Abstract
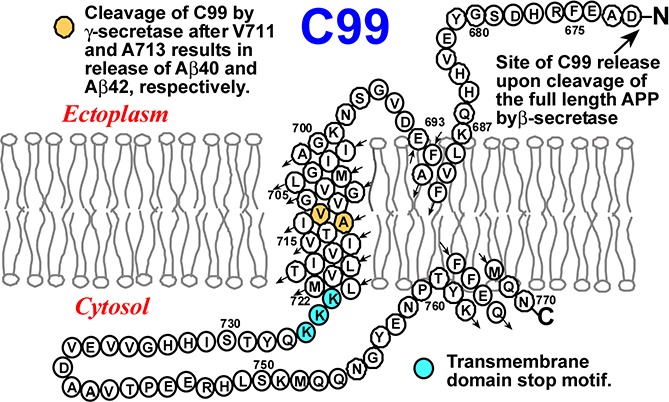
C99 (also known as β-CTF) is the 99 residue transmembrane C-terminal domain (residues 672–770) of the amyloid precursor protein and is the immediate precursor of the amyloid-β (Aβ) polypeptides. To test the dependence of the C99 structure on the composition of the host model membranes, NMR studies of C99 were conducted both in anionic lyso-myristoylphosphatidylglycerol (LMPG) micelles and in a series of five zwitterionic bicelle compositions involving phosphatidylcholine and sphingomyelin in which the acyl chain lengths of these lipid components varied from 14 to 24 carbons. Some of these mixtures are reported for the first time in this work and should be of broad utility in membrane protein research. The site-specific backbone 15N and 1H chemical shifts for C99 in LMPG and in all five bicelle mixtures were seen to be remarkably similar, indicating little dependence of the backbone structure of C99 on the composition of the host model membrane. However, the length of the transmembrane span was seen to vary in a manner that alters the positioning of the γ-secretase cleavage sites with respect to the center of the bilayer. This observation may contribute to the known dependency of the Aβ42-to-Aβ40 production ratio on both membrane thickness and the length of the C99 transmembrane domain.
The transmembrane (TM) C99 protein is a critical intermediate on the amyloidogenic pathway associated with the genesis of Alzheimer’s disease. C99 is the product of β-secretase cleavage of the full length amyloid precursor protein and is the substrate for cleavage by γ-secretase to release the amyloid-β polypeptides. The structure of monomeric C99 in anionic LMPG detergent micelles was determined by NMR1 and was seen to be composed of a disordered N-terminus (NTD, 672–687), followed by a surface-associated N-helix extending from 688 to 694, a flexible “N-loop” (695–699), a helical transmembrane domain (700–723), a disordered intracellular “C-loop”, and finally a surface associated “C-helix” (CTD, 762–770, see TOC graphic). The NMR structural work was followed by both EPR1 and computational2−4 studies of the protein under membrane conditions. However, the dependence of the C99 structure and membrane interactions as a function of variations in lipid composition has not been investigated. Here, we provide insight into this issue and illuminate previous studies showing that alteration of either the transmembrane span of C99 or of the bilayers in which it is solubilized impact cleavage of the protein by γ-secretase.5−13 For this purpose we used solution NMR to examine monomeric C99 in a series of detergent edge-stabilized lipid bilayers (bicelles), using the same detergent in all samples, but varying the lipid compositions.
We screened more than 40 potential bicelle compositions using either dihexanoylphosphatidylcholine (DHPC) or 3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1-propane-sulfonate (CHAPSO) as the detergent component mixed with a variety of neutral (cholesterol) and zwitterionic (phosphatidylcholine and sphingomyelin) lipids (Table S1). Sixteen of the mixtures appear to form bicelles. For this study we focused on DHPC-based bicelles, which yield better solution NMR spectra than CHAPSO-based bicelles. In addition, because C99 forms a 1:1 complex with cholesterol,1 which would complicate interpretation of results, we excluded all cholesterol-containing mixtures. We then selected a cross-section of five bicelle lipid compositions. Two of the chosen compositions have been previously reported: DHPC-dimyristoylphosphatidylcholine (DMPC) bicelles,14which have a relatively thin transbilayer span due to the C14 chains of DMPC, and DHPC-POPC (1-pamitoyl-2-oleoyl-phosphatidylcholine) bicelles,15,16which have an intermediate transbilayer span that includes an unsaturated acyl chain. The other three bicelle systems are novel but resemble previously described brain (mostly C18) sphingomyelin bicelles17 in that they are sphingolipid-based: DHPC-egg sphingomyelin (ESM) bicelles, in which the fatty amide chain composition is of intermediate (mostly C16) chain length (http://avantilipids.com), DHPC-milk sphingomyelin (MSM) bicelles, which have a fatty amide chain composition dominated (∼60%) by very long C22–C24 chains, and finally a DHPC-POPC/MSM (1:1 POPC:MSM) mixture. All bicelles used in this work contained a mole ratio of 2:1 DHPC:lipid (q = 0.5). Light scattering measurements confirmed that each of these five bicelle compositions form monodisperse assemblies of similar dimensions (Table S2).
C99 was reconstituted into the selected bicelles at a concentration low enough (<1:800 C99:{lipid+detergent}) to ensure that the protein is monomeric.181H,15N-TROSY NMR was acquired (Figures 1 and S1). Because peak positions in all cases were similar to those seen in LMPG micelles (Figure S1), it was possible to assign the bicelle spectra of C99 based on correlating peaks to the previously assigned19 peaks in LMPG. Based on these assignments, we then measured site-specific variation of backbone amide 1H and 15N chemical shifts from random coil values. Results for the 15N chemical shifts are shown in Figures 2 and S2, while results for 1H shifts are shown in Figures S3 and S4. For both data sets it can be seen that there are only very minor differences in site-specific shifts between LMPG micelle conditions and any of the five bicelle mixtures tested. This indicates that C99 has a very similar backbone conformation in all six mixtures. This is despite the facts that (i) micelles are morphologically distinct from bicelles, (ii) LMPG is anionic, whereas all the bicelles tested contained only zwitterionic lipid and detergent, and (iii) the lipid acyl chain lengths in the various mixtures varied dramatically, from C14 to C24 carbons. The similarity of the NMR data for C99 from such very different model membrane hosts suggests that the conformational and dynamic features of C99 are robust and tolerant of changes in membrane environment. Variations in cleavage of C99 by γ-secretase as a function of changes in lipid composition are unlikely to be due to composition-dependent changes in C99 conformation. We also repeated these measurement under reduced salt conditions (Figures S2 and S4–S6) and obtained nearly identical results, indicating that the C99 structure is also largely independent of ionic strength.
Figure 1.
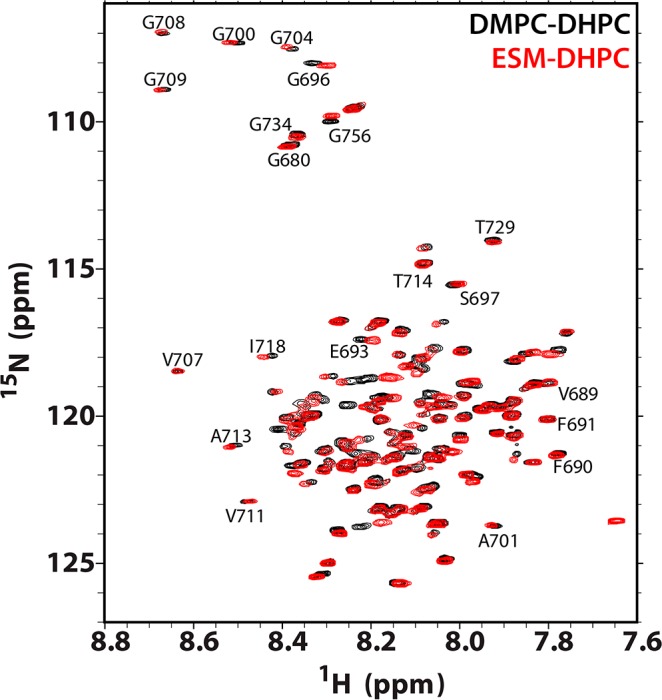
Examples of 900 MHz 1H–15N TROSY NMR spectra of U–15N-C99 in bicelles at 45 °C. Shown are spectra of the protein in ESM-DHPC bicelles (red) and the corresponding spectrum from conventional DMPC-DHPC bicelles (black). Bicelle samples contained 0.2–0.3 mM C99, 20% w/v bicelles, 250 mM imidazole, 1 mM EDTA, 10% D2O, and pH 4.5. In all cases the bicelle q ratio (lipid-to-detergent mol/mol) was 0.5. Selected resonance assignments are illustrated.
Figure 2.
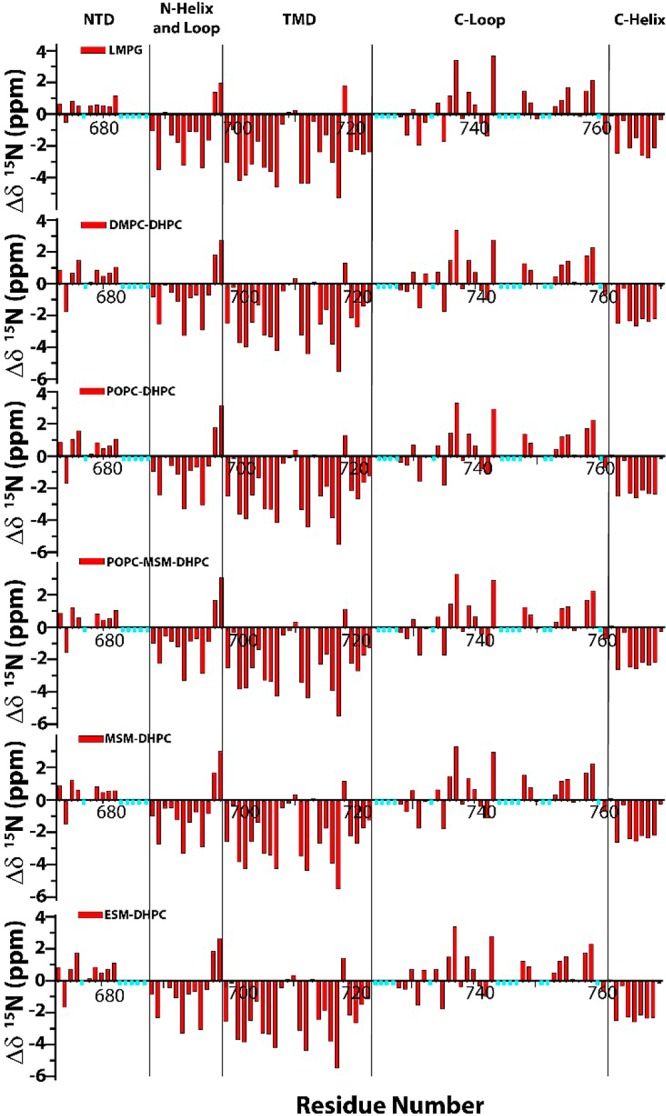
Residue-specific backbone amide 15N chemical shifts for C99 in 10% LMPG micelles and in various DHPC-based bicelles. The values reported here represent the difference between the measured chemical shift and the random coil chemical shift (estimated as described in the Supporting Information). The residues marked with cyan bars are either too broad to observe (even in the absence of a paramagnet) or lack peak assignments. The four vertical lines represent the boundaries of the disordered N-terminal cytosolic domain (NTD, 672–687), the combined N-helix and N-loop (688–699), the transmembrane domain (TMD, 700–723), the C-loop (724–761), and the distal C-terminal domain (C-helix, 762–770).1 All samples contained 0.2–0.3 mM C99, 20% w/v bicelles (q = 0.5), 250 mM imidazole, 1 mM EDTA, 10% D2O, and pH 4.5 (except for LMPG, which was pH 6.5). The temperature was 45 °C. The data for LMPG micelles were previously reported in Beel et al., 2008.19
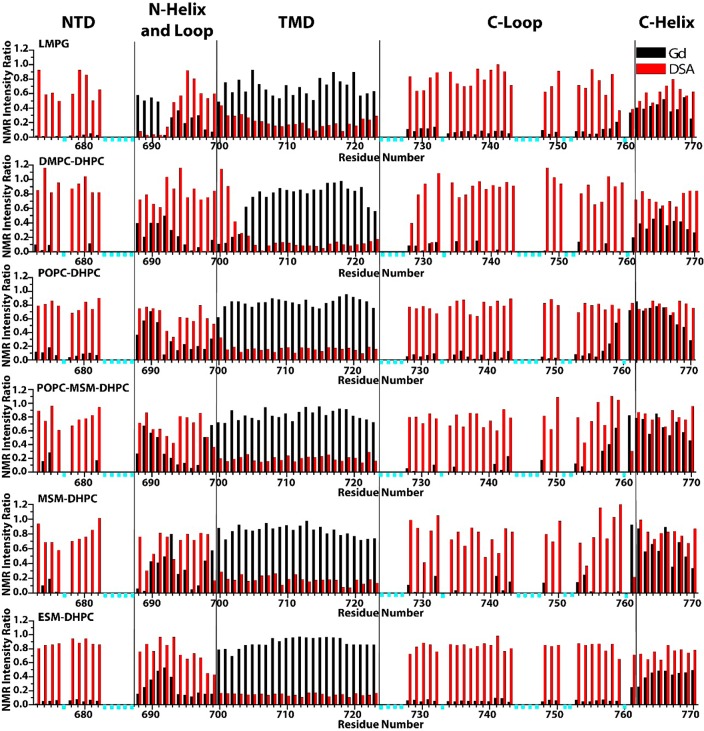
We next used water-soluble Gd(III)-diethylenetriamine-pentaacetate (Gd-DTPA) and lipophilic 16-doxylstearate (16-DSA) paramagnetic probes to examine whether the membrane topology of C99 varies as a function of host model membrane type and composition (cf. Figure S7). Figure 3 reveals no gross changes in membrane topology for C99 in all six mixtures tested: In all cases the three membrane-associated domains of C99 (the N-helix, TMD, and C-helix) remained membrane associated. Moreover, no new membrane interacting structural elements formed. However, some changes of a more modest nature are evident from Figure 3: (i) LMPG micelles are unique in that the N-helix peaks were completely broadened by the lipophilic probe, 16-DSA. This indicates that the N-helix is more deeply buried in the surface of LMPG micelles than in any of the five bicelle mixtures. This observation does not apply to the C-helix, which is protected to a similar degree from both probes in all mixtures examined. (ii) The N-terminal end of the TMD for C99 in DMPC-DHPC micelles is much more exposed to the polar Gd-DTPA probe than in either LMPG micelles or the other bicelle mixtures. This suggests that the C14 acyl chains of DMPC do not provide an adequate bilayer span to accommodate the 24 residue TM helix of C99. It is notable that the C-terminus of the C99 TMD remains largely protected from Gd-DTPA and exposed to 16-DSA in DHPC-DMPC bicelles. This means that adjustment of the TM span of C99 to thinner bilayers is not symmetric but is localized to the N-terminal end, probably because the C-terminal end is flanked by three consecutive Lys residues (724–726), which serve as a TMD termination motif. This implies that the γ-secretase cleavage sites in C99 are shifted in position with respect to center of the bilayer when bilayer thickness varies. This observation may shed light on previous results showing that the Aβ42:Aβ40 production ratio decreases as bilayer thickness increases9,10 and increases when additional hydrophobic residues are inserted to extend the C-terminal end of the TMD.11−13 Of course, bilayer composition and properties may also impact cleavage of C99 through other mechanisms, such as direct modulation of γ-secretase activity. (iii) The transmembrane span of C99 is longer by two N-terminal residues in the MSM-containing bicelle mixtures than in LMPG micelles and ESM bicelles. In POPC bicelles the TMD span is intermediate between the MSM-containing bicelles and ESM bicelles. (iv) The YEN segment (residues 757–759) that precedes the surface-associated C-helix of C99 is more deeply membrane buried (protected from Gd(III)-DTPA-induced line broadening) in POPC-containing bicelles than in the two sphingomyelin-only bicelles or in DMPC bicelles. This is possibly the consequence of the cis double bond present in the sn-2 chain of POPC, which will expand the bilayer surface area relative to bilayers with only saturated acyl chains.20 The YEN segment leads into the NPTY762 sequence, a known trafficking motif that is subject to tyrosine phosphorylation.21−23 One wonders whether access of this motif by tyrosine kinases and/or by trafficking adaptor proteins is dependent on lipid composition-dependent degree of membrane association of the preceding YEN segment.
Figure 3.
Paramagnetic probe-induced reductions in TROSY NMR peak intensities for backbone amide sites of C99 in LMPG micelles and in the five bicelle compositions examined in this work. Data were collected at 900 MHz and 45 °C as described in the Supporting Information and as exemplified by Figure S7. The reported intensity ratios are for peak height in a paramagnetic probe-containing sample divided by the corresponding peak intensity observed in a matched control sample. The water-soluble paramagnetic probe was Gd(III)-DTPA (black bars), while the lipophilic paramagnetic probe was 16-DSA (red bars). The residues marked with cyan bars are either invisible (even in the absence of a paramagnet) or lack peak assignments. Data were also collected for samples with low-salt content, yielding data very similar to that shown here (Figure S8).
The results of this work show C99 appears to have a robust conformational structure, a property that it potentially shares with many other membrane proteins.24 These results are interesting in light of considerable current interest in the question of to what degrees membrane protein structures determined under micellar conditions can be assumed to be native-like.24−27 While the C99 structure seems to vary little, it is significant that modest membrane topological adjustments were seen when C99 was reconstituted in bicelles containing lipids with very different chemistries (glycerol- vs sphingosine-based) or acyl chains of dramatically different lengths. These changes in topology may provide insight into how the cleavage sites of C99 by γ-secretase depend on membrane thickness.9−11 It will be interesting in future work to see if the known complex formation of C99 with cholesterol1 or dimerization of the protein4,6,18,28−31 alters the membrane topology of the protein (although the physiological relevance of dimerization has been questioned18). Finally, we note that this work also provides over a dozen new bicelle compositions that may be used in future studies of other membrane proteins.
Acknowledgments
We thank Dr. Martin Egli for providing access to light scattering instrumentation and Dr. Tarjani Thaker for instructing us in its use and related data analysis. This work was supported by NIH grants U54 GM094608, RO1 DC007416, RO1 DK083187, RO1 NS058815, and RO1 GM106672. K.F.M. was supported by NSF Predoctoral Research Fellowship DGE090667. The NMR instrumentation used in this work was supported by NIH grant S10 RR025677 and NSF grant DBI-0922862.
Supporting Information Available
Materials and methods, supporting tables, supporting references, and supporting figures and captions. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
§ These authors contributed equally.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Barrett P. J.; Song Y.; Van Horn W. D.; Hustedt E. J.; Schafer J. M.; Hadziselimovic A.; Beel A. J.; Sanders C. R. Science 2012, 336, 1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmin T.; Dimitrov M.; Fraering P. C.; Dal Peraro M. J. Biol. Chem. 2014, 10.1074/jbc.M113.470781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez L.; Meredith S. C.; Straub J. E.; Thirumalai D. J. Am. Chem. Soc. 2014, 136, 854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pester O.; Barrett P. J.; Hornburg D.; Hornburg P.; Pröbstle R.; Widmaier S.; Kutzner C.; Dürrbaum M.; Kapurniotu A.; Sanders C. R.; Scharnagl C.; Langosch D. J. Am. Chem. Soc. 2013, 135, 1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marenchino M.; Williamson P. T.; Murri S.; Zandomeneghi G.; Wunderli-Allenspach H.; Meier B. H.; Kramer S. D. Biophys. J. 2008, 95, 1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita N.; Straub J. E.; Thirumalai D. J. Am. Chem. Soc. 2009, 131, 17843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osenkowski P.; Ye W.; Wang R.; Wolfe M. S.; Selkoe D. J. J. Biol. Chem. 2008, 283, 22529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura K.; Farner K. C.; Nasser-Ghodsi N.; Jones P.; Berezovska O. Mol. Neurodegener. 2011, 6, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes O.; Paturi S.; Ye W.; Wolfe M. S.; Selkoe D. J. Biochemistry 2012, 51, 3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler E.; Kamp F.; Scheuring J.; Ebke A.; Fukumori A.; Steiner H. J. Biol. Chem. 2012, 287, 21326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler S. F.; Beher D.; Grimm H. S.; Wang R.; Shearman M. S.; Masters C. L.; Beyreuther K. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M. P.; Hickman L. J.; Eckman C. B.; Uljon S. N.; Wang R.; Golde T. E. J. Biol. Chem. 1999, 274, 11914. [DOI] [PubMed] [Google Scholar]
- Ousson S.; Saric A.; Baguet A.; Losberger C.; Genoud S.; Vilbois F.; Permanne B.; Hussain I.; Beher D. J. Neurochem. 2013, 125, 610. [DOI] [PubMed] [Google Scholar]
- Sanders C. R.; Schwonek J. P. Biochemistry 1992, 31, 8898. [DOI] [PubMed] [Google Scholar]
- Chou J. J.; Baber J. L.; Bax A. J. Biomol. NMR 2004, 29, 299. [DOI] [PubMed] [Google Scholar]
- Triba M. N.; Devaux P. F.; Warschawski D. E. Biophy. J. 2006, 91, 1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T.; Suzuki T.; Yasuda T.; Oishi T.; Matsumori N.; Murata M. Bioorg. Med. Chem. 2012, 20, 270. [DOI] [PubMed] [Google Scholar]
- Song Y.; Hustedt E. J.; Brandon S.; Sanders C. R. Biochemistry 2013, 52, 5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beel A. J.; Mobley C. K.; Kim H. J.; Tian F.; Hadziselimovic A.; Jap B.; Prestegard J. H.; Sanders C. R. Biochemistry 2008, 47, 9428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B. A.; Engelman D. M. J. Mol. Biol. 1983, 166, 211. [DOI] [PubMed] [Google Scholar]
- Radzimanowski J.; Simon B.; Sattler M.; Beyreuther K.; Sinning I.; Wild K. EMBO Rep. 2008, 9, 1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schettini G.; Govoni S.; Racchi M.; Rodriguez G. J. Neurochem. 2010, 115, 1299. [DOI] [PubMed] [Google Scholar]
- Tamayev R.; Zhou D.; D’Adamio L. Mol. Neurogen. 2009, 4, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders C. R.; Mittendorf K. F. Biochemistry 2011, 50, 7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews E. E.; Zoonens M.; Engelman D. M. Cell 2006, 127, 447. [DOI] [PubMed] [Google Scholar]
- Warschawski D. E.; Arnold A. A.; Beaugrand M.; Gravel A.; Chartrand E.; Marcotte I. Biochim. Biophys. Acta 2011, 1808, 1957. [DOI] [PubMed] [Google Scholar]
- Zhou H. X.; Cross T. A. Ann. Rev. Biophys. 2013, 42, 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadezhdin K. D.; Bocharova O. V.; Bocharov E. V.; Arseniev A. S. FEBS Lett. 2012, 586, 1687. [DOI] [PubMed] [Google Scholar]
- Sato T.; Tang T. C.; Reubins G.; Fei J. Z.; Fujimoto T.; Kienlen-Campard P.; Constantinescu S. N.; Octave J. N.; Aimoto S.; Smith S. O. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.; Barreyro L.; Provasi D.; Djemil I.; Torres-Arancivia C.; Filizola M.; Ubarretxena-Belandia I. J. Mol. Biol. 2011, 408, 879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botev A.; Munter L. M.; Wenzel R.; Richter L.; Althoff V.; Ismer J.; Gerling U.; Weise C.; Koksch B.; Hildebrand P. W.; Bittl R.; Multhaup G. Biochemistry 2011, 50, 828–835. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.