Abstract
DNA sequences with the potential to form secondary structures such as i-motifs (iMs) and G-quadruplexes (G4s) are abundant in the promoters of several oncogenes and, in some instances, are known to regulate gene expression. Recently, iM-forming DNA strands have also been employed as functional units in nanodevices, ranging from drug delivery systems to nanocircuitry. To understand both the mechanism of gene regulation by iMs and how to use them more efficiently in nanotechnological applications, it is essential to have a thorough knowledge of factors that govern their conformational states and stabilities. Most of the prior work to characterize the conformational dynamics of iMs have been done with iM-forming synthetic constructs like tandem (CCT)n repeats and in standard dilute buffer systems. Here, we present a systematic study on the consequences of epigenetic modifications, molecular crowding, and degree of hydration on the stabilities of an iM-forming sequence from the promoter of the c-myc gene. Our results indicate that 5-hydroxymethylation of cytosines destabilized the iMs against thermal and pH-dependent melting; contrarily, 5-methylcytosine modification stabilized the iMs. Under molecular crowding conditions (PEG-300, 40% w/v), the thermal stability of iMs increased by ∼10 °C, and the pKa was raised from 6.1 ± 0.1 to 7.0 ± 0.1. Lastly, the iM’s stability at varying degrees of hydration in 1,2-dimethoxyethane, 2-methoxyethanol, ethylene glycol, 1,3-propanediol, and glycerol cosolvents indicated that the iMs are stabilized by dehydration because of the release of water molecules when folded. Our results highlight the importance of considering the effects of epigenetic modifications, molecular crowding, and the degree of hydration on iM structural dynamics. For example, the incorporation of 5-methylycytosines and 5-hydroxymethlycytosines in iMs could be useful for fine-tuning the pH- or temperature-dependent folding/unfolding of an iM. Variations in the degree of hydration of iMs may also provide an additional control of the folded/unfolded state of iMs without having to change the pH of the surrounding matrix.
The discovery of DNA secondary structures, including quadruplexes in G-rich DNA (G4s) and iMs in C-rich DNA (iMs), has enabled the diversification of nucleic acid uses from their original roles in conventional biological processes to building blocks for nanoscale composite materials. Both G4s and iMs are four-stranded tetraplexes that can be formed by multiple individual single strands or by internal folding of a single-strand (ss) DNA.1,2 iMs are formed from cytosine repeats in C-rich DNA at slightly acidic pH, where unprotonated and protonated C–C+ base pairs intercalate and stabilize the structure.3,4 In genomic DNA, G- and C-rich regions capable of forming G4/iMs are enriched in regulatory elements of genes, particularly around the transcriptional start sites (TSS).4,5 G4s/iMs are associated with gene ontological (GO) terms like transcription factor activity, development, cell differentiation, and neurogenesis.6 DNA-based nanomaterials have found usefulness in arenas ranging from electronic circuit building to drug delivery systems.7−10
In mammalian cells, C-rich DNA located near the TSS has also been found to be susceptible to two different epigenetic modifications: methylation and hydroxymethylation.11,12 The occurrence of 5-hydroxymethylcytosine (5hmC) modifications has been discovered only recently and is generated by oxidation of 5-methylcytosines (5mC) by the ten eleven translocation (TET) family of oxygenases.13−20 Jin et al.20 demonstrated the role of 5hmCs as epigenetic regulators of gene expression, which is similar to the role of 5mC.21,22 Thus, 5mC, 5hmC, and iM formation have been studied individually as gene regulators. Although Dai et al. showed (via NMR) that methylation could adversely affect the integrity iMs,23 there have been no thorough studies that document the thermodynamic consequences of methylation or hydroxymethylation of cytosines to iMs.
The cellular environment likely also influences the pH- and temperature-dependent stability of G4s/iMs. Crowding agents and cosolutes together constitute about 20–40% of cellular volume.24 Cosolvents can alter the thermal/pH stability of DNA secondary structures via dehydration effects.25 The effects of cosolvents and crowding agents on the thermal stabilities of DNA triplexes, G-quadruplexes, and hairpin structures have been previously characterized by Chaires et al. and Sugimoto,26,27 but iMs were not among the secondary structures examined. This prior research showed duplexes are destabilized and G4s are stabilized under identical solution conditions where fewer water molecules associated with the DNA structure. Crowding is known to induce the pH-dependent stability in iMs, leading to their formation at near physiological pH,28,29 but there has been no detailed investigation of effects of dehydration on iMs. Because iMs and G4s are complementary structures that could be formed in gene regulatory regions, it is essential to understand the effects of hydration on iMs just as for G4s.
The biological function of G4s/iMs has yet to be completely established, as most of the experiments done on these structures have been in vitro. However, the consistency of G4/iM formation in the laboratory has created a new role for G4/iM DNA in nanomaterials. The iM structures have been used as proton-fueled nanomachines, which are reversibly actuated by cycling the solution conditions from acidic to basic and hence operate as a conformational switch to generate precise nanometer-scale motions.7,8 In these devices, iMs can act as pH-stimulated mechanical arms and/or as nanometer-height containers; in their folded form, they can nonspecifically trap small molecules and release these particles on controlled unfolding.30 When coupled with quantum dots and immobilized on gold electrodes, iMs work as photoelectric switches.31 iMs have been used as nanoprobes inside living cells to map spatial and temporal pH variations.32 Intracellular applications like this not only require characterization of iM probes under varying pH but also under intracellular crowding conditions that could affect the conformation of the probes and hence the signal output.
In both biological systems and nanomaterials, it is essential to have a thorough knowledge of the factors that affect iM formation and stability both in vivo and in vitro. Hence, here we present a systematic study on three important factors (epigenetic modification, molecular crowding, and hydration) that have thermodynamic consequences on the formation of the widely studied iM sequence from the c-myc gene.
Experimental Section
Methods: Genome-Wide Analysis of Colocalization of iMs and 5hmC
iM Density Calculations
To determine how widespread the phenomenon of 5hmC incorporation into putative iM structures in genomic DNA is, we first identified all of the putative unimolecular iMs using the Quadfinder tool developed by Scaria et al.33 This tool searches for sequences composed of CxNyCxNyCxNyCx motifs (for iMs on template strands) or GxNyGxNyGxNyGx motifs (for iMs on nontemplate strands), where x = 3–5 denotes the G/C stretch and y = 1–25 is the intervening loop length. The Quadfinder analyzes for, and lists all of, the probable motifs, including the overlapping ones, in a given DNA sequence. Promoters and intragenic regions of 15 760 reference sequence genes from the human GRCh37.p10 primary assembly were analyzed for the presence of iMs/G4s. The promoter region is defined as a 1 kb stretch upstream of the TSS,6 whereas the intragenic analyses covered a 1 kb stretch downstream of the TSS. To account for the iMs present on template and nontemplate strands, the total numbers of iMs were calculated by summing the G-motifs and C-motifs found in the template strand. To calculate the density of iMs, the 1 kb regions upstream and downstream of the TSS were divided into 100 bp segments for each gene, and each of these segments was analyzed with Quadfinder. The density of iMs per gene in any 100 bp was then calculated using the following equation.
The resulting plots (Figure 1) are similar to those in prior published reports.34
Figure 1.
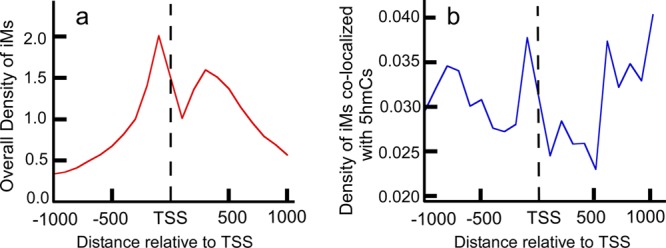
Very few 5hmC-modified iMs contribute to the overall density of iMs around the TSS. (a) Overall density of iM-forming genes relative to the TSS. (b) Density of iM-forming genes having 5hmCs colocalized within 100 bp of an iM.
Localization of 5-Hydroxymethylcytosine
We used the 5hmC sequencing data from H1 human ESC deposited to the Gene Expression Omnibus (accession GSE36173) by Yu et al.35 Their 5hmC sequencing was done using Tet-assisted bisulphite sequencing and was done on the UCSC hg18 build. These data were converted by us to GRCh37 using the liftOver genome tool by UCSC. (36) The 5hmC density calculation is similar to the iM density calculation and is shown in the following equation.
To visualize the enrichment of 5hmCs with the putative G4/iM forming genes, a contour plot of the enrichment of 5hmC content with varying GC content and G4/iM-forming potentials were plotted using JMP 10 statistical software (Figure 2).
Figure 2.
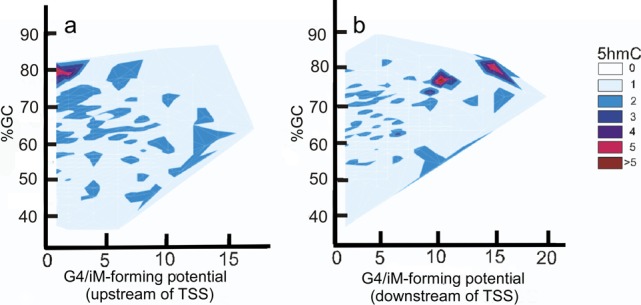
Relationship between iM potential and 5hmC density. (a) Contour plot for the sequences upstream of the TSS shows that 5hmC enrichment is associated with genes having low-iM-forming potential. (b) Contour plot for the downstream sequences shows that the 5hmC enrichment is associated with genes having high-iM-forming potentials.
Materials
As a representative iM, we have studied an iM-forming sequence from the nuclease-hypersensitive element (NHEIII) of the c-myc gene. The product of the c-myc gene is a proto-oncogene that is known to be overexpressed in cancer cells.11 The 22-mer sequence we used has been mutated from C to T at the sixth position to eliminate structural polymorphism and restrict its melting transition from folded iM to unfolded iM to two states.12 While evaluating the enrichment of 5hmC modification associated with iMs (Figure 2), we concluded that most of the iM-forming sequences show only single 5hmC modification; very infrequently, the 5hmC modification occurs more than once in an iM-forming sequence, regardless of whether it is located in intercalated C–C+ residues or in loop regions. Hence, for thermodynamic measurements, we have modified only a single cytosine to 5mC or 5hmC as a model of single-strand genomic DNA. Even though the C6T sequence predominantly exists as a single major conformer, a minor fraction can exist as a slip structure.12 However, in both of these conformations, the cytosine in the fourth position in the sequence always remains intercalated. Hence, we modified the cytosine in the fourth position to 5mC/5hmC to ensure that when the structure was folded, irrespective of the conformation, the modified cytosine could participate in intercalation. This approach facilitated considering the changed thermodynamic properties of fully intercalated iM residues as a function of changing epigenetic modifications alone. Although most methylation occurs in CpG islands (which do not occur in the myc promoter), the modification also occurs at significant numbers of CpC locations.2,37−39 Hence, the DNA oligonucleotides used have the following sequences
| C6T: | 5′-TTCCCTACCCTCCCCACCCTAA-3′ |
| 5mC-C6T: | 5′-TTC(mC)CTACCCTCCCCACCCTAA-3′ |
| 5hmC-C6T: | 5′-TTC(5hmC)CTACCCTCCCCACCCTAA-3′ |
where 5mC and 5hmC indicate 5-methyl- and 5-hydroxymethylcytidine, respectively. All oligos were purchased from Midland Certified Reagent Co. (Midland, TX, USA). For all experiments, the oligos were dissolved either in 20 mM sodium cacodylate buffer (for pH ranging from 5.3 to 7.4) or in 10 mM Tris base, 1 mM EDTA (for pH ranging from 7.6 to 8.0). All buffers were made from spectroscopic-grade reagents from Fisher Scientific. Poly(ethylene glycol) (PEG-300) was obtained from PCCA (Houston, TX, USA), and 2-methoxyethanol, ethylene glycol, glycerol, 1,3-propanediol, and 1,2-dimethoxyethane were purchased from Fisher Scientific.
Sample Preparation
Solutions of oligonucleotides were initially heated at 90 °C for 10 min and then cooled to room temperature to eliminate any possible duplex structures. For pKa determinations, concentrations of samples were matched via absorbance spectra when the samples were still hot. The circular dichroism (CD) spectra of the oligos at decreasing pH were obtained at 25 °C with DNA concentrations of 2 to 3 μM. The molecular crowding agents or cosolvents in 10, 20, 30, and 40 wt % (w/v) were added to the buffers and the pH adjusted as necessary for a range of 5.3 to 8.0.
Temperature- and pH-Dependent Circular Dichroism
CD experiments were performed in 1 cm cuvette using an Olis instrument (DSM 20 CD). Wavelength scans were collected from 225 to 350 nm. For melting experiments, temperature increments were either 1 or 2 °C per minute, and samples were allowed to equilibrate for 30 s before collecting each spectrum. Final CD spectra were obtained by averaging at least three scans for a given set of buffer conditions. All melting experiments were performed at pH 5.4 ± 0.1. To ensure that there were no intermolecular iMs, thermal melting experiments were performed at concentrations ranging from 0.3 to 20 μM. Plots of Tm versus concentration showed that all Tm values were identical, indicating that at all concentrations in this range the iMs were intramolecular. Furthermore, nondenaturing PAGE at pH 5.4 was used to look for formation of multiple structures. Concentrations of DNA < 30 μM traveled as a single band, suggesting that at pH 5.4 and at a concentration of 2 to 3 μM (those used in our studies) the sequences adopted an intramolecular iM structure. To obtain the thermodynamic parameters from the melting curve, curve fitting and data analyses were done using IGOR Pro (version 4.0, Wavemetrics). The values for Tm, ΔG°T, and ΔHm were obtained by fitting the data to a two-state model, where ΔG°T is a change in the free energy at a particular temperature and ΔHm is the enthalpy at the melting temperature, Tm.
The two-state model for DNA melting is described by
| 1 |
and
| 2 |
where K is the equilibrium constant for unfolding and [U] and [N] are the concentrations of unfolded and folded state, respectively.
The mole fraction of unfolded DNA, f(U), is given by
| 3 |
The free energy of unfolding at any given temperature, T, is given by eq 4(40)
| 4 |
The fraction folded was normalized from 0 to 1 prior to fitting, and the change in heat capacity (ΔCp) was assumed to be negligible. The fits yielded the values for Tm and ΔHm.
Hydration Effects on iM Stabilities
Water activities were obtained from osmolality measurements, which were done using a model 5520R vapor pressure osmometer (Wescor, Inc.). For higher concentrations of volatile cosolvents, osmolality was calculated. The water activity (aw) values were obtained from osmolality using eq 5
| 5 |
where Cosm is osmoles of cosolvent per kilogram of solvent.41
To assess the effects of hydration on the thermodynamic properties of the iM structure, we used the approach of Sugimoto42 that was previously used to study hairpins and G-quadruplex structures. The equilibrium between the folded and unfolded iM structure can be represented by
| 6 |
where U and N represent unfolded and native (folded iM) forms, CS represents cosolvent, and H+ represent protons. The equilibrium constant, K, is then given by
| 7 |
where (U/N) = Kobs.
At constant temperature and pressure, the first derivative of ln Kobs by ln aw is given by
| 8 |
In our plots of ln Kobs against ln aw, because of buffering, the difference in the number of protons, ΔnH+, between the folded and unfolded state is negligible. The change in the activity of cosolvents with respect to the change in activity of water is assumed to be insignificant over the small range tested. On the basis of the previously reported work on duplexes and triplexes,26,42 the linear slopes of the graph of eq 8 give the number of water molecules associated with each iM at the temperature studied, which, in our case, was physiologically relevant 37 °C.
Results
Colocalization of iMs with 5hmCs Showed That Most iM-Forming Sequences Have Only a Single 5hmC Modification
The density of iMs within 1 kb upstream and downstream was plotted for all genes (Figure 1a,b). The contour plots of 5hmC with respect to iM-forming potentials and GC content indicate that in the 1 kb region upstream relative to the TSS (Figure 2a) the 5hmC enrichment occurs around those sequences that have lower potential to form iMs. In contrast, in the 1 kb downstream region, the 5hmC enrichment occurs around the sequences with high-iM-forming potential (Figure 2b). Asymmetry in the distribution could be caused by intragenic regions that are more likely to be enriched in 5hmC content. In any case, most of the iMs do not have more than single 5hmC modification associated with them. The genes with putative iM-forming sequences were a subset of all 5hmC-modified genes in the database. Each individual putative iM-forming sequence was analyzed for the presence of 5hmC. Quantitatively, our analyses indicated that among all of the putative iM-forming sequences that could have cytosines modified to 5hmCs only a small fraction (less than 15% of sequences) had two or more 5hmCs located within an iM-forming sequence. Although many 5hmC residues were found in regions that would constitute loops in putative i-motifs, there were also a significant number of 5hmC associated with residues that could form the C–C+ intercalation bond that stabilizes the i-motif. Hence, as a representative of those sequences, we have used the constructs having only single 5mC and 5hmC modifications to evaluate whether a singly modified cytosine can affect the biophysical properties of iMs by altering the intercalated bases.
Single 5hmC and 5mC Modifications Do Not Inhibit iM Formation, but the Presence of 5hmC Alters pH-Dependent Folding
Initially, we evaluated whether the 5hmC and 5mC modifications affected the ability of the DNA to adopt the iM conformation. The conformations of 22-mers at decreasing pH were examined to determine the pKa for iM formation (defined as the pH at which 50% of the strands are folded). The characteristic CD signal maxima for iMs at 289 nm and minima at 254 nm was observed for modified strands, indicating that they formed iM structures. By measuring the change in the ellipticity at 289 nm, the pKa was determined (Figure 3). This showed that the pKa values were 6.1 ± 0.1 for C6T and 6.3 ± 0.1 for 5mC-C6T but were slightly lowered to 5.9 ± 0.1 for 5hmC-C6T. Thus, a single 5hmC modification reduced the pH-induced stability of the iM structure. However, the unfolding with respect to increase in pH for 5hmC modified construct was strikingly different from that of 5mC and unmodified DNA; for 5hmC modification, the unfolding was highly cooperative, transitioning from fully folded to fully unfolded over the small pH range of 5.6–6.2. We attribute this dramatic change in pH response to the 5hmC polarity, which likely makes it more favorable for water molecules to interact with ssDNA and hence results in an ease of unfolding of the structure at relatively low pH compared to C6T and 5mC-C6T.
Figure 3.
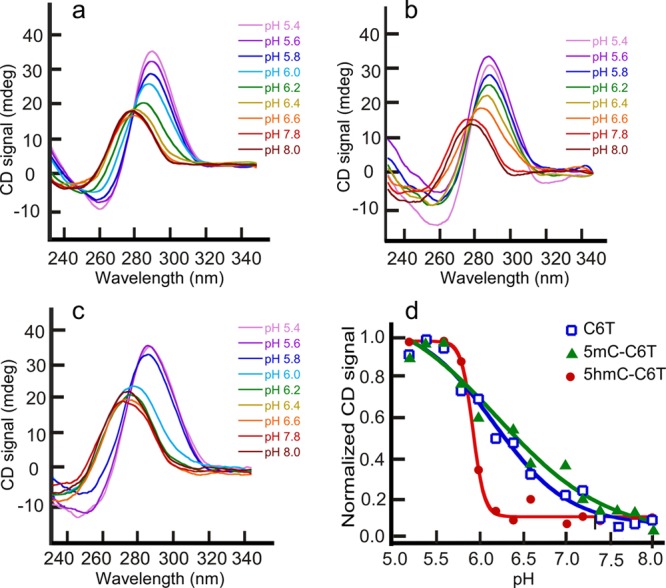
pH denaturation of (a) C6T, (b) 5mC-C6T, and (c) 5hmC-C6T. pKa increases slightly with 5mC modification and is lowered with 5hmC modification. The pH melting curve (d) shows substantial co-operativity when the iM contains 5hmC modification.
Macromolecular Crowding Agents Shift the pKa of iMs toward Physiological pH, whereas Smaller Cosolvents Have No Effect on the pKa
Physiologically, crowding agents and cosolutes occupy 20–40% of cellular volume.27 Hence, for evaluating the effects of cosolvents and crowding agents on the iM structure of C6T, we used 40% (w/v) of 1,2-dimethoxyethane, 2-methoxyethanol, ethylene glycol, 1,3-propanediol, glycerol, and PEG-300 as representative of the cellular milieu. The pKa measurements were repeated as described above. Addition of cosolvents did not perturb the maxima and minima in the CD spectra of iM structures. The low-molecular-weight cosolvents showed no effect on the pKa of iMs (±0.1). Thus, dehydration did not have any significant consequence on the pKa of iMs. However, PEG-300 stabilized the iMs at higher pH. The pKa of the C6T iM structure was raised to 7.0 ± 0.1 in the presence of PEG-300 (Figure 4), which is a considerable shift and is consistent with the previously published literature.28 Similarly, a rise in pKa was observed for 5mC-C6T as well as for 5hmC-C6T. However, under molecular crowding conditions, the pKa difference between the C6T and epigenetically modified DNA is rendered insignificant. The comparative studies conducted using the crowding agent PEG 300 and other smaller cosolvents lead to the conclusion that it is primarily crowding because of steric effects that is responsible for conferring pH-dependent stability in PEG 300. Previously, it has been reported that the direct interaction between ssDNA and macromolecules like PEG are thermodynamically unfavorable.40 However, Hänsel et al.43 reported on the differences between the effects of the two molecular crowding mimetics, Ficoll 70 and PEG 200, on G-quadruplex folding topology. Their results suggested that PEG may promote the formation of high-order parallel topologies in G-quadruplexes via a mechanism other than simple crowding. However, unlike the G-quadruplexes studied, our iM-forming sequence cannot adopt intramolecular parallel/antiparallel conformations. Also, our iM-forming sequence is conformationally restricted and cannot undergo significant structural polymorphism by intercalated residues. Hence, the increase in stability of the iM in the PEG solution we attribute to the resistance offered by macromolecular crowding to iM unfolding and not to an alternate physical mechanism or structure.
Figure 4.
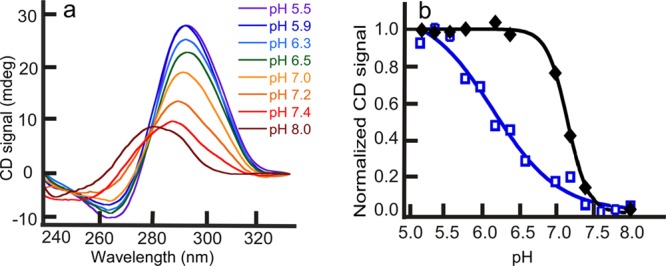
pH scans for C6T in the presence of PEG-300. (a) Melting scans of iM structure, under molecular crowding conditions, with increasing pH. (b) Fits showing that the pKa of C6T in the presence (black) of molecular crowding agents shifts toward neutral, whereas the pKa in the absence (blue) of crowding is in the acidic range. The other smaller cosolutes did not show any pKa shifts.
Epigenetic Modification by 5hmC Results in iM Thermal Destabilization
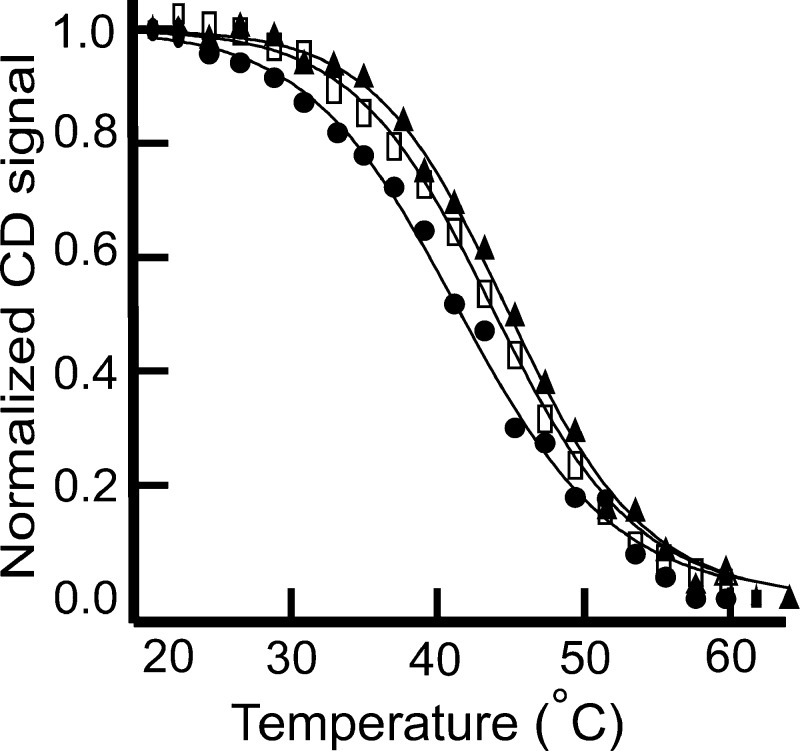
To characterize the thermodynamic differences between the modified and unmodified oligos, thermal denaturation experiments were conducted. The CD signal at 289 nm (maximum for iMs) was then plotted against temperature. The melting profiles are shown in Figure 5. All the three oligos melted in a fashion that could be described by a two-state transition but showed small yet significant (p value <0.05 using one-way ANOVA) differences in melting temperature of C6T when the cytosine was modified to 5hmC. 5hmC-C6T had the lowest melting temperature of 40.5 ± 0.6 °C, whereas 5mC-C6T and unmodified C6T have Tm of 43.7 ± 0.3 and 42.5 ± 0.9 °C, respectively. The results of the fits to the Tm profiles for the three oligos are shown in Table 1. The ΔG° of unfolding at 37 °C for 5hmC-C6T is substantially lower than the other two oligos, indicating the unfolded state is more favorable at physiological temperatures. With 5hmC, we again interpret the difference as being due to the additional −OH group, which likely leads to the increase in the water-accessible surface area upon denaturation. As with pH-dependent unfolding, the −OH group in thermally unfolded DNA can interact with water molecules, facilitating the unfolding of iMs at lower temperatures. Conversely, the methyl group of 5mC, being nonpolar, likely introduces an entropic penalty for formation of additional iceberg waters with the ssDNA form, making 5mC iMs slightly more difficult to thermally unfold. An alternative explanation for the lower melting temperature and pH response of 5hmC-C6T could be attributed to a different conformation of iM in 5hmC-modified DNA. However, this seems unlikely given the similarity of the CD spectra for each folded DNA, modified or not.
Figure 5.
CD melting trends for C6T (□), 5hmC-C6T (●), and 5mC-C6T (▲). 5hmC modification thermally destabilized the iM structure.
Table 1. Comparison of the Thermodynamic Parameters for Melting of C6T, 5mC-C6T, and 5hmC-C6Ta.
| ΔHm (kcal/mol) | Tm (°C) | ΔG° (kcal/mol, 37 °C) | |
|---|---|---|---|
| C6T | 42.0 ± 0.3 | 42.5 ± 0.9 | 0.80 ± 0.20 |
| 5mC-C6T | 44.4 ± 0.3 | 43.7 ± 0.3 | 0.94 ± 0.05 |
| 5hmC-C6T | 37.9 ± 0.1b | 40.5 ± 0.6b | 0.42 ± 0.07b |
Values are given ± standard deviations.
Indicates that the difference in the thermodynamic parameters between the epigenetically modified and unmodified DNA is significant. ANOVA (Turkey post hoc test) with p < 0.05 was considered significant.
Thermodynamics of iM Melting Systematically Change with the Total Number and Position of Hydroxyl Groups in the Cosolvents
The thermal stability of the C6T iM in water is greatly dependent on the presence and composition of cosolvents. Nonionic cosolvents like 1,2-dimethoxyethane (with no −OH groups), 2-methoxyethanol (with a single −OH group), 1,3-propanediol (with two −OH groups), ethylene glycol (with two vicinal −OH groups), and glycerol (with three vicinal −OH groups) were used from 10 to 40% (w/v). Increasing amounts of 1,2-dimethoxyethane and 2-methoxyethanol increased the thermal stability of the iM structure, glycerol lowered it significantly, and ethylene glycol and 1,3-propanediol had a negligible effect on the melting temperature. Hence, adding cosolvents did not always stabilize the iM structure; the composition of the cosolvents played an important role. The melting profiles for C6T in 1,2-dimethoxyethane, 2-methoxyethanol, glycerol, and PEG-300 are shown in Figure 6. The melting temperature was raised approximately 10 °C on addition of 1,2-dimethoxyethane or PEG-300 to the buffer. The thermodynamic parameters obtained from fitting all thermal melting data are given in Table 2. Below, we further analyze the hydration of iMs in the presence of the different cosolvents.
Figure 6.
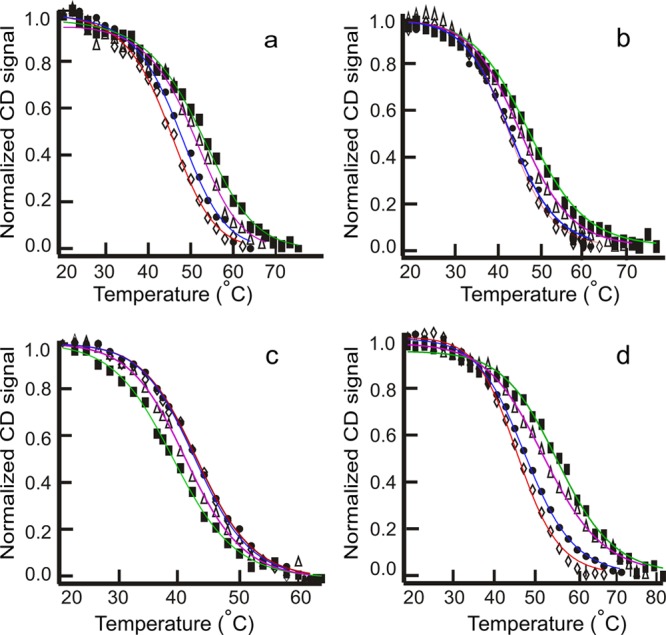
Thermal melting profiles for C6T in 10 (◊), 20 (●), 30 (▲), and 40% (■) cosolvents. Melting temperature increases with the addition of (a) dimethoxyethane, (b) 2-methoxyethanol, and (d) PEG-300. Melting temperature decreases with the addition of (c) glycerol.
Table 2. Comparison of the Thermodynamic Parameters of C6T in Varying Concentrations of the Cosolvents and PEG-300a.
| cosolvent/crowding agent | 40% (w/v) | 30% (w/v) | 20% (w/v) | 10% (w/v) | |
|---|---|---|---|---|---|
| dimethoxyethane | ΔHm (kcal/mol) | 33.9 ± 1.7 | 34.6 ± 2.0 | 37.3 ± 0.8 | 43.0 ± 1.8 |
| Tm (°C) | 52.0 ± 0.4 | 49.0 ± 1 | 46.5 ± 0.2 | 44.7 ± 0.3 | |
| ΔG° (kcal/mol, 37 °C) | 1.5 ± 0.2 | 1.3 ± 0.2 | 1.1 ± 0.1 | 1.04 ± 0.02 | |
| methoxyethanol | ΔHm (kcal/mol) | 32.0 ± 1.9 | 35.0 ± 2 | 38.0 ± 2.8 | 41.0 ± 1.8 |
| Tm (°C) | 47.0 ± 0.2 | 45.0 ± 2 | 44.0 ± 0.1 | 43.0 ± 1 | |
| ΔG° (kcal/mol, 37 °C) | 1.0 ± 0.2 | 0.9 ± 0.2 | 0.8 ± 0.1 | 0.77 ± 0.02 | |
| ethylene glycol | ΔHm (kcal/mol) | 38.0 ± 0.4 | 36.7 ± 0.4 | 38.0 ± 1.0 | 37.1 ± 0.4 |
| Tm (°C) | 40 ± 0.1 | 41.0 ± 0.1 | 42.2 ± 0.3 | 42.1 ± 0.6 | |
| ΔG° (kcal/mol, 37 °C) | 0.6 ± 0.1 | 0.5 ± 0.2 | 0.6 ± 0.2 | 0.6 ± 0.1 | |
| glycerol | ΔHm (kcal/mol) | 34.0 ± 2.8 | 41.0 ± 3.0 | 42.0 ± 2.0 | 42.5 ± 0.4 |
| Tm (°C) | 39.0 ± 0.2 | 41.0 ± 0.1 | 43.0 ± 0.5 | 43 ± 1 | |
| ΔG° (kcal/mol, 37 °C) | 0.23 ± 0.03 | 0.5 ± 0.1 | 0.8 ± 0.2 | 0.8 ± 0.2 | |
| PEG-300 | ΔHm (kcal/mol) | 31.1 ± 0.9 | 34.0 ± 2.0 | 37.0 ± 1.0 | 38.0 ± 1.0 |
| Tm (°C) | 52.0 ± 0.2 | 50.2 ± 0.3 | 47.2 ± 0.2 | 45.0 ± 0.9 | |
| ΔG° (kcal/mol, 37 °C) | 1.4 ± 0.1 | 1.40 ± 0.01 | 1.2 ± 0.2 | 0.9 ± 0.2 |
Values are given ± standard deviations.
Under Molecular Crowding Conditions or in the Presence of Cosolvents, the Epigenetic Modifications Lose Their Governance on the iM Stability
When pKa and thermal melting experiments were done with the modified DNA in the presence of cosolvents, all of the differences caused as a result of modifications were abolished (Table 3). Thus, epigenetic modification governs the iM stability only weakly compared to cosolvents. From these experiments, we conclude that in the cellular environment the iM stability would not vary significantly in the presence or absence of the modified cytosine residues. Table 3 gives the comparisons of melting temperatures of 5hmC-C6T and 5mC-C6T in various solvents, and these temperatures are similar to the melting temperatures of C6T (Table 2).
Table 3. Melting Temperatures of Epigenetically Modified DNA Strands in the Presence of Cosolventsa.
| 5mC-C6T (°C) | 5hmC-C6T (°C) | C6T (°C) | |
|---|---|---|---|
| 40% dimethoxyethane | 51.8 ± 0.2 | 52.4 ± 0.2 | 52.0 ± 0.4 |
| 20% dimethoxyethane | 46.2 ± 0.2 | 46.6 ± 0.2 | 46.5 ± 0.2 |
| 40% methoxyethanol | 46.6 ± 0.2 | 46.9 ± 0.4 | 47.0 ± 0.2 |
| 20% methoxyethanol | 43.4 ± 0.3 | 43.5 ± 0.2 | 44.0 ± 0.21 |
| 40% glycerol | 39.1 ± 0.2 | 39.0 ± 0.2 | 39.0 ± 0.2 |
| 20% glycerol | 42.9 ± 0.2 | 43.1 ± 0.2 | 43.0 ± 0.5 |
| 40% PEG-300 | 52.5 ± 0.4 | 52.6 ± 0.6 | 52.0 ± 0.2 |
| 20% PEG-300 | 47.4 ± 0.3 | 48.9 ± 0.9 | 47.2 ± 0.2 |
Values are given ± standard deviations.
Stability of iM Structure Is Affected by Both Hydration and Molecular Crowding
The stability of iMs is affected by the composition of the cosolvent, which governs the hydration state of the structure. The small cosolvents, such as 1,2-dimethoxyethane (MW = 90 g/mol), 2-methoxyethanol (MW = 76 g/mol), ethylene glycol (MW = 62 g/mol), 1,3-propanediol (MW =76 g/mol), and glycerol (MW = 92 g/mol), vary the degree of hydration associated with the iM structure. These smaller cosolvents have similar molecular weights and should contribute similarly to any crowding resulting from their steric bulk. Hence, the differences in the thermodynamic characteristics of iMs in the presence of these small cosolvents were attributed to differences in the degree of hydration of the iMs by these cosolvents. To be able to quantify the hydration associated with the DNA molecules at 37 °C, the logarithm of water activity is plotted against the logarithm of Kobs (Figure 7). From the slopes of the plots in Figure 7, we calculated that in the presence of glycerol there were six water molecules associated with the folded DNA structure, whereas ethylene glycol led to the uptake of 1.4 water molecules per iM structure. However, 1,2-dimethoxyethane and 2-methoxyethanol caused the release of eight and two water molecules, respectively. Likewise, in the folded state, PEG-300 facilitated release of 26 water molecules. Addition of 1,3-propanediol did not result in either the uptake or release of water molecules, and the total number of water molecules associated with iM-forming sequence remained unchanged between folded and unfolded states. From the Tm data, we demonstrated that the presence of ethylene glycol and glycerol lowered the Tm, whereas 1,2 dimethoxyethane, 2-methoxyethanol, and PEG-300 considerably increased the thermal stability of C6T. Together, these results lead to a very important inference that the iMs are stabilized by the release of water molecules. It has been previously reported that macromolecules like PEG do not interact with DNA directly because such interactions are thermodynamically unfavorable.42 Therefore, PEG-dependent stabilization of iMs are more likely a result of the molecular crowding phenomenon, which restricts the degrees of freedom available for iMs to unfold, thereby hindering the process of unfolding. In contrast to PEG, small molecules like 1,2-dimethoxyethane, 2-methoxyethanol, ethylene glycol, 1,3-propanediol, and glycerol can directly interact with both structured and random-coiled-single stranded DNA. These cosolvents can be either taken up or released during the secondary-structure formation along with water molecules. In our studies, glycerol, having three hydroxyl groups, increases the water molecules associated with the folded iMs and also destabilizes the iM form. In contrast, dimethoxymethane favors the release of water molecules associated with the iM and imparts stability to the iM structure. These data suggest that the entropic penalty for increased water participation during folding is the thermodynamic phenomenon that disfavors the folded form in selected cosolvents. Hence, cosolvent effects on iM formation in cells are likely to be a complex function of the cellular environment of genomic DNA.
Figure 7.
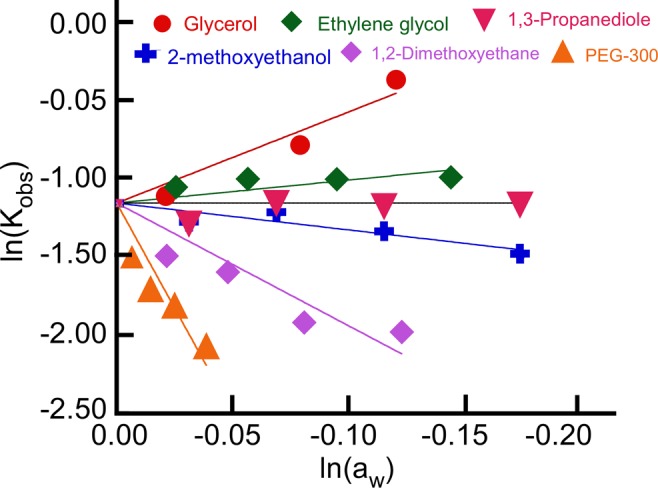
Changes in Kobs with respect to changing water activity (aw) for C6T at 37 °C in pH 5.4 solutions. Nearly identical results were obtained for 5hmC-C6T and 5mC-C6T, indicating that the solvent effects are indifferent to epigenetic modification.
Discussion
With the recent confirmation of the existence of G4s in vivo,44 the probability of biological roles for iMs has increased significantly. G4s/iMs are gene regulatory elements abundant near the TSS of several genes.34 G4s/iMs are known to promote expression of certain genes. As one example, the presence of G4s/iMs enhances myoD-dependent gene expression.45 In contrast, transcription of the c-myb gene is suppressed by G4/iM formation.46 These conflicting roles in regulating gene expression indicate a complex underlying mechanism of gene regulation. Hence, to develop a better understanding of the gene regulatory function of iMs, it is important to comprehensively study and account for the factors affecting the formation and stability of iMs in vivo.
One of the factors that might affect iM stability is epigenetic modification. The cytosines in mammalian DNA can undergo epigenetic modifications to 5mC and 5hmC.13,15 Our determination of the pKa of modified iMs indicated that the 5mC modification shifted the pKa toward physiological pH (from 6.1 to 6.3), whereas the 5hmC modification shifted the pKa to more acidic pH (from 6.1 to 5.9). The thermal melting data showed that a single cytosine modification to either 5mC or 5hmC did not inhibit the formation of iMs at 37 °C. The 5hmC modification had the lowest melting temperature of 40.5 ± 0.6 °C, whereas 5mC and unmodified C6T have Tm at 43.7 ± 0.3 and 42.5 ± 0.9 °C, respectively. The observed shifts in pKa and melting temperature indicate that 5mC and 5hmC modifications can alter the conformational stability of iMs, depending on experimental conditions. Therefore, these modifications may need to be accounted for when studying transcriptional regulation of iM-forming, epigenetically modified genes.
Macromolecules including nucleic acids, proteins, and polysaccharides occupy 20–40% of the intracellular volume, resulting in the phenomenon of molecular crowding.27 Crowding conditions are known to stabilize G4s and triplexes.26 Experiments done in dilute aqueous buffers often exclude volume and osmotic pressure effects found in the cellular environment. Hence, we used PEG-300 as a surrogate to introduce molecular crowding into our experiment on the stability of iMs. Concurring with the earlier work on crowding and stability of iMs,28 our results demonstrate that molecular crowding via PEG-300 significantly increased the pKa of iMs approximately to physiological pH (7.0). Also, addition of PEG-300 increased the Tm of iMs. Our data indicate that the increased Tm and pKa result from the resistance offered by the crowding agents to the unfolding process, making the folded form more favored. Hence, along with above-mentioned epigenetic modifications, crowding conditions should also be accounted for when studying conformational dynamics and energetic landscapes of folding/unfolding and gene regulatory mechanisms involving iMs.
Owing to their pH- and temperature-dependent conformational response, iMs are being used in a diverse range of nanotechnological applications.47 These applications include drug delivery systems, nanocircuitry, and mechanical motors. For example, Pu et al. have developed a supramolecular complex based on iMs that is capable of performing multiplex logical operations. The ability iMs/G4s to undergo conformational variation (from tetraplex to duplex) in response to temperature and/or pH stimuli is harnessed to operate these logic gates. To build logic gates operational over different temperature and pH ranges, Pu et al. have suggested altering the sequence and the length of iM-forming oligonucleotides. Alternatively, our studies show that introducing 5mC and 5hmC modifications in intercalating cytosines alter both the pKa and Tm of iMs. Thus, 5mC and 5hmC modifications could be used to fine-tune the pH- and temperature-dependent iM conformational switching of C-rich sequences. The pH- and temperature-dependent response of iMs can also be varied by changing the environment in which iMs are formed. For example, the presence of a cosolvent like glycerol lowers the Tm of iMs and hence glycerol could be used to design iM-based logic gates that operate over a narrower range of temperature changes. In contrast, crowding agents like PEG-300 raise the Tm of iMs, resulting in a molecular switch that undergoes a conformational change at higher temperatures. Overall, our work on the effects of epigenetic modifications, hydration, and crowding effects provides a systematic study of ways to modulate iM-based conformational changes.
Conclusions
In this work, we examined the stabilities of iMs for comparison with prior work on DNA G4s, duplexes, and triplexes.26,42 In dilute aqueous solutions, modification of cytosine to 5mC raised the pKa and Tm of iMs, whereas modification to 5hmC lowered the pKa and Tm. The introduction of molecular crowding by using PEG-300 stabilized the iMs versus both temperature and pH. However, under molecular crowding conditions, neither 5mC nor 5hmC modification could alter the stability of iMs. The degree of hydration of iMs changed their temperature-dependent stability. Depletion of water molecules associated with folded iMs stabilized them against thermal melting, indicating an important cosolvent dependence. Our methodical study of the effects of hydration and molecular crowding on iMs suggests that the microenvironment around the iMs should be accounted for when studying these structures. Thus, both epigenetic modification and the matrix surrounding the iMs affect their formation; hence, these factors could be used both for understanding the physiological roles of iMs and for fine tuning pH- and temperature-dependent responses of nanodevices based on iM structures.
Acknowledgments
We thank Dr. Christopher J. Fields from the University of Illinois–Urbana–Champaign, USA, for helping us with BioPerl. We thank Dr. Souvik Maiti and Vinod Scaria form the Institute of Genomics and Integrative Biology, India, for sharing the code for the Quadfinder program. We also acknowledge the valuable input of Samantha M. Reilly, a graduate student at the University of Mississippi, USA.
Glossary
Abbreviations
- G4
guanine quadruplex
- iM
i-motif
- TSS
transcription start site
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
This research was supported by National Science Foundation Mississippi EPSCoR grant EPS-0903787 and National Institutes of Health/National Cancer Institute grant 1R15CA173667-01A1 (T.A.B., PI; R.M.W., co-PI).
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Moehlig A. R.; Djernes K. E.; Krishnan V. M.; Hooley R. J. (2012) Cytosine derivatives form hemiprotonated dimers in solution and the gas phase. Org. Lett. 14, 2560–2563. [DOI] [PubMed] [Google Scholar]
- Choi J.; Kim S.; Tachikawa T.; Fujitsuka M.; Majima T. (2011) pH-induced intramolecular folding dynamics of i-motif DNA. J. Am. Chem. Soc. 133, 16146–16153. [DOI] [PubMed] [Google Scholar]
- Leroy J. L. (2009) The formation pathway of i-motif tetramers. Nucleic Acids Res. 37, 4127–4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy J.; Maizels N. (2008) Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes. Nucleic Acids Res. 36, 1321–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S.; Hurley L. H.; Neidle S. (2011) Targeting G-quadruplexes in gene promoters: A novel anticancer strategy?. Nat. Rev. Drug Discovery 10, 261–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert J. L. (2007) Four-stranded DNA: Cancer, gene regulation and drug development. Philos. Trans. R. Soc., A 365, 2969–2984. [DOI] [PubMed] [Google Scholar]
- Liu D.; Bruckbauer A.; Abell C.; Balasubramanian S.; Kang D. J.; Klenerman D.; Zhou D. (2006) A reversible pH-driven DNA nanoswitch array. J. Am. Chem. Soc. 128, 2067–2071. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Chouai A.; Degtyareva N. N.; Lutterman D. A.; Dunbar K. R.; Turro C. (2005) Chemical control of the DNA light switch: Cycling the switch ON and OFF. J. Am. Chem. Soc. 127, 10796–10797. [DOI] [PubMed] [Google Scholar]
- Cheng E.; Xing Y.; Chen P.; Yang Y.; Sun Y.; Zhou D.; Xu L.; Fan Q.; Liu D. (2009) A pH-triggered, fast-responding DNA hydrogel. Angew. Chem. 121, 7796–7799. [DOI] [PubMed] [Google Scholar]
- Allen T. M.; Cullis P. R. (2004) Drug delivery systems: Entering the mainstream. Science 303, 1818–1822. [DOI] [PubMed] [Google Scholar]
- Brooks T. A.; Hurley L. H. (2010) Targeting MYC expression through G-quadruplexes. Genes Cancer 1, 641–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J.; Hatzakis E.; Hurley L. H.; Yang D. (2010) I-motif structures formed in the human c-MYC promoter are highly dynamic–insights into sequence redundancy and I-motif stability. PloS One 5, e11647-1–e11647-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R.; Nagarajan A.; Wajapeyee N. (2010) Advances in genome-wide DNA methylation analysis. Biotechniques 49, iii–xi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra S. K.; Bettuzzi S. (2009) Epigenetic DNA-(cytosine-5-carbon) modifications: 5-Aza-2′-deoxycytidine and DNA-demethylation. Biochemistry (Moscow) 74, 613–619. [DOI] [PubMed] [Google Scholar]
- Matarese F.; Carrillo-de Santa Pau E.; Stunnenberg H. G. (2011) 5-Hydroxymethylcytosine: A new kid on the epigenetic block?. Mol. Syst. Biol. 7, 562-1–562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shock L. S.; Thakkar P. V.; Peterson E. J.; Moran R. G.; Taylor S. M. (2011) DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc. Natl. Acad. Sci. U.S.A. 108, 3630–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S.; Heintz N. (2009) The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl C.; Gronbaek K.; Guldberg P. (2011) Advances in DNA methylation: 5-Hydroxymethylcytosine revisited. Clin. Chim. Acta 412, 831–836. [DOI] [PubMed] [Google Scholar]
- Koh K. P.; Yabuuchi A.; Rao S.; Huang Y.; Cunniff K.; Nardone J.; Laiho A.; Tahiliani M.; Sommer C. A.; Mostoslavsky G.; Lahesmaa R.; Orkin S. H.; Rodig S. J.; Daley G. Q.; Rao A. (2011) Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 8, 200–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S. G.; Kadam S.; Pfeifer G. P. (2010) Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 38, e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim O.; Li R.; Hung J.-H.; Chen P. B.; Dong X.; Ee L.-S.; Weng Z.; Rando O. J.; Fazzio T. G. (2011) Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell 147, 1498–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus T. F. J.; Globisch D.; Wagner M.; Eigenbrod S.; Widmann D.; Münzel M.; Müller M.; Pfaffeneder T.; Hackner B.; Feiden W.; Schüller U.; Carell T.; Kretzschmar H. A. (2012) Low values of 5-hydroxymethylcytosine (5hmC), the sixth base, are associated with anaplasia in human brain tumors. Int. J. Cancer 131, 1577–1590. [DOI] [PubMed] [Google Scholar]
- Dai J.; Ambrus A.; Hurley L. H.; Yang D. (2009) A direct and nondestructive approach to determine the folding structure of the I-motif DNA secondary structure by NMR. J. Am. Chem. Soc. 131, 6102–6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann J. W.; Steffen D.; Gusella J.; Tabin C.; Bird S.; Cowing D.; Weinberg R. A. (1982) DNA methylation affecting the expression of murine leukemia proviruses. J. Virol. 44, 144–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhakal S.; Cui Y.; Koirala D.; Ghimire C.; Kushwaha S.; Yu Z.; Yangyuoru P. M.; Mao H. (2013) Structural and mechanical properties of individual human telomeric G-quadruplexes in molecularly crowded solutions. Nucleic Acids Res. 41, 3915–3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spink C. H.; Chaires J. B. (1999) Effects of hydration, ion release, and excluded volume on the melting of triplex and duplex DNA. Biochemistry 38, 496–508. [DOI] [PubMed] [Google Scholar]
- Miyoshi D.; Sugimoto N. (2008) Molecular crowding effects on structure and stability of DNA. Biochimie 90, 1040–1051. [DOI] [PubMed] [Google Scholar]
- Rajendran A.; Nakano S.; Sugimoto N. (2010) Molecular crowding of the cosolutes induces an intramolecular i-motif structure of triplet repeat DNA oligomers at neutral pH. Chem. Commun. 46, 1299–1301. [DOI] [PubMed] [Google Scholar]
- Cui J.; Waltman P.; Le V. H.; Lewis E. A. (2013) The effect of molecular crowding on the stability of human c-MYC promoter sequence i-motif at neutral pH. Molecules 18, 12751–12767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y.; Liu D.; Wang S.; Luo S.; Wang W.; Yang Y.; Ouyang Q.; Jiang L. (2007) Alternating-electric-field-enhanced reversible switching of DNA nanocontainers with pH. Nucleic Acids Res. 35, e33-1–e33-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng H.; Yang Y.; Chen Y.; Zhou Y.; Liu Y.; Chen X.; Ma H.; Tang Z.; Liu D.; Jiang L. (2009) Photoelectric conversion switch based on quantum dots with i-motif DNA scaffolds. Chem. Commun. 17, 2293–2295. [DOI] [PubMed] [Google Scholar]
- Modi S.; Swetha M. G.; Goswami D.; Gupta G. D.; Mayor S.; Krishnan Y. (2009) A DNA nanomachine that maps spatial and temporal pH changes inside living cells. Nat. Nanotechnol. 4, 325–330. [DOI] [PubMed] [Google Scholar]
- Scaria V.; Hariharan M.; Arora A.; Maiti S. (2006) Quadfinder: Server for identification and analysis of quadruplex-forming motifs in nucleotide sequences. Nucleic Acids Res. 34, W683–W685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.; Du Z.; Li N. (2007) Extensive selection for the enrichment of G4 DNA motifs in transcriptional regulatory regions of warm blooded animals. FEBS Lett. 581, 1951–1956. [DOI] [PubMed] [Google Scholar]
- Yu M.; Hon G. C.; Szulwach K. E.; Song C.-X.; Zhang L.; Kim A.; Li X.; Dai Q.; Shen Y.; Park B. (2012) Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149, 1368–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E. S.; Linton L. M.; Birren B.; Nusbaum C.; Zody M. C.; Baldwin J.; Devon K.; Dewar K.; Doyle M.; FitzHugh W. (2001) Initial sequencing and analysis of the human genome. Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- Robertson A. B.; Dahl J. A.; Vågbø C. B.; Tripathi P.; Krokan H. E.; Klungland A. (2011) A novel method for the efficient and selective identification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 39, e55-1–e55-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestor C. E.; Ottaviano R.; Reddington J.; Sproul D.; Reinhardt D.; Dunican D.; Katz E.; Dixon J. M.; Harrison D. J.; Meehan R. R. (2012) Tissue type is a major modifier of the 5-hydroxymethylcytosine content of human genes. Genome Res. 22, 467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandjean V.; Yaman R.; Cuzin F.; Rassoulzadegan M. (2007) Inheritance of an epigenetic mark: The CpG DNA methyltransferase 1 is required for de novo establishment of a complex pattern of non-CpG methylation. PLoS One 2, e1136-1–e1136-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruylants G.; Wouters J.; Michaux C. (2005) Differential scanning calorimetry in life science: Thermodynamics, stability, molecular recognition and application in drug design. Curr. Med. Chem. 12, 2011–2020. [DOI] [PubMed] [Google Scholar]
- Kiss G.; Hansson H. C. (2004) Application of osmolality for the determination of water activity and the modelling of cloud formation. Atmos. Chem. Phys. Discuss. 4, 7667–7689. [Google Scholar]
- Miyoshi D.; Karimata H.; Sugimoto N. (2006) Hydration regulates thermodynamics of G-quadruplex formation under molecular crowding conditions. J. Am. Chem. Soc. 128, 7957–7963. [DOI] [PubMed] [Google Scholar]
- Hänsel R.; Löhr F.; Foldynová-Trantírková S.; Bamberg E.; Trantírek L.; Dötsch V. (2011) The parallel G-quadruplex structure of vertebrate telomeric repeat sequences is not the preferred folding topology under physiological conditions. Nucleic Acids Res. 39, 5768–5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffi G.; Tannahill D.; McCafferty J.; Balasubramanian S. (2013) Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 5, 182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shklover J.; Weisman-Shomer P.; Yafe A.; Fry M. (2010) Quadruplex structures of muscle gene promoter sequences enhance in vivo MyoD-dependent gene expression. Nucleic Acids Res. 38, 2369–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbo S. L.; Memmott R. M.; Uribe D. J.; Krotova-Khan Y.; Hurley L. H.; Ebbinghaus S. W. (2008) A novel G-quadruplex-forming GGA repeat region in the c-myb promoter is a critical regulator of promoter activity. Nucleic Acids Res. 36, 1755–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimron S.; Magen N.; Elbaz J.; Willner I. (2011) pH-programmable DNAzyme nanostructures. Chem. Commun. 47, 8787–8789. [DOI] [PubMed] [Google Scholar]