Abstract
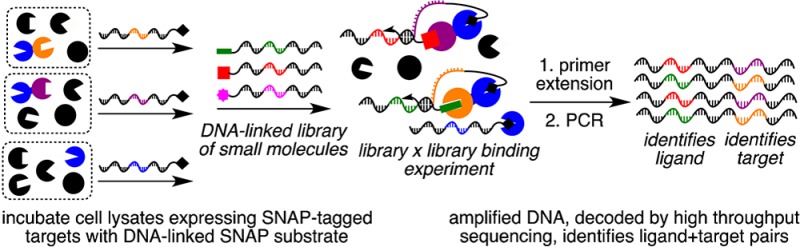
We describe the development and validation of interaction determination using unpurified proteins (IDUP), a method that selectively amplifies DNA sequences identifying ligand+target pairs from a mixture of DNA-linked small molecules and unpurified protein targets in cell lysates. By operating in cell lysates, IDUP preserves native post-translational modifications and interactions with endogenous binding partners, thereby enabling the study of difficult-to-purify targets and increasing the potential biological relevance of detected interactions compared with methods that require purified proteins. In IDUP, target proteins are associated with DNA oligonucleotide tags either non-covalently using a DNA-linked antibody or covalently using a SNAP-tag. Ligand–target binding promotes hybridization of a self-priming hairpin that is extended by a DNA polymerase to create a DNA strand that contains sequences identifying both the target and its ligand. These sequences encoding ligand+target pairs are selectively amplified by PCR and revealed by high-throughput DNA sequencing. IDUP can respond to the effect of affinity-modulating adaptor proteins in cell lysates that would be absent in ligand screening or selection methods using a purified protein target. This capability was exemplified by the 100-fold amplification of DNA sequences encoding FRB+rapamycin or FKBP+rapamycin in samples overexpressing both FRB and FKBP (FRB·rapamycin+FKBP, Kd ≈ 100 fM; FKBP·rapamycin+FRB, Kd = 12 nM). In contrast, these sequences were amplified 10-fold less efficiently in samples overexpressing either FRB or FKBP alone (rapamycin+FKBP, Kd ≈ 0.2 nM; rapamcyin+FRB, Kd = 26 μM). Finally, IDUP was used to process a model library of DNA-linked small molecules and a model library of cell lysates expressing SNAP-target fusions combined in a single sample. In this library×library experiment, IDUP resulted in enrichment of sequences corresponding to five known ligand+target pairs ranging in binding affinity from Kd = 0.2 nM to 3.2 μM out of 67,858 possible combinations, with no false positive signals enriched to the same extent as that of any of the bona fide ligand+target pairs.
Introduction
Advances in genomic and proteomic studies continue to reveal new targets for therapeutic intervention. The identification of ligands for such targets remains a major opportunity and challenge. To this end, a variety of target-oriented ligand-binding assays have been developed, including affinity selections on DNA-encoded chemical libraries,1,2 selection-like methods such as interaction-dependent PCR,3 and a wide variety of screening platforms.4 Selections offer substantially improved throughput and decreased time, cost, and material consumption compared to screens, but they generally rely on purified, heterologously expressed proteins in an artificial context that includes an immobilized1 or DNA-linked3 protein, the compound library, and buffer. Selections conducted in this manner can be incompatible with poorly soluble, aggregation-prone, difficult-to-purify, intrinsically disordered, or membrane-bound targets. Moreover, the results of selections on immobilized targets may lack biological relevance for proteins that adopt non-native conformations or lack binding partners or cofactors essential for their function when taken out of the cellular context.5 Although successful selections have been conducted using purified proteins,1 increasing the biological relevance of selection methods will significantly increase their effectiveness. Here we report the development and validation of interaction determination using unpurified proteins (IDUP), a method to rapidly identify ligand+target pairs from one-pot mixtures of DNA-linked ligands and unpurified protein targets in cell lysates (Figure 1A).
Figure 1.
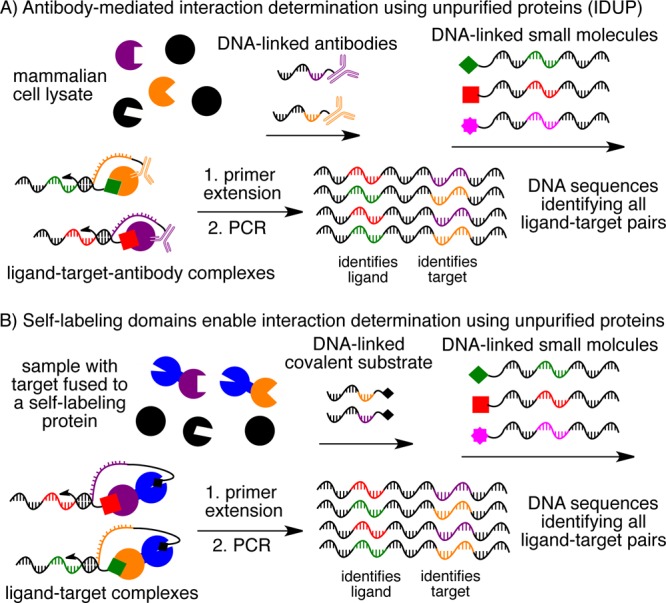
(A) Antibody-mediated interaction determination using unpurified proteins (IDUP) uses DNA-linked antibodies to recognize a target protein or epitope tag. (B) Alternatively, a covalent bond can be formed between the target and identifying DNA strand in IDUP by fusing the target to a self-labeling protein tag such as SNAP-tag, CLIP-tag, or HaloTag. After primer extension and PCR, the resulting DNA encodes all ligand+target combinations.
IDUP is triggered by the formation of a ternary complex involving a DNA-linked ligand, a target protein, and a DNA oligonucleotide that identifies the target protein. The association of the target protein with its corresponding DNA oligonucleotide can be established either non-covalently using a DNA-linked antibody (Figure 1A) or covalently using a self-labeling protein that reacts with a DNA-linked small molecule (Figure 1B). Formation of this ternary complex is dependent on ligand–target binding and promotes hybridization of short complementary regions on the target- and ligand-linked oligonucleotides. A DNA polymerase can then extend this hybridized region to generate a double-stranded product that contains DNA sequences identifying both the target and its bound ligand. This extension product contains two primer-binding sites and therefore can be amplified by PCR (Figure 1).3 By removing the requirement for purified protein targets, the IDUP approach enables ligand-binding “selections” to be performed on proteins that are free to undergo post-translational modification, interact with endogenous accessory proteins and metabolites, and access physiologically relevant conformational states.5
Results and Discussion
Pairs of DNA-linked antibodies have been used to measure the presence of proteins and protein–protein complexes by the proximity ligation assay6 and the proximity extension assay.7 On the basis of these concepts and our previous development of interaction-dependent PCR (IDPCR),3 we speculated that formation of a ternary complex of a DNA-linked antibody, a protein target, and a DNA-linked small molecule could promote hybridization of the linked oligonucleotides and enable primer extension by a DNA polymerase in a manner dependent on binding of the ligand to the target (Figure 1A). Such a system would offer the benefits of IDPCR but without the significant limitation of requiring a purified target protein conjugated to a DNA oligonucleotide.
To test this hypothesis, quantitative PCR (qPCR) was used to compare the amount of primer extension product from reactions containing a DNA-linked anti-streptavidin antibody (DNA-αSA), streptavidin protein (SA), and a DNA-linked ligand. The DNA-linked ligand was varied among four small molecules: DNA-linked hexylamine (DNA-amine, no significant affinity for SA), DNA-linked Gly-Leu-carboxybenzene sulfonamide (DNA-GLCBS, no significant affinity for SA), and either one of two ligands of SA, DNA-desthiobiotin (Kd = 2 nM)8 or DNA-biotin (Kd = 40 pM).9 Consistent with formation of an antibody+SA+biotin complex or an antibody+SA+desthiobiotin complex, samples containing DNA-αSA, SA, and either DNA-biotin or DNA-desthiobiotin were amplified much more quickly than those containing DNA-amine, containing DNA-GLCBS, or lacking SA, resulting in a qPCR cycle threshold (CT) difference of five cycles (ΔCT = 5, corresponding to a 32-fold difference in effective template availability; Supporting Information, Figure S1). Together, these results demonstrate the ability of an antibody+protein+ligand ternary complex to trigger the selective amplification of a DNA sequence identifying the protein and ligand.
Because potential applications of IDUP include selections on DNA-encoded chemical libraries, we next asked whether formation of an αSA+SA+biotin complex would result in selective amplification of the DNA sequence encoding SA+biotin when DNA-biotin was present in a mock library containing an excess of DNA-GLCBS. After primer extension, PCR, restriction digestion, and polyacrylamide gel electrophoresis (PAGE) of samples containing DNA-αSA, SA, and mixtures containing a 1:10, 1:100, or 1:1000 ratio of DNA-biotin/DNA-GLCBS, we found that the sequence corresponding to SA+biotin was enriched ∼10-fold (Figure S1). This relatively modest enrichment of the SA+biotin DNA sequence suggested that an improvement in the signal-to-noise ratio of IDUP would be required to enable enrichment of sequences corresponding to interactions with affinities weaker than that of SA+biotin.
To optimize the effectiveness of IDUP, we investigated key aspects of the primer extension step using the related and previously validated IDPCR system, in which the purified target protein is covalently preconjugated to an identifiable DNA oligonucleotide.3 In a series of model library×library experiments with 258 DNA-linked proteins and 260 DNA-linked ligands, we systematically varied parameters of the DNA extension step using the DNA polymerase Klenow exo– and found that varying primer extension conditions did not substantially improve enrichment factors for sequences corresponding to weaker interactions (Figures S2 and S3). We next performed model library×library experiments using other mesophilic polymerases, and consistent with a previous report describing the ability of polymerases with 3′-exonuclease activity to increase the signal-to-noise ratio of the proximity extension assay,7 we found that IDPCR with T4 DNA polymerase and a complementary region of 8-nt or 9-nt resulted in 4.5- to 140-fold improvements in the enrichment of DNA sequences corresponding to ligand+target pairs with binding affinities ranging from 40 nM to 13 μM (Figures S4–S6).
Next we tested whether these improvements would apply to IDUP and enable the enrichment of sequences corresponding to known binding interactions from a large excess of nonbinding entities (Figure 2A). We incubated mixtures containing 1:10, 1:100, or 1:1000 ratios of DNA-biotin/DNA-GLCBS with DNA-αSA and SA. Because we are interested in the ability of IDUP to detect protein+ligand binding in a complex mixture, such as a cell lysate, we also added HeLa cell lysate to the solution so that SA was present at 0.01 wt% relative to the total protein content of the HeLa cell lysate, an amount that is representative of the endogenous expression level of members of protein classes of interest such as MAP kinases, histone deacetylases, Ras-related proteins, and isocitrate dehydrogenases.10,11 After IDUP, restriction digestion, and PAGE analysis, we observed ∼1000-fold enrichment of the sequence corresponding to SA+biotin. Replacing Klenow exo– with T4 DNA polymerase and replacing a 6-nt complementary region with an 8-nt complementary region resulted in a 100-fold improvement in enrichment of the sequence encoding SA+biotin (Figures 2B,C and S1).
Figure 2.
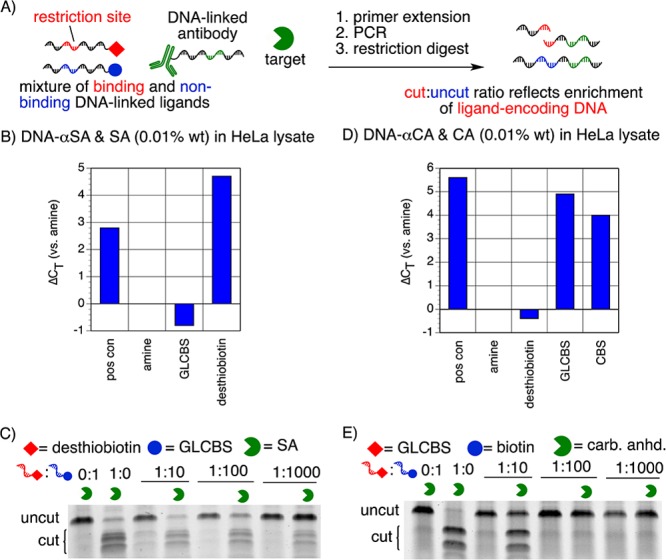
(A) The ability of IDUP to enrich the sequence corresponding to a particular interaction is evaluated by incubating mixtures of binding and nonbinding DNA-linked ligands with a target protein and a DNA-linked antibody. After primer extension and PCR, a restriction digest is used to determine the fraction of the amplified sequences corresponding to the target+ligand interaction. (B) IDUP with DNA-αSA and 0.01% SA in HeLa lysate shows selective amplification of a sequence corresponding to SA+desthiobiotin in qPCR (ΔCT = 4.7). (C) IDUP on a mock library containing mixtures of DNA-desthiobiotin and DNA-GLCBS shows ∼1000-fold enrichment of a sequence corresponding to SA+desthiobiotin. (D) When analyzed by qPCR, IDUP with DNA-αCAII and 0.01% CAII in HeLa lysate shows rapid amplification of sequences corresponding to CA+GLCBS and CA+CBS, but not CA+desthiobiotin or CA+amine (ΔCT = 4–5). (E) IDUP on a mock library containing mixtures of DNA-GLCBS and DNA-desthiobiotin shows ∼10-fold enrichment of a sequence corresponding to CAII+GLCBS.
To determine whether IDUP using T4 DNA polymerase could enrich DNA sequences corresponding to weaker ligand–target interactions, we similarly studied the interaction between carbonic anhydrase II (CAII) and its ligand GLCBS (Kd = 40 nM)12 and observed 10-fold enrichment of the sequence corresponding to CAII+GLCBS (Figure 2D,E), despite the observation that the polyclonal antibody used to generate DNA-αCAII appears to partially compete for ligand binding, likely reducing the enrichment obtained by IDUP using DNA-αCAII (Figure S7). Together, these results demonstrate that, for targets for which suitable antibodies exist, antibody-mediated IDUP provides a selection-like method for the detection and reporting of small molecule–protein interactions from cell lysates.
For some targets of interest, an antibody capable of selectively binding the target in solution without obscuring the ligand-binding site may be difficult to obtain. Because oligohistidine is a rare sequence among naturally occurring proteins,13 we expected that an antibody against the His6 epitope tag would be less likely to interfere with target protein function, including ligand binding. We therefore investigated the ability of an anti-His5 antibody (Qiagen, His6+αHis Kd = 1–50 nM)14 linked to DNA (DNA-αHis) to participate in IDUP with His6-tagged target proteins (Figure 3A). We performed IDUP with DNA-αHis and purified, C-terminally His6-tagged CAII (CAII-His6, 0.01% in HeLa cell lysate) and observed that using DNA-αHis resulted in 100-fold enrichment of DNA encoding CAII+GLCBS, representing a 10-fold improvement over the enrichment factor using DNA-αCAII (Figure S8). These results demonstrate the feasibility of IDUP mediated by an epitope tag-binding antibody instead of an antibody that directly binds the target protein’s coding sequence.
Figure 3.
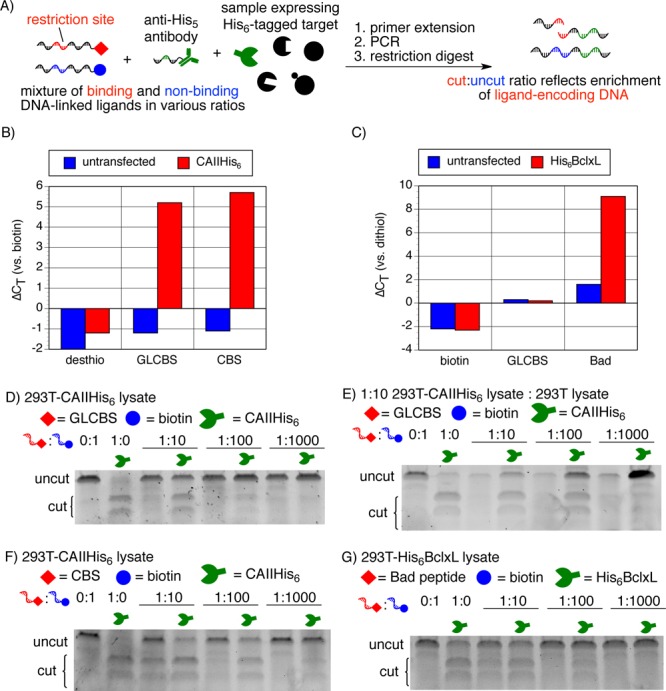
(A) IDUP using DNA-αHis and genetically encoded His6-tagged target proteins. (B) IDUP using DNA-αHis with 293T cell lysate expressing CAII-His6 shows rapid amplification of sequences corresponding to CA+GLCBS and CA+CBS but not CA+desthiobiotin (ΔCT = 5–6). (D) IDUP using a mock library shows ∼10-fold enrichment of the sequence corresponding to CAII+GLCBS and (F) ∼100-fold enrichment of a sequence corresponding to CAII+CBS. (E) When the transfected lysate was diluted 1:10 into untransfected lysate, the enrichment of the CAII+GLCBS sequence increased to ∼100-fold. (C) IDUP using DNA-αHis with 293T cell lysate expressing His6-BclxL shows rapid amplification of DNA-Bad but not DNA-GLCBS or DNA-biotin (ΔCT = 8) and ∼100-fold enrichment of a sequence corresponding to Bcl-xL+Bad (G).
To assess the compatibility of IDUP with unpurified, genetically encoded targets, we transiently transfected HEK-293T cells with a plasmid expressing CAII-His6. We performed an IDUP enrichment experiment in the resulting cell lysate using DNA-αHis and observed ∼10-fold enrichment of the sequence corresponding to CAII+GLCBS (Figure 3B,D). When we similarly performed IDUP using mixtures of DNA-CBS/DNA-biotin, we observed ∼100-fold enrichment of the sequence corresponding to CAII+CBS (Figure 3F). Enrichment by antibody-mediated IDUP depends on formation of a ternary complex of DNA–antibody+target+ligand–DNA. According to a recent model of three-body binding,15 for αHis-mediated IDUP (His+αHis, Kd = 10 nM), a target protein concentration of 30 nM is optimal for CAII+GLCBS (Kd = 40 nM),12 but the optimal target concentration is 190 nM for CAII+CBS (Kd = 3.2 μM).16,15 By Western blot, we determined that the concentration of CAII-His6 in the 293T cell lysate was ∼300 nM, corresponding to ∼130 nM in the IDUP assay (Figure S9). Consistent with the model of three-body binding,15 we observed optimal IDUP enrichment of the sequence corresponding to CAII+GLCBS (∼100-fold) when the 293T cell lysate transfected with a CAII-His6 expression plasmid was diluted 1:10 into untreated 293T cell lysate (Figures 3E and S9). The enrichment of the sequence corresponding to CAII+CBS decreased to ∼10-fold in this diluted lysate sample (Figure S9).
IDUP using DNA-αHis also resulted in ∼100-fold enrichment of a sequence corresponding to Bcl-xL+Bad (Kd = 0.6 nM)17 when performed in lysates of 293T cells transfected with a plasmid expressing His6-Bcl-xL (amino acids 1–212) (Figure 3G). Taken together, these results suggest that IDUP using the DNA-αHis antibody can enrich sequences corresponding to ligand+target combinations for unpurified targets in cell lysates.
Covalent protein–DNA linkages offer several potential advantages during IDUP compared to non-covalent antibody–target or antibody–tag associations. We anticipated that formation of a covalent bond between a target protein and its identifying DNA might increase the stability of the DNA–target entity and the sensitivity of IDUP for weaker small molecule–target binding interactions. In principle, replacing non-covalent antibody–target binding with a covalent linkage can be accomplished by expressing the target protein as a fusion to a self-labeling protein domain such as SNAP-tag,18,19 CLIP-tag,20 or HaloTag.21 Moreover, the use of a small-molecule-reactive tag removes the requirement for a non-covalent ternary complex to form, and therefore reduces the assay’s dependence on target protein concentration.15 Finally, the small size of self-labeling proteins compared to antibodies (∼30 kDa vs ∼150 kDa) suggests that the former are less likely to obscure ligand-binding sites or disrupt native protein–protein interactions. Self-labeling proteins have been used successfully in protein–ligand binding assays19,20 and in linking target proteins to DNA.22 We therefore speculated that self-labeling proteins expressed as fusions to target proteins might serve as effective reagents for linking targets to DNA during IDUP.
We transiently transfected 293T cells with vectors expressing N- or C-terminally SNAP-tagged CAII (SNAP-CAII or CAII-SNAP). The resulting lysates were individually incubated with a SNAP substrate, O6-benzylguanine (BG), linked to DNA (DNA-BG) for 15 min before incubation with mixtures of DNA-GLCBS/DNA-desthiobiotin or DNA-CBS/DNA-desthiobiotin. After IDUP and restriction digestion, we observed ∼100-fold enrichment of the sequences encoding CAII+GLCBS and CAII+CBS in samples expressing SNAP-CAII or CAII-SNAP, but not in untransfected samples or samples expressing SNAP-tag alone (Figures 4B,D and S10).
Figure 4.
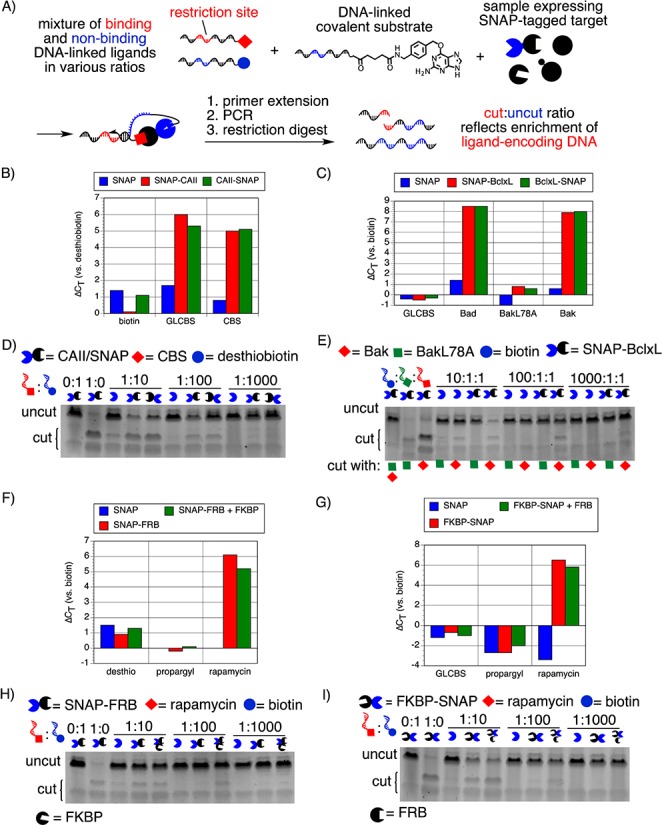
(A) IDUP in cell lysates expressing a SNAP-tagged target protein. DNA was rapidly amplified in samples corresponding to known interactions: (B) SNAP/CAII+(GL)CBS (ΔCT = 5–6), (C) SNAP/Bcl-xL+Bad or Bak (ΔCT = 8–9), (F) SNAP-FRB+rapamycin (ΔCT = 6), and (G) FKBP-SNAP+rapamycin (ΔCT = 6). (D) A sequence corresponding to CAII+CBS was enriched ∼100-fold in a sample expressing either SNAP-CAII (lane 7) or CAII-SNAP (lane 8). (E) In samples expressing SNAP-Bcl-xL, a sequence corresponding to the interaction between Bcl-xL+Bak was enriched ∼100-fold (lane 11), but no enrichment was observed for a sequence corresponding to BclxL+BakL78A, a weakly binding mutant of the Bak peptide (lane 9). (H) Overexpression of FKBP with SNAP-FRB increased the enrichment of a sequence encoding FRB+rapamycin by 10-fold compared to a sample transfected with SNAP-FRB alone. (I) Overexpression of FRB with FKBP-SNAP also increased the enrichment of a sequence corresponding to FKBP+rapamycin by 10-fold compared to a sample transfected with FKBP-SNAP alone.
Similarly, IDUP performed on 293T cell lysate expressing SNAP-Bcl-xL or Bcl-xL-SNAP with DNA-BG and mixtures of DNA-Bak/DNA-BakL78A/DNA-biotin resulted in ∼100-fold enrichment of a sequence corresponding to Bcl-xL+Bak (Kd = 340 nM)23 but no enrichment of the DNA sequence encoding an interaction between Bcl-xL and the closely related negative control peptide BakL78A (Kd = 270 μM).23 (Figures 4C,E and S10). Collectively, these results demonstrate the ability of protein targets fused to self-labeling domains to participate in IDUP. In contrast to our results with αHis-mediated IDUP, we noticed that both N- and C-terminally SNAP-tagged proteins resulted in roughly equivalent enrichment levels of DNA encoding known ligand–target pairs, suggesting that the SNAP-tag also offers increased generality compared to the His6-tag+αHis approach.
The ability of IDUP to evaluate ligand–protein binding in complex mixtures enables the detection of interactions that require exogenous factors. For example, the interaction of rapamycin with FRB, the rapamycin-binding domain of mTOR, is substantially increased in the presence of another rapamycin-binding protein, FKBP; the Kd of rapamcyin+FRB is 26 μM, while the Kd of FKBP·rapamycin+FRB is 12 nM.24 We wondered whether the FKBP-dependent modulation in the strength of the FRB+rapamycin interaction could be detected by IDUP. We conjugated azide-linked rapamycin24 to DNA using the Cu(I)-catalyzed azide–alkyne cycloaddition reaction.25 When we performed IDUP using 293T cell lysates overexpressing SNAP, SNAP-FRB, or SNAP-FRB and FKBP, we observed ∼10-fold enrichment of the sequence corresponding to FRB+rapamycin in the sample overexpressing SNAP-FRB and ∼100-fold enrichment in the sample overexpressing SNAP-FRB and FKBP (Figure 4F,H). Similar results were obtained with recombinant, preconjugated DNA-FRB (Figure S11). The rapamycin·FRB complex also has a higher affinity for FKBP (Kd ≈ 100 fM) than rapamycin alone (Kd ≈ 0.2 nM).24 293T cell lysate overexpressing FRB and FKBP-SNAP also showed ∼10-fold greater enrichment for a sequence encoding FKBP+rapamycin than a sample overexpressing FKBP-SNAP alone (Figure 4G,I). Together these results demonstrate that IDUP results can reflect the influence of accessory proteins on the target affinity of small-molecule ligands.
Because a key advantage of IDUP is the ability to simultaneously assay all interactions between combined libraries of targets and ligands in a single solution, we next tested the ability of IDUP to selectively enrich known target+ligand interactions from a model library containing 262 DNA-linked ligands (comprising DNA-linked GLCBS, CBS, rapamycin, Bad, Bak, BakL78A, and hexylamine linked to a set of 256 DNA sequences) and 259 DNA-linked targets. To generate a library of 259 DNA-linked targets, lysates from 293T cells previously transfected with vectors encoding SNAP-FKBP, SNAP-CA, SNAP-Bcl-xL, or SNAP were individually incubated with BG linked to unique DNA sequences (in the case of SNAP-FKBP, SNAP-CA, or SNAP-Bcl-xL) or to a library of 256 sequences (in the case of SNAP alone), quenched with a free BG derivative, and pooled to obtain an equimolar ratio of the 259 DNA-linked targets. As a control, aliquots of the same lysates were separately incubated with DNA sequences lacking conjugated BG. Both samples were incubated with the library of DNA-linked ligands and processed by primer extension, PCR, and high-throughput DNA sequencing using conditions identified in previous experiments (Figures S2 and S6). We divided the number of sequence counts for each protein+ligand sequence from the sample treated with DNA-BG by the corresponding number of counts from the sample treated with DNA alone and observed enrichment factors from 68.9 to 328.7 for sequences corresponding to all five known ligand–target binding interactions, including FKBP+rapamycin, CAII+GLCBS, CAII+CBS, Bcl-xL+Bad, and Bcl-xL+Bak (Figure 5). The mean enrichment for all 67,858 possible ligand+target sequences was 1.5. We observed strong enrichment of the sequences corresponding to all of the known target+ligand interactions, despite the fact that the corresponding dissociation constants vary over 4 orders of magnitude (Kd ≈ 0.2 nM–3.2 μM). No sequences corresponding to any presumed nonbinding interactions were enriched greater than 31-fold, and only 21 presumed false-positive sequences had enrichment factors greater than 20, a signal level less than one-third that of the weakest bona fide positive (Tables S1 and S2).
Figure 5.
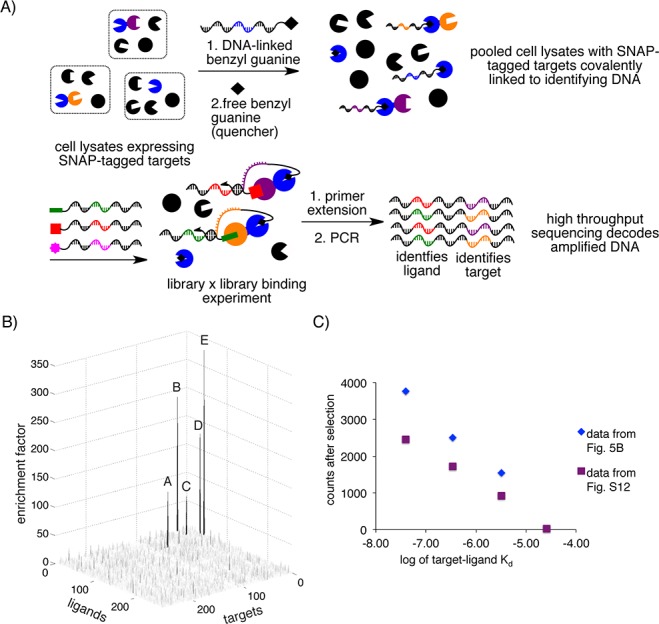
(A) Cell lysates expressing SNAP-CA, SNAP-Bcl-xL, and SNAP-FKBP were individually labeled with one of three DNA sequences and combined with a cell lysate expressing SNAP and labeled with 256 DNA sequences. The pooled lysates were combined with a library of 262 DNA-linked small molecules, including DNA-linked GLCBS, CBS, Bad, Bak, BakL78A, and rapamycin, for a model library×library IDUP “selection”. (B) IDUP using a library of cell lysates expressing SNAP-target fusions identified all five known target+ligand pairs, including A - FKBP+rapamycin, B - Bcl-xL+Bad, C - Bcl-xL+Bak, D - CAII+GLCBS, and E - CAII+CBS, despite having affinities from 0.2 nM to 3.2 μM. (C) For interactions with Kd = 40 nM–26 μM, we observed a strong relationship between the log of target–ligand Kd and the number of sequence counts after selection.
When we performed a similar IDUP experiment containing 260 DNA-linked targets, including both SNAP-FKBP and FRB-SNAP, we observed all of the known interactions except for that of FRB+rapamycin, likely due to both the relatively weak affinity of rapamycin for FRB (Kd = 26 μM)24 and the relatively low expression level of FRB-SNAP (Figure S12). In an IDUP experiment containing FRB-SNAP but lacking SNAP-FKBP, we observed 23.4-fold enrichment of the sequence corresponding to rapamycin+FRB, with only three presumed false positive sequences enriched as strongly (Figure S12). Together, these results validate the ability of IDUP to identify interactions between combined libraries of small molecules and SNAP-tagged target proteins in crude cell lysates.
The relationship between target–ligand affinity and sequence counts following IDUP should be governed by several factors, including concentrations of individual library members, differences in the expression levels of SNAP-tagged targets, and differences in the extent to which both DNA-linked ligands and SNAP-tagged targets are obscured by factors present in the cell lysate. For example, 293T cells have been shown to natively express FK506-binding proteins,28 mTor,29 CAII,26 and BclxL,27 and these untagged targets may compete for ligand binding. Despite these potential complications, for interactions with dissociation constants from 40 nM to 26 μM, we observed a surprisingly strong relationship between log(Kd) and the number of counts observed after selection (Figures 5C and S13). In principle, this relationship can be used to estimate the detection limit of the IDUP assay (here 30–60 μM; see Figure S13B) and to infer the affinities of newly detected interactions.
The relationship between affinity and enrichment factor was less strong than that between affinity and sequence counts. Enrichment factor values also depend on the number of sequence counts in the negative control sample, which are generally smaller (despite 12- to 18-fold sequence coverage) and more susceptible to variation caused by DNA sequence bias and sampling stochasticity during PCR or high-throughput sequencing. Plotting enrichment factors is, however, an effective way to distinguish true binding events from presumed false positives by eliminating sequences likely amplified due to PCR bias (Figure S13C,D). Interactions with Kd ≈ 0.2–0.6 nM did not follow the linear trend (Figure S13A). A plausible explanation for this observation is that the concentration of each library member during IDUP is 0.4 nM, and thus interactions with affinities in this range (here, FKBP+rapamycin and Bcl-xL+Bad) could approach binding saturation.
Conclusions
IDUP is a method for rapidly evaluating potential small-molecule–target interactions from mixtures in a single solution that is compatible with unpurified targets in biological samples. The ability to identify ligand+target pairs from complex samples including cell lysates offers significant advantages compared to other methods for evaluating DNA-encoded chemical libraries. Samples in cell lysates are able to undergo native post-translational modification and interact with accessory proteins and metabolites in ways that better reflect their relevant biological environment. Because IDUP is compatible with crude cell lysates, difficult-to-purify, poorly soluble, intrinsically unstable, and aggregation-prone targets may also be compatible with this method, without requiring truncation or other strategies used to promote heterologous expression. We demonstrated the ability of IDUP to selectively amplify DNA sequences corresponding to interactions between His6-tagged or SNAP-tagged target proteins and their ligands in cell lysates, and we have demonstrated that IDUP results reflect the ability of accessory proteins to modulate ligand–target affinity. We also demonstrated the ability of IDUP to selectively enrich DNA sequences corresponding to known protein+ligand interactions with affinities from 0.2 nM to 26 μM from a library of SNAP-target-expressing cell lysates and a library of DNA-linked small molecules. Moreover, we observed a relationship between sequence counts and ligand–target identity, suggesting that IDUP sequencing results may be able to not only identify new interactions but also estimate their affinities. We anticipate that IDUP will provide a general and highly efficient strategy to evaluate DNA-encoded libraries under conditions in which purified protein targets are unavailable or differ in important ways from their native cellular counterparts.
Acknowledgments
This work was supported by the Howard Hughes Medical Institute and the NIH/NIGMS (R01GM065865). L.M.M. gratefully acknowledges fellowships from the NSF GRFP and the Harvard Graduate Society. Plasmids expressing CAII and Bcl-xL were the kind gifts from Carol Fierke and Loren Walensky, respectively. We thank Rick McDonald and Ryan Hili for many helpful discussions.
Supporting Information Available
Additional information and experimental methods. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare the following competing financial interest(s): D.R.L. is a consultant for Ensemble Therapeutics, a company that uses DNA-templated synthesis and in vitro selection for drug discovery.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Reviewed in the following:; a Kleiner R. E.; Dumelin C. E.; Liu D. R. Chem. Soc. Rev. 2011, 40, 5707. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Mannocci L.; Leimbacher M.; Wichert M.; Scheuermann J.; Neri D. Chem. Commun. 2011, 47, 12747. [DOI] [PubMed] [Google Scholar]; c Clark M. A. Curr. Opin. Chem. Biol. 2010, 14, 396. [DOI] [PubMed] [Google Scholar]
- a Buller F.; Steiner M.; Frey K.; Mircsof D.; Scheuermann J.; Kalisch M.; Buhlmann P.; Supuran C. T.; Neri D. ACS Chem. Biol. 2011, 6, 336. [DOI] [PubMed] [Google Scholar]; b Kleiner R. E.; Dumelin C. E.; Tiu G. C.; Sakurai K.; Liu D. R. J. Am. Chem. Soc. 2010, 132, 11779. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Clark M. A.; Acharya R. A.; Arico-Muendel C. C.; Belyanskaya S. L.; Benjamin D. R.; Carlson N. R.; Centrella P. A.; Chiu C. H.; Creaser S. P.; Cuozzo J. W.; Davie C. P.; Ding Y.; Franklin G. J.; Franzen K. D.; Gefter M. L.; Hale S. P.; Hansen N. J. V.; Israel D. I.; Jiang J.; Kavarana M. J.; Kelley M. S.; Kollmann C. S.; Li F.; Lind K.; Mataruse S.; Medeiros P. F.; Messer J. A.; Myers P.; O’keefe H.; Oliff M. C.; Rise C. E.; Satz A. L.; Skinner S. R.; Svendsen J. L.; Tang L.; Van Vloten K.; Wagner R. W.; Yao G.; Zhao B.; Morgan B. A.. Nat. Chem. Biol. 2009, 5, 647. [DOI] [PubMed] [Google Scholar]; d Wrenn S. J.; Weisinger R. M.; Halpin D. R.; Harbury P. B. J. Am. Chem. Soc. 2007, 129, 13137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor L. M.; Gorin D. J.; Dumelin C. E.; Liu D. R. J. Am. Chem. Soc. 2010, 132, 15522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Inglese J.; Johnson R. L.; Simeonov A.; Xia M.; Zheng W.; Austin C. P.; Auld D. S. Nat. Chem. Biol. 2007, 3, 466. [DOI] [PubMed] [Google Scholar]; b Scott D. E.; Coyne A. G.; Hudson S. A.; Abell C. Biochemistry 2012, 51, 4990. [DOI] [PubMed] [Google Scholar]
- Good M. C.; Zalatan J. G.; Lim W. A. Science 2011, 332, 680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Fredriksson S.; Gullberg M.; Jarvius J.; Olsson C.; Pietras K.; Gustafsdottir S. M.; Ostman A.; Landegren U. Nat. Biotechnol. 2002, 20, 473. [DOI] [PubMed] [Google Scholar]; b Soderberg O.; Gullberg M.; Jarvius M.; Ridderstrale K.; Leuchowius K. J.; Jarvius J.; Wester K.; Hydbring P.; Bahram F.; Larsson L. G.; Landegren U. Nat. Methods 2006, 3, 995. [DOI] [PubMed] [Google Scholar]; c Hammond M.; Nong R. Y.; Ericsson O.; Pardali K.; Landegren U. PLoS One 2012, 7, e40405. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Gustafsdottir S. M.; Wennstrom S.; Fredriksson S.; Schallmeiner E.; Hamilton A. D.; Sebti S. M.; Landegren U. Clin. Chem. 2008, 54, 1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg M.; Eriksson A.; Tran B.; Assarsson E.; Fredriksson S. Nucleic Acids Res. 2011, 39, e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumelin C. E.; Scheuermann J.; Melkko S.; Neri D. Bioconjugate Chem. 2006, 17, 366. [DOI] [PubMed] [Google Scholar]
- Green N. M. Methods Enzymol. 1990, 184, 51. [DOI] [PubMed] [Google Scholar]
- Schwanhausser B.; Busse D.; Li N.; Dittmar G.; Schuchhardt J.; Wolf J.; Chen W.; Selbach M. Nature 2011, 437, 337. [DOI] [PubMed] [Google Scholar]
- Wang F.; Travins J.; DeLaBarre B.; Penard-Lacronique V.; Schalm S.; Hansen E.; Straley K.; Kernytsky A.; Liu W.; Gliser C.; Yang H.; Gross S.; Artin E.; Saada V.; Mylonas E.; Quivoron C.; Popovici-Muller J.; Saunders J. O.; Salituro F. G.; Yan S.; Murray S.; Wei W.; Gao Y.; Dang L.; Dorsch M.; Agresta S.; Schenkein D. P.; Biller S. A.; Su S. M.; de Botton S.; Yen K. E. Science 2013, 340, 622. [DOI] [PubMed] [Google Scholar]
- Mincione F.; Starnotti M.; Menabuoni L.; Scozzafava A.; Casini A.; Supuran C. T. Bioorg. Med. Chem. Lett. 2001, 11, 1787. [DOI] [PubMed] [Google Scholar]
- Fritze C. E.; Anderson T. R. Methods Enzymol. 2000, 327, 3. [DOI] [PubMed] [Google Scholar]
- QIAexpress Detection and Assay Handbook. http://www.qiagen.com (accessed September 19, 2012).
- Douglass E. F. Jr.; Miller C. J.; Sparer G.; Shapiro H.; Spiegel D. A. J. Am. Chem. Soc. 2013, 135, 6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West G. M.; Tang L.; Fitzgerald M. C. Anal. Chem. 2008, 80, 4175. [DOI] [PubMed] [Google Scholar]
- Petros A. M.; Nettesheim D. G.; Wang Y.; Olejniczak E. T.; Meadows R. P.; Mack J.; Swift K.; Matayoshi E. D.; Zhang H.; Thompson C. B.; Fesik S. W. Protein Sci. 2000, 9, 2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollwitz B.; Brunk E.; Schmitt S.; Pojer F.; Bannwarth M.; Schiltz M.; Rothlisberger U.; Johnsson K. Biochemistry 2012, 51, 986. [DOI] [PubMed] [Google Scholar]
- Chidley C.; Haruki H.; Pedersen M. G.; Muller E.; Johnsson K. Nat. Chem. Biol. 2011, 7, 375. [DOI] [PubMed] [Google Scholar]
- Haruki H.; Gonzalez M. R.; Johnsson K. PLoS One 2012, 7, e37598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encell L. P.; Friedman Ohana R.; Zimmerman K.; Otto P.; Vidugiris G.; Wood M. G.; Los G. V.; McDougall M. G.; Zimprich C.; Karassina N.; Learish R. D.; Hurst R.; Hartnett J.; Wheeler S.; Stecha P.; English J.; Zhao K.; Mendez J.; Benink H. A.; Murphy N.; Daniels D. L.; Slater M. R.; Urh M.; Darzins A.; Klaubert D. H.; Bulleit R. F.; Wood K. V. Curr. Chem. Genomics 2012, 6, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Niemeyer C. M. Angew. Chem., Int. Ed. 2010, 49, 1200. [DOI] [PubMed] [Google Scholar]; b Sacca B.; Meyer R.; Erkelenz M.; Kiko K.; Arndt A.; Schroeder H.; Rabe K. S.; Niemeyer C. M. Angew. Chem., Int. Ed. 2010, 49, 9378. [DOI] [PubMed] [Google Scholar]
- Sattler M.; Liang H.; Nettesheim D.; Meadows R. P.; Harlan J. E.; Eberstadt M.; Yoon H. S.; Shuker S. B.; Chang B. S.; Minn A. J.; Thompson C. B.; Fesik S. W. Science 1997, 275, 983. [DOI] [PubMed] [Google Scholar]
- Banaszynski L. A.; Liu C. W.; Wandless T. J. J. Am. Chem. Soc. 2005, 127, 4715. [DOI] [PubMed] [Google Scholar]
- El-Sagheer A. H.; Brown T. Chem. Soc. Rev. 2010, 39, 1388. [DOI] [PubMed] [Google Scholar]
- Sterling D.; Brown N. J.; Supuran C. T.; Casey J. R. Am. J. Physiol. Cell Physiol. 2002, 283, C1522. [DOI] [PubMed] [Google Scholar]
- Pradhan A. K.; Mohapatra A. D.; Nayak K. B.; Chakraborty S. PLoS One 2011, 6, e25370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman J. S.; Sironi J.; Berezniuk I.; Dasgupta S.; Castro L. M.; Gozzo F. C.; Ferro E. S.; Fricker L. D. PLoS One 2013, 8, e53263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov D. D.; Ali S. M.; Sengupta S.; Sheen J. H.; Hsu P. P.; Bagley A. F.; Markhard A. L.; Sabatini D. M. Mol. Cell 2006, 22, 159. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


