Abstract
Sublancin 168 is a member of a small group of glycosylated antimicrobial peptides known as glycocins. The solution structure of sublancin 168, a 37-amino-acid peptide produced by Bacillus subtilis 168, has been solved by nuclear magnetic resonance (NMR) spectroscopy. Sublancin comprises two α-helices and a well-defined interhelical loop. The two helices span residues 6–16 and 26–35, and the loop region encompasses residues 17–25. The 9-amino-acid loop region contains a β-S-linked glucose moiety attached to Cys22. Hydrophobic interactions as well as hydrogen bonding are responsible for the well-structured loop region. The three-dimensional structure provides an explanation for the previously reported extraordinary high stability of sublancin 168.
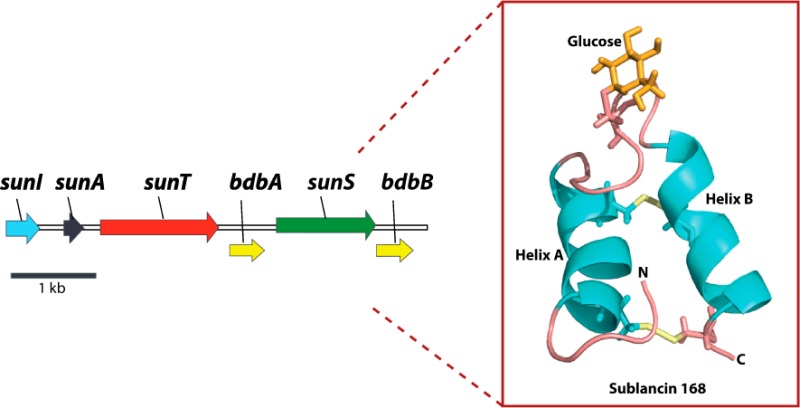
Glycosylation is an important post-translational modification in eukaryotic and bacterial cells. Typically, the carbohydrate group is covalently linked to a peptide or protein through the hydroxyl group of serine/threonine residues (O-linked) or through the side chain nitrogen of asparagine (N-linked).1 A rare form of glycosylation involves a carbohydrate covalently linked to the sulfur atom of a cysteine residue (S-linked). To date, two ribosomally synthesized S-glycosylated peptides have been characterized.2,3 Both peptides have antimicrobial activity, and hence the name glycocins has been introduced to describe this group.3 Glycocins belong to the family of ribosomally synthesized and post-translationally modified peptide natural products (RiPPs).4 One such glycocin is sublancin 168, a 37-amino-acid peptide produced by Bacillus subtilis 168 (Figure 1).2,5
Figure 1.
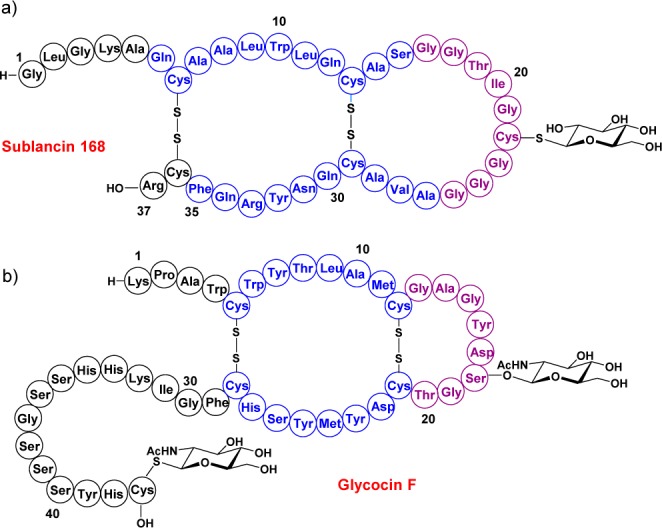
Glycocin structures. (a) Sublancin 168. (b) Glycocin F. Helical regions are depicted in blue, and loop regions in magenta.
Like other RiPPs, sublancin is biosynthesized as a precursor peptide, SunA. Sublancin’s biosynthetic gene cluster encodes an S-glycosyltransferase (SunS), an immunity protein (SunI), a transporter protein that also removes a leader peptide (SunT), and two thiol-disulfide oxidoreductases (BdbAB).2,5−7 SunS, BdbAB, and SunT convert SunA into sublancin 168. To understand glycocin peptides in more depth and obtain insight into their potential mode of action, it is important to elucidate their three-dimensional structures at high resolution. Glycocin F (GccF), a 43-amino-acid peptide produced by Lactobacillus plantarum KW30, is the second currently known member of the glycocin family (Figure 1).3 Pascal and co-workers reported the NMR structure of GccF,8 which contains both S- and O-linked N-acetyl glucosamine (Figure 1). The primary sequences of sublancin 168 and glycocin F display similarities as well as some key differences. Both peptides contain two disulfides and a loop region, but the loop in sublancin is substantially longer than that in glycocin F. Furthermore, the C-terminus of sublancin is shorter by 13 residues, sublancin contains one glycosylation compared to two in GccF, and the S-linked sugar is located in the loop region rather than at the C-terminal residue. Moreover, the primary sequences of the two peptides are quite different (Figure 1). Because of the unknown role(s) of the glycosylations in glycocins, we determined the three-dimensional structure of sublancin 168 to compare it to the structure of GccF.
Results and Discussion
Structure Determination
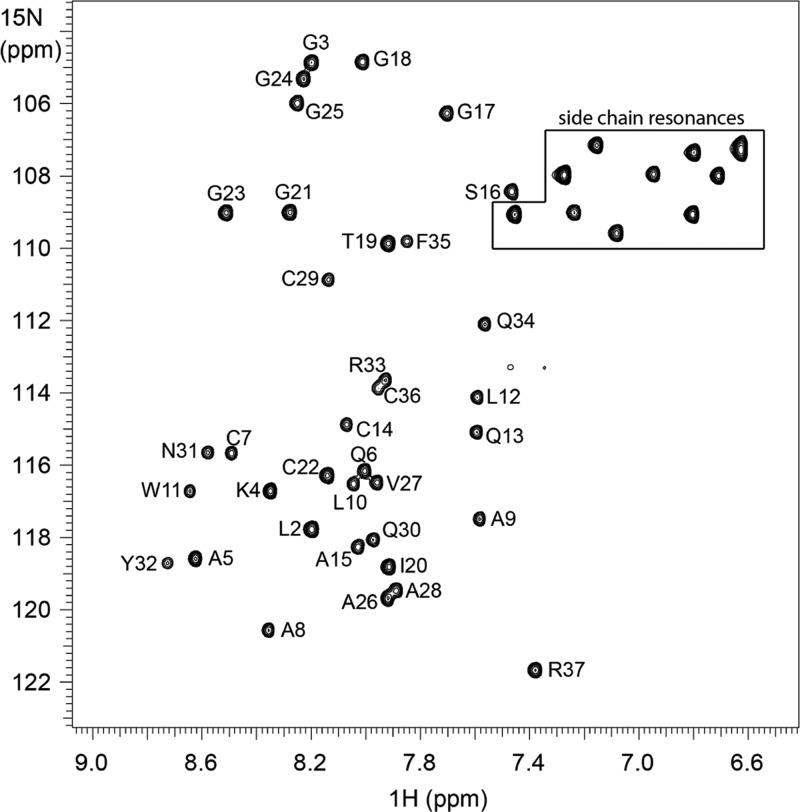
The 1H–15N HSQC spectrum of sublancin produced 36 amide backbone signals and 10 side chain nitrogen resonances (Figure 2). All resonances were assigned (Supplementary Table S1), and the structure of sublancin 168 was solved with 628 structural constraints derived from 3D 1H–13C–15N HNCA, 3D 1H–13C–15N CBCA(CO)NH, 2D 1H–15N HSQC, 2D 1H–13C HSQC, 3D 15N HSQC-NOESY, 3D 15N HSQC-TOCSY, 2D 1H–1H TOCSY, 2D 1H–1H NOESY, 3D 15N HNHA, and DQCOSY experiments. Molecular Operating Environment (MOE) software9 was used to computationally attach the carbohydrate moiety to Cys22. The details of the structure can be found at PDB ID 2MIJ and BMRB ID 19683.
Figure 2.
HSQC spectrum of sublancin. The cross peaks correspond to the backbone amide region of 1H–15N correlations in the HSQC spectrum recorded at 25 °C. Specific amino acid assignments are indicated. Side chain nitrogen resonances are located within the boxed region.
An ensemble of 15 sublancin structures with the lowest NMR constraint violations and lowest XPLOR energies10,11 were used for detailed analysis, with the structural statistics given in Table 1. All experimental NMR constraints were satisfied in the structures, with all nuclear Overhauser effect (NOE) violations below 0.5 Å, J-coupling violations below 1 Hz, and dihedral angle constraints below the 5° violation limit.
Table 1. Structural Statistics for Sublancin 168.
| total restraints used | 628 |
| total NOE restraints | 531 |
| intraresidue | 184 |
| sequential (|i – j| = 1) | 185 |
| medium (1 < |i – j| ≤ 4) | 100 |
| long range (|i – j| > 4) | 62 |
| dihedral | |
| ϕ angles | 23 |
| φ angles | 16 |
| disulfide bridges (Cys7–Cys36, Cys14–Cys29) | 2 |
| hydrogen bonds | 56 |
| rmsd from experimental distance restraints | |
| bonds (Å) | 0.006 ± 0.000 |
| bond angles (deg) | 0.655 ± 0.037 |
| improper torsions (deg) | 0.470 ± 0.037 |
| average pairwise rmsd (Å) | |
| backbone atoms in helical regions (6–16) | 0.52 |
| backbone atoms in helical regions (26–35) | 0.34 |
| all backbone atoms (residues 1–37) | 0.79 |
| all heavy atoms in helical regions (6–16) | 0.87 |
| all heavy atoms in helical regions (26–35) | 1.21 |
| all heavy atoms (residues 1–37) | 1.36 |
| procheck analysis | |
| residues in most favorable regions (%) | 89.3 |
| residues in additional allowed regions (%) | 0.0 |
| residues in generously allowed regions (%) | 10.7 |
| residues in disallowed regions (%) | 0.0 |
Procheck analysis of an ensemble of 15 conformers shows Cys22 in a generously allowed region of the Ramachandran plot (Supplementary Figure S1). This finding is not uncommon for Cys. Previous studies have shown that Ser followed by Cys are the residues with the highest propensity for location in generously allowed and disallowed regions.12 Two additional residues, Leu2 and Ala5, are also found in the generously allowed regions (Supplementary Figure S1). The average pairwise root-mean-square deviation (rmsd) for backbone heavy atoms (C′, Cα, N) of all residues (1–37) is 0.79 Å, with smaller values in well-structured regions (Table 1).
The restraints that have the greatest weight in a structure calculation are those of medium- and long-range NOEs.13 The three-dimensional structure of sublancin was obtained from over 160 medium- and long-range restraints, resulting in well-defined α helices as well as loop regions. Overall, an average of more than 17 NMR constraints per residue was used for the structural calculation.
Description of the Structure
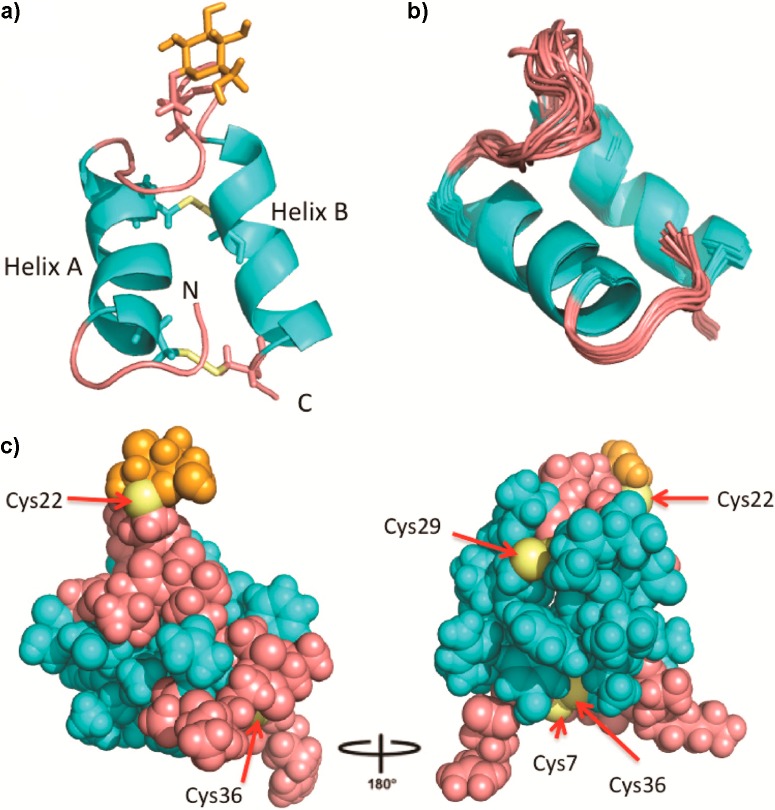
The ribbon diagram of a representative structure of sublancin is shown in Figure 3a. As predicted by analysis of the primary sequence and on the basis of backbone dihedral angles and characteristic NOEs, the solution structure shows two well-defined α helices encompassing residues 6–16 and 26–35. Superposition of the backbone Cα atoms of the 15 lowest energy conformers is shown in Figure 3b illustrating the highly ordered structure. Helices A and B are not parallel but rather offset from one another at an approximately 25° angle. The two helices are connected by an extended loop region.
Figure 3.
Three-dimensional solution structure of sublancin 168. (a) Ribbon diagram of the lowest energy conformer with helices colored in cyan, the loop regions, N- and C-termini colored pink, cysteine residues in yellow, and the carbohydrate moiety in orange. (b) Ensemble of the 15 lowest energy conformers depicting all backbone atoms. (c) Ball and stick representation of the lowest energy conformer with the loop region and termini in pink coming together to seal off one face of the helices. Exposed sulfur atoms are labeled.
This interhelical loop (residues 17–25) is relatively flexible but has a defined conformation, with the loop folded on top of both helices giving sublancin a compact conformation. The loop does not significantly open up in the 15 structures but does display flexibility as expected with 6 out of 9 residues being glycine (Figure 3b). Comparison of sublancin’s interhelical loop to those in the loop conformation database ArchDB did not provide any hits.14 A search of the database for a similar sequence of a loop located between two helices resulted in 17 examples. Inspection of the structures of these hits revealed only one example with a similar conformation in which the loop folded back over the helices (Supplementary Figure S2).15
In addition to the relatively ordered interhelical loop, the N-terminal pentapeptide of sublancin is also well-defined. The ordered conformation of the interhelix and N-terminal loops is evidenced by 24% of all long-range NOEs coming from these regions. The N-terminal amino group is not entirely solvent-accessible but instead is inserted between the two helices (Figure 3a).
The superposition of the structures shows that the sugar moiety is the least well-defined section of the structure (Supplementary Figure S3), also indicated by the lack of long-range NOE restraints between the glucose unit and the amino acids of sublancin. We did observe an NOE restraint between one of the β protons of Cys22 to the H1 proton of the glucose moiety, which supports the β-linkage of the glucose to the sulfur atom of Cys22 reported previously.2 No obvious hydrophobic interactions were observed between the hydrophobic face of the glucose and amino acids of the peptide.
The structure of the C-terminal portion of sublancin is also well-defined as indicated by the observed medium and long-range NOE restraints. The very compact structure of sublancin (Figure 3b,c) provides a possible explanation for the extraordinary high chemical stability of sublancin reported previously.5
Hydrogen Bond Interactions
The ensemble of the 15 lowest energy conformers calculated without hydrogen bond restraints was analyzed using Chimera software to predict tentative hydrogen bond interactions between residues in the interhelical loop and the helices (Supplementary Figure S4). Then, hydrogen bond interactions were experimentally investigated with a series of deuterium exchange 1H–15N HSQC experiments with a 15N-labeled sublancin sample. Within the first few minutes, we observed protection from exchange of 27 backbone amide protons as well as the side chain protons of Asn31 (Supplementary Table S2). As expected, most of the hydrogen bond donors belonged to residues involved in helix formation, but several hydrogen bond interactions were located in the loop region, including the amide hydrogens of Gly17, Gly18, and Ile20 (Figure 4a). On the basis of hydrogen bond distance restrictions, the amide protons from Gly17 and Gly18 interact with the carbonyl oxygen of the side chain of Gln13. In addition, the amide proton of Ile20 hydrogen bonds with the side chain oxygen atom from Thr19. The well-defined turn is reinforced by additional hydrogen bonds between the N–H of Ser16, the last residue in helix A, and the side chain carbonyl of Gln13 (Figure 4a). The other end of the loop is held in place by hydrogen bonds involving the amide protons of Ala26, Val27, and Ala28, located in helix B, which interact with the carbonyl oxygen atoms of Gly24 and Gly25 (Figure 4b). These hydrogen bond interactions located at the beginning and end of the loop offer a plausible explanation as to why the loop is folded on top of the two helices, although the flexibility of the loop seen in Figure 3b is also reflected in exchange of most of the protons involved in hydrogen bond interactions of the loop residues by 14 min (Supplementary Table S2). After 45 h, 8 amide protons were still protected from exchange (Supplementary Table S2). Of these, 5 protons are located in helix A (Leu10, Trp11, Leu12, Gln13, Cys14) and 3 in helix B (Tyr32, Phe35, and Cys36).
Figure 4.
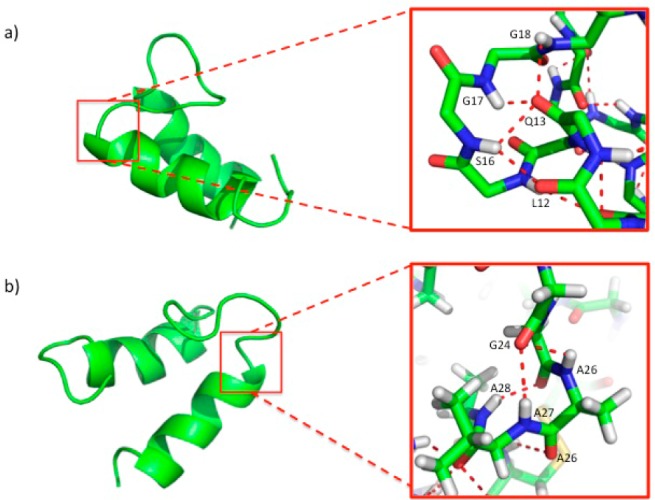
Hydrogen bond interactions in the interhelical loop region. Ribbon diagram showing the hydrogen bonds between residues in the loop and the helices. Hydrogen bond interactions are depicted in red.
Hydrophobic Interactions
The compact and well-defined structure of sublancin is also enforced in part by hydrophobic interactions. As shown in Figure 5, a hydrophobic core consisting of Leu2, Leu10, Trp11, Ala28, Tyr32, and Phe35 helps maintain the structure, including a hydrophobic interaction between Ile20 in the loop and Leu10 in helix A. Use of the program IsoCleft Finder16 identified Gly1, Gln6, Leu10, Ile20, Gly21, Gly23, Val27, Ala28, Asn31, and Phe35 as residues involved in creating a small hydrophobic cleft (Supplementary Figure S5) that may be important for target binding.
Figure 5.
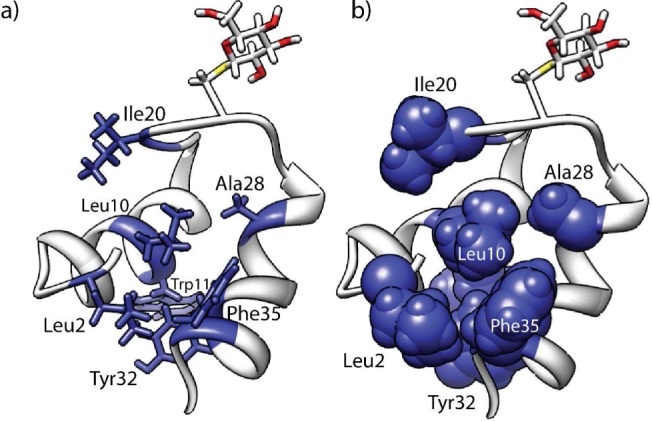
Hydrophobic interactions. Ribbon diagram of sublancin depicting the hydrophobic residues in (a) sticks or (b) spheres.
Glycocin Structural Comparison
The three-dimensional structures of GccF and sublancin were superimposed to visualize structural similarities and differences of these two glycocins (Figure 6). The orientation of the helices match very closely, with both structures displaying the 25° offset, even though the primary sequences of the amino acids involved in helix formation are vastly different. As expected on the basis of the primary sequences, the helices in sublancin are longer. The location and orientation of the sugar moiety in the interhelix loop is quite different in GccF and sublancin 168, and the N-and C-terminal segments are also oriented quite differently (Figure 6).
Figure 6.
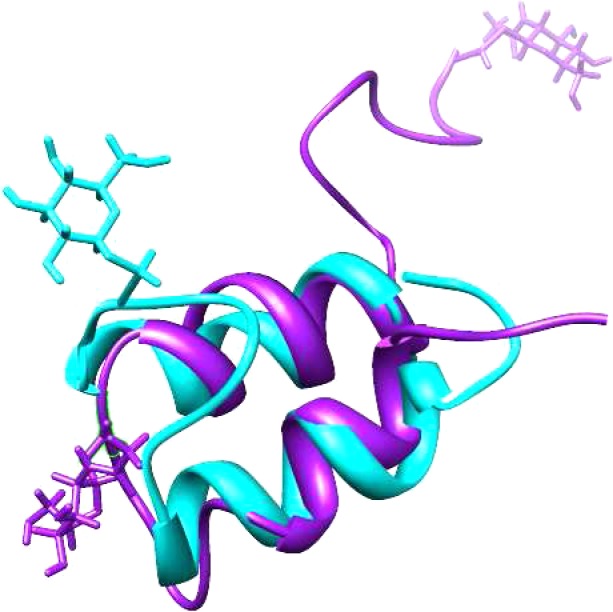
Superposition of sublancin 168 and glycocin F. Sublancin is depicted in cyan, and glycocin F in magenta.
The lack of defined contacts between the glucose and the peptide in the sublancin structure do not provide any evidence for a role of the sugar in folding of the peptide as is often seen for glycopeptides,17−21 although an interaction with the biosynthetic thiol-disulfide oxidoreductases6 cannot be ruled out. The conformational flexibility of the sugar does provide a possible explanation for the ease by which the S-linked glucose in sublancin could be substituted by several other hexoses in previous work.2 The previous observation that the stereochemistry of the hexose in these analogues was not critical for antimicrobial activity of sublancin 168,2 combined with the findings that the Cys22Ser analogue displayed antimicrobial activity22 and that the glucose in the structure shown here is conformationally flexible, suggests that the sugar in sublancin 168 may not be installed for target recognition. These findings are consistent with the previous suggestion that the sugar in sublancin might fulfill a role in self-protection in the producing strain.22
In summary, we have obtained a high-resolution three-dimensional structure of sublancin 168. The structure is highly compact with well-defined N- and C-terminal regions, two nearly antiparallel helices, and a conformationally highly structured interhelix loop that carries the glucose. Further studies are required to better understand the mode of action of the glycocin group of antimicrobial peptides.
Methods
Materials
All chemicals were purchased from Fisher Scientific or Aldrich unless noted otherwise.
Sample Preparation
Wild type sublancin was isolated and purified from Bacillus subtilis 168 as described previously5 and analyzed by mass spectrometry. Uniformly 15N-labeled sublancin and 13C/15N-labeled sublancin were produced by culturing B. subtilis 168 in M9 minimal media enriched with 15NH4Cl as the sole nitrogen source and with 15NH4Cl and [13C]-glucose (Cambridge Isotope Laboratories) as the sole nitrogen and carbon source, respectively. The peptides were purified by reversed phase chromatography and their identity assessed by MALDI-TOF MS (Supplementary Figures S7–S10). NMR samples contained 2.0 mM peptide in 90% H2O/10% D2O or 100% D2O.
NMR Spectroscopy
All NMR experiments were performed at 25 °C on Varian INOVA 500, 600, and 750 MHz spectrometers equipped with a 5 mm triple resonance (1H–13C–15N) triaxial gradient probe and pulse-shaping capabilities. The VNMRJ 2.1B software with the BioPack suite of pulse sequences was used. The spectra were processed with NMRPipe software23 and analyzed by Sparky24 and VNMRJ (Agilent Technology).
Peptide Chemical Shift Assignments
Backbone resonance assignments (15N, 13Ca, and 13Cb) were obtained from 3D 1H–13C–15N HNCA spectra recorded with a spectral width of 14 ppm (2048 points), 36 ppm (32 points), and 36 ppm (32 points) in 1H, 13C, and 15N dimensions, respectively; 3D 1H–13C–15N CBCA(CO)NH spectra were recorded with a spectral width of 14 ppm (2048 points), 96 ppm (32 points), and 36 ppm (32 points) for 1H, 13C, and 15N dimensions, respectively; and 1H–15N HSQC spectra were recorded with a spectral width of 14 ppm (2048 points) and 36 ppm (256 points) in the 1H and 15N dimensions. The proton signals from the amino acid side chains were assigned by analysis of 3D 15N HSQC-NOESY (150 ms NOESY mixing time) and 3D 15N HSQC-TOCSY (80 ms TOCSY mixing time) spectra and two-dimensional 1H–1H TOCSY (80 ms mixing time) and 1H–1H NOESY spectra (200 ms mixing time).
The dihedral angle restraints were obtained based on 3JHN-Hα coupling constants measured in an HNHA experiment using 15N-labeled sublancin and were obtained from the Torsion Angle Likelihood Obtained from Shift and Sequence similarity (TALOS)23,25 program based on backbone chemical shifts.
Hydrogen Bond Identification by Deuterium Exchange
A sample lyophilized from 90% H2O/10% D2O was dissolved in 100% D2O. A series of 1H–15N HSQC spectra with a 7 min duration each were collected. Hydrogen bonding donors were identified within the first 7 min with 27 backbone and 1 side chain NH observed, with 3 signals belonging to residues located in the loop region (Gly17, Gly18, and Ile20). After the first 30 min, 15 signals were still present; by 60 min, 12 signals were present; and by 45 h, 8 signals could still be observed (see Supporting Information).
Sugar Chemical Shift Assignments
Sugar assignments were obtained by analysis of TOCSY (80 ms mixing time), 1H–13C HSQC, and DQCOSY spectra for identification of neighboring protons.
Structure Calculations
The three-dimensional structure of the peptide was calculated on the basis of both distance and angle restraints by using the simulated annealing protocol in the NIH version of X-PLOR10,11 3.1, and the quality of the NMR structures was evaluated using the program PROCHECK.26−28 Distance restraints were derived from NOE peak heights in the 15N HSQC NOESY with a 150 ms mixing time and from two- dimensional NOESY spectra with a 200 ms mixing time collected on unlabeled material. The distance restraints were grouped by classifying the NOE cross-peak heights into ranges of 2.5, 3.5, 5.0, and 6.0 Å (strong, medium, weak, and very weak, respectively). The peptide backbone restraints extracted from JNHα and TALOS were used as dihedral phi and psi angle restraints. A list of the number of NMR distances and angle restraints used for structural calculations is given in Table 1. In total 150 structures were calculated. An ensemble of 15 structures with the lowest total energy was chosen for structural analysis.
Acknowledgments
The authors thank M. Brothers for technical assistance. This work was supported by the U.S. National Institutes of Health (GM 58822 to W.A.v.d.D.) and partially supported by an NIGMS-NIH Chemistry-Biology Interface Training Grant (5T32-GM070421 to C.V.G.D.G). Mass spectra were recorded in part on an instrument purchased with grant S10RR027109-01 from the U.S. National Institutes of Health.
Supporting Information Available
Description of all molecular biology procedures, protein purifications, and supporting figures. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Spiro R. G. (2002) Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 12, 43R–56R. [DOI] [PubMed] [Google Scholar]
- Oman T. J.; Boettcher J. M.; Wang H.; Okalibe X. N.; van der Donk W. A. (2011) Sublancin is not a lantibiotic but an S-linked glycopeptide. Nat. Chem. Biol. 7, 78–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepper J.; Shastri S.; Loo T. S.; Preston J. C.; Novak P.; Man P.; Moore C. H.; Havlicek V.; Patchett M. L.; Norris G. E. (2011) Cysteine S-glycosylation, a new post-translational modification found in glycopeptide bacteriocins. FEBS Lett. 585, 645–650. [DOI] [PubMed] [Google Scholar]
- Arnison P. G.; Bibb M. J.; Bierbaum G.; Bowers A. A.; Bugni T. S.; Bulaj G.; Camarero J. A.; Campopiano D. J.; Challis G. L.; Clardy J.; Cotter P. D.; Craik D. J.; Dawson M.; Dittmann E.; Donadio S.; Dorrestein P. C.; Entian K. D.; Fischbach M. A.; Garavelli J. S.; Goransson U.; Gruber C. W.; Haft D. H.; Hemscheidt T. K.; Hertweck C.; Hill C.; Horswill A. R.; Jaspars M.; Kelly W. L.; Klinman J. P.; Kuipers O. P.; Link A. J.; Liu W.; Marahiel M. A.; Mitchell D. A.; Moll G. N.; Moore B. S.; Muller R.; Nair S. K.; Nes I. F.; Norris G. E.; Olivera B. M.; Onaka H.; Patchett M. L.; Piel J.; Reaney M. J.; Rebuffat S.; Ross R. P.; Sahl H. G.; Schmidt E. W.; Selsted M. E.; Severinov K.; Shen B.; Sivonen K.; Smith L.; Stein T.; Sussmuth R. D.; Tagg J. R.; Tang G. L.; Truman A. W.; Vederas J. C.; Walsh C. T.; Walton J. D.; Wenzel S. C.; Willey J. M.; van der Donk W. A. (2013) Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik S. H.; Chakicherla A.; Hansen J. N. (1998) Identification and characterization of the structural and transporter genes for, and the chemical and biological properties of, sublancin 168, a novel lantibiotic produced by Bacillus subtilis 168. J. Biol. Chem. 273, 23134–23142. [DOI] [PubMed] [Google Scholar]
- Dorenbos R.; Stein T.; Kabel J.; Bruand C.; Bolhuis A.; Bron S.; Quax W. J.; Van Dijl J. M. (2002) Thiol-disulfide oxidoreductases are essential for the production of the lantibiotic sublancin 168. J. Biol. Chem. 277, 16682–16688. [DOI] [PubMed] [Google Scholar]
- Dubois J. Y.; Kouwen T. R.; Schurich A. K.; Reis C. R.; Ensing H. T.; Trip E. N.; Zweers J. C.; van Dijl J. M. (2009) Immunity to the bacteriocin sublancin 168 Is determined by the SunI (YolF) protein of Bacillus subtilis. Antimicrob. Agents Chemother. 53, 651–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal H.; Edwards P. J.; Schwalbe M.; Claridge J. K.; Libich D. S.; Stepper J.; Loo T.; Patchett M. L.; Norris G. E.; Pascal S. M. (2011) Structural, dynamic, and chemical characterization of a novel S-glycosylated bacteriocin. Biochemistry 50, 2748–2755. [DOI] [PubMed] [Google Scholar]
- Molecular Operating Environment (MOE) 2013.08 (2013) Chemical Computing Group Inc., 1010 Sherbooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7. [Google Scholar]
- Schwieters C. D.; Kuszewski J. J.; Tjandra N.; Clore G. M. (2003) The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160, 65–73. [DOI] [PubMed] [Google Scholar]
- Schwieters C. D.; Kuszewski J. J.; Tjandra N.; Clore G. M. (2006) Using Xplor-NIH for NMR molecular structure determination. Prog. NMR Spectrosc. 48, 47–62. [Google Scholar]
- Pal D.; Chakrabarti P. (2002) On residues in the disallowed region of the Ramachandran map. Biopolymers 63, 195–206. [DOI] [PubMed] [Google Scholar]
- Wüthrich K. (1986) NMR of Proteins and Nucleic Acids, Wiley, New York. [Google Scholar]
- Bonet J.; Planas-Iglesias J.; Garcia-Garcia J.; Marín-López M. A.; Fernandez-Fuentes N.; Oliva B. (2013) ArchDB 2014: structural classification of loops in proteins. Nucleic Acids Res. 42, D315–D319 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromme J. C.; Verdine G. L. (2003) DNA lesion recognition by the bacterial repair enzyme MutM. J. Biol. Chem. 278, 51543–51548. [DOI] [PubMed] [Google Scholar]
- Kurbatova N.; Chartier M.; Zylber M. I.; Najmanovich R. (2013) IsoCleft Finder - a web-based tool for the detection and analysis of protein binding-site geometric and chemical similarities. F1000Research 10.12688/f1000research.2-117.v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.; Eufemi M.; Turano C.; Giartosio A. (1996) Influence of the carbohydrate moiety on the stability of glycoproteins. Biochemistry 35, 7299–7307. [DOI] [PubMed] [Google Scholar]
- Helenius A.; Aebi M. (2004) Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73, 1019–1049. [DOI] [PubMed] [Google Scholar]
- Chen M. M.; Bartlett A. I.; Nerenberg P. S.; Friel C. T.; Hackenberger C. P.; Stultz C. M.; Radford S. E.; Imperiali B. (2010) Perturbing the folding energy landscape of the bacterial immunity protein Im7 by site-specific N-linked glycosylation. Proc. Natl. Acad. Sci. U.S.A. 107, 22528–22533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culyba E. K.; Price J. L.; Hanson S. R.; Dhar A.; Wong C. H.; Gruebele M.; Powers E. T.; Kelly J. W. (2011) Protein native-state stabilization by placing aromatic side chains in N-glycosylated reverse turns. Science 331, 571–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson S. R.; Culyba E. K.; Hsu T. L.; Wong C. H.; Kelly J. W.; Powers E. T. (2009) The core trisaccharide of an N-linked glycoprotein intrinsically accelerates folding and enhances stability. Proc. Natl. Acad. Sci. U.S.A. 106, 3131–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.; van der Donk W. A. (2011) Substrate selectivity of the sublancin S-glycosyltransferase. J. Am. Chem. Soc. 133, 16394–16397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F.; Grzesiek S.; Vuister G. W.; Zhu G.; Pfeifer J.; Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- Lee W.; Westler W. M.; Bahrami A.; Eghbalnia H. R.; Markley J. L. (2009) PINE-SPARKY: graphical interface for evaluating automated probabilistic peak assignments in protein NMR spectroscopy. Bioinformatics 25, 2085–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornilescu G.; Delaglio F.; Bax A. (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13, 289–302. [DOI] [PubMed] [Google Scholar]
- Laskowski R. A.; MacArthur M. W.; Moss D. S.; Thornton J. M. (1993) PROCHECK - a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291. [Google Scholar]
- Laskowski R. A.; Rullmannn J. A.; MacArthur M. W.; Kaptein R.; Thornton J. M. (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486. [DOI] [PubMed] [Google Scholar]
- Morris A. L.; MacArthur M. W.; Hutchinson E. G.; Thornton J. M. (1992) Stereochemical quality of protein structure coordinates. Proteins 12, 345–364. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.