli synthesis, inducing RydC expression reduces the csgD mRNA steady-state levels down to half when compared with the isogenic strain. There is about a 5 h gap between the peaks of csgD transcription and translation that might be ascribed to previously reported or unknown regulators of csgD expression acting at the post-transcriptional level. Additional time points between 10 and 15 h would be required to further investigate this observation. Compared with an isogenic strain, the lack of endogenous levels of RydC increases csgD mRNA synthesis about three-fold after 8 h of curli formation on YESCA plates (Figure 3C). This result demonstrates the negative influence of RydC on csgD mRNA steady-state levels in vivo. RydC reduces biofilm formation by impairing curli synthesis through lowering of CsgD protein and mRNA levels, in turn decreasing CsgA mRNA and CsgA and CsgB protein levels.
Figure 3.
RydC lowers csgD mRNA and protein levels and the absence of endogenous levels of RydC increases csgD mRNA synthesis during curli formation. (A) Immunoblots with anti-CsgD antibodies monitoring CsgD protein expression between 8 to 48 h curli formation on YESCA agar plates at 28°C in an E. coli strain harbouring pUC18-rydC versus an isogenic strain containing the empty plasmid (E. coli+pUC18). The asterisk indicates an aspecific protein revealed by the antibody. The graph shows CsgD protein quantification in the two isogenic strains (E. coli+pUC18 is blue; E. coli+pUC18-rydC is pink, Arbitrary Units, AU) relative to the amount of the aspecific protein. (B) Northern blot analysis of the csgD and csgDEF mRNAs in the two strains during curli formation at time points, as in panel A. The blots were also probed for tmRNA as loading internal controls. The graph shows csgD mRNA quantification in the strains relative to tmRNA (similar colour code as in panel A, Arbitrary Units, AU). (C) The qPCR comparison of csgD mRNA expression in E. coli (white) and E. coli-ΔrydC (dark grey) strains during curli formation for 8 h on YESCA plates, normalized against the tmrna reference gene (Arbitrary Units, AU). The downregulation of csgD mRNA by RydC occurs after 4 h of incubation.
CsgD expression reduction by RydC interaction with csgD mRNA and the influence of the mRNA 5′-UTR in complex formation
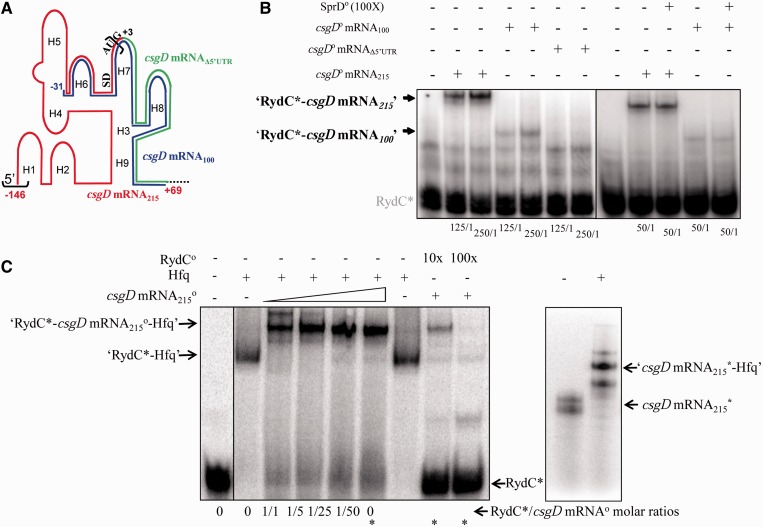
RydC controls csgD mRNA expression either indirectly via dedicated regulators or directly by antisense pairings with the mRNA. The CsgDEFG mRNA transcriptional start site was mapped by primer extension analysis and is located 146 nt upstream from the CsgD initiation codon (22). Gel retardation assays were used to analyse duplex formation between RydC and a 215 nt-long csgD mRNA fragment (mRNA215). The mRNA215 contains the 5′-UTR sequence (146 nt) followed by 69 nt corresponding to the first 23 codons from its coding sequence (Figure 4A). An ‘RydC-csgD mRNA215’ duplex was detected (Figure 4B and Supplementary Figure S1) and its binding is specific, as a 100-fold molar excess of unrelated RNA (SprD) does not remove the csgD mRNA215 from its preformed ‘RydC–csgD mRNA215’ complex. To test the importance of the csgD mRNA 5′-UTR in the binding of RydC, a csgD mRNA deletion mutant lacking the 5′-UTR and starting at G+3 was constructed (mRNAΔ5’UTR). The mRNAΔ5′-UTR does not interact with RydC (Figure 4B), demonstrating that the csgD mRNA 5′-UTR is essential for binding. Is the entire 5′-UTR of csgD mRNA required? To see if this is so, a second mutant (mRNA100) was made containing 31 nt from the csgD mRNA 5′-UTR including the TIS, followed by 69 nt from its coding sequence. A ‘RydC-csgD mRNA100’ duplex was detected (Figure 4B and Supplementary Figure S1), and the binding was specific, as a 100-fold molar excess of unrelated RNA (SprD) did not remove the csgD mRNA100 from its preformed ‘RydC–csgD mRNA100’ complex. The binding ability of mRNA100 with RydC was lower than that with mRNA215 (Figure 4B and Supplementary Data), suggesting structural differences between these mRNAs (see later in the text), or a lower affinity between RydC and mRNA100 as compared with that of mRNA215. Our results demonstrate that RydC forms a stable complex with the csgD mRNA in vitro and that at least a section of its 5′-UTR, including the TIS, is required for binding.
Figure 4.
Direct interaction between RydC and the csgD mRNA; ternary complex formation between RydC Hfq and its mRNA target. (A) Schematic representation of the csgD mRNA 5′-domain emphasizing three RNA constructs. The csgD mRNA215 corresponds to the 215 nt from the 5′-end of the mRNA (red), emphasizing the SD and AUG translation initiation signals. In the csgD mRNA100 variant (blue), 115 nt from the csgD mRNA 5′-end were deleted. The 5′-UTR of the csgD mRNA (the sequence between the black brackets) was deleted in mutant csgD mRNAΔ5′-UTR, therefore starting at G+3 (green). (B) Complex formation between RydC and each of the three csgD mRNA constructs. Native gel retardation assays of purified labelled RydC with increasing amounts of purified unlabelled csgD mRNA215, csgD mRNA100 or csgD mRNAΔ5′-UTR are shown. The csgD mRNA construct/RydC molar ratios are indicated below each lane. Competition assays were performed with a 100-fold molar excess of unrelated purified SprD RNA (18) in the presence of each of the csgD mRNA215 and csgD mRNA100 constructs. (C) Ternary complex formation between RydC, csgD mRNA215 and Hfq. Left panel: Native gel retardation assays show complex formation between labelled RydC and increasing amounts of unlabelled csgD mRNA215 (at a 1- to 50-fold excess as compared with RydC) in the presence or absence of purified Hfq. Hfq is at a 1:1 molar ratio with RydC. The asterisks indicate the ‘RydC*/csgD mRNA’ molar ratio used to perform the competition assays with a 10- to 100-fold molar excess of unlabelled RydC. Right panel: csgD mRNA215 interacts with Hfq in the absence of RydC in vitro. Hfq is at a 20:1 molar ratio with the mRNA. The csgD mRNA adopts two conformations on a native gel.
Hfq facilitates the interaction between RydC and the csgD mRNA
RydC interacts with Hfq in vitro, and the protein considerably enhances RydC stability in vivo (4). Therefore, Hfq may facilitate the pairings between RydC and csgD mRNA. To test this, gel retardation assays were performed between labelled RydC, purified E. coli Hfq (1:1 molar ratio relative to RydC) and increasing concentrations of unlabelled csgD mRNA215. An ‘RydC–Hfq–csgD mRNA215’ ternary complex is detected (Figure 4C, left), and nearly all of the RydC is in the complex at a one-to-one molar ratio with csgD mRNA215. In the absence of Hfq, to obtain about half the amount of RydC in complex with its target, there is a need for a 1000-fold molar excess of csgD mRNA215 versus RydC (Supplementary Figure S1). This also indicates that RydC and csgD mRNA215 can simultaneously interact with Hfq. In the absence of RydC, Hfq interacts with the csgD mRNA215in vitro (Figure 4C, right). Hfq facilitates the interaction between RydC and the csgD mRNA, improving the efficiency of the regulation.
The csgD mRNA ribosome binding site is sequestered by RydC and by Hfq to prevent translation initiation
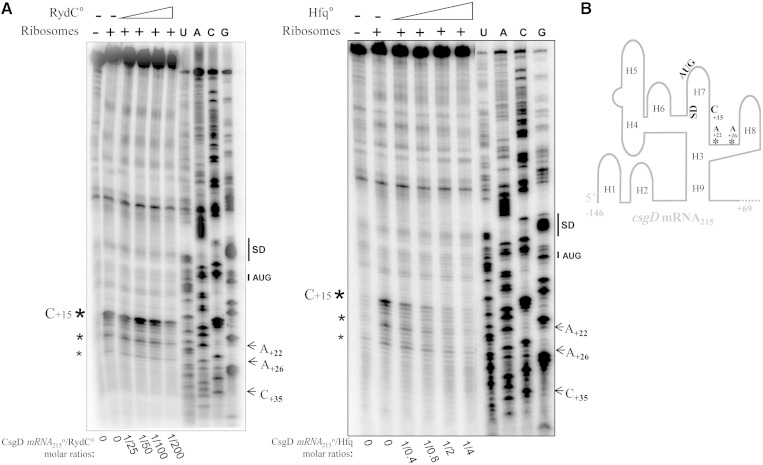
Because the interaction of RydC with the csgD mRNA requires the 31 nt upstream from the initiation codon that contain the TIS, RydC could prevent ribosome loading onto the csgD mRNA. To test this, toeprint assays were performed on ternary initiation complexes, including purified ribosomes, initiator tRNAfMet and the csgD mRNA215. A strong ribosome toeprint was detected at position C+15 on csgD mRNA215, 14 nt downstream from A+1 of the initiation codon (Figure 5A, left). Minor toeprints were also detected upstream, at positions A+22 and A+26, suggesting some degree of freedom in the positioning of the ribosome onto csgD mRNA215, or else a structural rearrangement of the mRNA on ribosome binding. In the absence of Hfq, RydC reduced ribosome loading onto the csgD mRNA in a concentration-dependent manner, requiring elevated amounts of sRNA for the regulation (Figure 5A, left). In the absence of RydC, low amounts of purified Hfq also prevent csgD mRNA translation initiation (Figure 5A, right). Thus, both Hfq alone or elevated amounts of RydC have the ability to reduce CsgD translation initiation in vitro.
Figure 5.
Hfq and RydC both prevent ribosome loading onto the csgD mRNA. (A) Ribosome toeprint assays performed on csgD mRNA215 in the presence of increasing amounts of RydC or purified Hfq. Left panel: 25- to 200-fold excess RydC as compared with csgD mRNA215. Right panel: 0.4- to 4-fold excess purified Hfq as compared with csgD mRNA215. The experimentally proven toeprints are indicated with asterisks, with their sizes reflecting the intensity of the toeprints. Plus/minus indicates the presence of purified ribosomes with the csgD mRNA; U, A, G and C: indications of the csgD mRNA215 sequencing ladders. The SD sequence and AUG initiation codon of the csgD mRNA215 are also indicated. (B) Schematic representation of the csgD mRNA 5′-domain, emphasizing the location of the ribosome toeprints (marked with asterisks) induced by either Hfq or RydC.
Monitoring the CsgD mRNA conformation by structural probes
As a prerequisite, conformations of free csgD mRNA215 and free csgD mRNA100 were investigated in solution. Both transcripts were end labelled and their solution structures were probed by RNase V1, which cleaves double-stranded RNAs or stacked nucleotides, and by nuclease S1 and lead, which both cleave accessible single-stranded RNAs. The reactivity towards these structural probes was monitored for each nucleotide (Supplementary Figure S2 for csgD mRNA215 and Supplementary Figure S3 for csgD mRNA100). The data are summarized onto the supporting model of csgD mRNA215 (Figure 6A) and csgD mRNA100 (Supplementary Figure S3). For csgD mRNA215, the data showed the existence of nine folded helices (H1–H9, with V1 cuts and without lead or S1 cleavages), all of which except H3 and H9 are capped by loops (presenting S1 and lead cleavages but no V1 cuts). An internal bulge between H4 and H5 was revealed by numerous S1 cuts at G−62-U−65. Structural analysis of the csgD mRNA is consistent with a previous RNase T1 and lead analysis (13) that proposed the existence of SL1 (H4–H5) and SL2 (H7). However, our data suggest the existence of additional helices (H1–H3, H6, H8–H9; Figure 6A) that may not be conserved (13). Probing data indicate that the beginning of the csgD mRNA coding sequence is tightly folded and embedded within four helices (H3, H7–H9). The conformation of csgD mRNA100 was monitored by structural probes (Supplementary Figure S3), and these data were compatible with the existence of H7 and H8. In that shorter mRNA fragment, however, the conformation of its 5′- and 3′-ends is different than that of csgD mRNA215: it lacks H6 and H9 but has an additional helix (H10) that bridges the 5′- and 3′-ends (Supplementary Figure S3). H7, H8 and H10 are joined by three accessible single-stranded RNAs (U−25-A−14, A+22-A+26 and C+50-U+60).
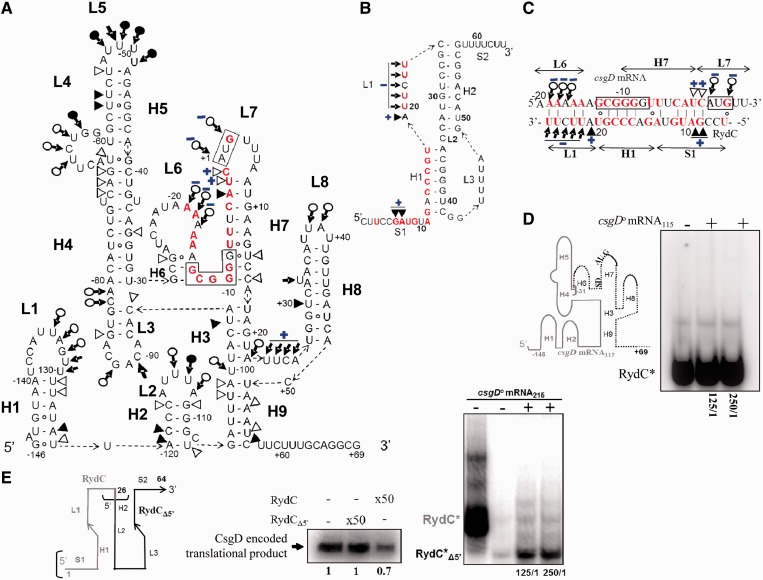
Figure 6.
Structural probing and deletion analysis of the interaction between RydC and csgD mRNA. (A) Secondary structure of the csgD mRNA215 5′-end (−146 to +69 nt) from E. coli. This is based on structural probes in solution (Supplementary Figure S2), which provide experimental support for the proposed structure. Triangles are V1 cuts; arrows capped by a circle are S1 cuts; plain arrows are lead cleavages. Cleavage intensity is shown with filled (strong cuts) or open (weaker) symbols. Structural domains (H1–H9, L1–L8) are indicated. The structural changes in the csgD mRNA induced by RydC are blue. Most of these changes are clustered onto L6-H6 and H7-L7. Nucleotides from the csgD mRNA proposed to interact with RydC are in red (for details, see Figure 7). (B) RydC secondary structure (4) emphasizing the nucleotides from its 5′-domain (red) interacting with the csgD mRNA. The structure is based on probing (Supplementary Figure S3) and mutational analysis (panel E and Figure 7). Structural changes induced by the binding of csgD mRNA with RydC are clustered on S1-H1-L1. Only the structural changes induced by duplex formation are indicated here, using the same indicators as in section A. (C) Proposed antisense pairing between RydC and the csgD mRNA leads to the sequestration of the mRNA ribosome binding site (outlined) by the RydC 5′-domain. Pairing interactions between RydC and the csgD mRNA are based on native gel retardation assays, deletion analysis and structural mapping of RydC in complex with the csgD mRNA. Only the structural data concerning the RNA duplex conformation is indicated, with the same symbols and colours as Figure 6A. The blue plus (+) and minus (−) signs indicate the appearance or disappearance of cleavages induced by structural probes when the two RNAs are, respectively, in duplex (D). RydC binding does not require 115 nt from the csgD mRNA 5′-end. Schematic representation of the csgD mRNA 5′-domain, emphasizing the csgD mRNA115 construct (in grey), which lacks the H6–H9 domains (dotted lines). The grey bracket delineates the shorter csgD mRNA115 construct. Native gel retardation assays of purified labelled RydC with increasing amounts of csgD mRNA115 (125- to 250-fold excess relative to RydC) show that the first 115 nt from the csgD mRNA 5′-end are unable to interact with RydC. This result is in agreement with the probing data, which reveal a lack of structural changes in this area of the mRNA when in complex with RydC. (E) Native gel retardation and in vitro translation evidence that the RydC 5-domain interacts with and controls CsgD translation. Left panel: Schematic representation of RydC, emphasizing the RydCΔ5′ construct (in black), which lacks the S1-H1-L1 5′-domains (in grey). Right panels: a 250-fold excess of synthetic purified RydCΔ5′ is unable to bind with csgD mRNA215, whereas wild-type RydC can (Figure 4B). In vitro translation of csgD mRNA215 in the presence of RydCΔ5′ at a 50-fold molar ratio with the csgD mRNA, showing that the RydC 5′-domain S1-H1-L1 is essential in lowering CsgD translation. The lack of RydCΔ5′ activity is due to its incapacity to interact with the csgD mRNA, as evidenced by the absence of complex formation (upper panel).
Monitoring the ‘RydC–CsgD mRNA’ complex by structural probes
Structural changes induced by RydC complex formation were examined by subjecting the ‘RydC–csgD mRNA215’ and ‘RydC–csgD mRNA100’ complexes to nuclease S1, RNases V1 and lead statistical digestions. Binding of RydC induced a cluster of structural changes located in a similar restricted region within the two csgD mRNA constructs encompassing the SD and AUG sequences, from A−20 to G+3 (Figure 6A, Supplementary Figures S2 and Supplementary Data). When RydC interacts with csgD mRNA215, the sRNA pseudoknot undergoes structural changes at its 5′-end that includes S1, H1 and L1 (Figure 6B and Supplementary Figure S2), as a result of which S1 and L1 should become double stranded. The structural data support a model of interaction between csgD mRNA and RydC in which ‘L6-H7-L7’ from the mRNA (including the TIS) pairs with ‘S1-H1-L1’ from RydC (Figure 6C). To provide additional experimental evidence for the proposed pairing model, a csgD mRNA mutant lacking H6–H9 was engineered and produced (csgD mRNA115, Figure 6D). Based on the probing data and pairing model, it should not be able to bind RydC. When csgD mRNA215 was in complex with RydC, there was no structural modifications at the first 115 nt from the csgD mRNA 5′-end (Figure 6A and Supplementary Figure S2). CsgD mRNA115 did not interact with RydC, even when at a 250-fold excess (Figure 6D), indicating that the recognition domains of csgD mRNA for binding RydC are not in the first 115 nt from the mRNA leader region. Conversely, a RydC mutant lacking ‘S1-H1-L1’ was constructed (RydCΔ5′, Figure 6E), and gel retardation assays with csgD mRNA215 revealed the absence of complex formation between the two RNAs (Figure 6E, right). Translation assays provide direct experimental evidence that, unlike wt RydC, RydCΔ5′ was unable to reduce csgD mRNA translation (Figure 6E, right).
Hfq induces a conformational rearrangement of the csgD mRNA
Structural changes induced by complex formation between Hfq and the csgD mRNA were examined by subjecting an ‘Hfq-csgD mRNA215’ complex to nuclease S1, RNases V1 and lead statistical digestions (Supplementary Figure S4). Binding of Hfq induced a cluster of structural changes on the csgD mRNA at loops L4, L4-5, L5 and L6, all of which became protected against lead and S1 cuts (Figure 7A and Supplementary Data). This provides direct evidence for structural modifications of the csgD mRNA 5′-UTR. Hfq also induced reactivity changes within the csgD mRNA coding sequence, especially within helices H7 and H8 (Figure 7A). This indicates that Hfq induced a significant conformational rearrangement of the csgD mRNA 5′-UTR, including part of its actual coding sequence.
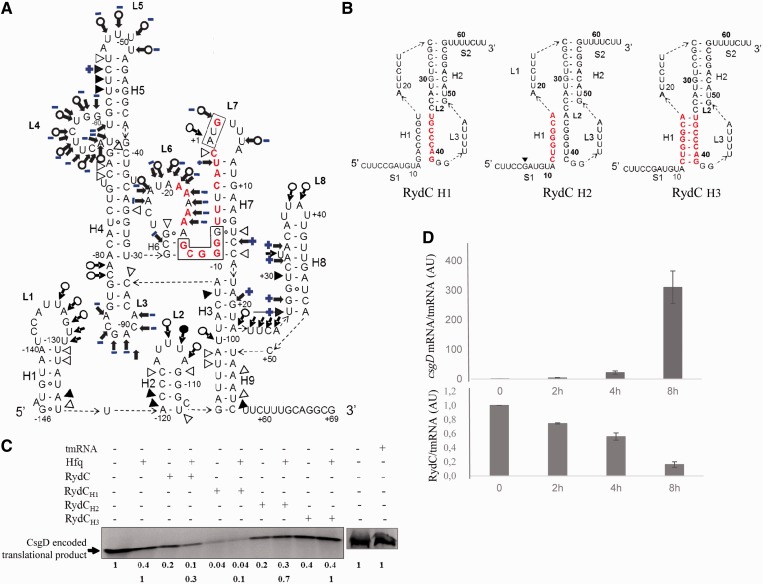
Figure 7.
The interaction between Hfq and csgD mRNA, the role of Hfq in translational regulation and the inverse correlation between RydC and csgD mRNA expression during curli formation in vivo. (A) Secondary structure of csgD mRNA215, with the structural changes induced by Hfq on the csgD mRNA conformation in blue. This model is based on structural probing of the RNA-protein complex in solution (Supplementary Figure S4). The blue plus (+) and minus (−) signs indicate the appearance or disappearance of cleavages induced by the structural probes when the protein is in complex with the mRNA. (B). RydC mutants with mutated nucleotides in red: disrupted stem H1 (RydCH1), stem H1 and the interacting sequence with the csgD mRNA (RydCH2) and a compensatory mutant that restores the H1 structure (RydCH3). (C)In vitro translation of csgD mRNA503 in the presence of various RydC mutants, with and without a 2-fold molar excess of Hfq. The translation products arbitrarily set to 1 were quantified relative to csgD mRNA503 translation in the absence of RydC and Hfq (upper lane). To explore the effect of RydC in the presence of Hfq, the translation products were also set to 1 and quantified relative to CsgD translation in the presence of Hfq (lower lane); tmRNA was used as an internal negative control. (D) qPCR monitoring of csgD mRNA and RydC expression in E. coli cells during curli formation on YESCA plates, normalized against the tmrna reference gene.
Evaluation of the involvement of the RydC structure and pairings in regulation of csgD mRNA translation
According to the probing data and the RNA deletion mutants (Figure 6), one can predict that the RydC domains S1-H1-L1 will interact with the csgD mRNA. Specific mutations were generated within the central element of the pairing interaction, stem H1 (Figure 7). These disrupted the pseudoknot fold (RydCH1), removed its csgD mRNA binding site (RydCH2) or restored stem H1 (RydCH3). Mutant RydCH1 disrupts stem H1 and therefore unfolds the pseudoknot while maintaining its csgD mRNA binding site, resulting in increased efficacy and translation blockage in the absence of Hfq (Figure 7C). This shows that unfolding the RydC pseudoknot greatly enhances its translational control of csgD mRNA. Mutant RydCH2 had a similar effect on CsgD translation, implying that pairings between S1, L1 and the csgD mRNA TIS are necessary and sufficient for translational control. In the regulation triggered by RydCH2, in which the pseudoknot was unfolded, the addition of Hfq was not beneficial. Finally, compensatory mutant RydCH3 was only half as active as RydC in reducing csgD mRNA translation, and Hfq had no effect on the translation regulation induced by RydCH3. Because RydCH3 is ∼10-fold less active than RydCH1 for reducing CsgD translation, it suggests that an unfolded state of the RydC pseudoknot significantly increases its capacity to reduce CsgD translation.
RydC and Hfq control of csgD mRNA translation
In vitro translation assays were done to provide direct experimental evidence that RydC, Hfq or ‘RydC–Hfq’ complex represses csgD mRNA protein synthesis. These assays were performed on a csgD mRNA503 construct encoding the first 119 amino acids of the CsgD protein. Without RydC and Hfq, a 13-kDa polypeptide was detected (Figure 7C). Hfq reduced CsgD translation down to 40%. This is in agreement with the substantial reduction of the ribosome toeprints induced by Hfq (Figure 5A, right), and the 20% reduction of translation by RydC (Figure 7C). When RydC and Hfq acted together, CsgD translation dropped down to 10%. Hfq or elevated amounts of RydC by themselves reduced CsgD translation by impairing ribosome binding, but the presence of an ‘Hfq–RydC’ complex significantly amplified the regulation. As an internal negative control, similar concentrations of tmRNA did not impact CsgD translation when compared with RydC (Figure 7C), demonstrating the specificity of the RydC-induced CsgD translation reduction.
DISCUSSION
In this report, we show that RydC expression affects biofilm formation and cell adhesion in two enterobacteria: S. enterica and E. coli. RydC is an important negative regulator of curli synthesis in vivo, as its endogenous expression gradually decreases over time while csgD mRNA expression progressively increases and triggers curli synthesis and biofilm formation (Figure 7D). In addition, the lack of endogenous levels of RydC augments csgD mRNA synthesis (Figure 3C). This tiny 64 nt-long sRNA also regulates the expression of a membrane transporter involved in nutrient and antibiotic uptake (5,6). Escherichia coli RydC possesses at least two direct targets (yejABEF and csgD mRNAs), which suggests physiological links between these encoded proteins. In Salmonella, RydC regulates bacterial membrane integrity through mRNA stabilization of cyclopropane fatty acid synthase (8). Thus, RydC acts both as a target activator/repressor and as a sensor for nutrient uptake, membrane remodelling and biofilm formation (Figure 8A). Interestingly, the RydC pairings with both cfa and csgD mRNAs involve accessible nucleotides at the RydC 5′-end, although in the case of the csgD mRNA, the pairing interaction is longer and spreads deeper into the sRNA pseudoknot. When food supplies are available and enter the bacteria, RydC expression is turned on to enable nutrient uptake and membrane stabilization. It also prevents unwanted biofilm formation, avoiding this survival mode triggered in hostile environments such as under feeding limitations. Previous observations (4) are in agreement with our conclusions, as RydC expression is activated during the exponential growth phase and ‘switched off’ at the stationary phase. However, when enteric bacteria are in ‘curli’ and ‘biofilm’ modes, RydC expression gradually decreases over time (Figure 7D), probably due to unknown regulators. RydC-induced reduction of biofilm formation and cell adhesion results from a drop-off in curli synthesis via the direct downregulation of CsgD expression at both the RNA and protein levels, which in turn lowers CsgA and CsgB curli structural proteins levels. The csgD mRNA is a direct target of RydC and Hfq, reducing translation initiation by blocking the mRNA TIS through direct pairings.
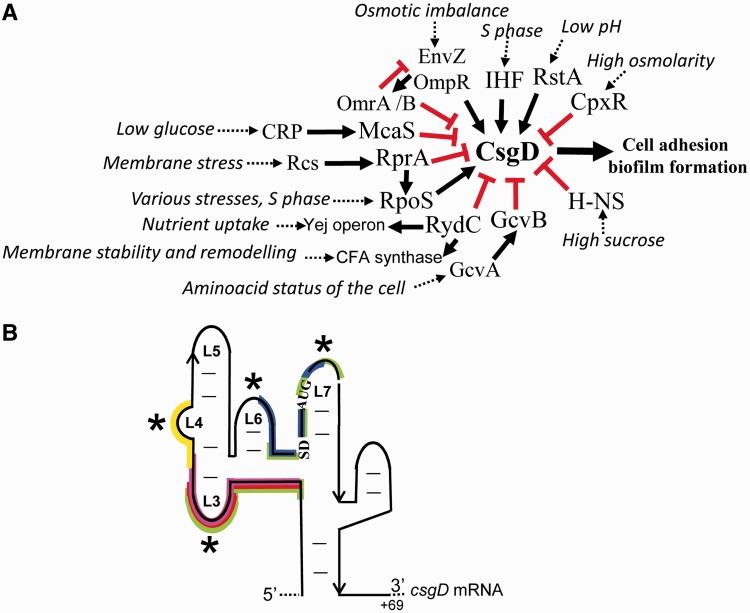
Figure 8.
Schematic integration of protein and RNA regulators of CsgD expression, colocalization of the binding sites of the six sRNAs regulating this expression and the Hfq-induced mRNA structural changes. (A) The proteins and sRNAs (16) that control CsgD expression in response to various specific environmental changes that trigger cell adhesion and biofilm formation. The black arrows and red bars indicate positive and negative regulations, respectively. Endogenous levels of RydC induce positive regulations of the Yej operon (4) and of CFA synthase expression (8). The various environmental triggers that influence and initiate these regulations are in italics, the ultimate effector molecule being the CsgD transcription factor. (B) The binding sites of the six sRNAs that reduce CsgD translation initiation are indicated on the csgD mRNA 5′ platform. OmrA/B is yellow, GcvB is pink, McaS is red, RprA is green and RydC is blue. The asterisks indicate the csgD mRNA domains from that are subjected to reactivity changes in the presence of Hfq, which strikingly match the sRNA binding sites.
In E. coli, RydC is the sixth Hfq-dependent sRNA that negatively controls CsgD transcription factor expression, and all of these sRNAs impair translation initiation. With the help of Hfq, OmrA, OmrB, RprA, McaS, GcvB and RydC regulate CsgD expression by pairing at the csgD mRNA 5′-UTR (13–17). Each of these six possesses specific binding sites on the csgD 5′-UTR, some with binding overlaps (Figure 8B). Interestingly, most of the structural changes induced by Hfq on the csgD mRNA overlap with the binding sites of these sRNA regulators (Figure 8B). Hfq modifies the conformation of the csgD mRNA at and around the binding sites of each of these sRNAs, probably to facilitate pairing between the mRNA target and its RNA regulators. Hfq can, however, repress csgD mRNA translation in the absence of sRNA, as recently observed in the translation inhibition of the cirA mRNA involved in iron uptake (26).
Interestingly, RydC is the only sRNA from the group that pairs exclusively at the csgD mRNA TIS rather than upstream (RprA interacts at both the TIS and upstream). In fact, RydC binding still occurred after the removal of 115 nt at the csgD mRNA 5′-end (Figure 4). In addition, RydC reduces cellular levels of csgD mRNA (Figure 3), implying that the regulation occurs at both the post-transcriptional and translational levels, as is usually the case for ‘Hfq-dependent’ sRNAs (27). In bacteria, transcription and translation are simultaneous, but we detected ∼5-h delay between csgD mRNA and protein synthesis (Figure 3). This is attributable to previously reported or unknown regulators of csgD expression acting at the post-transcriptional level. As reported for other sRNAs that interact with Hfq, RydC-Hfq-induced CsgD translation inhibition could promote target mRNA turnover, stimulating endonucleolytic cleavages and decay (28).
The CsgD 5′-UTR structure, inferred from structural probes, is highly folded and includes a portion of the TIS (Figure 6A). This implies unfolding both when translation initiates and when initiation is blocked through the joint action of Hfq and the six sRNAs that bind at various locations within the csgD 5′-UTR (Figure 8B). In this latter situation, each sRNA acts as a specific external stimulus sensor (Figure 8A). Hfq facilitates interactions between an sRNA and its targets by binding both RNAs or by restructuring one or both RNAs (3). We previously reported that Hfq binds RydC and restructures its conformation (4), presumably to facilitate pairing with its mRNA targets. Based on previous probing data collected on a RydC–Hfq complex (4), the protein induces reactivity changes at the two connecting single-stranded loops within the RydC pseudoknot, triggering pairing rearrangements within H1. Hfq modifies RydC structure, thus destabilizing H1 (4) but also changing csgD mRNA conformations. These particular RydC domains are those with which our structural and mutational evidence indicates csgD mRNA interacts. Hfq interacts with csgD mRNA (Figure 4C) and reduces its translation in the absence of sRNAs (Figure 5A). As previously reported for sodB mRNA (28), Hfq remodels both the conformations of RydC and csgD mRNA to improve translational control. In the absence of Hfq, the ribosomal toeprint on the csgD mRNA requires a large amount of RydC (Figure 5A). Accordingly, CsgD translation decreases only when RydC is in excess (Figure 7C). Interestingly, RydC is considerably lowered in the presence of Hfq. This implies that Hfq is required in vivo to regulate RydC-induced csgD translation initiation. Hfq orientation and proximity to the complementary target site may facilitate RydC unfolding and the annealing between the two RNAs (29). Hfq could also assist in the exchange of RNA strands between the interacting RNAs.
For the most part, single strands accessible within the scaffolds of sRNAs pair with their mRNA targets, occasionally requiring conformational activations. The interaction between RydC and csgD mRNA is striking because it is the first time that an interaction between an mRNA target and an sRNA pseudoknot, which requires chaperone-induced restricted unfolding, is reported. These observations come from structural and mutational analysis of ‘sRNA-mRNA’ duplexes, which indicate that the RydC 5′-end is involved in pairings with the csgD mRNA TIS. For pairing, helices H1 from RydC and H7 from the csgD mRNA should unfold. This is probably facilitated by Hfq, which interacts with both RydC (4) and csgD mRNA, to form a ternary complex with the two RNAs (Figure 4). The 5′-seeding between RydC 5′-accessible nucleotides and the csgD mRNA AUG codon is involved in pairing. Demonstrated previously by probing (4), RydC pseudoknot ‘breathing’ in solution opens helix H1 to promote pairing with the csgD mRNA, a transition facilitated by Hfq.
Pseudoknots are ingenious dynamic structural modules that can be temporarily unfolded (here with the assistance of a chaperone) to allow for antisense seed pairing and subsequent propagation. Two pseudoknots have already been detected and experimentally validated in another bacterial sRNA (29). In that case, they both contained an internal open reading frame that can only be translated under specific conditions. Bacterial sRNAs can act as antitoxic components in toxin–antitoxin systems, and an RNA pseudoknot was recently reported to inhibit and antagonize a harmful protein in the toxin–antitoxin pair (30). Antisense RNAs can modulate mRNA pseudoknot formation to control plasmid replication (31), indicating that pseudoknot structural plasticity can also be manipulated by chaperoned RNAs to control gene expression. In addition to their essential roles as cis-regulatory modules within mRNAs (31), including riboswitches (32), regulatory sRNAs pseudoknots are, when assisted by RNA chaperones, ingenious tools for efficient and reversible gene regulation processes in living organisms.
FUNDING
Agence Nationale pour la Recherche [ANR-09-MIEN-030-01 to B.F.]; Institut National de la Santé Et de la Recherche Médicale (INSERM); Ministère de l'Enseignement supérieur et de la Recherche. Funding for open access charge: INSERM.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are thankful to Prof. M. Chapman (University of Michigan) for the gift of the polyclonal antibodies to CsgA and CsgB, to Dr Hajnsdorf (IBPC, Paris) for providing plasmid pTE607 plasmid for Hfq expression and purification, to Dr M. Guillier (IBPC, Paris) and Dr M. Hallier (our laboratory) for critical reading of the manuscript and comments and to Dr S. Chabelskaya and A. Eyraud from our laboratory for providing SprD and purified ribosomes, respectively.
REFERENCES
- 1.Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell. 2011;43:880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sobrero P, Valverde C. The bacterial protein Hfq: much more than a mere RNA-binding factor. Crit. Rev. Microbiol. 2012;38:276–299. doi: 10.3109/1040841X.2012.664540. [DOI] [PubMed] [Google Scholar]
- 3.Soper TJ, Doxzen K, Woodson SA. Major role for mRNA binding and restructuring in sRNA recruitment by Hfq. RNA. 2011;17:1544–1550. doi: 10.1261/rna.2767211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antal M, Bordeau V, Douchin V, Felden B. A small bacterial RNA regulates a putative ABC transporter. J. Biol. Chem. 2005;280:7901–7908. doi: 10.1074/jbc.M413071200. [DOI] [PubMed] [Google Scholar]
- 5.Novikova M, Metlitskaya A, Datsenko K, Kazakov T, Kazakov A, Wanner B, Severinov K. The Escherichia coli Yej transporter is required for the uptake of translation inhibitor microcin C. J. Bacteriol. 2007;189:8361–8365. doi: 10.1128/JB.01028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eswarappa SM, Panguluri KK, Hensel M, Chakravortty D. The yejABEF operon of Salmonella confers resistance to antimicrobial peptides and contributes to its virulence. Microbiology. 2008;154:666–678. doi: 10.1099/mic.0.2007/011114-0. [DOI] [PubMed] [Google Scholar]
- 7.Ortega AD, Gonzalo-Asensio J, Garcia-del Portillo F. Dynamics of Salmonella small RNA expression in non-growing bacteria located inside eukaryotic cells. RNA Biol. 2012;9:469–488. doi: 10.4161/rna.19317. [DOI] [PubMed] [Google Scholar]
- 8.Frohlich KS, Papenfort K, Fekete A, Vogel J. A small RNA activates CFA synthase by isoform-specific mRNA stabilization. EMBO J. 2013;32:2963–2979. doi: 10.1038/emboj.2013.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 10.Romling U. Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell Mol. Life Sci. 2005;62:1234–1246. doi: 10.1007/s00018-005-4557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ogasawara H, Yamada K, Kori A, Yamamoto K, Ishihama A. Regulation of the Escherichia coli csgD promoter: interplay between five transcription factors. Microbiology. 2010;156:2470–2483. doi: 10.1099/mic.0.039131-0. [DOI] [PubMed] [Google Scholar]
- 12.Pesavento C, Becker G, Sommerfeldt N, Possling A, Tschowri N, Mehlis A, Hengge R. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev. 2008;22:2434–2446. doi: 10.1101/gad.475808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holmqvist E, Reimegard J, Sterk M, Grantcharova N, Romling U, Wagner EG. Two antisense RNAs target the transcriptional regulator CsgD to inhibit curli synthesis. EMBO J. 2010;29:1840–1850. doi: 10.1038/emboj.2010.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomason MK, Fontaine F, De Lay N, Storz G. A small RNA that regulates motility and biofilm formation in response to changes in nutrient availability in Escherichia coli. Mol. Microbiol. 2012;84:17–35. doi: 10.1111/j.1365-2958.2012.07965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mika F, Busse S, Possling A, Berkholz J, Tschowri N, Sommerfeldt N, Pruteanu M, Hengge R. Targeting of csgD by the small regulatory RNA RprA links stationary phase, biofilm formation and cell envelope stress in Escherichia coli. Mol. Microbiol. 2012;84:51–65. doi: 10.1111/j.1365-2958.2012.08002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jorgensen MG, Nielsen JS, Boysen A, Franch T, Moller-Jensen J, Valentin-Hansen P. Small regulatory RNAs control the multi-cellular adhesive lifestyle of Escherichia coli. Mol. Microbiol. 2012;84:36–50. doi: 10.1111/j.1365-2958.2012.07976.x. [DOI] [PubMed] [Google Scholar]
- 17.Boehm A, Vogel J. The csgD mRNA as a hub for signal integration via multiple small RNAs. Mol. Microbiol. 2012;84:1–5. doi: 10.1111/j.1365-2958.2012.08033.x. [DOI] [PubMed] [Google Scholar]
- 18.Kikuchi T, Mizunoe Y, Takade A, Naito S, Yoshida S. Curli fibers are required for development of biofilm architecture in Escherichia coli K-12 and enhance bacterial adherence to human uroepithelial cells. Microbiol. Immunol. 2005;49:875–884. doi: 10.1111/j.1348-0421.2005.tb03678.x. [DOI] [PubMed] [Google Scholar]
- 19.Wagner EG, Altuvia S, Romby P. Antisense RNAs in bacteria and their genetic elements. Adv. Genet. 2002;46:361–398. doi: 10.1016/s0065-2660(02)46013-0. [DOI] [PubMed] [Google Scholar]
- 20.Sakellaris H, Hannink NK, Rajakumar K, Bulach D, Hunt M, Sasakawa C, Adler B. Curli loci of Shigella spp. Infect. Immun. 2000;68:3780–3783. doi: 10.1128/iai.68.6.3780-3783.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barnhart MM, Chapman MR. Curli biogenesis and function. Annu. Rev. Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hammar M, Arnqvist A, Bian Z, Olsen A, Normark S. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol. Microbiol. 1995;18:661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x.. [DOI] [PubMed] [Google Scholar]
- 23.Collinson SK, Clouthier SC, Doran JL, Banser PA, Kay WW. Salmonella enteritidis agfBAC operon encoding thin, aggregative fimbriae. J. Bacteriol. 1996;178:662–667. doi: 10.1128/jb.178.3.662-667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammar M, Bian Z, Normark S. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc. Natl Acad. Sci. USA. 1996;93:6562–6566. doi: 10.1073/pnas.93.13.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arnqvist A, Olsen A, Normark S. Sigma S-dependent growth-phase induction of the csgBA promoter in Escherichia coli can be achieved in vivo by sigma 70 in the absence of the nucleoid-associated protein H-NS. Mol. Microbiol. 1994;13:1021–1032. doi: 10.1111/j.1365-2958.1994.tb00493.x. [DOI] [PubMed] [Google Scholar]
- 26.Salvail H, Caron MP, Belanger J, Masse E. Antagonistic functions between the RNA chaperone Hfq and an sRNA regulate sensitivity to the antibiotic colicin. EMBO J. 2013;32:2764–2778. doi: 10.1038/emboj.2013.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gottesman S, Storz G. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb. Perspect. Biol. 2011;3:pii: a003798. doi: 10.1101/cshperspect.a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr. Opin. Microbiol. 2007;10:134–139. doi: 10.1016/j.mib.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 29.Panja S, Woodson SA. Hfq proximity and orientation controls RNA annealing. Nucleic Acids Res. 2012;40:8690–8697. doi: 10.1093/nar/gks618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sayed N, Jousselin A, Felden B. A cis-antisense RNA acts in trans in Staphylococcus aureus to control translation of a human cytolytic peptide. Nat. Struct. Mol. Biol. 2012;19:105–112. doi: 10.1038/nsmb.2193. [DOI] [PubMed] [Google Scholar]
- 31.Brantl S. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr. Opin. Microbiol. 2007;10:102–109. doi: 10.1016/j.mib.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 32.Babitzke P, Baker CS, Romeo T. Regulation of translation initiation by RNA binding proteins. Annu. Rev. Microbiol. 2009;63:27–44. doi: 10.1146/annurev.micro.091208.073514. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.