Abstract
Protein kinases are dynamic molecular switches that have evolved to be only transiently activated. Kinase activity is embedded within a conserved kinase core, which is typically regulated by associated domains, linkers and interacting proteins. Moreover, protein kinases are often tethered to large macromolecular complexes to provide tighter spatiotemporal control. Thus, structural characterization of kinase domains alone is insufficient to explain protein kinase function and regulation in vivo. Recent progress in structural characterization of cyclic AMP-dependent protein kinase (PKA) exemplifies how our knowledge of kinase signalling has evolved by shifting the focus of structural studies from single kinase subunits to macromolecular complexes.
As we strive to understand the biological complexity of cell signalling, it becomes increasingly clear that we cannot study signalling molecules in isolation. Instead, we must try to understand how individual molecules function as part of large polyvalent and often highly dynamic signalling scaffolds. In some cases, these scaffolds are stably assembled at the cytoplasmic surface of transporters and/or at channels that are located at the plasma membrane, whereas in other instances intracellular scaffolds are assembled as a consequence of an extracellular ligand binding to a receptor. In either case, these scaffolds are regulated by the dynamic turnover of phosphates through the activity of colocalized kinases and phosphatases and by oscillating second messengers such as calcium and cyclic AMP that are regulated by the activity of cyclases and phosphodiesterases (PDEs).
Protein kinases constitute one of the largest protein families that regulates a plethora of biological processes and that is associated with many diseases (BOX 1). cAMP-dependent protein kinase (PKA) serves in many ways as a prototype for the entire superfamily of eukaryotic protein kinases. Historically, PKA had a central role in the elucidation of the mechanisms that regulate protein phosphorylation. It serves as the prototypical protein kinase not only for the large AGC subfamily, which includes PKA, PKG, AKT (also known as PKB), S6 kinase (S6K) and phosphoinositide-dependent protein kinase 1 (PDK1), but also for the entire kinome. Although phosphorylase kinase was the first regulatory protein kinase to be discovered1, with PKA being the second, PKA was the first to be sequenced by conventional methods2. When the transforming protein SRC from Rous sarcoma virus was fist cloned, its similarity to PKA defined the boundaries and complexity of this large superfamily3. Moreover, the catalytic subunit of PKA was the first protein kinase to be crystallized, and this structure immediately explained the role of the conserved motifs and revealed how all protein kinases are organized.
Box 1. Discovery of PKA signalling and the protein kinase superfamily.
Protein phosphorylation was first described as a regulatory mechanism in 1958 through the pioneering studies of Krebs and Fischer1,68. They showed for the first time that the activity of an enzyme, phosphorylase kinase, was activated by the covalent addition of a phosphate moiety. Whereas G protein-coupled receptors (GPCRs) are probably the largest gene family, protein kinases account for ~2% of most genomes and include over 500 separate genes in the human genome69. Because protein kinases are very important for biology and are linked to many diseases, they are major drug targets. Thus, the rather comprehensive structural kinome includes structures that were solved by academia, industry and by structural genomics consortia (see the figure, part a). The protein kinases are clustered into seven major subfamilies (as indicated). Kinase structures are indicated by black dots, whereas the historically important kinases, phosphorylase kinase (PHK), cyclic AMP-dependent protein kinase (PKA) and SRC are indicated by red dots.
About the same time as the discovery of protein phosphorylation, Sutherland discovered that cAMP was a second messenger for hormone action in mammalian cells70. In the intervening decades, G proteins were discovered as a mechanism for transducing the information from the hormone receptor to adenylyl cyclases71. It is now recognized that protein kinases, like the G proteins, are widespread biological switches that regulate most events in eukaryotic cells. The discovery of a second protein kinase in 1968 (REF. 72), PKA, and later the elucidation of the mechanism for its regulation by cAMP73–75 (which is produced by adenylyl cyclase), brought together these two major signalling mechanisms (see the figure, part b). PKA has a special role and is ubiquitous in every mammalian cell. This kinase functions both in the cytoplasm and in the nucleus to regulate a plethora of biological events, including memory, differentiation, proliferation and metabolism. It is conserved in fungi and even in single-cell pathogens such as Plasmodium falciparum and Trypanosomes76. Historically, the PKA catalytic subunit remains also as a frame of reference for every protein kinase.
The PKA signalling system includes the regulatory (R) and catalytic (C) subunits, PKA substrates and targeting proteins (such as A kinase anchoring proteins (AKAPs)) that localize PKA to specific sites in the cell in close proximity to dedicated substrates (see the figure, part b and c).
CAMK, calcium/calmodulin-dependent protein kinase; CK1, casein kinase 1; CMGC, CDK, MAPK, GSK3 and CLK; GPCR, G protein-coupled receptor; PDE, phosphodiesterase; STE, sterile 7, sterile 11 and sterile 20 kinase; TK, Tyr kinase; TKL, Tyr kinase-like.

The structure and in vitro and in vivo kinetic properties of free PKA catalytic subunits have been extensively studied. However, these kinases do not usually function in isolation; instead, they are part of larger signalling systems. In eukaryotic cells, PKA catalytic subunits assemble with regulatory subunits into a holoenzyme complex that is sequestered in an inactive state in the absence of cAMP. In mammals, the ‘PKA signalling system’ comprises four genes that encode functionally non-redundant regulatory subunits and three genes that encode catalytic subunits (BOX 1). The PKA holoenzyme consists of a regulatory subunit dimer and two catalytic subunits. The regulatory subunits are the receptors for cAMP, and structural and functional studies revealed that the cyclic nucleotide-binding domains (CNB domains) are conserved across all phyla4. The structural analysis of the individual subunits was useful for the understanding of the structure and function of protein kinases and cAMP receptors. However, these studies shed no light onto the mechanism of inhibition of the catalytic subunit by the regulatory subunits, and they did not reveal the molecular mechanism by which cAMP activates the holoenzyme. In addition, the structures of full-length holoenzymes had to be elucidated to understand the mechanisms underlying the functional non-redundancy of regulatory subunits.
Importantly, the specificity of PKA signalling is not only determined by the cell type-specific expression of the different regulatory and catalytic PKA isoforms and the different PKA substrates, but also by the subcellular localization of PKA. Targeting of PKA to specific sites within the cell is largely achieved by A kinase anchoring proteins (AKAPs)5,6. The hallmark motif in AKAPs is an amphipathic helix that docks with high affinity to the PKA regulatory subunits7,8. Most protein kinases assemble into signalling scaffolds, and a layer of complexity is added by the existence of multiple kinase isoforms and/or splice variants. However, we are just beginning to appreciate how functionally unique each protein can be in terms of its expression, regulation and targeting. Perhaps it is the increased number of splice variants, especially in the brain and in immune-responsive cells, that explains the enhanced complexity for PKA signalling in higher eukaryotes and distinguishes humans from mice and flies, in which the actual number of genes is surprisingly similar.
In this Review, we discuss the functional insights that have been gained from the structures of signalling proteins and how our knowledge evolved as these structural studies moved to higher levels of complexity. By using PKA as an example, we emphasize why it is so important to elucidate structures not only of single domains and subunits but also of full-length proteins and macromolecular signalling complexes to fully appreciate the beauty and elegance of signalling pathways in the cell. Although PKA is used as an example, the lessons extend well beyond this kinase.
Functional insights from single subunit structures
A wealth of information has been obtained from initial structural studies of the individual regulatory and catalytic subunits of PKA. The structure of the catalytic subunit in particular took the field of kinase research instantly from the one-dimensional sequence to a three-dimensional template9,10. This solved structure provided a mechanistic understanding of this enzyme family and laid the foundation for all the drug discovery enterprises that have become so fundamental to present day therapeutics.
The PKA catalytic subunit
When the sequence of PKA was compared with the sequence of newly cloned SRC, the eukaryotic protein kinase superfamily was enlarged to include enzymes that phosphorylate Ser, Thr and Tyr residues3. With the advent of DNA cloning, it soon became clear that protein kinases constitute a large enzyme family that shares a conserved catalytic core comprising an amino-terminal N-lobe and a carboxy-terminal C-lobe (FIG. 1a,b). Based solely on the alignment of a few protein kinase sequences, many conserved motifs were identified and classified as subdomains11 (FIG. 1c). The subsequent elucidation of the structure of PKA showed for the first time how these sequence motifs and subdomains converge to form an active enzyme9. This single structure helped define all the fundamental features that are conserved in every protein kinase. The PKA catalytic subunit core is a bilobal protein in which the conserved residues cluster mostly around the active site cleft that lies between the two lobes. Subsequent structures of the catalytic subunit helped identify the flexibility of the core, specifically how the cleft opens and closes as the enzyme traverses through a catalytic cycle, and how substrates and inhibitors dock. These studies established PKA as a template for the function of the whole protein kinase superfamily.
Figure 1. The catalytic subunit of PKA as a prototype for the protein kinase superfamily.
a | The cyclic AMP-dependent protein kinase (PKA) catalytic core, which contains an N-lobe and a C-lobe, is shared by all eukaryotic protein kinases. b | In contrast to the conserved core, the flanking regions (that is, the amino-terminal tail (N-tail) and the carboxy-terminal tail (C-tail)) vary among protein kinases. The regulatory subunits can have a separate function (as in PKA) or be part of the kinase (as for PKG). Post-translational modifications include phosphorylation (indicated in red) and myristylation (indicated in blue). c | The sequence motifs that span the entire core are highly conserved, and the core can be segregated into 12 subdomains11. d | By comparing many structures, the core hydrophobic spine architecture that is shared by all eukaryotic protein kinases can be defined14. The regulatory spine (R-spine) is typically dynamically assembled following phosphorylation of the activation loop, whereas the catalytic spine (C-spine) is completed by the adenine ring of bound ATP. Both spines are anchored to the unusual hydrophobic αF-helix. e | In the C-tails of AGC kinases, which include PKA, PKC, PKG, AKT, S6 kinase (S6K) and p90 ribosomal protein S6K (RSK), there are at least three highly conserved motifs: Pro-x-x-Pro (PXXP) in the C-lobe tether, Phe-Asp-Asp-Tyr (FDDY) in the active site tether and Phe-x-x-Phe (FxxF) in the N-lobe tether. This region contains numerous motifs that contribute to catalysis and to interactions with other proteins such as SMAD3 (wich interacts with AKT), SRC (which associates via its SRC homology 3 (SH3) domain with AKT), heat shock protein 90 (HSP90; which interacts via its SH3 domain with PKC) and the prolyl isomerase PIN1 (which binds to PKC). The C-tail thus serves as a cis-regulatory element that is essential for the activity of every AGC kinase and also as a trans-regulatory element that allows communication with other proteins21. The C-tail is highly regulated by phosphorylation. The activation segment contains an essential phosphorylation site (Thr197) that is conserved in most kinases. f | The N-tail is unique to PKA and also serves multiple functions. This flanking region undergoes post-translational modifications that include myristylation, phosphorylation and deamidation. Like the C-tail, it wraps around both the C-lobe and the N-lobe and is essential for activity. The myristyl moiety becomes mobilized in the type II holoenzymes and serves as a membrane anchor77 (not shown). The N-tail also contributes to localization and trafficking through its interactions with a novel A kinase interacting protein 1 (AKIP1)25. CDK2, cyclin-dependent kinase 2; CNB, cyclic nucleotide-binding domains; D/D, dimerization and docking domain; EGFR, epidermal growth factor receptor.
Because of their importance for the understanding of biological processes and for the design of therapeutic interventions targeting kinases that have a role in disease, many kinase structures have now been solved, and these structures confirm the universality of the conserved residues and motifs. By comparing the structures of protein kinase cores, the architecture of the internal core, which includes a set of hydrophobic motifs (referred to as a regulatory spine (R-spine) and a catalytic spine (C-spine)12–14), was defined (FIG. 1d). The spine residues are from both the N-lobe and the C-lobe and provide a mechanism for fluid communication between the two lobes in the active enzyme. These spines are docked onto the unusual hydrophobic αF-helix, which spans the C-lobe. The active conformation of the R-spine is typically assembled transiently as a consequence of activation loop phosphorylation, allowing tight regulation of protein kinase activity15,16.
Protein kinases are typically not efficient catalysts but instead have evolved to be highly regulated molecular switches, controlled typically by phosphorylation. In addition to the conserved core, PKA catalytic subunits contain two flanking tails and two phosphorylation sites (FIG. 1e, f).The first phosphorylation site near the C terminus (Ser338) is modified by cis autophosphorylation while the protein is still bound to the ribosome. The second phosphorylation site (Thr197), which is located at the activation loop, is phosphorylated in trans either by another PKA enzyme or by PDK1 (3-phosphoinositide- dependent protein kinase 1) after the catalytic subunit leaves the ribosome17,18. This phosphorylation event is essential for full activity of the kinase19,20 (FIG. 1e). Unlike most other protein kinases, once the fully phosphorylated catalytic subunit is synthesized the phosphates are very resistant to phosphatases13,14. The activity of the assembled holoenzyme solely depends on cAMP levels and not on the turnover of the activation loop phosphate. Most kinase structures that have subsequently been solved are unlikely to represent fully active structures, as they comprise only the conserved, often dephosphorylated kinase core. By contrast, the PKA catalytic subunit is autophosphorylated, even when expressed in Escherichia coli, and therefore this structure identified not only the conserved fold but also the fully active enzyme and highlighted the importance of phosphorylation for kinase activity.
The crucial importance of linkers and tails was also apparent from that first structure. The N-terminal and the C-terminal tails (N-tail and C-tail) of the catalytic subunit are an integral part of the active enzyme and function as cis-regulatory elements21,22. For example, most of the ATP-binding site consists of core residues, but two hydrophobic residues in the C-tail, referred to as the Phe-Asp-Asp-Tyr (FDDY) motif, are an essential part of the ATP-binding site in PKA. The C-tail wraps around both lobes of the kinase core and is conserved in the entire AGC subfamily of protein kinases21 (FIG. 1e). In contrast to the C-tail, the N-tail is unique to the mammalian PKA catalytic subunit. It is myristylated at its N terminus and consists mostly of a long and stable αA-helix that docks onto the surface of both lobes23 (FIG. 1f). The αA-helix is important for catalysis24 and also binds to A kinase interacting protein 1 (AKIP1), which is involved in PKA shuttling between the cytoplasm and the nucleus25 and in nuclear translocation of nuclear factor-κB (NF-κB)26.
The PKA regulatory subunits
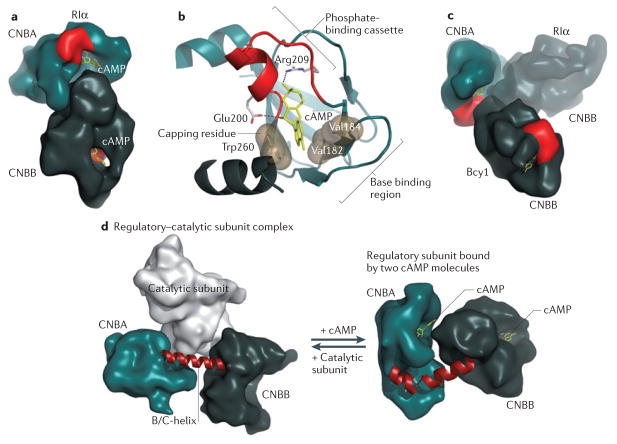
The regulatory subunits of PKA control its activity depending on the levels of the second messenger cAMP. PKA regulatory subunits are the primary receptors for cAMP in mammalian cells. In the absence of cAMP, the regulatory subunits bind to the catalytic subunits and suppress their activity. Mammalian cells possess four regulatory subunit isoforms (RIα, RIβ, RIIα and RIIβ), and knockout studies show that these isoforms are functionally non-redundant (see also below). The domain organization is conserved in all regulatory subunit isoforms and includes a dimerization and docking domain (D/D domain) located at the N terminus, followed by a flexible linker and two CNB domains in tandem (CNBA and CNBB) at the C terminus (FIG. 2a). The linker contains an inhibitor site, which resembles a peptide substrate and docks to the active site cleft of the catalytic subunit in the holoenzyme (FIG. 2b). Because RII subunits possess a Ser residue at their inhibitor site, they are also substrates for the catalytic subunit, whereas RI subunits have an Ala or Gly residue at their P-site and are therefore pseudosubstrates — that is, they mimic the substrate and thus inhibit PKA activity but are not phosphorylated by PKA (FIG. 2a). This is a fundamental difference between the two classes of regulatory subunits27,28. Crystallization of the two cAMP-binding domains of RI and RII subunits confirmed that the CNB domain is a universal docking motif for cAMP (FIG. 3a), which is conserved from bacteria to humans. The binding of cAMP to a CNB domain is thus an ancient signalling mechanism that allows cAMP to convert an extracellular signal such as glucose deprivation into a biological response. A small phosphate-binding cassette (PBC) is the signature motif of the CNB domain, whereas the adenine ring is capped by a hydrophobic residue that is recruited to the cAMP-binding site29 (FIG. 3b). Although CNBA and CNBB resemble the CNB domain of catabolite gene activator protein (CAP) in bacteria30, the orientation of the two CNB domains relative to one another in the various isoforms of the PKA regulatory subunit is surprisingly different from each other and compared with the orientation seen in CAP (FIG. 3a,c). These different domain interfaces could not be readily predicted from homology modelling and only became apparent when different crystal structures were solved.
Figure 2. Assembly of full length tetrameric holoenzymes is isoform specific and involves ordering of the intrinsically disordered linker.
a | The organization of the RIα and RIIβ regulatory subunits and the sequence of the linker regions for all four isoforms are shown. The amino terminus contains a dimerization and docking domain (D/D domain). In the absence of a catalytic subunit, the linker is disordered. The inhibitor site is embedded in the middle of the linker, followed by two cyclic nucleotide-binding domains (termed CNBA and CNBB). b | Following binding to the catalytic subunit, the inhibitor site docks to the active site cleft of the catalytic subunit (indicated by a red box) and the carboxy-terminal segment of the linker (C-linker) becomes ordered. The remaining portion of the linker (N-linker) has an important role in defining the quaternary structure of each holoenzyme. The linker becomes ordered differently in RIα and RIβ following their binding to the catalytic subunit. The binding of the inhibitor site and the C-linker is conserved, whereas the N-linker is positioned differently and contributes in unique ways to the organization of each tetrameric holoenzyme.
Figure 3. Molecular basis for regulation of PKA by cAMP.
The cyclic nucleotide-binding domains (CNB domains) of cyclic AMP-dependent protein kinase (PKA), which are conserved throughout all species, are defined by a unique fold. a | The two CNB domains (CNBA and CNBB) in RIα in the cAMP-bound state create an extensive network of allosteric communication. b | The conserved features of each CNB domain are summarized. The phosphate-binding cassette (PBC), shown in red, is the signature motif of the CNB domain. The adenine ring of bound cAMP is capped by a hydrophobic residue from within the CNB domain or from associated domains or motifs29. The base-binding region stabilizes the adenine ring by binding on the other side of cAMP. c | Although each CNB domain is highly conserved, the two domains of RIα and Bcy1 (the yeast homologue of RIα) are oriented in distinctly different ways. The overlay of the two structures is shown. d | The remarkable malleability of the regulatory subunit was first recognized when the structure of a regulatory–catalytic subunit complex was solved. On the left is the conformation of the CNB domains (amino acids 91–379 of the regulatory subunit) in the holoenzyme complex, and on the right are the CNB domains bound to two molecules of cAMP. The conformational change is mediated by the B/C-helix in CNBA, which is kinked in the cAMP-bound state and extended into a single long helix in the holoenzyme.
Inhibition of the kinase activity through an independently expressed inhibitory subunit as opposed to inhibition by a contiguous inhibitory domain or motif is a novel mechanism of regulation for protein kinases. This mechanism of regulation has led the catalytic subunits of mammalian cells to acquire, during evolution, novel features that allow them to function as potent sensors of low cAMP levels. For example, when the catalytic subunit is phosphorylated at its C-terminal tail and at its activation loop, these phosphates are very resistant to removal by phosphatases31. This is in contrast to Tyr kinases and to most other eukaryotic protein kinases, as their activity is controlled by the rapid turnover of a phosphate on the essential activation loop. When the mature enzyme has been synthesized, the catalytic subunit and the regulatory subunit of PKA are assembled into an inactive tetrameric holoenzyme18, and the activity of the holoenzyme depends solely on cAMP levels.
As described above, the structures of the individual regulatory and catalytic PKA subunits defined the dissociated and fully active conformations and provided insights into protein kinase evolution4,32 and CNB domain proteins32,33. However, these findings did not explain how PKA activity is regulated. This required the study of multisubunit complexes.
Regulatory–catalytic subunit heterodimers
In addition to understanding the regulation of cAMP synthesis and degradation, two fundamental questions of PKA regulation remained to be addressed: how is the catalytic subunit inhibited by the regulatory subunit, and how is PKA regulated by cAMP? These questions were answered by resolving the structure of a regulatory–catalytic subunit complex rather than of each independent subunit. This approach was completely analogous to the one that was used to decipher the mechanisms involved in the regulation of the activity of cyclin-dependent kinases (CDKs), which required the analysis of the structure of a CDK–cyclin complex34. The first structures of a PKA regulatory–catalytic subunit heterodimer used truncated forms of RIα that contained the inhibitor site and either one or two CNB domains. The resolved structures provided a new dimension to the understanding of PKA signalling35,36 (FIG. 3d), as they revealed for the first time how the inhibitor site of the regulatory subunit was docked into the active site cleft of the catalytic subunit, thereby blocking access to other substrates. These structures also demonstrated the remarkable flexibility of the regulatory subunits.
In contrast to the catalytic subunit, which functions as a very stable docking scaffold, the regulatory subunit undergoes major conformational changes as it releases cAMP and binds to the catalytic subunit. In the cAMP-bound state, a large allosteric interface exists between the two CNB domains of the regulatory subunit, whereas in the heterodimers the major interface is located between the regulatory and catalytic subunits. In the heterodimer complex the two CNB domains are separated by a long helix that is formed by the extension of the kinked B/C-helix in CNBA36,37 (FIG. 3d). In many ways this conformational change is reminiscent of the conformational change that occurs in calmodulin following calcium binding, during which a central helix becomes extended and separates the two calcium-binding domains at each end38. One unanticipated consequence of this conformational switch is that the cAMP-binding sites in PKA are substantially altered. Thus, two very different conformations of the regulatory subunits exist, one having high affinity for cAMP and the other one having high affinity for the catalytic subunit. This conformational flexibility of CNB domains is now recognized to be a conserved feature of these domains, as it is also observed in other CNB domain-containing proteins, such as the guanine nucleotide- binding protein EPAC (exchange protein directly activated by cAMP)39,40.
Holoenzymes give novel insights
The solved structures of the catalytic subunits bound to monomeric regulatory subunits revealed how enzymatic activity is inhibited in the absence of cAMP and how interfaces were broken and new interfaces were formed. However, the regulatory–catalytic subunit heterodimers are remarkably similar for the different isoforms (RIα, RIIα and RIIβ)36,37,41, and the heterodimers did not explain why the isoforms are functionally non-redundant. Therefore, resolving the structures of full-length holoenzymes, which also reflect the physiological state of the enzyme, would be required to fully understand the regulation of PKA activity. Although we focus here on PKA as an example, it is likely that we will not understand the full complexity and regulation of any protein kinase until the entire protein assembly including the flanking domains and/ or regulatory subunits is revealed. The kinase core alone is only a starting point.
Disordered regions drive the assembly of tetramers
The linkers that join the very stable D/D domain to the well-folded CNB domains are classified as intrinsically disordered regions (IDRs)42. IDRs are commonly found in kinases, for example the segment that joins the membrane- spanning helix to the kinase domain in epidermal growth factor receptor (EGFR) and the linkers that flank the inhibitor sequence in PKA (FIG. 2b). IDRs have inherent dynamic properties that allow them to ‘scan’ for different binding partners depending on the activation state of the cell. These flexible linkers pose the greatest challenge for the structural biologist in resolving a larger macromolecular complex and not just the single kinase, as IDRs typically interfere with crystallization by favouring a more dynamic state. Thus, the tendency is to simply delete these regions to conduct structural studies, even though IDRs are extremely important for biological function and regulation of kinases.
The portion of the linker that extends from the inhibitory site to the CNBA domain was always disordered in structures of single regulatory subunits (FIG. 2), as it becomes ordered only following binding of the regulatory subunit to the catalytic subunit. However, the remaining part of the linker, referred to as the N-linker, remained a mystery, as it was absent from all initial PKA structures. Interestingly, the N-linkers exhibit the highest sequence variability among the four isoforms of PKA regulatory subunits.
Generating RIα monomers that contained the extended N-linker offered the first glimpse of a tetrameric holoenzyme. Surprisingly, the stabilization of the N-linker in this crystal structure was achieved by interaction of this linker with the opposite heterodimer. This structure showed for the first time how two heterodimers can interact to create a novel twofold symmetry. The holoenzyme is thus a dimer of two heterodimers43 (FIG. 4). Based on this structure, a full-length RIα holoenzyme was modelled (FIG. 4c). Small angle X-ray scattering (SAXS) and small angle neutron scattering (SANS), which provide low resolution models of the general shape and maximum diameter of a protein or a protein complex in solution, validated the reconstructed model of the full-length RIα holoenzyme44. SAXS indicated early on that the RIα holoenzyme was Y-shaped in solution, whereas SANS showed that the two catalytic subunits were far apart, and these data were completely consistent with our model of full length RIα43. Moreover, the SAXS studies revealed that holoenzymes formed with the different regulatory subunits are all distinct45,46, even though the structures of all PKA heterodimers are remarkably similar. This work has contributed to the realization that the structural diversity of the PKA isoforms could be appreciated only by solving structures of tetrameric holoenzymes.
Figure 4. Assembly of tetrameric holoenzymes.
Here we show how complexity is built up from regulatory–catalytic subunit heterodimers to tetrameric holoenzymes. The strategic positioning of the linker is shown in red. a–c | The solved structure of RIα regulatory–catalytic subunit heterodimer showed for the first time how the catalytic subunit was inhibited and how the complex was activated by cyclic AMP35,36. The complex of the catalytic subunit bound to the RIα regulatory subunit defines the conformational flexibility of the two cyclic nucleotide-binding domains (CNB domains; termed CNBA and CNBB) that are located within the RIα subunit. By extending the amino-terminal segment of the linker (N-linker) it was possible to get a first glimpse of a tetrameric holoenzyme (c). d | The structure of the complex shown in (b) was used to model the full-length tetrameric holoenzyme on the basis of small angle X ray scattering (SAXS) and small angle neutron scattering (SANS) analyses as well as on the basis of previous cyclic AMP-dependent protein kinase (PKA) crystal structures. The model was validated by mutagenesis. Rotation of the model allows one to appreciate for the first time the twofold axis of symmetry that is created by the interaction of the two regulatory–catalytic subunit heterodimers. e | A cartoon of the tetramer structure shown in (d) highlights how the N-linker from one heterodimer is positioned in close proximity to the CNBA domain of the opposite dimer.
The RI tetrameric PKA holoenzymes
Although RIα and RIβ are close homologues that share over 85% sequence identity, they are functionally non-redundant. On the one hand, genetic deletion of the gene encoding RIα is embryonic lethal47. RIα mRNA levels are tightly controlled by nonsense-mediated mRNA decay (NMD), and haploinsufficiency leads to a wide range of disease states, including Carney complex disease and numerous endocrine disorders48. On the other hand, deletion of RIβ is not embryonic lethal but leads to defects in long-term depression (LTD) and memory and to dysregulated long-term potentiation (LTP)49,50. RIα and RIβ also differ in terms of their subcellular localization. Unlike RIα, which is typically diffused in the cytoplasm in the absence of stimuli51, RIβ is localized to mitochondria52. Tissue expression also distinguishes these two isoforms. RIα is constitutively expressed by every cell type, whereas RIβ expression is enriched in the brain and in neurons53,54. Finally, as mentioned above, SAXS data indicated that the RIα holoenzymes are much more compact than the RIβ holoenzymes52.
Although both RIα and RIβ require ATP and two magnesium ions to form a high affinity holoenzyme complex55, the quaternary structures of the two holoenzymes are surprisingly different on the basis of the crystal structures (FIG. 5). Consistent with the findings from SAXS studies, the RIβ holoenzyme is much more elongated and rod-shaped. In contrast to the RIα holoenzyme, in which the two catalytic subunits are well separated, in the RIβ holoenzyme the two heterodimers interact directly through an interface between the two catalytic subunits in a ‘head to head’ configuration between the two N-lobes. This leaves the two CNBB domains at opposite ends of the structure, where they are predicted to interact with an as yet unknown class II PDZ domain protein through their C-terminal PDZ-binding motifs.
Figure 5. Specific motifs define isoform-specific interfaces between heterodimers.
a | The conformation of the regulatory–catalytic subunit heterodimer (RIα is shown here) is similar in all isoforms, but each heterodimer assembles into distinctive quaternary structures. The structure is shown on the left and the model on the right. Two motifs, in addition to the amino-terminal linker (N-linker) motif (red) are exposed to solvent in the regulatory–catalytic subunit heterodimers, but have important isoform-specific roles in the assembly of each tetramer. The β4–β5 loop (purple) is located within the cyclic nucleotide-binding A domain (CNBA domain) of the regulatory subunits. The Phe-Asp-Asp-Tyr (FDDY) motif (yellow) resides in the carboxy-terminal tail (C-tail) of the catalytic subunit and is an integral part of the ATP-binding site. b–d | The quaternary structures of RIα-, RIβ- and RIIβ-containing cyclic AMP-dependent protein kinase (PKA) holoenzymes, shown on the top, highlighting the differences in the overall architecture of each tetramer and also indicating the differences in the positioning of the dimerization and docking domain (D/D domain; shown by arrows and visualized only in RIβ (c; in brown)). The linkers are shown in red. The twofold axis of symmetry is also indicated on the lower panels. In each lower panel, rotation allows one to appreciate the twofold symmetry that is found in each tetramer and also how the two motifs, the β4–β5 loops and the FDDY motifs, contribute in novel ways to the assembly of the tetramer.
As discussed above, the highest sequence variability in the isoforms of the regulatory subunits is seen in the linker region between the cAMP-binding domains and the D/D domain, in particular in the N-linker, which joins the D/D domain to the inhibitor site. Each N-linker region docks in a different way onto the surface of the catalytic subunit, whereas the structure and the position of the inhibitor site and the C-linker of all isoforms is similar43 (FIG. 2b). In the RIβ tetramer, the N-linker is directed in a different way and this forces the D/D domain into a completely different location. This variability in the linker region seems to be a major factor for promoting different inter-domain arrangements that lead to remarkably distinct quaternary structures (FIG. 5).
In principle, the D/D domain can function either as an independent docking motif or as an integral part of the holoenzyme by interacting with either the catalytic subunit or the CNB domains. The positioning of the D/D domain in RIβ suggests that it has the potential to be an integral part of the complex (FIG. 5c). The structure also suggests that the interaction of the D/D domain with AKAP would be sensed by the entire complex. It may also explain how RI subunits can serve as redox sensors, as the D/D domains of both RI subunits can be oxidized to form two interchain disulphide bonds56, and there is evidence that these disulphide bonds may be induced by stress57.
The RII tetrameric PKA holoenzyme
RIIβ is the pre-dominant PKA regulatory subunit isoform in white and brown adipose tissue and in the brain47,58, and RIIβ knockout mice have a lean phenotype and are not susceptible to diet-induced insulin resistance59,60. Deletion of RIIβ even reverses the obesity syndrome of agouti mice61. RII subunits are usually anchored to membrane proteins through high affinity binding to AKAPs. Interestingly, SAXS studies indicated significant differences not only between RI- and RII-containing PKA holoenzymes but also between the RIIα and RIIβ holoenzymes. Based on SAXS data, the RIIα and RIIβ dimers are both very extended and almost shaped like a dumb-bell. Surprisingly, however, the two holoenzymes are very different46. Although the RIIα holoenzyme remains extended, similarly to the RIIα dimer, the RIIβ holoenzyme is compact and almost globular. Confirming the SAXS predictions, the holoenzyme containing two RIIβ and two catalytic subunits (RIIβ2–C2) was shown to have a compact doughnut shape with extensive interactions between the twofold symmetry-related regulatory–catalytic subunit heterodimers62 (FIG. 5d). The axis of symmetry of this quaternary structure passes through the central cavity. Although all three holoenzymes, RIα, RIβ and RIIβ, have a twofold axis of symmetry, the geometry is very different for each complex.
A striking feature of the RIIβ tetramer is the closed conformation of the inhibited catalytic subunit even in the absence of ATP, and this finding has important implications for the regulation of the formation of the RIIβ PKA holoenzyme. Formation of a tight RIα regulatory- catalytic subunit heterodimer or a RIα2–C2 tetramer (that is, the holoenzyme) requires two magnesium ions and ATP binding to the active site cleft of each catalytic subunit55. This leads to the closed conformation of the catalytic subunit that is observed in all the solved RI holoenzyme structures35,36,43. Similarly to the RIα isoform, PKI is a pseudosubstrate of PKA and also requires two magnesium ions and ATP to form a tight complex, in which the PKA catalytic subunit is in a fully closed conformation10. The formation of the holoenzyme with RII subunits, however, does not require ATP. For example, in the RIIα–C heterodimer37 the catalytic subunit is in an open conformation with an empty ATP-binding pocket, a disordered C-tail and a dynamic N-lobe. The catalytic subunit retains high affinity binding to the regulatory subunit (that is, 0.1 nM) even in the absence of ATP. The β4–β5 loop in the CNBA domain of RIIβ promotes a closed conformation for the catalytic subunit in the absence of ATP by interacting with the C-tail of the neighbouring catalytic subunit. Following diffusion of magnesium and ATP into crystals of the apo form of the RIIβ2–C2 holoenzyme, ADP and the phosphorylated RIIβ subunit, but not ATP, were detected62. This suggests that the dissociation and reassociation cycling of PKA (which is mediated through dephosphorylation of the RII subunits) may be accomplished most efficiently in the presence of calcineurin, a calcium-activated phosphatase that is often tethered on the same AKAP in close proximity to PKA63. Such a complex that has a fully closed conformation in the absence of nucleotide has not been observed for other protein kinases.
The apparent activation constant (Ka(cAMP)) for activation of the RIIβ tetramer by cAMP is 584 nM62, and this is significantly higher than the Ka(cAMP) for the RIIβ heterodimer (which is 65 nM). This can be explained by the extensive interfaces between the two RIIβ–C heterodimers in the holoenzyme. The solved structure of the RIIβ tetramer also provides a mechanistic explanation for the highly cooperative allosteric activation of the RIIβ holoenzyme by cAMP, as it reveals that the CNBA domain from one heterodimer binds directly to the C-linker of the regulatory subunit and the C-tail of the catalytic subunit from the other heterodimer, thus providing a direct crosstalk between the two heterodimers62 (FIG. 5d). The unique quaternary structure of the RIIβ holoenzyme is also confirmed by single particle electron microscopy reconstruction data, which are also consistent with the RIα and RIIβ holoenzyme structures (F. Ye, M. Ginsburg, P. Z. and S. T., unpublished observations).
Achieving specificity
In general, kinases signal through the dynamic assembly and disassembly of macromolecular complexes. Although inherent conformational dynamics are an intrinsic feature of every kinase core, specificity is achieved through the evolution of multiple kinase isoforms, multiple regulatory domains or proteins and multiple scaffolds. In this way each kinase isoform creates a distinct signalling hub that can respond to local levels of second messengers such as cAMP or calcium. This combinatorial diversity creates a myriad of opportunities for achieving specificity but also demands the independent characterization of each of the kinases in a specific context as their properties are likely to be different. PKA specificity cannot be understood by studying individual subunits. Instead, one needs to focus on each ‘PKA signalling system’ as a whole that allows the cell to create localized sites for PKA signalling (FIG. 6).
Figure 6. PKA is anchored to scaffolds that assemble macromolecular complexes and define foci for PKA signalling.
a | Cardiomyocytes are used as an example to illustrate how cyclic AMP-dependent protein kinase (PKA) can regulate different functions in a cell. PKA signalling is mediated by complexes with different subcellular localizations. Targeting and assembly of these PKA-containing complexes is mediated by A kinase anchoring proteins (AKAPs). Most AKAPs are specific for the RII regulatory subunit (blue ovals), a few have dual specificity and can bind to either RI or RII (yellow oval and dashed R subunits) and a few, such as SKIP (which functions as an AKAP), are RI regulatory subunit-specific (red oval). By binding to different regulatory subunits, AKAPs assemble complexes that regulate diverse functions. For example, in cardiomyocytes, different PKA-containing complexes can regulate Ca2+ uptake by regulating the activity of L-type Ca2+ channels located at the plasma membrane, promote the storage of Ca2+ in the sarcoplasmic reticulum by activating Ca2+ uptake pumps or its release by activating Ca2+-release channels (which are coupled to the junction–triadin–calsequestrin complex that binds Ca2+). PKA-containing complexes can also regulate cardiomyocyte contraction when localized to the cytoskeleton, where these enzyme regulate mitochondrial activity necessary for contraction. b | Assembly of PKA at the voltage gated Ca2+ channel 1.2 (CaV1.2) is mediated by AKAP5 (also known as AKAP79 in humans or AKAP150 in mice), which interacts with the plasma membrane and brings PKA in close proximity to the tail of the channel (shown in red) that harbours a target phosphorylation site and calcineurin (CaN). PKA phosphorylates and activates the channel. After that, Ca2+ enters the cell and binds to CaN and anchored calmodulin (CaM). CaN is activated in two ways by Ca2+: by binding to the calmodulin-like B-subunit of CaN and to a separate Ca2+-bound CaM. Activated CaN can then dephosphorylate the channel, decreasing its activity. Furthermore CaN dephosphorylates the RIIβ regulatory subunit of PKA, promoting its binding to the catalytic subunit and inactivation of PKA. Dephosphorylation can also be accomplished by the phosphatase PP2A, which binds directly to the channel. Targeting of the PKA holoenzyme is mediated by an amphipathic helix in the AKAP, referred to as the A kinase-binding (AKB) motif that binds with high affinity to the dimerization and docking domain (D/D domain) of RII regulatory subunits.
The role of macromolecular complexes in cAMP signalling
Specificity is achieved in part by cell type-specific expression of PKA signalling proteins. These include, in addition to PKA isoforms, more than 30 different G protein- coupled receptors that couple PKA signalling to Gαs activation through the generation of cAMP, the cyclases that generate cAMP and to the PDEs that break it down. There are multiple isoforms for each of the signalling proteins that are expressed in a cell type-specific manner. Specificity is also achieved by the differential expression of scaffold proteins such as AKAPs, which sequester families of PKA signalling proteins in close proximity of a dedicated substrate such as an ion channel, a receptor or a transporter. In this way the cell creates discrete foci of cAMP signalling. PKC, a close homologue of PKA, is another example of different isoforms being recruited to discrete foci, where they signal in synchrony with calcium and diacylglycerol64. The many different PDE isoforms are also assembled and regulated in unique ways65.
Spatiotemporal control of cAMP signalling
It is known from genetic research that the PKA isoforms are functionally non-redundant, and it has become apparent that the structures of tetrameric holoenzymes are different from one another. Another contributing factor to PKA signalling specificity is the remarkable resistance of the active catalytic subunit to phosphatases. This feature renders the kinase responsive only to cAMP and not to the dynamic turnover of an activation loop phosphate as is the case for so many other protein kinases18. By sequestering a family of PKA signalling proteins (which also includes phosphodiesterases) to a specific site, the cell ensures that cAMP signals are local and do not diffuse as a wave across the cell. The addition of forskolin (which stimulates most cyclases) and IBMX (which is a PDE inhibitor) results in the generation of high concentrations of cAMP, which can drive PKA holoenzyme dissociation but, in case of RIα, not its full dissociation27,28. Obviously under these conditions, which are typically used to induce PKA signalling in cells, all specificity is lost. By contrast, PKA signalling under physiological conditions is likely to result from localized pulses of cAMP. By assembling foci of PKA signalling, the cell creates oscillating systems between kinases and phosphatases, cyclases and PDEs and between second messengers such as calcium and cAMP66. In this setting, negative feedback loops can be rate-limiting. The fact that most signalling events occur in a rapid time frame (less than a minute) emphasizes even more the importance of localized signalling environments. It is essential that PKA signalling system are considered as a multicomponent entity as opposed to individual subunits.
Interdisciplinary approaches to monitor the regulation of cAMP signalling
Visualizing the structures of larger kinase complexes resulted in the appreciation of the importance of these macromolecular complexes. Recent reports showing that Gαi interacts with the RIIβ subunit indicates further levels of crosstalk between these signalling proteins that are clustered in close proximity67. To understand these macromolecules beyond the holoenzymes will require the elucidation the macromolecular complexes by using cryo-electron microscopy and X-ray crystallography. In addition, single molecule technologies, fluorescence techniques, nuclear magnetic resonance (NMR), SAXS and SANS, which allow monitoring of signalling molecules in solution and in live cells, will help to appreciate the dynamic features of PKA signalling systems. Obviously one also needs advanced computational tools not only to analyse the complexes but also to model cell signalling processes. Thus, the elucidation of protein kinase regulation and function in living cells is an exciting interdisciplinary challenge that requires the combination of skills and expertise of multiple specialists to span the many scales of biology.
Perspectives and conclusions
By tracing how the understanding of PKA signalling has evolved, one can appreciate the impact of structural biology. At each step along the way, knowledge gained from the structures has enhanced the perspective of biological function. Much of the detailed understanding of the protein kinase superfamily has come from studies of the PKA catalytic subunit, and by comparing many protein kinases the underlying features such as the spines that define this family have been discovered. Importantly structural studies of PKA holoenzymes have revealed that the characterization of kinase signalling at the molecular level relies on the study of the assembly of highly specific signalling foci at discrete sites in close proximity to a dedicated substrate. In particular, it is important to elucidate how the holoenzymes communicate with their neighbours in these polyvalent assemblies. Multiscale tools that allow the spatiotemporal imaging of signalling will be essential to unravel how this process is orchestrated by macromolecular complexes of signalling proteins and their substrates and, essentially, to understand how this process is altered in disease.
Acknowledgments
Work in the authors’ laboratory is supported by the Howard Hughes Medical Institute and grants from the US National Institutes of Health (NIH) (GM19301, GM34921) and DK54441 to S. S. T.
Glossary
- Phosphodiesterases (PDEs)
A family of proteins that break down cyclic AMP to AMP
- AGC subfamily
A group of about 60 Ser/Thr kinases that share several sequence, structural and functional similarities. The name is derived from three main representatives of this kinase family, namely cyclic AMP-dependent protein kinase (PKA), PKG and PKC
- Cyclic nucleotide-binding domains (CNB domains)
Conserved protein domains that bind a single molecule of cyclic AMP. Binding of cAMP causes a substantial change of the domain tertiary structure, thus providing a sensor for cAMP levels in the solution
- A kinase anchoring proteins (AKAPs)
A diverse family of scaffolding proteins that have a binding site for the dimerization and docking domain in cyclic AMP-dependent protein kinase regulatory subunits
- Cyclin-dependent kinases (CDKs)
Kinases that regulate the cell cycle. They are activated by a cyclin molecule, which is a small protein that is generated inside the nucleus during mitosis
- EPAC (Exchange protein directly activated by cyclic AMP)
A protein that regulates the activity of RAP1, which is an important cellular regulator. EPAC has a crucial role in cell proliferation and survival
- Intrinsically disordered regions (IDRs)
Parts of protein sequences that cannot accept stable secondary or tertiary structures. Disordered regions are very flexible and often serve as binding sites for other proteins
- Small-angle X-ray scattering (SAXS)
Experimental technique that studies scattering of X-rays by proteins in solution (or protein solutions). Scattering profiles in the small angle range (0.1–10 degrees) provide information about general, low-resolution (50–250Å) forms of the protein
- Small-angle neutron scattering (SANS)
Experimental technique that studies scattering of neutron beams by proteins in solution or protein solutions. It is similar to small-angle X-ray scattering (SAXS) but allows the use of isotope labelling of the protein sample, thus enhancing the technique
- Nonsense-mediated mRNA decay (NMD)
A cellular process that controls the correct synthesis of mRNA. It detects nonsense mutations that insert an erroneous stop codon in the mRNA and thus prevents its folding through degradation
- Long-term depression (LTD)
Weakening of the ability of a neuron to transmit a signal. LTD can last for hours or longer and is one of the most fundamental processes in neurophysiology
- Long-term potentiation (LTP)
Strengthening of the ability of a neuron to transmit signal. This form of synaptic plasticity is thought to underlie memory formation and is characterized by synapses using leads for their long-term strengthening. LTP can last for hours or longer
- PDZ domain
Common domain in a family of signalling proteins. It is named after the three first discovered representatives of this family (Postsynaptic density of 95 kDa, Discs large and zonula occludens 1)
- Agouti mice
Heterozygous mice that carry a mutation in the gene encoding agouti. This mutation leads to the yellow obese mouse syndrome, which is characterized by obesity, insulin resistance, hyperglycaemia, increased growth and yellow coat colour
- Apparent activation constant K−a(cAMP)
The concentration of cAMP that is required for 50% activation of cyclic AMP-dependent protein kinase
Footnotes
Competing interests statement
The authors declare no competing financial interests.
FURTHER INFORMATION
Susan S. Taylor’s homepage: http://susantaylorlab.ucsd.edu
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Krebs EG, Graves DJ, Fischer EH. Factors affecting the activity of muscle phosphorylase B kinase. J Biol Chem. 1959;234:2867–2873. [PubMed] [Google Scholar]
- 2.Shoji S, et al. Complete amino acid sequence of the catalytic subunit of bovine cardiac-muscle cyclic AMP-dependent protein-kinase. Proc Natl Acad Sci USA. 1981;78:848–851. doi: 10.1073/pnas.78.2.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker WC, Dayhoff MO. Viral src gene products are related to the catalytic chain of mammalian cAMP-dependent protein kinase. Proc Natl Acad Sci USA. 1982;79:2836–2839. doi: 10.1073/pnas.79.9.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kannan N, et al. Evolution of allostery in the cyclic nucleotide binding module. Genome Biol. 2007;8:R264. doi: 10.1186/gb-2007-8-12-r264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pidoux G, Tasken K. Specificity and spatial dynamics of protein kinase A signaling organized by A-kinase-anchoring proteins. J Mol Endocrinol. 2010;44:271–284. doi: 10.1677/JME-10-0010. [DOI] [PubMed] [Google Scholar]
- 6.Wong W, Scott JD. AKAP signalling complexes: focal points in space and time. Nature Rev Mol Cell Biol. 2004;5:959–970. doi: 10.1038/nrm1527. [DOI] [PubMed] [Google Scholar]
- 7.Newlon MG, et al. The molecular basis for protein kinase A anchoring revealed by solution NMR. Nature Struct Biol. 1999;6:222–227. doi: 10.1038/6663. [DOI] [PubMed] [Google Scholar]
- 8.Sarma GN, et al. Structure of D-AKAP2:PKA RI complex: insights into AKAP specificity and selectivity. Structure. 2010;18:155–166. doi: 10.1016/j.str.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knighton DR, et al. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:407–414. doi: 10.1126/science.1862342. [DOI] [PubMed] [Google Scholar]
- 10.Knighton DR, et al. Structure of a peptide inhibitor bound to the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:414–420. doi: 10.1126/science.1862343. [DOI] [PubMed] [Google Scholar]
- 11.Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 12.Kornev AP, Haste NM, Taylor SS, Ten Eyck LF. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc Natl Acad Sci USA. 2006;103:17783–17788. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kornev AP, Taylor SS, Ten Eyck LF. A helix scaffold for the assembly of active protein kinases. Proc Natl Acad Sci USA. 2008;105:14377–14382. doi: 10.1073/pnas.0807988105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor SS, Kornev AP. Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem Sci. 2011;36:65–77. doi: 10.1016/j.tibs.2010.09.006. Comprehensive review on eukaryotic protein kinases, their conserved core and kinase-specific regions. Compares eukaryotic protein kinases to their evolutionary predecessors, which are eukaryote-like kinases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson LN, Lewis RJ. Structural basis for control by phosphorylation. Chem Rev. 2001;101:2209–2242. doi: 10.1021/cr000225s. [DOI] [PubMed] [Google Scholar]
- 16.Nolen B, Taylor S, Ghosh G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol Cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 17.Iyer GH, Garrod S, Woods VL, Jr, Taylor SS. Catalytic independent functions of a protein kinase as revealed by a kinase-dead mutant: study of the Lys72His mutant of cAMP-dependent kinase. J Mol Biol. 2005;351:1110–1122. doi: 10.1016/j.jmb.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 18.Keshwani MM, et al. Cotranslational cis-phosphorylation of the COOH-terminal tail is a key priming step in the maturation of cAMP-dependent protein kinase. Proc Natl Acad Sci USA. 2012 Apr 9; doi: 10.1073/pnas.1202741109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steichen JM, et al. Global consequences of activation loop phosphorylation on protein kinase A. J Biol Chem. 2010;285:3825–3832. doi: 10.1074/jbc.M109.061820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steichen JM, et al. Structural basis for the regulation of protein kinase A by activation loop phosphorylation. J Biol Chem. 2012;287:14672–14680. doi: 10.1074/jbc.M111.335091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kannan N, Haste N, Taylor SS, Neuwald AF. The hallmark of AGC kinase functional divergence is its C-terminal tail, a cis-acting regulatory module. Proc Natl Acad Sci USA. 2007;104:1272–1277. doi: 10.1073/pnas.0610251104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romano RA, Kannan N, Kornev AP, Allison CJ, Taylor SS. A chimeric mechanism for polyvalent trans-phosphorylation of PKA by PDK1. Protein Sci. 2009;18:1486–1497. doi: 10.1002/pro.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bastidas AC, et al. Role of N-terminal myristylation in the structure and regulation of cAMP-dependent protein kinase. J Mol Biol. 2012;422:215–29. doi: 10.1016/j.jmb.2012.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herberg FW, Zimmermann B, McGlone M, Taylor SS. Importance of the A-helix of the catalytic subunit of cAMP-dependent protein kinase for stability and for orienting subdomains at the cleft interface. Protein Sci. 1997;6:569–579. doi: 10.1002/pro.5560060306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sastri M, Barraclough DM, Carmichael PT, Taylor SS. A-kinase-interacting protein localizes protein kinase A in the nucleus. Proc Natl Acad Sci USA. 2005;102:349–354. doi: 10.1073/pnas.0408608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.King CC, Sastri M, Chang P, Pennypacker J, Taylor SS. The rate of NF-κB nuclear translocation is regulated by PKA and A kinase interacting protein 1. PLoS ONE. 2011;6:e18713. doi: 10.1371/journal.pone.0018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diskar M, Zenn HM, Kaupisch A, Prinz A, Herberg FW. Molecular basis for isoform-specific autoregulation of protein kinase A. Cell Signal. 2007;19:2024–2034. doi: 10.1016/j.cellsig.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 28.Martin BR, Deerinck TJ, Ellisman MH, Taylor SS, Tsien RY. Isoform-specific PKA dynamics revealed by dye-triggered aggregation and DAKAP1α-mediated localization in living cells. Chem Biol. 2007;14:1031–1042. doi: 10.1016/j.chembiol.2007.07.017. Using live cells, this study shows that two PKA isoforms have different localization and reaction to cAMP. [DOI] [PubMed] [Google Scholar]
- 29.Berman HM, et al. The cAMP binding domain: an ancient signaling module. Proc Natl Acad Sci USA. 2005;102:45–50. doi: 10.1073/pnas.0408579102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weber IT, Steitz TA, Bubis J, Taylor SS. Predicted structures of cAMP binding domains of type I and II regulatory subunits of cAMP-dependent protein kinase. Biochemistry. 1987;26:343–351. doi: 10.1021/bi00376a003. [DOI] [PubMed] [Google Scholar]
- 31.Humphries KM, Deal MS, Taylor SS. Enhanced dephosphorylation of cAMP-dependent protein kinase by oxidation and thiol modification. J Biol Chem. 2005;280:2750–2758. doi: 10.1074/jbc.M410242200. [DOI] [PubMed] [Google Scholar]
- 32.Kannan N, Taylor SS, Zhai Y, Venter JC, Manning G. Structural and functional diversity of the microbial kinome. PLoS Biol. 2007;5:e17. doi: 10.1371/journal.pbio.0050017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kornev AP, Taylor SS, Ten Eyck LF. A generalized allosteric mechanism for cis-regulated cyclic nucleotide binding domains. PLoS Comput Biol. 2008;4:e1000056. doi: 10.1371/journal.pcbi.1000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeffrey PD, et al. Mechanism of CDK activation revealed by the structure of a cyclinA–CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 35.Kim C, Xuong NH, Taylor SS. Crystal structure of a complex between the catalytic and regulatory (RIα) subunits of PKA. Science. 2005;307:690–696. doi: 10.1126/science.1104607. [DOI] [PubMed] [Google Scholar]
- 36.Kim C, Cheng CY, Saldanha SA, Taylor SS. PKA-I holoenzyme structure reveals a mechanism for cAMP-dependent activation. Cell. 2007;130:1032–1043. doi: 10.1016/j.cell.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 37.Wu J, Brown SH, von Daake S, Taylor SS. PKA type IIα holoenzyme reveals a combinatorial strategy for isoform diversity. Science. 2007;318:274–279. doi: 10.1126/science.1146447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crivici A, Ikura M. Molecular and structural basis of target recognition by calmodulin. Annu Rev Biophys Biomol Struct. 1995;24:85–116. doi: 10.1146/annurev.bb.24.060195.000505. [DOI] [PubMed] [Google Scholar]
- 39.Bos JL. Epac: a new cAMP target and new avenues in cAMP research. Nature Rev Mol Cell Biol. 2003;4:733–738. doi: 10.1038/nrm1197. [DOI] [PubMed] [Google Scholar]
- 40.Li S, et al. Mechanism of intracellular cAMP sensor Epac2 activation: cAMP-induced conformational changes identified by amide hydrogen/deuterium exchange mass spectrometry (DXMS) J Biol Chem. 2011;286:17889–17897. doi: 10.1074/jbc.M111.224535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown SH, Wu J, Kim C, Alberto K, Taylor SS. Novel isoform-specific interfaces revealed by PKA RIIβ holoenzyme structures. J Mol Biol. 2009;393:1070–1082. doi: 10.1016/j.jmb.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dunker AK, et al. Intrinsically disordered protein. J Mol Graph Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 43.Boettcher AJ, et al. Realizing the allosteric potential of the tetrameric protein kinase A RIα holoenzyme. Structure. 2011;19:265–276. doi: 10.1016/j.str.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heller WT, et al. C subunits binding to the protein kinase A RI α dimer induce a large conformational change. J Biol Chem. 2004;279:19084–19090. doi: 10.1074/jbc.M313405200. [DOI] [PubMed] [Google Scholar]
- 45.Vigil D, et al. Conformational differences among solution structures of the type Iα, IIα and IIβ protein kinase A regulatory subunit homodimers: role of the linker regions. J Mol Biol. 2004;337:1183–1194. doi: 10.1016/j.jmb.2004.02.028. [DOI] [PubMed] [Google Scholar]
- 46.Vigil D, Blumenthal DK, Taylor SS, Trewhella J. Solution scattering reveals large differences in the global structures of type II protein kinase A isoforms. J Mol Biol. 2006;357:880–889. doi: 10.1016/j.jmb.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 47.Amieux PS, et al. Compensatory regulation of RIa protein levels in protein kinase A mutant mice. J Biol Chem. 1997;272:3993–3998. doi: 10.1074/jbc.272.7.3993. [DOI] [PubMed] [Google Scholar]
- 48.Veugelers M, et al. Comparative PRKAR1A genotype–phenotype analyses in humans with Carney complex and prkar1a haploinsufficient mice. Proc Natl Acad Sci USA. 2004;101:14222–14227. doi: 10.1073/pnas.0405535101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brandon EP, et al. Hippocampal long-term depression and depotentiation are defective in mice carrying a targeted disruption of the gene encoding the RIβ subunit of cAMP-dependent protein kinase. Proc Natl Acad Sci USA. 1995;92:8851–8855. doi: 10.1073/pnas.92.19.8851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang YY, et al. A genetic test of the effects of mutations in PKA on mossy fiber LTP and its relation to spatial and contextual learning. Cell. 1995;83:1211–1222. doi: 10.1016/0092-8674(95)90146-9. [DOI] [PubMed] [Google Scholar]
- 51.Skalhegg BS, et al. Location of cAMP-dependent protein kinase type I with the TCR–CD3 complex. Science. 1994;263:84–87. doi: 10.1126/science.8272870. [DOI] [PubMed] [Google Scholar]
- 52.Ilouz R, et al. Localization and quaternary structure of PKA RIb holoenzyme. Proc Natl Acad Sci USA. 2012;109:12443–12448. doi: 10.1073/pnas.1209538109. Shows how small intrinsically disordered linkers in the full length PKA RIβ holoenzyme can drive packing of large unique macromolecular complexes by using almost identical building blocks. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cadd G, McKnight GS. Distinct patterns of cAMP-dependent protein kinase gene expression in mouse brain. Neuron. 1989;3:71–79. doi: 10.1016/0896-6273(89)90116-5. [DOI] [PubMed] [Google Scholar]
- 54.Clegg CH, Cadd GG, McKnight GS. Genetic characterization of a brain-specific form of the type I regulatory subunit of cAMP-dependent protein kinase. Proc Natl Acad Sci USA. 1988;85:3703–3707. doi: 10.1073/pnas.85.11.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Herberg FW, Taylor SS. Physiological inhibitors of the catalytic subunit of cAMP-dependent protein kinase: effect of MgATP on protein–protein interactions. Biochemistry. 1993;32:14015–14022. doi: 10.1021/bi00213a035. [DOI] [PubMed] [Google Scholar]
- 56.Bubis J, Vedvick TS, Taylor SS. Antiparallel alignment of the two protomers of the regulatory subunit dimer of cAMP-dependent protein kinase I. J Biol Chem. 1987;262:14961–14966. [PubMed] [Google Scholar]
- 57.Brennan JP, et al. Oxidant-induced activation of type I protein kinase A is mediated by RI subunit interprotein disulfide bond formation. J Biol Chem. 2006;281:21827–21836. doi: 10.1074/jbc.M603952200. [DOI] [PubMed] [Google Scholar]
- 58.McConnachie G, Langeberg LK, Scott JD. AKAP signaling complexes: getting to the heart of the matter. Trends Mol Med. 2006;12:317–323. doi: 10.1016/j.molmed.2006.05.008. Reviews large signalling complexes formed with different AKAPs that provide spatial and temporal specificity of cAMP signals. [DOI] [PubMed] [Google Scholar]
- 59.Cummings DE, et al. Genetically lean mice result from targeted disruption of the RIIβ subunit of protein kinase A. Nature. 1996;382:622–626. doi: 10.1038/382622a0. [DOI] [PubMed] [Google Scholar]
- 60.Schreyer SA, Cummings DE, McKnight GS, LeBoeuf RC. Mutation of the RIIβ subunit of protein kinase A prevents diet-induced insulin resistance and dyslipidemia in mice. Diabetes. 2001;50:2555–2562. doi: 10.2337/diabetes.50.11.2555. [DOI] [PubMed] [Google Scholar]
- 61.Czyzyk TA, Sikorski MA, Yang L, McKnight GS. Disruption of the RIIβ subunit of PKA reverses the obesity syndrome of agouti lethal yellow mice. Proc Natl Acad Sci USA. 2008;105:276–281. doi: 10.1073/pnas.0710607105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang P, et al. Structure and allostery of the PKA RIIβ tetrameric holoenzyme. Science. 2012;335:712–716. doi: 10.1126/science.1213979. Shows the first structure of the full length PKA tetrameric holoenzyme. This structure demonstrates how the tertiary structure can define the cooperative features of PKA activation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oliveria SF, Dell’Acqua ML, Sather WA. AKAP79/150 anchoring of calcineurin controls neuronal L-type Ca2+ channel activity and nuclear signaling. Neuron. 2007;55:261–275. doi: 10.1016/j.neuron.2007.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Violin JD, Zhang J, Tsien RY, Newton AC. A genetically encoded fluorescent reporter reveals oscillatory phosphorylation by protein kinase C. J Cell Biol. 2003;161:899–909. doi: 10.1083/jcb.200302125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Conti M, Beavo J. Biochemistry and physiology of cyclic nucleotide phosphodiesterases: essential components in cyclic nucleotide signaling. Annu Rev Biochem. 2007;76:481–511. doi: 10.1146/annurev.biochem.76.060305.150444. [DOI] [PubMed] [Google Scholar]
- 66.Ni Q, et al. Signaling diversity of PKA achieved via a Ca2+–cAMP–PKA oscillatory circuit. Nature Chem Biol. 2011;7:34–40. doi: 10.1038/nchembio.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stefan E, et al. PKA regulatory subunits mediate synergy among conserved G-protein-coupled receptor cascades. Nature Commun. 2011;2:598. doi: 10.1038/ncomms1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krebs EG, Fischer EH. The phosphorylase-b to bhosphorylase-a converting enzyme of rabbit skeletal muscle. Biochim Biophys Acta. 1956;20:150–157. doi: 10.1016/0006-3002(56)90273-6. [DOI] [PubMed] [Google Scholar]
- 69.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 70.Rall TW, Sutherland EW. Formation of a cyclic adenine ribonucleotide by tissue particles. J Biol Chem. 1958;232:1065–1076. [PubMed] [Google Scholar]
- 71.Northup JK, et al. Purification of the regulatory component of adenylate cyclase. Proc Natl Acad Sci USA. 1980;77:6516–6520. doi: 10.1073/pnas.77.11.6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Walsh DA, Perkins JP, Krebs EG. An adenosine 3′,5′-monophosphate-dependant protein kinase from rabbit skeletal muscle. J Biol Chem. 1968;243:3763–3765. [PubMed] [Google Scholar]
- 73.Brostrom CO, Corbin JD, King CA, Krebs EG. Interaction of the subunits of adenosine 3′:5′-cyclic monophosphate-dependent protein kinase of muscle. Proc Natl Acad Sci USA. 1971;68:2444–2447. doi: 10.1073/pnas.68.10.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gill GN, Garren LD. A cyclic-3′,5′-adenosine monophosphate dependent protein kinase from the adrenal cortex: comparison with a cyclic AMP binding protein. Biochem Biophys Res Commun. 1970;39:335–343. doi: 10.1016/0006-291x(70)90581-4. [DOI] [PubMed] [Google Scholar]
- 75.Tao M, Salas ML, Lipmann F. Mechanism of activation by adenosine 3′:5′-cyclic monophosphate of a protein phosphokinase from rabbit reticulocytes. Proc Natl Acad Sci USA. 1970;67:408–414. doi: 10.1073/pnas.67.1.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Haste NM, et al. Exploring the Plasmodium falciparum cyclic-adenosine monophosphate (cAMP)-dependent protein kinase (PfPKA) as a therapeutic target. Microbes Infect. 2012;14:838–850. doi: 10.1016/j.micinf.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gangal M, et al. Mobilization of the A-kinase N-myristate through an isoform-specific intermolecular switch. Proc Natl Acad Sci USA. 1999;96:12394–12399. doi: 10.1073/pnas.96.22.12394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gisler SM, et al. PDZK1: II. an anchoring site for the PKA-binding protein D-AKAP2 in renal proximal tubular cells. Kidney Int. 2003;64:1746–1754. doi: 10.1046/j.1523-1755.2003.00267.x. [DOI] [PubMed] [Google Scholar]