Abstract

Cells in vivo exist within the context of a multicellular tissue, where their behavior is governed by homo- and heterotypic cell–cell interactions, the material properties of the extracellular matrix, and the distribution of various soluble and physical factors. Most methods currently used to study and manipulate cellular behavior in vitro, however, sacrifice physiological relevance for experimental expediency. The fallacy of such approaches has been highlighted by the recent development and application of three-dimensional culture models to cell biology, which has revealed striking phenotypic differences in cell survival, migration, and differentiation in genetically identical cells simply by varying culture conditions. These perplexing findings beg the question of what constitutes a three-dimensional culture and why cells behave so differently in two- and three-dimensional culture formats. In the following review, we dissect the fundamental differences between two- and three-dimensional culture conditions. We begin by establishing a basic definition of what “three dimensions” means at different biological scales and discuss how dimensionality influences cell signaling across different length scales. We identify which three-dimensional features most potently influence intracellular signaling and distinguish between conserved biological principles that are maintained across culture conditions and cellular behaviors that are sensitive to microenvironmental context. Finally, we highlight state-of-the-art molecular tools amenable to the study of signaling in three dimensions under conditions that facilitate deconstruction of signaling in a more physiologically relevant manner.
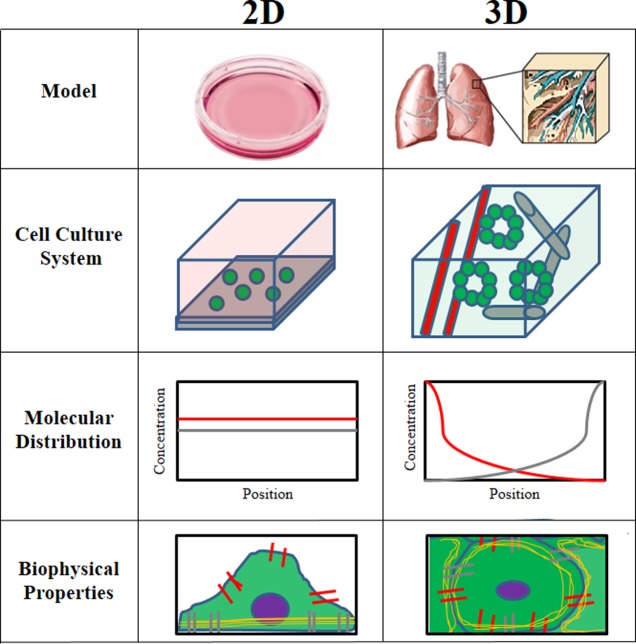
It is important to begin this discussion of signaling in three dimensions (“3D”) by defining what constitutes 3D as compared to a 2D environment. “2D” most frequently refers to a monolayer culture of cells plated on polystyrene or glass surfaces. On these conventional 2D substrates, the cell interacts with a basal extracellular “matrix” and with neighboring cells via lateral cell–cell junctions. Because of equal exposure to the bulk culture medium, a uniform nutrient distribution is assumed. The term 3D was initially used to contrast a 3D culture from a monolayer culture. In one definition, a 3D culture consists of a cell embedded within and surrounded by an extracellular matrix, such that the cell is able to encounter its extracellular microenvironment within a 3D volume of space.1 Another definition suggests 3D refers to the specific topological features and 3D organization of the extracellular matrix.2,3 One common feature of these systems is the exposure of the cell to matrix and/or cell–cell interactions in all directions. Regardless, early studies using these various 3D culture systems demonstrated dramatic differences in cell behavior and signaling between cells grown in 2D versus 3D. Cells in 3D respond differently to exogenous growth factors and are highly resistant to apoptosis.4,5 Consistently, gene expression in a variety of cells types, including glomerular, endothelial, and melanoma cells, is altered in a 3D culture.6−8 Moreover, cell–matrix adhesions in 3D can be composed of proteins different from those in 2D, resulting in altered metabolic activity and nuclear architecture.2,9,10 These observations suggest that cells can undergo a profound and systematic rewiring when they undergo the transition from a 2D to a 3D context. However, while these experimental observations emphasize how cellular context can profoundly modify a plethora of cellular behaviors, the precise mechanism by which dimensionality induces these changes is unclear. Indeed, 3D can mean different things at different biological scales. Within a cell, 3D is felt through changes in cell shape and volume, in the organization of the cytoskeleton, and in the distribution of signaling components throughout the cell. At the cellular level, 3D means that the cell is exposed to stimuli on all sides, whether from the ECM or neighboring cells. On a multicellular level, 3D manifests in the structural organization of units like lobules, ducts, and vessels. These factors all influence the manner in which signaling takes place in distinct ways, as shown in overview in Figure 1. In the following review, we discuss how signaling is influenced by dimensionality at each of these scales and highlight novel tools for the further elucidation of these relationships.
Figure 1.
Comparison of 2D vs 3D. The environment in which the cell is cultured differs dramatically between 2D and 3D.
1. Signaling in 3D within the Cell: Intracellular Localization
Conventionally, signaling events within the cell are modeled as concentration-dependent biochemical reactions. This paradigm assumes that signaling molecules are freely diffusing and uniformly distributed in the cytoplasm and has resulted in a “whole-cell” perspective on signaling, where the cell is considered the reaction vessel in which given concentrations of signaling molecules react with each other. Conversely, a large body of evidence has arisen to suggest that the localization of a specific signaling molecule within a cell greatly impacts the magnitude and the effect of the resultant signaling event.11,12 Signaling molecules can be constrained in space by barriers to free diffusion, scaffolded into multiprotein complexes, or tethered to 2D membranes. Each of these situations significantly alters the diffusivity of signaling molecules, which in turn changes the frequency, speed, and duration of biochemical reactions.
Barriers to Diffusion: Scaffolding Proteins
Scaffolding proteins can nucleate large, multiprotein complexes that accelerate reaction kinetics. By bringing multiple components of a signaling cascade together, these proteins alter the reaction kinetics governing a specific signaling cascade. β-arrestin, for example, can bind several components of the MAPK pathway, including RAF, MEK, and ERK.13,14 By clustering these proteins, β-arrestin eliminates RAF–MEK binding as a rate-limiting step. Following MEK sequestration by β-arrestin, MEK phosphorylation by RAF occurs at a constant rate.13 Physiologically, β-arrestin enhances ERK activation downstream of growth factor receptor signaling by prolonging the RAF–MEK–ERK interaction time in the cytoplasm.13,15
Similarly, paxillin serves as a scaffold for proteins at sites of integrin-mediated cell–matrix adhesions. These adhesion complexes connect the cytoskeleton to proteins within the extracellular matrix (ECM) and are critical sites for the transduction of mechanical inputs to intracellular signaling.16 Notably, activated paxillin increases the frequency of interaction between protein kinases such as focal adhesion kinase and Src, which can control downstream Rac activity and increase cell motility.17,18 Thus, paxillin mediates mechanotransduction by connecting adhesion proteins with signaling pathways governing motility and proliferation.
Scaffolds can also serve to reduce the extent of signaling in specific pathways. LKB-1 interacting protein 1 (LIP-1) is a scaffolding protein that sequesters the transcription factor Smad4 from binding to TGFβ or BMP promoter sequences, thereby acting as a tumor suppressor.19 Depending on availability, scaffolding proteins may serve either enhancing or inhibitory functions. An elegant example of this may be found in the MAPK pathway. The kinase suppressor of Ras-1 (KSR1) can scaffold members of the MAPK cascade.20,21 Titrating KSR1 concentration reveals its cooperative role in the pathway until a certain threshold is reached, after which the concentration of scaffolding protein exceeds that of “signaling-competent complexes” and KSR1 sequesters individual proteins from interacting with others in the cascade.14 In general there is no doubt scaffolding proteins influence signal transduction kinetics by altering the interaction dynamics of signaling components.
Barriers to Diffusion: The Cytoskeleton
When cells are in a 3D environment, the cytoskeleton is usually nonuniform in shape and composition. For example, cells in collagen matrices often exhibit actin cytoskeleton alignment with fibers within the ECM.22,23 Changes in cytoskeletal organization, in turn, influence intracellular signaling by acting as barriers to diffusion. The cytoskeleton can be a passive barrier, where an increased level of cross-linking represents more obstacles to free diffusion. Computational molecular models have predicted that components of the MAPK, protein kinase A (PKA), and PI3K–Akt–mTOR pathways have reduced rates of diffusion because of increased levels of molecular crowding.11 Alternatively, cytoskeletal proteins can act as scaffolds to promote or inhibit interaction of signaling molecules. Nanoclustering of epidermal growth factor receptor (EGFR) at the cell periphery can be due to high actin activity at these sites.24 Altered clustering and diffusion of receptor tyrosine kinases has been proposed as a molecular mechanism employed by cells in 3D environments to resist drug therapies, such as resistance to HER2 targeting agents in breast cancer via increased HER2 dimerization in 3D.25,26 Thus, external stimuli leading to changes in actin density can alter the spatiotemporal behavior of intracellular signaling.
Altered Dimensionality: Membrane Sequestration
In some cases, transition from 3D diffusion in the cytoplasm to 2D diffusion in a membrane can enhance cellular signaling. An example of this is given by the BCL-2 family of proteins, which mediate caspase-driven apoptosis by regulating the release of cytochrome c from mitochondria.27 Bax, a BCL-2 protein typically found freely diffusing in the cytoplasm, activates mitochondrial membrane permeabilization via oligomerization. When cytokine activation of apoptosis occurs, Bax is recruited to the mitochondrial membrane surface via upstream signaling events.28 When Bax is in the cytoplasm, diffusing in a 3D environment, the probability that it will oligomerize with itself is quite low to negligible. However, once it is limited to 2D diffusion in the mitochondrial membrane, the likelihood of interaction with other Bax proteins also anchored in the membrane greatly increases.28 This leads to Bax oligomerization, release of cytochrome c, and progression of apoptotic signaling.
Novel Tools for Characterization of Spatial Control of Signaling
As can be seen, the spatial organization of signaling molecules can greatly impact the dynamics of intracellular signaling. Moreover, there are many mechanisms by which molecular localization of these proteins can be affected. Traditional biochemical assays like Western blots and enzyme-linked immunosorbent assays (ELISA) lack the subcellular resolution to capture this information (Table 1A). Electron microscopy has long been used to characterize the molecular organization of the cytoskeleton, membranes, and organelles in 2D and 3D, including changes in epithelial cell structure after hormone stimulation or altered mechanical properties of the ECM (Table 1B).29−31 However, 3D samples must be fixed, dehydrated, and sectioned, prohibiting studies of spatial or temporal dynamics of cells in a 3D environment. Light microscopy, on the other hand, is highly conducive to characterizing how protein localization influences cellular signaling. To empirically determine the diffusion coefficients and dissociation constants of signaling molecules, fluorescence recovery after photobleaching (FRAP) can be used. FRAP involves tagging a protein of interest with a fluorescent molecule, taking an image of the basal fluorescence level, photobleaching a specific area of interest with high-intensity laser illumination, and then quantifying the recovery of fluorescence intensity as unbleached molecules switch places in the area of interest with the bleached molecules (Table 1D). FRAP is a technique that can be used to measure the ensemble change in protein behavior at a region of interest and has been used to great advantage to determine the binding kinetics of transcription factors and the mobility of receptors in the plasma membrane (Table 1D).32,33
Table 1. Quantitative Techniques for Determining Reaction Kinetics.

To study protein–protein interactions, techniques like Förster resonance energy transfer (FRET) and fluorescence lifetime imaging microscopy (FLIM) can be used. FRET imaging involves using the energy transfer between fluorophores as a proxy for the distance between fluorescently tagged molecules; it is sensitive within the 5–10 nm range and is often used as an indication of protein–protein binding (Table 1E). FRET has been used to study receptor–ligand binding at the membrane as well as protein conformation.34,35 Grashoff et al. demonstrated the use of an intramolecular FRET probe that allowed detection of vinculin stretching under cytoskeletal tension.36 FLIM, on the other hand, takes advantage of the sensitivity of a fluorescent molecule’s exponential rate of decay of its fluorescence (lifetime) to its environment to characterize protein–protein interactions. Unlike FRET and FRAP, which require genetically encoded probes or fluorescent antibodies, FLIM can use the inherent fluorescence of specific molecules such as NADPH and is a promising technique for analysis of unmodified and untagged human cancer cells in the future (Table 1F).37
Summary
The impact of the 3D subcellular distribution of signaling molecules on intracellular signaling is not often discussed despite the fact that localization can profoundly affect cell and tissue behavior and fate. Nonetheless, as new technologies that allow us to peer into the cell emerge, the need to understand the role of protein localization in signaling cannot be ignored.
2. The Cell in 3D: Cell–Matrix Interactions
As we zoom out to examine the cell, we must consider its natural context. Within the body, cells interact in 3D with the extracellular matrix (ECM) and neighboring cells. Through these interactions, cells gather information about their surroundings, which is integrated to determine cell behavior and fate. In addition, a cell’s 3D environment affects the organization of intracellular components and thus the context in which signaling occurs. This section will feature how intracellular signaling is dependent on the context of cell shape, barriers to intracellular diffusion, and protein localization.
Adhesions and Junctions: Gathering Information from the Environment
The adhesion of a cell to its external environment can dictate cell shape, growth, proliferation, and apoptosis signaling.38,39 Cells attach to neighboring cells and to ECM molecules, including secreted proteins (i.e., collagens and laminins), proteoglycans (heparin and chondroitin sulfate), and glycosaminoglycans (hyaluronic acid). Cells attach to these extracellular molecules via a plethora of adhesion receptors, including integrins, cadherins, selectins, discoidin receptors, and syndecans.40−42 These adhesion receptors bridge the connection between the extracellular environment and the intracellular cytoskeletal and signaling machinery.
Engagement of the ECM in all three dimensions, compared to only at the basal side of cells as in a conventional culture, significantly influences cell behavior. Depending on whether cells are cultured in 3D or 2D, a cell’s response to a given signal can be significantly altered.10,43,44 Mammary epithelial cells with intact cell–cell junctions in 3D hydrogels are more resistant to apoptosis-inducing factors like tumor necrosis factor α (TNF-α) than cells in 2D or cells in 3D lacking cell–cell junctions (Figure 2A).10 This resistance highlights the importance of cell–cell interactions in 3D and implies a connection between adhesion-dependent signaling and intracellular signaling directing cell fate.10 Culture in 2D versus 3D also alters cell spreading and traction force.44 When fibroblasts are plated onto closely spaced small pillars (5 μm), cells form adhesions on the top and side of the pillars, simulating a 3D environment.44 Under these conditions, fibroblasts pull with high levels of force on the external environment (Figure 2B).44 Conversely, if the cells are on pillars with an increased surface area (15 μm) that are widely spaced, simulating a 2D environment, cells do not form adhesions on the sides of the pillars and the extent of traction on the external environment is decreased (Figure 2B).44
Figure 2.
Changes in cell shape and 3D organization modulate intracellular signaling. (A) Transitions from nonmalignant S1s to malignant T4-2 epithelial cells show a reversal in cell polarity and 3D organization.10 (B) Cells on closely spaced pillars behave as though on a 2D environment, whereas those on widely spaced pillars exhibit 3D-like behaviors.44 (C) Actin organization in cells on pillar-based 2D substrates vs. inside 3D microwell scaffolds.43
The dimensionality, rigidity, and topology of the external environment have been implicated as regulating factors in cytoskeleton assembly and metabolism.43 Through the use of polymeric hydrogels, researchers can control rigidity by altering the cross-linker:monomer ratio in the gel solution. When fibroblasts were seeded in 3D silicone microwells of varying stiffness, they exhibited a range of novel behaviors compared to the behavior of those in 2D environments. In soft 3D environments, fibroblasts downregulate actin filament assembly and upregulate mitochondrial activity, in contrast to the activities in compliant 2D, stiff 2D, or stiff 3D environments (Figure 2C).43
An Altered Extracellular Context Leads to Cytoskeletal and Membrane Reorganization
Many of the affected cell behaviors associated with 3D culture have been attributed to altered cell shape. Altered cell shape leads to reorganization of the cytoskeleton as well as changes to membrane tension, curvature, and composition, changing the context in which intracellular signaling takes place.
Cell shape directly influences the organization of the cytoskeleton, which can have profound effects on intracellular signaling. In mesenchymal stem cells (MSC), cell spreading leads to RhoA activation of Rho-activated kinase (ROCK), myosin light chain kinase phosphorylation, elevated myosin contractility, increased traction forces to the external environment, and differentiation to an osteoblast phenotype.45 Conversely, if MSCs are rounded or express a dominant negative RhoA, these cells differentiate into an adipocyte phenotype.45,46
Cell shape, in combination with ECM stiffness, governs membrane properties like curvature and tension.47,48 These characteristics of the plasma membrane, in turn, determine membrane composition, membrane protein distribution, and intracellular trafficking rates.48,49 Several families of proteins, including the FERM (4.1 protein, Ezrin, Radixin, and Moesin) and BAR (Bin/amphiphysin/Rvs) domain proteins, have molecular motifs that are sensitive to membrane curvature. Proteins containing these domains, which include the ARF (adenosine-ribosylation factor) family,50,51 small GTPases like Rac,50,52 and guanine exchange factors,53 can assemble multiprotein complexes and preferentially target these complexes toward a curved membrane, thereby affecting protein localization and signaling. Similarly, membrane tension is a key regulator of endo- and exocytosis.48,49 Increased membrane tension decreases the probability of vesicular budding and favors exocytic merging of vesicles with the plasma membrane.49 This affects the overall balance of vesicular trafficking within the cell, which can influence such diverse signaling pathways as growth factor receptor signaling, reactive oxygen species production, and phagocytosis.
The composition and organization of the plasma membrane are different between 2D and 3D, including cholesterol and sphingomyelin content and organization.54 This membrane reorganization can alter the distribution of lipids and lipid rafts in the membrane.55,56 These differences can then affect lipid-dependent signaling, such as signaling through the PI3K–Akt–mTOR pathway, which regulates cell metabolism, migration, and apoptosis.57 The PI3K–Akt–mROR pathway is initiated by the recruitment of phosphatidylinositol 3-kinase (PI3K) to the membrane and the generation of lipid rafts with phosphatidylinositol 3,4,5-trisphosphates [PtdIns(3,4,5)P3] and is attenuated by phosphatase and tensin homologue (PTEN).58 PI3K and PTEN are usually sequestered in distinct lipid groups in the membrane that display limited diffusion and mixing between lipid rafts.59 This lipid raft-induced segregation of PI3K and PTEN is exacerbated in spread cells, in which an increased number of adhesions leads to lipid raft formation in the plasma membrane via microtubules and Arf6 recycling.60 When cells are rounded via cytoskeletal disruption or via micropatterned surfaces, lipid raft formation is decreased, the level of PI3K and PTEN interactions is increased, and PI3K signaling is attenuated.61,62 When cells are spread and PtdIns(3,4,5)P3 lipid rafts are present, downstream Akt activity leads to differences in BCL-2-mediated apoptosis and cell migration.62,63
When cells transit from a rounded to spread morphology, the Rnd family of proteins is targeted to lipid rafts via the KERPA (Lys-Glu-Arg-Pro-Ala) sequence at their N-termini.64 Rnd proteins recruit p190RhoGAP to lipid rafts at the plasma membrane, leading to increases in Rho activity that are mediated by filamin cross-linking of actin.64,65 This behavior can be ablated through inhibition of actin polymerization, filamin-dependent actin cross-linking, and cell spreading.65
The pattern of cell–ECM and cell–cell adhesions can dictate the landscape in which other signaling molecules may interact on the cell membrane. Ephrin signaling, for example, is sensitive to the spatial organization of ephrin receptors on the cell membrane.66 Salaita et al. restricted ephrin receptor diffusion in the membrane and showed obstacles to receptor oligomerization dramatically influence intracellular signaling. Specifically, limiting ephrinA1 movement led to changes in cytoskeletal organization and metalloprotease secretion in cancer cells.67 As many membrane-bound proteins act by forming protein complexes, the organization of adhesions on the cell membrane can greatly influence the dynamics of these signaling events.
Adhesions and Junctions: Mediating the Response of the Cell to Dynamic Input
In addition to transducing static information about its environment, adhesion molecules can mediate dynamic cellular responses to external input. In 3D environments like the lumen of a blood vessel, signaling at endothelial cell–cell and cell–ECM junctions regulates vessel integrity and the extravasation of leukocytes from the vasculature. Upon application of fluid shear stress in vitro, endothelial cells in culture undergo filamentous actin and focal adhesion remodeling in the direction of flow in a VE-cadherin-dependent manner (Figure 3B).68,69 Downstream, this results in Rho GTPase activation, which works in conjunction with vascular endothelial growth factor receptor 2 (VEGFR2) to activate p38 MAPK pathway activity.70,71 Thus, endothelial cells sense dynamic changes in the environment and feed this information into existing intracellular signaling to influence cell behavior.
Figure 3.
Tools for studying cell and matrix mechanical interactions in 3D. (A) Peptide gels with consistent pore sizes but variable stiffnesses.76 (B) Microfluidic chambers allow application of flow, chemical gradients, and mechanical forces.68,69 (C) In hydrogels, different materials lead to differential cell behavior.77 (D) Example of cellular organization on collagen gels with different stiffnesses.78
The need to process information about the extracellular environment is even more relevant for migrating cells. The transit of leukocytes from the vasculature to the subendothelial matrix, for example, relies upon mechanically activated adhesion receptors gathering information about the vascular endothelium upon which leukocytes travel. Endothelial presentation of selectins may be recognized by white blood cells flowing through the blood. Ligation of P-selectin with P-selectin glycoprotein ligand-1 (PSGL-1) on the surface of a rapidly moving leukocyte can result in strengthening of the P-selectin–PSGL-1 catch bond (force induced non-covalent bond), leading to leukocyte deceleration.72,73 While P-selectin and PSGL-1 bonds are transient on the scale of ∼0.5 s, they allow stronger, longer-lasting integrin–ECM bonds to form.72,73 These cell–ECM adhesions then lead to leukocyte polarization, accumulation of the membrane lipid PtdIns(3,4,5)P3, activation of Rac1 and Akt, cytoskeletal remodeling, and transmigration through the endothelium.72,74
Novel Tools for Studying Signaling in 3D Matrices
To study cell adhesion in 3D environments, many techniques have been developed to encapsulate cells in hydrogels, hydrated polymer networks that behave as viscoelastic solids (Table 2). Commonly used polymers in hydrogels include naturally derived materials (including collagen, a proteoglycan such as hyaluronic acid, fibrin gels, and cellulose) and synthetic polymers such as polyethylene glycol (PEG), poly(vinyl alcohol) (PVA), poly-lactic-glycolic-acid (PLGA), and engineered peptide-based biomaterials. Peptide hydrogels are a useful system for studying cell signaling in 3D as they allow tuning of individual environmental parameters, including matrix elasticity, cell adhesion binding sites, and degradation.75 Examples of different peptides used include elastin and silklike polypeptides, or novel repetitive peptide sequences that form fibrils in ionic environments like KFE ((acetyl)-FKFEFKFE-CONH2.75,76 Via modification of these gels with RGD (Arg-Gly-Asp) sequences for integrin binding and changing the concentration of peptide, KFE gels can be fabricated to allow tuning of cell binding, matrix compliance, and pore size (Figure 3A).76 When these gels are optimized, they allow formation of cell–ECM and cell–cell connections that promote endothelial tube formation, facilitating the study of 3D endothelial cell signaling in an in vitro system that more accurately resembles a capillary bed than a 2D culture does (Figure 3A).76
Table 2. Summary of 3D Culture Systems.

The 3D microenvironment can be specifically controlled in vitro by embedding cells in a hydrogel with a specific molecular composition and concentration. Individual cells can be placed in self-assembling peptide gels, cross-linked collagen matrices, or reconstituted basement membrane hydrogels such as Matrigel (Figure 3C).77 Breast cancer cells such as MDA-MB-231, for example, alter their morphology and adhesion in response to altered ECM properties, while MCF10a cells form stable spheroids in compliant but not stiff collagen matrices.77,78 Furthermore, cell behavior can be influenced by controlling ECM properties like stiffness, which, in the case of collagen gels, can be modulated by increasing the collagen concentration or via inhibition of lysyl oxidase-mediated collagen cross-linking (Figure 3D).79,80
Summary
The effect of altered cell–matrix and cell–cell interactions on intracellular signaling is undeniable. The cell is continuously gathering information about its surroundings and incorporating this information into its decision-making circuits. Culturing cells in 2D versus 3D, or one ECM component versus another, results in altered signaling at the cellular and subcellular level. As such, researchers need to be particularly aware of their phenomena of interest and how cellular context can introduce confounding factors into their studies.
3. 3D Signaling at the Multicellular Level
Cells in the body exist in a three-dimensional environment. They interact on all sides with other cells, the extracellular matrix, and interstitial fluid. These interactions provide inputs that the cell integrates to determine its behavior and fate. At the tissue level, two major factors influence cell behavior: local gradients in signaling molecules and multicellular structures. The following section highlights molecular gradients in development and ductlike multicellular structures.
Molecular Gradients in Multicellular Structures
Sources and sinks for signaling molecules, combined with interstitial pressure, serve to establish molecular gradients that can differentially influence cells within a tissue based on their location. These gradients are established through a combination of cell secretion, protein diffusion, proteoglycan-mediated stabilization, and endocytic depletion in neighboring cells.81−83 Some source, or collection of cells secreting the morphogen, acts as the focal point from which diffusion distributes the morphogen. Proteoglycans in the extracellular matrix, like glycosaminoglycans (GAGs), sequester and maintain local supplies of the morphogen.84 Finally, depletion of the morphogen from interstitial fluid occurs via receptor binding, endocytosis, and degradation.83,85
Many examples of molecular gradients can be found in development. In the early Drosophila embryo, before nuclei are separated by cell membranes, a Bicoid (Bcd) gradient governs the expression of gap genes.86,87 These genes dictate the anteroposterior patterning of the embryo and, if mutated, can lead to a loss of continuous segments in the developed organism.86,87 At later stages of development, Decapentaplegic (Dpp) and Wingless (Wg) gradients in the wing imaginal disc have been linked to proper segmentation and wing development.83,88 In the imaginal disc, the Dpp gradient arises through a combination of changes to Dpp secretion, diffusion, stabilization, and depletion. Dpp is captured at the surface of a cell via low-affinity interactions with heparin sulfate proteoglycans, decreasing the rate of diffusion of Dpp. The improved availability of Dpp leads to an increased number of interactions with its receptor, resulting in amplified intracellular signaling.89,90
Similarly, gradients of the vertebrate Dpp homologue, bone morphogenetic protein (BMP), are vital for dorsal–ventral patterning.91,92 High local BMP levels specify ventral tissues, while low BMP signaling levels lead to development of dorsal tissues.91 Sonic hedgehog (Shh), transforming growth factor-β (TGFβ), and fibroblast growth factor gradients have similar effects on developing cells.93,94 In all these cases, progenitors have concentration-dependent responses to morphogens. In chicks, for example, the duration of the responses of neural cells to Shh is directly proportional to its local concentration.81 This Shh response controls the expression of important transcription factors, which in turn direct differentiation into specific neuronal subtypes.94
While simple model organisms like Drosophila melanogaster and Caenorhabditis elegans make possible investigation of the effects of these morphogen gradients, translation and extension of this work into more complex specimens are major challenges. The deterministic pattern of development in C. elegans and the small number of cells in the developed organism, for example, facilitate reliable lineage tracing, where transfection of specific cells with genetic markers allows identification of their progeny.95 This can be combined with secondary markers that indicate the relative expression of genes responsible for driving different cell fates.95 The resilience of D. melanogaster permits the use of simple knockout systems to identify the role of genes like Dpp and Wg. These and other methods have led to enormous strides in understanding the processes governing development. However, the same techniques are difficult to apply in more complex systems.90 Many knockouts are embryonic lethal in mammals, precluding analysis of their effects on development. Further, many of these model organisms are transparent and thus particularly amenable to imaging approaches. Most tissues in mammals are not transparent and preclude the use of common visualization techniques.95 The increased genomic complexity of higher-order organisms makes systematic screening an often untenable challenge. These factors have hampered researchers’ investigation and left an important facet of biological function largely unexplored. While hints of the role of molecular gradients have arisen in studies of human embryonic development, little is known about their importance in normal tissue function. Given the fact that many of the same mechanisms that apply to morphogen gradients apply in normal tissues, however, it is probable that gradients play an equally important role in signaling in tissues.
Transport Phenomena in Tissues
A key indication that molecular gradients exist in tissues is the myriad convective transport phenomena known to be at play. These transport processes generate interstitial flow that can induce local gradients of signaling factors. A major driver of convective flow is interstitial pressure. Interstitial pressure results from differences in hydrostatic and osmotic pressure among the vasculature, interstitium, and lymphatics.96 Positive pressure gradients from blood vessels, combined with leaky capillaries, drive fluid and soluble factors into the tissue. Negative pressure gradients between the tissue and lymphatic system are maintained by active drainage into lymphatic capillaries.96 Tissue stretching and compression due to regular movement can also cause transient interstitial flow. These patterns are altered in tumors, where leaky vasculature and increased interstitial pressure inhibit leakage from capillaries into the interstitium. This altered transport can contribute to oxygen deprivation within the tumor, creating hypoxic conditions that activate HIF-1 (hypoxia-inducible factor 1), a transcription factor implicated in the control of metabolism, invasion, and apoptosis.97 HIF-1 upregulation, in turn, has been strongly implicated in tumor aggression and therapeutic resistance.98,99
In addition to pressure differences, ciliary movement in the lung and intestines can also drive interstitial flow. These small convective flows have been demonstrated to govern normal branching morphogenesis in the embryonic lung by directing points at which splitting should occur.100 Interstitial flow has been demonstrated to allow generation of pericellular gradients, where directional flow around a cell secreting some signaling factor leads to an asymmetrical distribution of that factor around the cell, thereby facilitating generation of an autologous chemotactic gradient.101 On a larger scale, immune cells use molecular gradients established by interstitial flow to home to the lymphatic system. While these chemotactic behaviors are well-documented, there is no doubt that nonmotile cells are similarly affected by gradients established by interstitial flow. Gradients of extracellular signaling factors within a multicellular structure are, therefore, relevant aspects to include if we are to gain a full picture of the dynamic nature of signaling in vivo.
Cell Organization and Signaling in Multicellular Tissues
Cellular localization is an important aspect of signaling at the multicellular level, affecting the cell’s exposure to other cells, the ECM, and soluble signaling molecules within the interstitial fluid. In contrast to a monolayer culture, where all cells have essentially the same relationship with one another and the nutrient source, multicellular structures can dramatically alter the environment in which a cell receives signals. In human breast ducts, for example, luminal and myoepithelial cells encounter dramatically different environments. Whereas the inner luminal epithelial cells are exposed to the hollow duct and lined with myoepithelial cells, the myoepithelial cells are sandwiched between the luminal epithelial cells and the basal lamina. Thus, myoepithelial cells are subject to more ECM signals than the luminal epithelial cells. This complexity is not captured in conventional 2D culture of these cells, which could lead to artifactual observations that do not translate to in vivo behavior. For example, culturing transformed mammary epithelial cells in a 3D matrix can suppress proliferation and reestablish cell polarity, while oral squamous cell carcinoma cells exhibited higher angiogenic potentials under 3D conditions.102,103
Similarly, the importance of paracrine signaling from neighboring cell types has been highlighted by the necessity for feeder cultures of stem cells. Since the early 1950s, maintenance of the undifferentiated state of embryonic stem cells in vitro has been achieved via coculture with fibroblasts.104,105 Recently, this has been substituted with a culture of conditioned media containing fibroblast-secreted factors like BMPs and leukemia inhibitory factor (LIF) or a 3D culture of stem cells.106−108 As cells are rarely in isolation in the body, there is no doubt that many other such mechanisms of cell control have been lost in the transition to an in vitro culture.
A functional example of the importance of multicellular structures may be found in the brain, where neuron–glia networks cooperate to transduce signals.109−111 Neurotransmitters secreted by neurons can initiate signaling in glial cells, which then secrete neuromodulatory factors that act on neurons within the synapse, including glutamate and thrombospondin.109,111−113 Glial cell morphology allows them to contact multiple neuronal cell bodies and extend into thousands of synapses, making them uniquely positioned to act as an integrator and modulator of neuronal activity.111 As such, the specific positional relationships among neurons and glial cells within a 3D network can be key to how that network responds to and propagates stimuli.
In addition, multicellular structures often optimize the access of cells to nutrients. In particular, hepatocytes are known for their sensitivity to in vitro culture. Compared to the dense populations found in vivo, hepatocytes lose viability or transdifferentiate at high densities when cultured in a monolayer.114−116 This can be partially addressed by culturing them in a microfluidic device with a geometry that maximizes the access of cells to a central fluidics chamber that mimics the hepatic artery.117 This enhanced proximity to nutrients via an altered organization allowed a high-density hepatocyte culture closer to in vivo conditions.114
Cells do not signal in isolation within the body. They are parts of multicellular structures exposed to gradients of signaling molecules and physical forces. Their positions within a highly organized structure dictate their exposure to a specific set of signaling inputs, which may differ from those of neighboring cells. This organization and coordination permits these cells to work as a unit to perform a biological function. Accordingly, a full understanding of cell signaling requires studying the cell as part of a larger unit.
Novel Techniques for Studying Multicellular Structures in 3D
Many of the conditions that may govern signaling in 3D have eluded researchers because of inadequacies in culture methods. Recent recognition of the importance of the context in which signaling occurs, including molecular gradients, cell–ECM interactions, and multicellular organization, has led to the development of novel culture techniques that seek to recapitulate 3D conditions seen in vivo (Table 2).
The most basic of these is 3D encapsulation culture. This involves embedding cells in a hydrogel composed of ECM components to better mimic the immersive environment of the body. This technique has spawned the development of many biomimetic or biosynthetic materials, including Matrigel, hyaluronic acid, collagen, and alginate.118 However, except in cases of self-organization, this method still fails to recapitulate the complex organization of cells and the ECM in vivo.
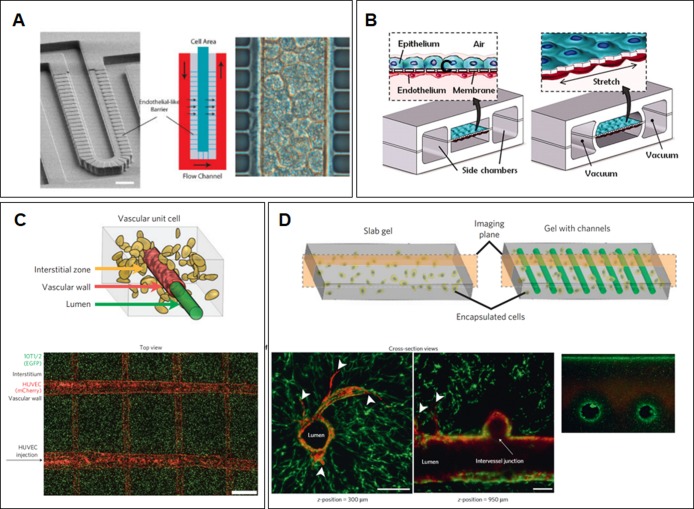
Soft lithography, on the other hand, is proving to be a boon for researchers seeking more precise control of their culture conditions (Figure 4). This family of methods adapts approaches used in manufacturing microelectronic chips to fabricate or replicate nanometer scale structures and patterns on elastomeric materials like polydimethylsiloxane (PDMS).119 These methods offer unprecedented control over features in a culture system, including geometry, patterns of ECM protein functionalization, and application of flow.119,120 Soft lithography has been used to generate features that mimic the layout of a blood vessel or facilitate a multicell-type culture (Figure 4C,D).121,122 The open design aspect of soft lithography also allows researchers to model in vivo multicellular organization, leading to fabrication of devices that recapitulate the shape and layout of major organ subunits within the liver and lung (Figure 4A,B).117,123,124 In addition, microfluidic culture permits introduction of flows, in contrast to conventional static culture. This allows recapitulation of both vascular flow and interstitial flow, facilitating the establishment of molecular gradients (Figure 4B,C).125,126 3D printing confers even greater control over culture conditions, allowing layer-by-layer construction of an extracellular matrix and cells. Biodegradable carbohydrate scaffolds can be used to create architecturally complex “organs”. Miller et al. used this method to create 3D cylindrical networks of cells within an ECM network (Figure 4C,D).122
Figure 4.
Methods for mimicking in vivo conditions of a 3D environment. (A) Imposing tissuelike organization through microfabrication. This example mimics liver ducts in a hepatocyte culture.117 (B) Flow application allows recapitulation of regular lung stretching in vitro.123 (C and D) 3D printing allows construction of complex matrices and cell seeding. In this case, vascular endothelial cells were encapsulated to form hollow vessels surrounded by an extracellular matrix. (C) The top panel is a schematic of the relationship of vascular endothelial cells (red) with matrix-encapsulated fibroblasts (yellow) in the interstitium. The bottom panel is a fluorescence image of a printed vascular system, with endothelial cells (mCherry) and fibroblasts (EGFP).122 (D) The top panel is a schematic contrasting culture systems. With the novel casting system, randomly encapsulated cells can be combined with regular vascular networks. The bottom panels shows cross-sectional views of the vascular network to illustrate both the cleared lumen without cells and the ability to generate intervessel junctions to study the effect of more complicated flow patterns on cell behavior.122
While these novel microfabrication approaches are both powerful and versatile, in many ways, they are still in their infancy. The elastomeric materials used in soft lithography have a limited range of mechanical properties that may not reflect in vivo conditions. Further, while proof-of-concept cell studies usually accompany technical descriptions of these systems, extensive analysis of intracellular signaling in these culture settings has not been performed. Finally, application of these systems requires a level of commitment and investment that may not be feasible for all laboratories. Nonetheless, the field is rapidly growing and has exciting potential.
Summary
Studying multicellular structures in vitro has only recently become a reality. While there remain many unsolved mysteries within the cell that would not benefit from the introduction of multicellular context, the application of these novel approaches could be vital to researchers interested in translational and clinically relevant results. As single-cell level studies cannot capture the emergent properties of an organized tissue, researchers should test their hypotheses in higher-order systems to gain a more thorough understanding of functions within the human body and to design therapeutics in an informed manner.
4. Conclusion
As researchers’ understanding of cellular behavior in 3D progresses, studying signaling in the appropriate context has become essential. From intracellular localization of signaling molecules to molecular gradients at the tissue level, it is apparent that signaling is influenced by a myriad of factors and is a process far more complex than the simple interaction of reactants to form an end product. This recognition has spurred the invention of new technologies to facilitate investigation of signaling in 3D contexts. At the cellular level, novel superresolution imaging tools allow unprecedented characterization of protein localization and interaction dynamics. Bioengineered materials serve as in vitro mimics of complex tissue ECM, helping to elucidate the role of cell–matrix interactions in governing intra- and intercellular signaling. Finally, microfabrication and 3D printing permit recapitulation of multicellular structures, bringing multiple cell types together in physiologically relevant ways and revealing emergent properties at the tissue level. Results from the application of these tools have demonstrated that cell behavior can change dramatically based on the conditions under which they are studied. As such, it is not surprising that the biomedical field has encountered unprecedented failure rates in translating hard-earned discoveries into clinical progress. Therefore, it has never been more urgent for both basic science and translational research to be conducted in the appropriate context. With the adoption of new methods, findings from the laboratory will become more transferable to improve human health.
Author Contributions
M.G.R. and G.O. contributed equally to this work.
The work was supported by a Department of Defense National Defense Science and Engineering Graduate Fellowship to M.G.R., the National Science Foundation Graduate Research Fellowship Program to G.O., and a BCRP Scholar Expansion Award BC122990 and National Institutes of Health/National Cencer Institute Grants PSOC U54CA143836-01, R01 CA138818-01A1, RO1 CA085492-11A1, U01 ES019458-01, TMEN U54CA163155-01, and U01 CA151925-01 to V.M.W.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Yamada K. M.; Cukierman E. (2007) Modeling tissue morphogenesis and cancer in 3D. Cell 130, 601–610. [DOI] [PubMed] [Google Scholar]
- Cukierman E.; Pankov R.; Stevens D. R.; Yamada K. M. (2001) Taking cell-matrix adhesions to the third dimension. Science 294, 1708–1712. [DOI] [PubMed] [Google Scholar]
- Sutherland R. M.; Durand R. E. (1976) Radiation response of multicell spheroids: An in vitro tumour model. Curr. Top. Radiat. Res. Q. 11, 87–139. [PubMed] [Google Scholar]
- Zahir N.; Weaver V. M. (2004) Death in the third dimension: Apoptosis regulation and tissue architecture. Curr. Opin. Genet. Dev. 14, 71–80. [DOI] [PubMed] [Google Scholar]
- Wang F.; Weaver V. M.; Petersen O. W.; Larabell C. A.; Dedhar S.; Briand P.; Lupu R.; Bissell M. J. (1998) Reciprocal interactions between β1-integrin and epidermal growth factor receptor in three-dimensional basement membrane breast cultures: A different perspective in epithelial biology. Proc. Natl. Acad. Sci. U.S.A. 95, 14821–14826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S. E.; Mavila A.; Salazar R.; Bayless K. J.; Kanagala S.; Maxwell S. A.; Davis G. E. (2001) Differential gene expression during capillary morphogenesis in 3D collagen matrices: Regulated expression of genes involved in basement membrane matrix assembly, cell cycle progression, cellular differentiation and G-protein signaling. J. Cell Sci. 114, 2755–2773. [DOI] [PubMed] [Google Scholar]
- Tsuboi N.; Yoshida H.; Kawamura T.; Furukawa Y.; Hosoya T.; Yamada H. (2000) Three-dimensional matrix suppresses E2F-controlled gene expression in glomerular mesangial cells. Kidney Int. 57, 1581–1589. [DOI] [PubMed] [Google Scholar]
- Ghosh S.; Spagnoli G. C.; Martin I.; Ploegert S.; Demougin P.; Heberer M.; Reschner A. (2005) Three-dimensional culture of melanoma cells profoundly affects gene expression profile: A high density oligonucleotide array study. J. Cell. Physiol. 204, 522–531. [DOI] [PubMed] [Google Scholar]
- Lelièvre S.; Weaver V. M.; Bissell M. J. (1996) Extracellular Matrix Signaling from the Cellular Membrane Skeleton to the Nuclear Skeleton: A Model of Gene Regulation. Recent Prog. Horm. Res. 51, 417–432. [PMC free article] [PubMed] [Google Scholar]
- Weaver V. M.; Lelièvre S.; Lakins J. N.; Chrenek M. A.; Jones J. C. R.; Giancotti F.; Werb Z.; Bissell M. J. (2002) β4 integrin-dependent formation of polarized three-dimensional architecture confers resistance to apoptosis in normal and malignant mammary epithelium. Cancer Cell 2, 205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla U. S. (2004) Signaling in small subcellular volumes. I. Stochastic and diffusion effects on individual pathways. Biophys. J. 87, 733–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehmelt L.; Bastiaens P. I. H. (2010) Spatial organization of intracellular communication: Insights from imaging. Nat. Rev. Mol. Cell Biol. 11, 440–452. [DOI] [PubMed] [Google Scholar]
- DeWire S. M.; Ahn S.; Lefkowitz R. J.; Shenoy S. K. (2007) β-Arrestins and cell signaling. Annu. Rev. Physiol. 69, 483–510. [DOI] [PubMed] [Google Scholar]
- Kolch W. (2005) Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat. Rev. Mol. Cell Biol. 6, 827–837. [DOI] [PubMed] [Google Scholar]
- Kholodenko B. N.; Hancock J. F.; Kolch W. (2010) Signalling ballet in space and time. Nat. Rev. Mol. Cell Biol. 11, 414–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb D. J.; Donais K.; Whitmore L. A.; Thomas S. M.; Turner C. E.; Parsons J. T.; Horwitz A. F. (2004) FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat. Cell Biol. 6, 154–161. [DOI] [PubMed] [Google Scholar]
- Calalb M. B.; Zhang X.; Polte T. R.; Hanks S. K. (1996) Focal adhesion kinase tyrosine-861 is a major site of phosphorylation by Src. Biochem. Biophys. Res. Commun. 228, 662–668. [DOI] [PubMed] [Google Scholar]
- Ishibe S.; Joly D.; Liu Z.-X.; Cantley L. G. (2004) Paxillin serves as an ERK-regulated scaffold for coordinating FAK and Rac activation in epithelial morphogenesis. Mol. Cell 16, 257–267. [DOI] [PubMed] [Google Scholar]
- Morén A.; Raja E.; Heldin C.-H.; Moustakas A. (2011) Negative regulation of TGFβ signaling by the kinase LKB1 and the scaffolding protein LIP1. J. Biol. Chem. 286, 341–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison D. K. (2001) KSR: A MAPK scaffold of the Ras pathway?. J. Cell Sci. 114, 1609–1612. [DOI] [PubMed] [Google Scholar]
- Kortum R. L.; Lewis R. E. (2004) The Molecular Scaffold KSR1 Regulates the Proliferative and Oncogenic Potential of Cells. Mol. Cell. Biol. 24, 4407–4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubow K. E.; Conrad S. K.; Horwitz A. R. (2013) Matrix microarchitecture and myosin II determine adhesion in 3D matrices. Curr. Biol. 23, 1607–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroll W. M.; Billiar K.; Starke J.; Maaser K.; Wehrle-Haller B.; Friedl P. (2013) Mechanotransduction of mesenchymal melanoma cell invasion into 3D collagen lattices: Filopod-mediated extension–relaxation cycles and force anisotropy. Exp. Cell Res. 319, 2424–2433. [DOI] [PubMed] [Google Scholar]
- Chung I.; Akita R.; Vandlen R.; Toomre D.; Schlessinger J.; Mellman I. (2010) Spatial control of EGF receptor activation by reversible dimerization on living cells. Nature 464, 783–787. [DOI] [PubMed] [Google Scholar]
- Pickl M.; Ries C. H. (2009) Comparison of 3D and 2D tumor models reveals enhanced HER2 activation in 3D associated with an increased response to trastuzumab. Oncogene 28, 461–468. [DOI] [PubMed] [Google Scholar]
- Weigelt B.; Lo A. T.; Park C. C.; Gray J. W.; Bissell M. J. (2010) HER2 signaling pathway activation and response of breast cancer cells to HER2-targeting agents is dependent strongly on the 3D microenvironment. Breast Cancer Res. Treat. 122, 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle R. J.; Strasser A. (2008) The BCL-2 protein family: Opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 9, 47–59. [DOI] [PubMed] [Google Scholar]
- Lovell J. F.; Billen L. P.; Bindner S.; Shamas-Din A.; Fradin C.; Leber B.; Andrews D. W. (2008) Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell 135, 1074–1084. [DOI] [PubMed] [Google Scholar]
- De Jonge N.; Ross F. M. (2011) Electron microscopy of specimens in liquid. Nat. Nanotechnol. 6, 695–704. [DOI] [PubMed] [Google Scholar]
- Vic P.; Vignon F.; Derocq D.; Rochefort H. (1982) Effect of Estradiol on the Ultrastructure of the MCF7 Human Breast Cancer Cells in Culture. Cancer Res. 42, 667–673. [PubMed] [Google Scholar]
- Loessner D.; Stok K. S.; Lutolf M. P.; Hutmacher D. W.; Clements J. A.; Rizzi S. C. (2010) Bioengineered 3D platform to explore cell-ECM interactions and drug resistance of epithelial ovarian cancer cells. Biomaterials 31, 8494–8506. [DOI] [PubMed] [Google Scholar]
- Stenoien D. L.; Patel K.; Mancini M. G.; Dutertre M.; Smith C. L.; O’Malley B. W.; Mancini M. A. (2001) FRAP reveals that mobility of oestrogen receptor-α is ligand- and proteasome-dependent. Nat. Cell Biol. 3, 15–23. [DOI] [PubMed] [Google Scholar]
- Chan S. W.; Lim C. J.; Loo L. S.; Chong Y. F.; Huang C.; Hong W. (2009) TEADs mediate nuclear retention of TAZ to promote oncogenic transformation. J. Biol. Chem. 284, 14347–14358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaborit N.; Larbouret C.; Vallaghe J.; Peyrusson F.; Bascoul-Mollevi C.; Crapez E.; Azria D.; Chardès T.; Poul M.-A.; Mathis G.; Bazin H.; Pèlegrin A. (2011) Time-resolved fluorescence resonance energy transfer (TR-FRET) to analyze the disruption of EGFR/HER2 dimers: A new method to evaluate the efficiency of targeted therapy using monoclonal antibodies. J. Biol. Chem. 286, 11337–11345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardaga J.-P.; Bünemann M.; Krasel C.; Castro M.; Lohse M. J. (2003) Measurement of the millisecond activation switch of G protein-coupled receptors in living cells. Nat. Biotechnol. 21, 807–812. [DOI] [PubMed] [Google Scholar]
- Grashoff C.; Hoffman B. D.; Brenner M. D.; Zhou R.; Parsons M.; Yang M. T.; McLean M. A.; Sligar S. G.; Chen C. S.; Ha T.; Schwartz M. A. (2010) Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature 466, 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezin M. Y.; Achilefu S. (2010) Fluorescence lifetime measurements and biological imaging. Chem. Rev. 110, 2641–2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel V.; Sheetz M. (2006) Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell Biol. 7, 265–275. [DOI] [PubMed] [Google Scholar]
- Chen C. S. (1997) Geometric Control of Cell Life and Death. Science 276, 1425–1428. [DOI] [PubMed] [Google Scholar]
- Zamir E.; Katz B. z.; Aota S.; Yamada K. m.; Geiger B.; Kam Z. (1999) Molecular diversity of cell-matrix adhesions. J. Cell Sci. 112, 1655–1669. [DOI] [PubMed] [Google Scholar]
- Aplin A. E. (2003) Cell adhesion molecule regulation of nucleocytoplasmic trafficking. FEBS Lett. 534, 11–14. [DOI] [PubMed] [Google Scholar]
- Juliano R. L. (2002) Signal Transduction by Cell Adhesion Receptors and the Cytoskeleton: Functions of Integrins, Cadherins, Selectins, and Immunoglobulin-Superfamily Members. Annu. Rev. Pharmacol. Toxicol. 42, 283–323. [DOI] [PubMed] [Google Scholar]
- Ochsner M.; Textor M.; Vogel V.; Smith M. L. (2010) Dimensionality controls cytoskeleton assembly and metabolism of fibroblast cells in response to rigidity and shape. PLoS One 5, e9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghibaudo M.; Di Meglio J.-M.; Hersen P.; Ladoux B. (2011) Mechanics of cell spreading within 3D-micropatterned environments. Lab Chip 11, 805–812. [DOI] [PubMed] [Google Scholar]
- McBeath R.; Pirone D. M.; Nelson C. M.; Bhadriraju K.; Chen C. S. (2004) Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev. Cell 6, 483–495. [DOI] [PubMed] [Google Scholar]
- Engler A. J.; Sen S.; Sweeney H. L.; Discher D. E. (2006) Matrix Elasticity Directs Stem Cell Lineage Specification. Cell 126, 677–689. [DOI] [PubMed] [Google Scholar]
- Raucher D.; Sheetz M. P. (2000) Cell Spreading and Lamellipodial Extension Rate Is Regulated by Membrane Tension. J. Cell Biol. 148, 127–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheetz M. P.; Dai J. (1996) Modulation of membrane dynamics and cell motility by membrane tension. Trends Cell Biol. 6, 85–89. [DOI] [PubMed] [Google Scholar]
- Dai J.; Sheetz M. P. (1995) Regulation of Endocytosis, Exocytosis, and Shape by Membrane Tension. Cold Spring Harbor Symp. Quant. Biol. 60, 567–571. [DOI] [PubMed] [Google Scholar]
- Tarricone C.; Xiao B.; Justin N.; Walker P. A.; Rittinger K.; Gamblin S. J.; Smerdon S. J. (2001) The structural basis of Arfaptin-mediated cross-talk between Rac and Arf signalling pathways. Nature 411, 215–219. [DOI] [PubMed] [Google Scholar]
- Williger B.-T.; Ostermann J.; Exton J. H. (1999) Arfaptin 1, an ARF-binding protein, inhibits phospholipase D and endoplasmic reticulum/Golgi protein transport. FEBS Lett. 443, 197–200. [DOI] [PubMed] [Google Scholar]
- Van Aelst L.; Joneson T.; Bar-Sagi D. (1996) Identification of a novel Rac1-interacting protein involved in membrane ruffling. EMBO J. 15, 3778–3786. [PMC free article] [PubMed] [Google Scholar]
- Kubo T.; Yamashita T.; Yamaguchi A.; Sumimoto H.; Hosokawa K.; Tohyama M. (2002) A Novel FERM Domain Including Guanine Nucleotide Exchange Factor Is Involved in Rac Signaling and Regulates Neurite Remodeling. J. Neurosci. 22, 8504–8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanova N.; Staneva G.; Petkova D.; Lupanova T.; Pankov R.; Momchilova A. (2009) Cell culturing in a three-dimensional matrix affects the localization and properties of plasma membrane cholesterol. Cell Biol. Int. 33, 1079–1086. [DOI] [PubMed] [Google Scholar]
- Caswell P. T.; Vadrevu S.; Norman J. C. (2009) Integrins: Masters and slaves of endocytic transport. Nat. Rev. Mol. Cell Biol. 10, 843–853. [DOI] [PubMed] [Google Scholar]
- Lingwood D.; Simons K. (2010) Lipid rafts as a membrane-organizing principle. Science 327, 46–50. [DOI] [PubMed] [Google Scholar]
- McAuliffe P. F.; Meric-Bernstam F.; Mills G. B.; Gonzalez-Angulo A. M. (2010) Deciphering the role of PI3K/Akt/mTOR pathway in breast cancer biology and pathogenesis. Clin. Breast Cancer 10(Suppl. 3), S59–S65. [DOI] [PubMed] [Google Scholar]
- Vara J. Á. F.; Casado E.; de Castro J.; Cejas P.; Belda-Iniesta C.; González-Barón M. (2004) PI3K/Akt signalling pathway and cancer. Cancer Treat. Rev. 30, 193–204. [DOI] [PubMed] [Google Scholar]
- Gao X.; Lowry P. R.; Zhou X.; Depry C.; Wei Z.; Wong G. W.; Zhang J. (2011) PI3K/Akt signaling requires spatial compartmentalization in plasma membrane microdomains. Proc. Natl. Acad. Sci. U.S.A. 108, 14509–14514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian N.; Scott D. W.; Castle J. D.; Casanova J. E.; Schwartz M. A. (2007) Arf6 and microtubules in adhesion-dependent trafficking of lipid rafts. Nat. Cell Biol. 9, 1381–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mañes S.; Ana Lacalle R.; Gómez-Moutón C.; Martínez-A C. (2003) From rafts to crafts: Membrane asymmetry in moving cells. Trends Immunol. 24, 319–325. [DOI] [PubMed] [Google Scholar]
- Matsuoka T.; Yashiro M.; Nishioka N.; Hirakawa K.; Olden K.; Roberts J. D. (2012) PI3K/Akt signalling is required for the attachment and spreading, and growth in vivo of metastatic scirrhous gastric carcinoma. Br. J. Cancer 106, 1535–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flusberg D. A.; Numaguchi Y.; Ingber D. E. (2001) Cooperative control of Akt phosphorylation, bcl-2 expression, and apoptosis by cytoskeletal microfilaments and microtubules in capillary endothelial cells. Mol. Biol. Cell 12, 3087–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oinuma I.; Kawada K.; Tsukagoshi K.; Negishi M. (2012) Rnd1 and Rnd3 targeting to lipid raft is required for p190 RhoGAP activation. Mol. Biol. Cell 23, 1593–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammoto A.; Huang S.; Ingber D. E. (2007) Filamin links cell shape and cytoskeletal structure to Rho regulation by controlling accumulation of p190RhoGAP in lipid rafts. J. Cell Sci. 120, 456–467. [DOI] [PubMed] [Google Scholar]
- Nikolov D. B.; Xu K.; Himanen J. P. (2013) Eph/ephrin recognition and the role of Eph/ephrin clusters in signaling initiation. Biochim. Biophys. Acta 1834, 2160–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salaita K.; Nair P. M.; Petit R. S.; Neve R. M.; Das D.; Gray J. W.; Groves J. T. (2010) Restriction of receptor movement alters cellular response: Physical force sensing by EphA2. Science 327, 1380–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek A. M.; Izumo S. (1996) Mechanism of endothelial cell shape change and cytoskeletal remodeling in response to fluid shear stress. J. Cell Sci. 109(Part 4), 713–726. [DOI] [PubMed] [Google Scholar]
- Chung S.; Sudo R.; Vickerman V.; Zervantonakis I. K.; Kamm R. D. (2010) Microfluidic platforms for studies of angiogenesis, cell migration, and cell-cell interactions. Ann. Biomed. Eng. 38, 1164–1177. [DOI] [PubMed] [Google Scholar]
- Gee E.; Milkiewicz M.; Haas T. L. (2010) p38 MAPK activity is stimulated by vascular endothelial growth factor receptor 2 activation and is essential for shear stress-induced angiogenesis. J. Cell. Physiol. 222, 120–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalali S.; Li Y. S.; Sotoudeh M.; Yuan S.; Li S.; Chien S.; Shyy J. Y. (1998) Shear stress activates p60src-Ras-MAPK signaling pathways in vascular endothelial cells. Arterioscler., Thromb., Vasc. Biol. 18, 227–234. [DOI] [PubMed] [Google Scholar]
- McEver R. P.; Zhu C. (2010) Rolling cell adhesion. Annu. Rev. Cell Dev. Biol. 26, 363–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin L. (2010) The impact of the extracellular matrix on inflammation. Nat. Rev. Immunol. 10, 712–723. [DOI] [PubMed] [Google Scholar]
- Srinivasan S.; Wang F.; Glavas S.; Ott A.; Hofmann F.; Aktories K.; Kalman D.; Bourne H. R. (2003) Rac and Cdc42 play distinct roles in regulating PI(3,4,5)P3 and polarity during neutrophil chemotaxis. J. Cell Biol. 160, 375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow D.; Nunalee M. L.; Lim D. W.; Simnick A. J.; Chilkoti A. (2008) Peptide-based Biopolymers in Biomedicine and Biotechnology. Mater. Sci. Eng., R 62, 125–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson M. D.; Piristine H.; Hogrebe N. J.; Nocera T. M.; Boehm M. W.; Reen R. K.; Koelling K. W.; Agarwal G.; Sarang-Sieminski A. L.; Gooch K. J. (2013) A self-assembling peptide matrix used to control stiffness and binding site density supports the formation of microvascular networks in three dimensions. Acta Biomater. 9, 7651–7661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi K.; Wang G.; Liu Z.; Feng Z.; Huang B.; Zhao X. (2009) Influence of a self-assembling peptide, RADA16, compared with collagen I and Matrigel on the malignant phenotype of human breast-cancer cells in 3D cultures and in vivo. Macromol. Biosci. 9, 437–443. [DOI] [PubMed] [Google Scholar]
- Miroshnikova Y. A.; Jorgens D. M.; Spirio L.; Auer M.; Sarang-Sieminski A. L.; Weaver V. M. (2011) Engineering strategies to recapitulate epithelial morphogenesis within synthetic three-dimensional extracellular matrix with tunable mechanical properties. Phys. Biol. 8, 026013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental K. R.; Yu H.; Kass L.; Lakins J. N.; Egeblad M.; Erler J. T.; Fong S. F. T.; Csiszar K.; Giaccia A.; Weninger W.; Yamauchi M.; Gasser D. L.; Weaver V. M. (2009) Matrix Crosslinking Forces Tumor Progression by Enhancing Integrin Signaling. Cell 139, 891–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willits R. K.; Skornia S. L. (2004) Effect of collagen gel stiffness on neurite extension. J. Biomater. Sci., Polym. Ed. 15, 1521–1531. [DOI] [PubMed] [Google Scholar]
- Ashe H. L.; Briscoe J. (2006) The interpretation of morphogen gradients. Development 133, 385–394. [DOI] [PubMed] [Google Scholar]
- Lander A. D.; Nie Q.; Wan F. Y. (2002) Do Morphogen Gradients Arise by Diffusion?. Dev. Cell 2, 785–796. [DOI] [PubMed] [Google Scholar]
- Bollenbach T.; Pantazis P.; Kicheva A.; Bökel C.; González-Gaitán M.; Jülicher F. (2008) Precision of the Dpp gradient. Development 135, 1137–1146. [DOI] [PubMed] [Google Scholar]
- Häcker U.; Nybakken K.; Perrimon N. (2005) Heparan sulphate proteoglycans: The sweet side of development. Nat. Rev. Mol. Cell Biol. 6, 530–541. [DOI] [PubMed] [Google Scholar]
- Yu S. R.; Burkhardt M.; Nowak M.; Ries J.; Petrášek Z.; Scholpp S.; Schwille P.; Brand M. (2009) Fgf8 morphogen gradient forms by a source-sink mechanism with freely diffusing molecules. Nature 461, 533–536. [DOI] [PubMed] [Google Scholar]
- Jaeger J.; Surkova S.; Blagov M.; Janssens H.; Kosman D.; Kozlov K. N.; Manu; Myasnikova E.; Vanario-Alonso C. E.; Samsonova M.; Sharp D. H.; Reinitz J. (2004) Dynamic control of positional information in the early Drosophila embryo. Nature 430, 368–371. [DOI] [PubMed] [Google Scholar]
- Houchmandzadeh B.; Wieschaus E.; Leibler S. (2002) Establishment of developmental precision and proportions in the early Drosophila embryo. Nature 415, 798–802. [DOI] [PubMed] [Google Scholar]
- Teleman A. A.; Cohen S. M. (2000) Dpp Gradient Formation in the Drosophila Wing Imaginal Disc. Cell 103, 971–980. [DOI] [PubMed] [Google Scholar]
- Belenkaya T. Y.; Han C.; Yan D.; Opoka R. J.; Khodoun M.; Liu H.; Lin X. (2004) Drosophila Dpp morphogen movement is independent of dynamin-mediated endocytosis but regulated by the glypican members of heparan sulfate proteoglycans. Cell 119, 231–244. [DOI] [PubMed] [Google Scholar]
- Rogers K. W.; Schier A. F. (2011) Morphogen gradients: From generation to interpretation. Annu. Rev. Cell Dev. Biol. 27, 377–407. [DOI] [PubMed] [Google Scholar]
- Tucker J. A.; Mintzer K. A.; Mullins M. C. (2008) The BMP signaling gradient patterns dorsoventral tissues in a temporally progressive manner along the anteroposterior axis. Dev. Cell 14, 108–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan B. L. (1996) Bone morphogenetic proteins: Multifunctional regulators of vertebrate development. Genes Dev. 10, 1580–1594. [DOI] [PubMed] [Google Scholar]
- Yu S. R.; Burkhardt M.; Nowak M.; Ries J.; Petrásek Z.; Scholpp S.; Schwille P.; Brand M. (2009) Fgf8 morphogen gradient forms by a source-sink mechanism with freely diffusing molecules. Nature 461, 533–536. [DOI] [PubMed] [Google Scholar]
- Dessaud E.; Yang L. L.; Hill K.; Cox B.; Ulloa F.; Ribeiro A.; Mynett A.; Novitch B. G.; Briscoe J. (2007) Interpretation of the sonic hedgehog morphogen gradient by a temporal adaptation mechanism. Nature 450, 717–720. [DOI] [PubMed] [Google Scholar]
- Kretzschmar K.; Watt F. M. (2012) Lineage Tracing. Cell 148, 33–45. [DOI] [PubMed] [Google Scholar]
- Swartz M. A.; Fleury M. E. (2007) Interstitial Flow and Its Effects in Soft Tissues. Annu. Rev. Biomed. Eng. 9, 229–256. [DOI] [PubMed] [Google Scholar]
- Pouysségur J.; Volmat V.; Lenormand P. (2002) Fidelity and spatio-temporal control in MAP kinase (ERKs) signalling. Biochem. Pharmacol. 64, 755–763. [DOI] [PubMed] [Google Scholar]
- Vaupel P.; Mayer A. (2007) Hypoxia in cancer: Significance and impact on clinical outcome. Cancer Metastasis Rev. 26, 225–239. [DOI] [PubMed] [Google Scholar]
- Harris A. L. (2002) Hypoxia: A key regulatory factor in tumour growth. Nat. Rev. Cancer 2, 38–47. [DOI] [PubMed] [Google Scholar]
- Warburton D.; Bellusci S.; De Langhe S.; Del Moral P.-M.; Fleury V.; Mailleux A.; Tefft D.; Unbekandt M.; Wang K.; Shi W. (2005) Molecular mechanisms of early lung specification and branching morphogenesis. Pediatr. Res. 57, 26R–37R. [DOI] [PubMed] [Google Scholar]
- Fleury M. E.; Boardman K. C.; Swartz M. A. (2006) Autologous Morphogen Gradients by Subtle Interstitial Flow and Matrix Interactions. Biophys. J. 91, 113–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach C.; Kong H. J.; Hsiong S. X.; Evangelista M. B.; Yuen W.; Mooney D. J. (2009) Cancer cell angiogenic capability is regulated by 3D culture and integrin engagement. Proc. Natl. Acad. Sci. U.S.A. 106, 399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.; Radisky D. C.; Wang F.; Bissell M. J. (2004) Polarity and proliferation are controlled by distinct signaling pathways downstream of PI3-kinase in breast epithelial tumor cells. J. Cell Biol. 164, 603–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khademhosseini A.; Ferreira L.; Blumling J. III; Yeh J.; Karp J. M.; Fukuda J.; Langer R. (2006) Co-culture of human embryonic stem cells with murine embryonic fibroblasts on microwell-patterned substrates. Biomaterials 27, 5968–5977. [DOI] [PubMed] [Google Scholar]
- Ludwig T. E.; Levenstein M. E.; Jones J. M.; Berggren W. T.; Mitchen E. R.; Frane J. L.; Crandall L. J.; Daigh C. A.; Conard K. R.; Piekarczyk M. S.; Llanas R. A.; Thomson J. A. (2006) Derivation of human embryonic stem cells in defined conditions. Nat. Biotechnol. 24, 185–187. [DOI] [PubMed] [Google Scholar]
- Amit M., and Itskovitz-Eldor J. (2006) in Feeder-Free Culture of Human Embryonic Stem Cells (Klimanskaya I., and Lanza R., Eds.) pp 37–49, Academic Press, New York. [DOI] [PubMed] [Google Scholar]
- Xu C.; Inokuma M. S.; Denham J.; Golds K.; Kundu P.; Gold J. D.; Carpenter M. K. (2001) Feeder-free growth of undifferentiated human embryonic stem cells. Nat. Biotechnol. 19, 971–974. [DOI] [PubMed] [Google Scholar]
- Ludwig T. E.; Bergendahl V.; Levenstein M. E.; Yu J.; Probasco M. D.; Thomson J. A. (2006) Feeder-independent culture of human embryonic stem cells. Nat. Methods 3, 637–646. [DOI] [PubMed] [Google Scholar]
- Alvarez-Maubecin V.; García-Hernández F.; Williams J. T.; Van Bockstaele E. J. (2000) Functional Coupling between Neurons and Glia. J. Neurosci. 20, 4091–4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H.; Kamm R. D.; Lee R. T. (2004) Cell mechanics and mechanotransduction: Pathways, probes, and physiology. Am. J. Physiol. 287, C1–C11. [DOI] [PubMed] [Google Scholar]
- Halassa M. M.; Haydon P. G. (2010) Integrated Brain Circuits: Astrocytic Networks Modulate Neuronal Activity and Behavior. Annu. Rev. Physiol. 72, 335–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydon P. G. (2000) Neuroglial networks: Neurons and glia talk to each other. Curr. Biol. 10, R712–R714. [DOI] [PubMed] [Google Scholar]
- Ullian E. M.; Christopherson K. S.; Barres B. A. (2004) Role for glia in synaptogenesis. Glia 47, 209–216. [DOI] [PubMed] [Google Scholar]
- Zhang M. Y.; Lee P. J.; Hung P. J.; Johnson T.; Lee L. P.; Mofrad M. R. K. (2008) Microfluidic environment for high density hepatocyte culture. Biomed. Microdevices 10, 117–121. [DOI] [PubMed] [Google Scholar]
- Powers M. J.; Janigian D. M.; Wack K. E.; Baker C. S.; Stolz D. B.; Griffith L. G. (2002) Functional Behavior of Primary Rat Liver Cells in a Three-Dimensional Perfused Microarray Bioreactor. Tissue Eng. 8, 499–513. [DOI] [PubMed] [Google Scholar]
- Powers M. J.; Domansky K.; Kaazempur-Mofrad M. R.; Kalezi A.; Capitano A.; Upadhyaya A.; Kurzawski P.; Wack K. E.; Stolz D. B.; Kamm R.; Griffith L. G. (2002) A microfabricated array bioreactor for perfused 3D liver culture. Biotechnol. Bioeng. 78, 257–269. [DOI] [PubMed] [Google Scholar]
- Lee P. J.; Hung P. J.; Lee L. P. (2007) An artificial liver sinusoid with a microfluidic endothelial-like barrier for primary hepatocyte culture. Biotechnol. Bioeng. 97, 1340–1346. [DOI] [PubMed] [Google Scholar]
- Tibbitt M. W.; Anseth K. S. (2009) Hydrogels as extracellular matrix mimics for 3D cell culture. Biotechnol. Bioeng. 103, 655–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitesides G. M.; Ostuni E.; Takayama S.; Jiang X.; Ingber D. E. (2001) Soft lithography in biology and biochemistry. Annu. Rev. Biomed. Eng. 3, 335–373. [DOI] [PubMed] [Google Scholar]
- Kane R. S.; Takayama S.; Ostuni E.; Ingber D. E.; Whitesides G. M. (1999) Patterning proteins and cells using soft lithography. Biomaterials 20, 2363–2376. [DOI] [PubMed] [Google Scholar]
- Sung J. H.; Kam C.; Shuler M. L. (2010) A microfluidic device for a pharmacokinetic–pharmacodynamic (PK–PD) model on a chip. Lab Chip 10, 446–455. [DOI] [PubMed] [Google Scholar]
- Miller J. S.; Stevens K. R.; Yang M. T.; Baker B. M.; Nguyen D.-H. T.; Cohen D. M.; Toro E.; Chen A. A.; Galie P. A.; Yu X.; Chaturvedi R.; Bhatia S. N.; Chen C. S. (2012) Rapid casting of patterned vascular networks for perfusable engineered three-dimensional tissues. Nat. Mater. 11, 768–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh D.; Matthews B. D.; Mammoto A.; Montoya-Zavala M.; Hsin H. Y.; Ingber D. E. (2010) Reconstituting Organ-Level Lung Functions on a Chip. Science 328, 1662–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao Y.; Kimura H.; Sakai Y.; Fujii T. (2011) Bile canaliculi formation by aligning rat primary hepatocytes in a microfluidic device. Biomicrofluidics 5, 022212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J. W.; Khetani S. R.; Bhatia S. N. (2005) In Vitro Zonation and Toxicity in a Hepatocyte Bioreactor. Toxicol. Sci. 84, 110–119. [DOI] [PubMed] [Google Scholar]
- Jang K.-J.; Suh K.-Y. (2010) A multi-layer microfluidic device for efficient culture and analysis of renal tubular cells. Lab Chip 10, 36–42. [DOI] [PubMed] [Google Scholar]
- Griffith L. G.; Swartz M. A. (2006) Capturing complex 3D tissue physiology in vitro. Nat. Rev. Mol. Cell Biol. 7, 211–224. [DOI] [PubMed] [Google Scholar]
- Lee G. Y.; Kenny P. A.; Lee E. H.; Bissell M. J. (2007) Three-dimensional culture models of normal and malignant breast epithelial cells. Nat. Methods 4, 359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vlierberghe S.; Dubruel P.; Schacht E. (2011) Biopolymer-based hydrogels as scaffolds for tissue engineering applications: A review. Biomacromolecules 12, 1387–1408. [DOI] [PubMed] [Google Scholar]
- Patterson J.; Martino M. M.; Hubbell J. A. (2010) Biomimetic materials in tissue engineering. Mater. Today 13, 14–22. [Google Scholar]
- Huh D.; Hamilton G. A.; Ingber D. E. (2011) From 3D cell culture to organs-on-chips. Trends Cell Biol. 21, 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]