Abstract
Although an X-ray crystal structure of lactose permease (LacY) has been presented with bound galactopyranoside, neither the sugar nor the residues ligating the sugar can be identified with precision at ∼3.5 Å. Therefore, additional evidence is important for identifying side chains likely to be involved in binding. On the basis of a clue from site-directed alkylation suggesting that Asn272, Gly268, and Val264 on one face of helix VIII might participate in galactoside binding, molecular dynamics simulations were conducted initially. The simulations indicate that Asn272 (helix VIII) is sufficiently close to the galactopyranosyl ring of a docked lactose analogue to play an important role in binding, the backbone at Gly268 may be involved, and Val264 does not interact with the bound sugar. When the three side chains are subjected to site-directed mutagenesis, with the sole exception of mutant Asn272 → Gln, various other replacements for Asn272 either markedly decrease affinity for the substrate (i.e., high KD) or abolish binding altogether. However, mutant Gly268 → Ala exhibits a moderate 8-fold decrease in affinity, and binding by mutant Val264 → Ala is affected only minimally. Thus, Asn272 and possibly Gly268 may comprise additional components of the galactoside-binding site in LacY.
The lactose permease of Escherichia coli (LacY) specifically binds and transports d-galactose and disaccharides containing a d-galactopyranosyl ring with a H+ (galactoside/H+ symport) and does not recognize d-glucose or d-glucopyranosides, which differ in the orientation of the C4-OH group only. By utilizing the free energy released from the energetically downhill movement of H+ in response to the electrochemical H+ gradient (Δμ̃H+, interior negative and/or alkaline), LacY catalyzes the uphill (active) transport of galactosides against a concentration gradient. Because coupling between sugar and H+ translocation is obligatory, in the absence of Δμ̃H+, LacY can also transduce the energy released from downhill sugar transport to drive the uphill H+ transport with the generation of Δμ̃H+, the polarity of which depends upon the direction of the sugar gradient (reviewed in refs (1) and (2)).
X-ray crystal structures of wild-type (WT) LacY,3 the conformationally restricted mutant C154G,4,5 and a single-Cys mutant with a covalently bound inactivator6 have been determined in the same inward-facing conformation. In each structure, 12 transmembrane α-helices are arranged in two six-helix pseudosymmetrical bundles linked by a long cytoplasmic loop between helices VI and VII. The two six-helix bundles surround a deep hydrophilic cavity that is tightly sealed on the periplasmic face and open to the cytoplasmic side only (an inward-facing conformation). Although the crystal structures suggest that LacY is rigid, a wealth of biochemical–spectroscopic data7−15 demonstrate that the molecule can open alternatively to either side of the membrane upon sugar binding, thereby providing evidence of an alternating-access model for symport (reviewed in refs (16) and (17)).
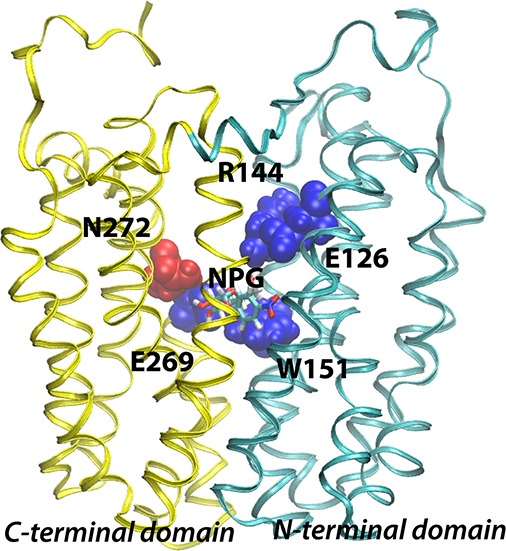
The initial X-ray structure of LacY was obtained with a density at the apex of the central cavity, but at ∼3.5 Å resolution, side-chain interactions with bound sugar cannot be identified clearly.4,5 However, biochemical–biophysical studies show that LacY contains a single galactoside-binding site and that the residues involved in sugar binding are located at or near the apex of the central cavity in the molecule (Figure 1). Although the specificity of LacY is strongly directed toward the C4-OH group of the d-galactopyranosyl ring, the C2-OH, C3-OH, and C6-OH groups are also important in the following order: C4-OH ≫ C6-OH > C3-OH > C2-OH.18,19 The residues responsible for sugar binding are located in helices IV, V, and VIII. Cys-scanning mutagenesis, site-directed alkylation, thiol cross-linking, and direct binding assays show that Glu126 (helix IV) and Arg144 (helix V) are critical for substrate binding and probably charge paired.20−23 Trp151 (helix V), two turns removed from Arg144, stacks aromatically with the galactopyranosyl ring.24,25 Glu269 (helix VIII), another irreplaceable residue,26,27 is also essential for sugar recognition and binding and cannot even be replaced with Asp without dramatically decreasing the affinity.28,29 Furthermore, mass spectrometry studies suggest that Glu269 may interact with the C3-OH group of the galactopyranosyl ring.30
Figure 1.
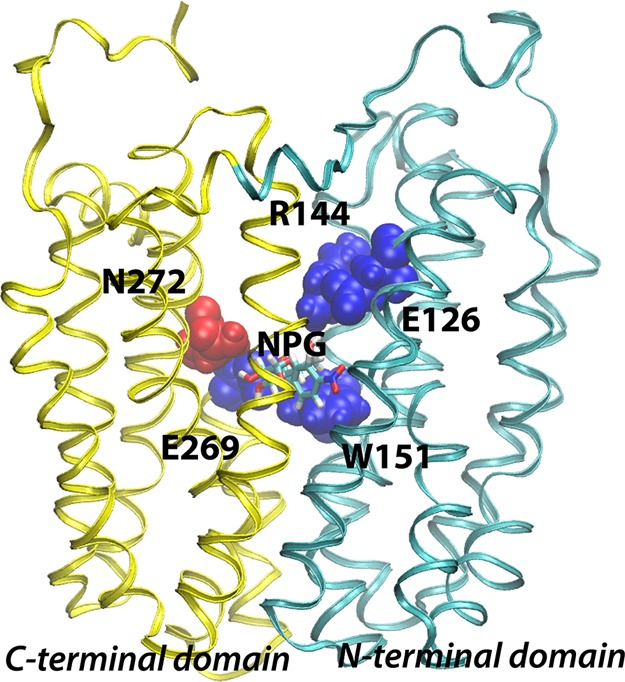
Backbone structure of LacY. The structure of wild-type LacY (PDB entry 2V8N) viewed from the side with the N-terminal six-helix bundle on the right and the C-terminal six-helix bundle on the left. Already known sugar-binding residues are depicted using blue van der Waal spheres, while Asn272 identified in this study is colored red. The NPG sugar is shown as sticks at the apex of the central cavity.
Cys-scanning mutagenesis31 and site-directed alkylation studies32 suggest that Asn272, Gly268, and Val264 on the same face of helix VIII might also be components of the sugar-binding site, and to gain further insight, we conducted a set of molecular dynamics (MD) simulations. The simulations indicate that Asn272, one turn removed from critical Glu269 in helix VIII, is very likely a component of the sugar-binding site, while the backbone at Gly268 or Val264 is unlikely to be directly involved in binding. Consistently, binding studies with purified mutant LacY molecules show that of a dozen replacements for Asn272 only mutant N272Q binds NPG with an affinity similar to that of the WT. All other replacements exhibit either markedly decreased affinity or no significant binding. Interestingly, mutant G268A also exhibits a significant decrease in affinity, suggesting that it may be peripherally involved in binding, and mutant V264A binds sugar with good affinity, demonstrating that it is not a component of the sugar-binding site.
Materials and Methods
Materials
Oligonucleotides for site-directed mutagenesis were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA). A QuikChange II mutagenesis kit was obtained from Agilent Technologies, Inc. (Santa Clara, CA). Restriction enzymes were purchased from New England Biolabs Inc. (Ipswich, MA). Lactose and p-nitrophenyl α-d-galactopyranoside (NPG) were from Sigma-Aldrich (St. Louis, MO). Melibiose was obtained from Acros Organics. [d-glucose-14C(U)]Lactose was from Moravek Biochemicals, Inc. (Brea, CA). The penta-His antibody–horseradish peroxidase (HRP) conjugate was from Qiagen (Hilden, Germany). Supersignal West Pico Chemiluminescent substrate kits for Western blotting were from Pierce (Rockford, IL). Micro BCA protein assay kits were from Thermo Scientific (Rockford, IL).
Molecular Dynamics (MD) Simulations
The Multipurpose Atom-Typer for CHARMM (MATCH)33,34 was used to obtain parameters for NPG. The crystal structure of WT, substrate-free LacY, in an inward-facing conformation, and including N-terminal Met1 to C-terminal Ala417 (PDB entry 2V8N), was inserted into a phosphatidylethanolamine lipid bilayer by aligning the centers of mass of the protein TM domain and the POPE bilayer, respectively, and removing lipids to avoid protein–lipid steric clashes. The system was solvated by explicit water molecules, and counterions were added to achieve electrical neutrality. The three simulation systems were relaxed using a 10000-step conjugate gradient energy minimization followed by gradual heating from 0 to 310 K over 120 ps at a constant temperature (310 K) and volume (NVT ensemble). Equilibrated positions of lipids, water molecules, and the protein were obtained by a series of consecutive 500 ps simulations, where the harmonic restraints on these groups were successively released at a constant temperature (310 K) and pressure (1 atm) (NPT ensemble). To introduce the NPG sugar molecule, WT LacY with β-d-galactopyranosyl 1-thio-β-d-galactopyranoside (TDG) coordinates (PDB entry 1PV7) was superimposed on the 1.5 ns NPT-equilibrated LacY structure followed by alignment of NPG with TDG. Water molecules were removed to eliminate water–NPG steric clashes, and the equilibration protocol was repeated, i.e., energy minimization and equilibration of water molecules and protein atoms, respectively, followed by unrestrained simulation of the wild-type, G268C, and V264C systems for 206, 79, and 89 ns, respectively.
The MD simulations were conducted with the NAMD 2.7 software package,35 and the CHARMM22 and CHARMM36 force fields36,37 were used for the protein and lipids, respectively. The TIP3P model was used for the water molecules.38 A reversible multiple-time step (MTS) algorithm39 was used to integrate the equations of motion with time steps of 1 fs for bonded forces, 2 fs for short-range, nonbonded forces, and 4 fs for long-range, electrostatic forces. The smooth particle mesh Ewald method40,41 was used to calculate electrostatic interactions. The short-range interactions were cut off at 12 Å. All bond lengths involving hydrogen atoms were held fixed using the SHAKE42 and SETTLE43 algorithms. A Langevin dynamics scheme was used for thermostating. Nosé–Hoover–Langevin pistons were used for pressure control.44,45 Molecular graphics and simulation analyses were generated with VMD version 1.9.1.46
Construction of Mutants
All mutants were constructed by site-directed mutagenesis using plasmid pT7-5 containing the cassette lacY gene with a C-terminal six-His tag as a template. All mutations were verified by sequencing of the entire lacY gene and the restriction sites.
Lactose Transport
E. coli T184 [lacI+O+Z–Y– (A) rpsL met–thr–recA hsdM hsdR/F′ lacIq O+ ZD118 (Y+A+)]47,48 cells were transformed with plasmid pT7-5 encoding WT LacY or a given mutant and grown in LB broth containing 0.1 mg/mL ampicillin at 37 °C overnight. The culture was diluted into 30 mL of LB broth and induced with 1 mM (final concentration) IPTG at an OD600 of 0.6. After being induced for 1 h, the cells were assayed for transport of [1-14C]lactose (10 mCi/mmol, final concentration of 0.4 mM) by rapid filtration.49
Western Blotting
The cells used in the lactose transport assays were lysed by sonication. A membrane fraction was obtained by ultracentrifugation (2.2 × 105 × g, 4 °C for 20 min) and suspended in 0.1 M KPi (pH 7.5) and 10 mM MgSO4. An aliquot of the sample containing 2 μg of membrane protein was used for Western blotting and probed with the penta-His antibody–HRP conjugate to indentify LacY.50
LacY Purification
E. coli XL-1 blue cells transformed with plasmid pT7-5 encoding a given mutant were grown in 1 L of LB broth containing 0.1 mg/mL ampicillin at 37 °C overnight. A 10-fold dilution of the culture was grown in a fermenter and induced with 0.3 mM (final concentration) IPTG at an OD600 of 0.6. After being induced for 3 h, the cells were harvested and lysed with a French press. His-tagged LacY in the cell lysate was purified as described previously.51 Purified LacY was solubilized in 50 mM NaPi (pH 7.6) and 0.01% DDM, flash-frozen in liquid nitrogen, and stored at −80 °C until use. The protein concentration was determined by a Micro BCA protein assay.
NPG Binding
NPG binding measurements were made with an SLM-Aminco 8100 spectrofluorometer as described previously52 with minor modifications. In a 1 cm × 1 cm cuvette, purified WT or a given mutant was diluted in 50 mM NaPi (pH 7.5) and 0.01% DDM to a final concentration of 1 μM in a volume of 2 mL. α-NPG was added to a given concentration, and then 30 mM (final concentration) melibiose was added to displace NPG. Changes in fluorescence resulting from Trp → α-NPG FRET were recorded while the sample was being constantly stirred and corrected for dilution caused by addition of the ligand. KD values were determined with GraFit version 6 (Erithacus Software, London, U.K.) using “1Site-Ligand Binding” equation
Results
MD Simulations
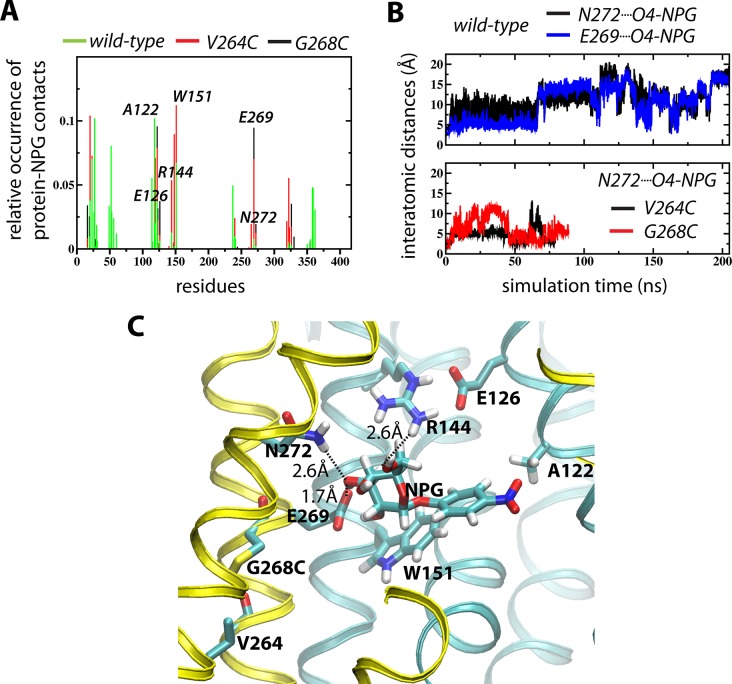
With the LacY WT crystal structure (PDB entry 2V8N) as a starting point, NPG was docked according to coordinates from the WT structure with β-d-galactopyranosyl 1-thio-β-d-galactopyranoside (TDG) coordinates (PDB entry 1PV7) (see Materials and Methods). A set of three MD simulations of WT, G268C, and V264C LacY disclose one major pose of NPG in which Asn272 contributes directly to sugar binding (Figure 2). As observed by contact analyses reporting <3.5 Å protein–NPG interactions, the sugar molecule interacts with residues thought to be involved in sugar binding (Figure 2A). A guanidino group at position 144 (Arg), a carboxyl group at position 126 (preferably Glu), an aromatic residue at position 151 (preferably Trp), and a carboxyl group at position 269 (Glu) are obligatory for sugar binding (reviewed in ref (1)). The nitro group in NPG is proximal to Ala122 in all three simulations, which is consistent with previous observations.53 Rather than being fixed in a single position with respect to the surrounding protein, the NPG molecule is highly dynamic, although with one clearly defined preferred binding pose. This is particularly evident in the wild-type simulation, where NPG visits the alternative locations in the wide, water-filled cytoplasmic cleft (Figure 2A).
Figure 2.
MD simulations of NPG binding. (A) Contact analyses of the WT (green), G268C (black), and V264C (red) simulations reporting <3.5 Å protein–NPG interactions. (B) Evolution of the interatomic distances between NPG O4 and N272 NH (black) and E269 O (blue) for the WT (top) and the NPG O4···N272 NH distances for the G268C (red) and V264C (black) simulations. (C) NPG binding pose displayed by the final frame of the G268C simulation.
Residues Asn272 and Glu269 on helix VIII make contacts with NPG (Figure 2A). In all three simulations, sugar binding dynamics between Asn272 and the sugar-interacting residue Glu269 are synchronized (Figure 2B). However, neither the backbone at position 268 nor that at position 264 appears to make contact with the galactopyranosyl ring of NPG (Figure 2A). In events where Asn272 and Glu269 are in the proximity of the galactopyranosyl moiety, known sugar-binding residues Glu126, Arg144, and Trp151 are also exposed to NPG (Figure 2C).
Lactose Transport
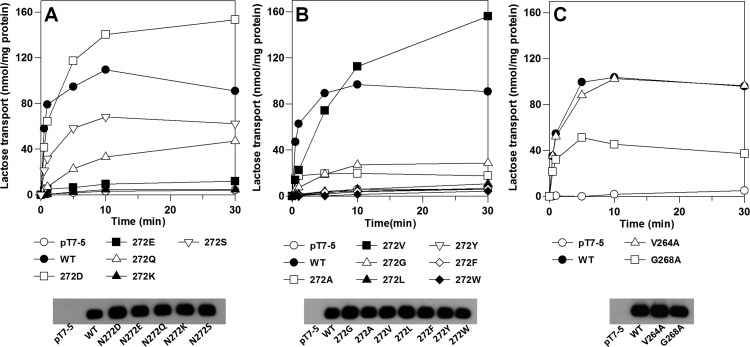
Of 12 Asn272 replacement mutants tested, only four (N272D, N272V, N272S, and N272Q) transport lactose to a steady-state level of accumulation that is ≥30% of the steady state established by WT LacY in 30 min (in decreasing order, N272D, N272V, N272S, and N272Q). Mutants N272G and N272A accumulate to levels that are ∼25 and ∼20% of that of the WT, respectively, and the activities of the remaining mutants are negligible (Figure 3A,B). The initial rate of transport by mutant N272D approximates that of the WT, but the initial rates of transport observed with mutants N272V, N272Q, and N272S are drastically lower than that of the WT. Mutant V264A accumulates lactose as well as WT. Although the initial rate of transport by mutant G268A is similar to that of the WT, the steady-state level of accumulation is only ∼40% of that of the WT.
Figure 3.
Lactose transport. Transport of [14C]lactose (10 mCi/mmol) of E. coli T184 expressing WT LacY, mutant N272D, N272E, N272K, N272Q, N272S, N272A, N272V, N272G, N272L, N272Y, N272F, N272W, V264A, or G268A, or no permease was measured at 0.4 mM lactose for given times as described in Materials and Methods. Expression of WT LacY and each mutant as determined by Western blotting.
Western blotting with the anti-His antibody reveals that each mutant is expressed at approximately the same levels as WT LacY (Figure 3A–C, bottom panels). Therefore, the differences in transport activity are not due to variations in the expression of the mutants.
NPG Binding
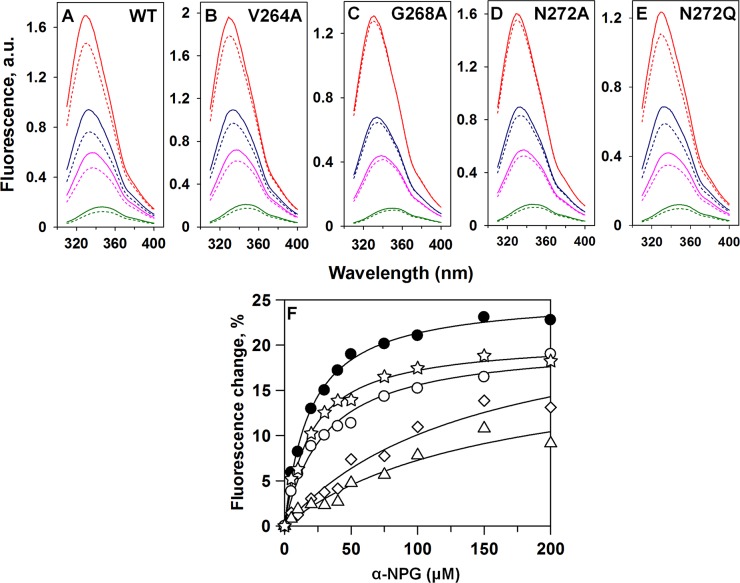
NPG is a high-affinity sugar analogue of lactose, and previous studies52 show that the distance between Trp151 in the binding site and the nitrophenyl group of NPG (∼12 Å) is a favorable distance for Förster resonance energy transfer (FRET). Because the analogue has a broad absorption spectrum with a maximum at 306 nm (not shown), NPG affects Trp fluorescence by two simultaneous processes: (1) by serving as a nonfluorescent FRET acceptor from Trp151 in the binding site and (2) by acting as an inner filter and absorbing irradiated excitation light at 295 nm, as well as fluorescence emission of Trp. To discriminate between the two processes, another lactose analogue, melibiose, which is not fluorescent and does not absorb light over the range of wavelengths studied, was used. Addition of saturating concentrations of melibiose in the absence of NPG causes little or no change in the emission spectrum of Trp. However, when melibiose is added after incubation with NPG, an increase in Trp fluorescence is observed because of displacement of NPG from the binding site. Thus, the increase in Trp fluorescence upon addition of melibiose represents a specific FRET effect, and the remainder of the fluorescence change that is not restored by melibiose represents the nonspecific inner filter effect caused by NPG in solution. The apparent affinity for NPG is estimated from the concentration dependence of the specific fluorescence change after addition of excess melibiose at various NPG concentrations (Figure 4A–E). The calculated KD of 19 ± 1 μM obtained for WT LacY (Figure 4F) is the same as that obtained previously.52
Figure 4.
NPG binding. (A–E) Trp fluorescence emission spectra at 20 (red), 50 (blue), 75 (pink), and 150 mM (green) NPG are shown for purified WT LacY (A), V264A (B), G268A (C), N272A (D), and N272Q (E). Broken lines are spectra after the addition of α-NPG. Solid lines are spectra after the addition of 30 mM melibiose. (F) Binding of α-NPG to purified WT LacY (●), V264A (○), G268A (△), N272A (◇), and N272Q (☆). The changes in fluorescence induced by addition of melibiose are plotted as a function of NPG concentration. Differences obtained with mutants N272D, N272E, N272F, N272S, and N272V were too small for accurate measurement.
In addition to WT LacY, significant fluorescence changes are observed with mutant N272Q (Figure 4E) and to a lesser extent with mutant N272A (Figure 4D), and KD values of 20 ± 2 and 143 ± 39 μM, respectively, are obtained (Figure 4F and Table 1). In contrast, the fluorescence changes observed with mutants N272D, N272V, N272E, N272F, and N272S were too small to allow estimates of binding constants. Significant fluoresecence changes are also observed with mutant V264A, which yield a KD of 30 ± 4 μM (Figure 4B and Table 1). However, mutant G268A exhibits smaller changes in fluorescence and an affinity for NPG (∼8-fold) lower than that of WT LacY (Figure 4C and Table 1).
Table 1. KD Values for NPG Binding of LacY Mutants.
| KDapp (μM)a | standard error | |
|---|---|---|
| WT | 19.2 | 1.1 |
| N272A | 142.6 | 39.4 |
| N272D | –b | –b |
| N272E | –b | –b |
| N272F | –b | –b |
| N272Q | 20.0 | 1.8 |
| N272S | –b | –b |
| N272V | –b | –b |
| V264A | 30.1 | 4.1 |
| G268A | 154.7 | 57.3 |
Values determined as described in the legend of Figure 4.
Binding is too poor to determine a KD.
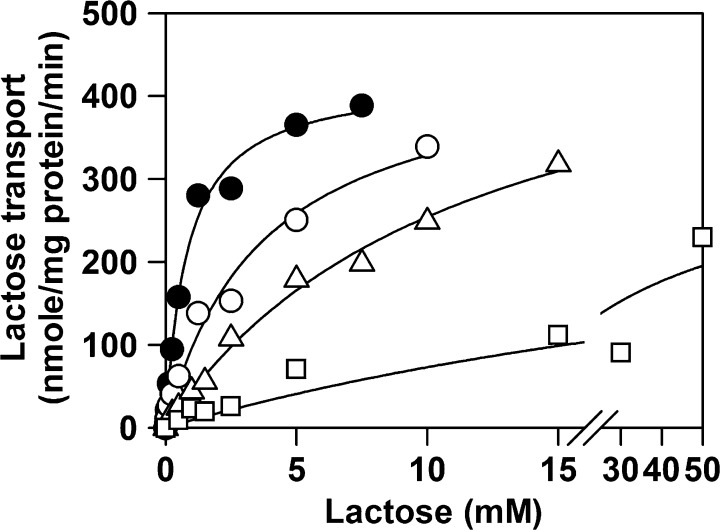
Kinetics of Selected Mutants
The kinetics of lactose transport for WT LacY and mutants N272D, N272Q, N272V, V264A, and G268A were measured to determine whether there is a correlation between the measured KD values and the kinetic constants (Figure 5 and Table 2). The Km obtained for WT LacY is ∼0.8 ± 0.1 mM with a Vmax of 442 ± 14 nmol (mg of protein)−1 min–1, in reasonable agreement with previous findings.54 The Km obtained for mutant N272D is ∼5 times higher than that of the WT, and the Vmax is approximately the same as that of the WT. Replacing Asn272 with Val leads to an ∼15-fold increase in Km, with a Vmax significantly higher than that of the WT. Remarkably, although WT LacY and mutant N272Q have essentially the same KD for NPG (Table 2), the Km obtained for lactose transport is more than 40 times higher for N272Q than for the WT, with a lower Vmax. Finally, replacement of Val264 or Gly268 with Ala has little or no effect on either Km or Vmax (data not shown).
Figure 5.
Table 2. Kinetics of Selected Forms of LacY.
| Km (mM)a | standard error | Vmax [nmol (mg of protein)−1 min–1]a | standard error | |
|---|---|---|---|---|
| WT | 0.8 | 0.1 | 422 | 14 |
| N272D | 3.7 | 0.8 | 450 | 40 |
| N272Q | 36 | 28 | 336 | 139 |
| N272V | 12 | 2 | 547 | 47 |
Values determined as described in the legend of Figure 5.
Discussion
The specificity of LacY for the substrate is directed exclusively toward the galactopyranosyl ring of the substrate. Thus, the monosaccharide d-galactose is the most specific sugar for LacY, although it has a very low affinity.18 Remarkably, substitutions on the anomeric carbon, particularly if they are hydrophobic, can lead to a marked increase in affinity (up to ∼3 orders of magnitude) with no change in specificity.55
Asn272 (helix VIII), positioned one turn from Glu269 toward the cytoplasmic side, is proximal to the galactopyranosyl ring. Cys-scanning mutagenesis of helix VIII reveals that mutant N272C transports lactose only 30% as well as the WT,31 and site-directed alkylation demonstrates that mutants N272C, G268C, and V264C are protected against alkylation by the substrate, suggesting that all three positions might be in the vicinity of the sugar-binding site.32 Moreover, sequence alignment data also show that Asn272 is conserved in bacterial galactoside/H+ symporters.56 Because the interactions of LacY with the substrate are based primarily upon biochemical findings (reviewed in refs (1) and (2)), to investigate further a possible role of Asn272, Gly268, and Val264 in sugar binding, we initially utilized molecular dynamics (MD) simulations.
The simulations reveal a highly dynamic NPG substrate that adopts a similar docking position in the WT, G268C, and V264C simulations. It was possible to monitor substrate entering and leaving this docking pose by tracking interactions between residues Glu269 and Asn272 and the C4-OH group on the NPG galactopyranosyl ring (Figure 2B). It is particularly evident in the >200 ns WT simulation that the sugar visits alternative positions in the cytoplasmic cleft. In both mutant simulations, NPG remains in a binding pose that also involves interaction between the guanidino group of Arg144 and the C6-OH group of the galactopyranosyl ring. While Glu126 is relatively far removed, Trp151 is stacked with the under side of the galactopyranosyl ring and the nitrophenyl group is directed toward Ala122 (Figure 2C).53 It is clear that backbone positions 264 and 268, which have been implicated in sugar binding, do not interact directly with NPG. Of all atoms in these positions, the backbone carbonyl oxygen at position 268 is closest (14.4 ± 3.2 Å), which corroborates an 8-fold decrease in the affinity of the G268A mutant. Because mutations G268C and V264C are also relatively far from the NPG-binding site, it is likely that the three simulations sample the same fundamental protein–sugar interactions.
Mutagenesis shows that except for mutant N272D, in which significant NPG binding cannot be measured, all other replacement mutants tested exhibit significantly decreased initial rates of lactose transport. Furthermore, only mutants N272D and N272V accumulate lactose to a steady state comparable to that of the WT (Figure 3A,B). Mutant N272Q exhibits low transport activity, but transport activity in the other mutants is essentially moribund. The transport defect in mutants N272F, N272Y, and N272W may be due to the bulky aromatic side chain at position 272, which could sterically block binding. The low activity of mutants N272G and N272A suggests that the amide group of Asn272 is probably important for binding.
Mutant N272Q with the most conservative replacement for Asn has a KD that is essentially the same as that of the WT, and replacing the amide group with virtually any other amino acid residue except Ala essentially abolishes NPG binding. Even with Ala in place of Asn272, the NPG affinity decreases by almost 8-fold. Although it is tempting to try to relate the transport data presented to the binding observations, it should be emphasized that the transport studies are conducted with lactose, which binds with very low affinity, while the binding data are obtained with the high-affinity homologue NPG. Thus, although the 45-fold increase in Km for mutant N272Q relative to that of the WT may explain the relatively low lactose transport activity, the KD for NPG is the same as that of the WT. In a similar vein, there appears to be no correlation whatsoever among decreased NPG affinity, increased Km, and transport activity.
In any case, primarily on the basis of the MD simulations and the NPG binding data in conjunction with the earlier site-directed alkylation experiments,31,32 it seems highly probable that Asn272 is a component of the sugar-binding site in LacY. Previous site-directed alkylation shows that two additional mutants, V264C and G268C, are also protected by sugar against NEM labeling. In this study, replacement of Val264 with Ala does not affect either NPG binding or the kinetics of lactose transport, indicating that Val264 is not a component of the sugar-binding site. However, mutant G268A exhibits an ∼8-fold increase in its KD (i.e., a moderate decrease in affinity), but lactose transport remains unchanged. Although the results are somewhat ambiguous, they imply that the backbone at the position of Gly268 might interact with bound sugar and that the moderate decrease in affinity exhibited by mutant G268A may be due to the steric hindrance. Another possibility is that a Gly residue at position 268 provides flexibility near the binding site.
Acknowledgments
We are indebted to Irina Smirnova, Vladimir Kasho, and Junichi Sugihara for helpful discussions and for technical assistance with the binding experiments.
Glossary
Abbreviations
- DDM
n-dodecyl β-d-maltopyranoside
- FRET
Förster resonance energy transfer
- IPTG
isopropyl β-d-1-thiogalactopyranoside
- KPi
potassium phosphate
- NaPi
sodium phosphate
- NEM
N-ethylmaleimide
- NPG
p-nitrophenyl α-d-galactopyranoside
- PDB
Protein Data Bank
- TDG
β-d-galactopyranosyl 1-thio-β-d-galactopyranoside
- SDS–PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis.
Author Present Address
§ M.A.: Science for Life Laboratory, Department of Theoretical Physics, Swedish e-Science Research Center, KTH Royal Institute of Technology, SE-171 21 Solna, Sweden.
The authors declare no competing financial interest.
The research was supported by National Institutes of Health (NIH) Grants DK051131, DK069463, GM073210, and GM074929 to H.R.K. The research was also supported in part by NIH Grants GM074637 and GM086685 to S.H.W. M.A. was supported in part by a Senior Postdoctoral Fellowship from the Branches Cost-Sharing Fund through the Institute of Complex Adaptive Matter ICAM, which was itself supported in part by the National Science Foundation, and by a Marie Curie Career Integration Grant (FP7-MC-CIG-618558).
While this paper was in review, an X-ray crystal structure of a LacY mutant was published which exhibits an almost occluded, slightly outward-open conformation with a clearly bound lactose analogue. In confirmation of the work presented here, Asn272 is ∼3.7 Å from the C4-OH of the galactopyranosyl ring [Kumar, H., Smirnova, I. et al (2014) Proc. Natl. Acad. Sci. USA 111, 1784−1788.
Funding Statement
National Institutes of Health, United States
References
- Guan L.; Kaback H. R. (2006) Lessons from lactose permease. Annu. Rev. Biophys. Biomol. Struct. 35, 67–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madej M. G., and Kaback H. R. (2014) The Life and Times of Lac Permease: Crystals Ain’t Enough, but They Certainly do Help. In Membrane transporter function: To structure and beyond (Ziegler C., and Kraemer R., Eds.) Springer Series in Biophysics: Transporters, Springer, Berlin. [Google Scholar]
- Guan L.; Mirza O.; Verner G.; Iwata S.; Kaback H. R. (2007) Structural determination of wild-type lactose permease. Proc. Natl. Acad. Sci. U.S.A. 104, 15294–15298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramson J.; Smirnova I.; Kasho V.; Verner G.; Kaback H. R.; Iwata S. (2003) Structure and mechanism of the lactose permease of Escherichia coli. Science 301, 610–615. [DOI] [PubMed] [Google Scholar]
- Mirza O.; Guan L.; Verner G.; Iwata S.; Kaback H. R. (2006) Structural evidence for induced fit and a mechanism for sugar/H+ symport in LacY. EMBO J. 25, 1177–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaptal V.; Kwon S.; Sawaya M. R.; Guan L.; Kaback H. R.; Abramson J. (2011) Crystal structure of lactose permease in complex with an affinity inactivator yields unique insight into sugar recognition. Proc. Natl. Acad. Sci. U.S.A. 108, 9361–9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaback H. R.; Dunten R.; Frillingos S.; Venkatesan P.; Kwaw I.; Zhang W.; Ermolova N. (2007) Site-directed alkylation and the alternating access model for LacY. Proc. Natl. Acad. Sci. U.S.A. 104, 491–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova I.; Kasho V.; Choe J. Y.; Altenbach C.; Hubbell W. L.; Kaback H. R. (2007) Sugar binding induces an outward facing conformation of LacY. Proc. Natl. Acad. Sci. U.S.A. 104, 16504–16509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar D. S.; Smirnova I.; Kasho V.; Nir E.; Kong X.; Weiss S.; Kaback H. R. (2007) Single-molecule FRET reveals sugar-induced conformational dynamics in LacY. Proc. Natl. Acad. Sci. U.S.A. 104, 12640–12645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y.; Guan L.; Freites J. A.; Kaback H. R. (2008) Opening and closing of the periplasmic gate in lactose permease. Proc. Natl. Acad. Sci. U.S.A. 105, 3774–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y.; Nie Y.; Kaback H. R. (2009) Residues Gating the Periplasmic Pathway of LacY. J. Mol. Biol. 394, 219–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova I.; Kasho V.; Sugihara J.; Kaback H. R. (2009) Probing of the rates of alternating access in LacY with Trp fluorescence. Proc. Natl. Acad. Sci. U.S.A. 106, 21561–21566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Y.; Kaback H. R. (2010) Sugar binding induces the same global conformational change in purified LacY as in the native bacterial membrane. Proc. Natl. Acad. Sci. U.S.A. 107, 9903–9908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X.; Nie Y.; Kaback H. R. (2011) Site-Directed Alkylation Studies with LacY Provide Evidence for the Alternating Access Model of Transport. Biochemistry 50, 1634–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova I.; Kasho V.; Sugihara J.; Kaback H. R. (2011) Opening the periplasmic cavity in lactose permease is the limiting step for sugar binding. Proc. Natl. Acad. Sci. U.S.A. 108, 15147–15151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaback H. R.; Smirnova I.; Kasho V.; Nie Y.; Zhou Y. (2011) The alternating access transport mechanism in LacY. J. Membr. Biol. 239, 85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova I.; Kasho V.; Kaback H. R. (2011) Lactose permease and the alternating access mechanism. Biochemistry 50, 9684–9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin-Tóth M.; Akhoon K. M.; Runner J.; Kaback H. R. (2000) Ligand recognition by the lactose permease of Escherichia coli: Specificity and affinity are defined by distinct structural elements of galactopyranosides. Biochemistry 39, 5097–5103. [DOI] [PubMed] [Google Scholar]
- Sahin-Tóth M.; Lawrence M. C.; Nishio T.; Kaback H. R. (2001) The C-4 hydroxyl group of galactopyranosides is the major determinant for ligand recognition by the lactose permease of Escherichia coli. Biochemistry 40, 13015–13019. [DOI] [PubMed] [Google Scholar]
- Frillingos S.; Gonzalez A.; Kaback H. R. (1997) Cysteine-scanning mutagenesis of helix IV and the adjoining loops in the lactose permease of Escherichia coli: Glu126 and Arg144 are essential. Biochemistry 36, 14284–14290. [DOI] [PubMed] [Google Scholar]
- Sahin-Toth M.; le Coutre J.; Kharabi D.; le Maire G.; Lee J. C.; Kaback H. R. (1999) Characterization of Glu126 and Arg144, two residues that are indispensable for substrate binding in the lactose permease of Escherichia coli. Biochemistry 38, 813–819. [DOI] [PubMed] [Google Scholar]
- Venkatesan P.; Kaback H. R. (1998) The substrate-binding site in the lactose permease of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 95, 9802–9807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin C. D.; Kaback H. R. (2000) Thiol cross-linking of transmembrane domains IV and V in the lactose permease of Escherichia coli. Biochemistry 39, 6130–6135. [DOI] [PubMed] [Google Scholar]
- Guan L.; Hu Y.; Kaback H. R. (2003) Aromatic stacking in the sugar binding site of the lactose permease. Biochemistry 42, 1377–1382. [DOI] [PubMed] [Google Scholar]
- Vazquez-Ibar J. L.; Guan L.; Svrakic M.; Kaback H. R. (2003) Exploiting luminescence spectroscopy to elucidate the interaction between sugar and a tryptophan residue in the lactose permease of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 100, 12706–12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujwal M. L.; Sahin-Toth M.; Persson B.; Kaback H. R. (1994) Role of glutamate-269 in the lactose permease of Escherichia coli. Mol. Membr. Biol. 11, 9–16. [DOI] [PubMed] [Google Scholar]
- Frillingos S.; Sahin-Toth M.; Wu J.; Kaback H. R. (1998) Cys-scanning mutagenesis: A novel approach to structure function relationships in polytopic membrane proteins. FASEB J. 12, 1281–1299. [DOI] [PubMed] [Google Scholar]
- He M. M.; Kaback H. R. (1997) Interaction between residues Glu269 (helix VIII) and His322 (helix X) of the lactose permease of Escherichia coli is essential for substrate binding. Biochemistry 36, 13688–13692. [DOI] [PubMed] [Google Scholar]
- Smirnova I.; Kasho V.; Sugihara J.; Choe J. Y.; Kaback H. R. (2009) Residues in the H+ translocation site define the pKa for sugar binding to LacY. Biochemistry 48, 8852–8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinglass A. B.; Whitelegge J. P.; Hu Y.; Verner G. E.; Faull K. F.; Kaback H. R. (2003) Elucidation of substrate binding interactions in a membrane transport protein by mass spectrometry. EMBO J. 22, 1467–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frillingos S.; Ujwal M. L.; Sun J.; Kaback H. R. (1997) The role of helix VIII in the lactose permease of Escherichia coli: I. Cys-scanning mutagenesis. Protein Sci. 6, 431–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frillingos S.; Kaback H. R. (1997) The role of helix VIII in the lactose permease of Escherichia coli: II. Site-directed sulfhydryl modification. Protein Sci. 6, 438–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadyvaloo V.; Smirnova I. N.; Kasho V. N.; Kaback H. R. (2006) Conservation of residues involved in sugar/H+ symport by the sucrose permease of Escherichia coli relative to lactose permease. J. Mol. Biol. 358, 1051–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yesselman J. D.; Price D. J.; Knight J. L.; Brooks C. L. III (2012) MATCH: An atom-typing toolset for molecular mechanics force fields. J. Comput. Chem. 33, 189–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips J. C.; Braun R.; Wang W.; Gumbart J.; Tajkhorshid E.; Villa E.; Chipot C.; Skeel R. D.; Kale L.; Schulten K. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauda J. B.; Venable R. M.; Freites J. A.; O’Connor J. W.; Tobias D. J.; Mondragon-Ramirez C.; Vorobyov I.; MacKerell A. D. Jr.; Pastor R. W. (2010) Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J. Phys. Chem. B 114, 7830–7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKerell A. D.; Bashford D.; Bellott M.; Dunbrack R.; Evanseck J.; Field M. J.; Fischer S.; Gao J.; Guo H.; Ha S. A. (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616. [DOI] [PubMed] [Google Scholar]
- Jorgensen W.; Chandrasekhar J.; Madura J.; Impey R.; Klein M. (1983) Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935. [Google Scholar]
- Grubmüller H.; Heller H.; Windemuth A.; Schulten K. (1991) Generalized Verlet Algorithm for Efficient Molecular Dynamics Simulations with Long-range Interactions. Mol. Simul. 6, 121–142. [Google Scholar]
- Darden T.; York D.; Pedersen L. (1993) Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092. [Google Scholar]
- Essmann U.; Perera L.; Berkowitz M.; Darden T.; Lee H.; Pedersen L. (1995) A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8593. [Google Scholar]
- Ryckaert J.-P.; Ciccotti G.; Berendsen H. (1977) Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 23, 327–341. [Google Scholar]
- Miyamoto S.; Kollman P. (1992) Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 13, 952–962. [Google Scholar]
- Feller S.; Zhang Y.; Pastor R.; Brooks B. (1995) Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 103, 4613–4621. [Google Scholar]
- Martyna G.; Tobias D.; Klein M. (1994) Constant pressure molecular dynamics algorithms. J. Chem. Phys. 101, 4177–4189. [Google Scholar]
- Humphrey W.; Dalke A.; Schulten K. (1996) VMD: Visual molecular dynamics. J. Mol. Graphics 14, 27–38. [DOI] [PubMed] [Google Scholar]
- Teather R. M.; Bramhall J.; Riede I.; Wright J. K.; Furst M.; Aichele G.; Wilhelm U.; Overath P. (1980) Lactose carrier protein of Escherichia coli. Structure and expression of plasmids carrying the Y-gene of the lac operon. Eur. J. Biochem. 108, 223–231. [DOI] [PubMed] [Google Scholar]
- Teather R. M.; Müller-Hill B.; Abrutsch U.; Aichele G.; Overath P. (1978) Amplification of the lactose carrier protein in Escherichia coli using a plasmid vector. Mol. Gen. Genet. 159, 239–248. [DOI] [PubMed] [Google Scholar]
- Kaback H. R. (1974) Transport in isolated bacterial membrane vesicles. Methods Enzymol. 31, 698–709. [DOI] [PubMed] [Google Scholar]
- Smirnova I. N.; Kaback H. R. (2003) A mutation in the lactose permease of Escherichia coli that decreases conformational flexibility and increases protein stability. Biochemistry 42, 3025–3031. [DOI] [PubMed] [Google Scholar]
- Sugihara J.; Sun L.; Yan N.; Kaback H. R. (2012) Dynamics of the l-fucose/H+ symporter revealed by fluorescence spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 109, 14847–14851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova I. N.; Kasho V. N.; Kaback H. R. (2006) Direct Sugar Binding to LacY Measured by Resonance Energy Transfer. Biochemistry 45, 15279–15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan L.; Sahin-Toth M.; Kaback H. R. (2002) Changing the lactose permease of Escherichia coli into a galactose-specific symporter. Proc. Natl. Acad. Sci. U.S.A. 99, 6613–6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson D. E.; Kaczorowski G. J.; Garcia M. L.; Kaback H. R. (1980) Active transport in membrane vesicles from Escherichia coli: The electrochemical proton gradient alters the distribution of the lac carrier between two different kinetic states. Biochemistry 19, 5692–5702. [DOI] [PubMed] [Google Scholar]
- Sahin-Tóth M.; Gunawan P.; Lawrence M. C.; Toyokuni T.; Kaback H. R. (2002) Binding of hydrophobic d-galactopyranosides to the lactose permease of Escherichia coli. Biochemistry 41, 13039–13045. [DOI] [PubMed] [Google Scholar]
- Kasho V. N.; Smirnova I. N.; Kaback H. R. (2006) Sequence alignment and homology threading reveals prokaryotic and eukaryotic proteins similar to lactose permease. J. Mol. Biol. 358, 1060–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]