Abstract
Carbon nanomaterials have been widely explored for diverse biosensing applications including bacterial detection. However, covalent functionalization of these materials can lead to the destruction of attractive electronic properties. To this end, we utilized a new graphene derivative, holey reduced graphene oxide (hRGO), functionalized with Magainin I to produce a broad-spectrum bacterial probe. Unlike related carbon nanomaterials, hRGO retains the necessary electronic properties while providing the high percentage of available oxygen moieties required for effective covalent functionalization.
Keywords: graphene, antimicrobial peptides, semiconductors, nanotechnology, nanomaterials
Introduction
The novel electronic properties1−3 and mechanical strength2,4 of graphene may render this carbon-based nanomaterial integral in future generations of electronics, batteries, sensors, and composites.1,2,5−8 Because of the ambipolar nature of graphene,2 however, lithographic9 or chemical10 techniques have been employed to produce graphene nanoribbons, which demonstrate semiconducting properties at room temperature when their widths are less than 10 nm as a result of quantum confinement and edge effects.11−13 The creation of holes in the basal plane of graphene results in an interconnected nanoribbon-like semiconducting nanomaterial; therefore, various techniques have been employed to create nanometer-sized holes on individual sheets of graphene.14−25
Single-walled nanotubes (SWNTs) and graphene, because of their nanometer-scale sizes and unique electronic properties, are considered to be ideal materials for biosensing applications.26−30 SWNT field-effect-transistor (FET) biosensors have been functionalized with antibodies or aptamers for the fast detection of multiple bacterial species,31,32 and electrochemical sensors using SWNT– and graphene–aptamer composites were reported to quickly detect ultralow concentrations of bacteria.33,34 Additionally, graphene-based FET devices functionalized with antibodies17 or antimicrobial peptides (AMPs)35 have been employed for the electronic detection of E. coli. Many of these reports rely on the noncovalent functionalization of either SWNTs or graphene. While this technique has demonstrated the capability of detecting analytes with selectivity and sensitivity, the primary limitation arises when using short peptide chains and molecules less capable of forming van der Waals interactions with the surface of carbon-based nanomaterials. Interactions can be improved by utilizing pyrene- or porphyrin-based conjugates capable of π–π stacking on a graphitic surface, which requires additional chemistry. While covalent attachment of molecules can overcome these issues, this approach requires the introduction of functional groups onto the nanomaterial, thereby reducing the overall efficiency and stability of the device.27
Results and Discussion
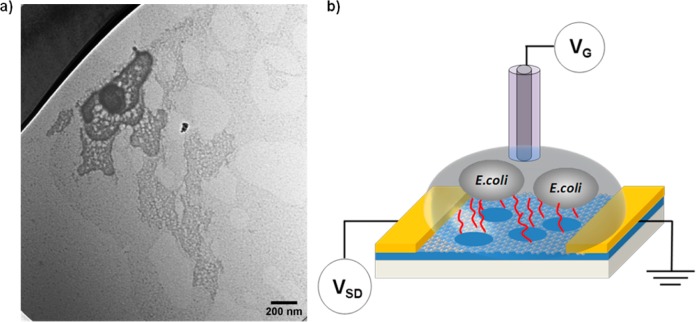
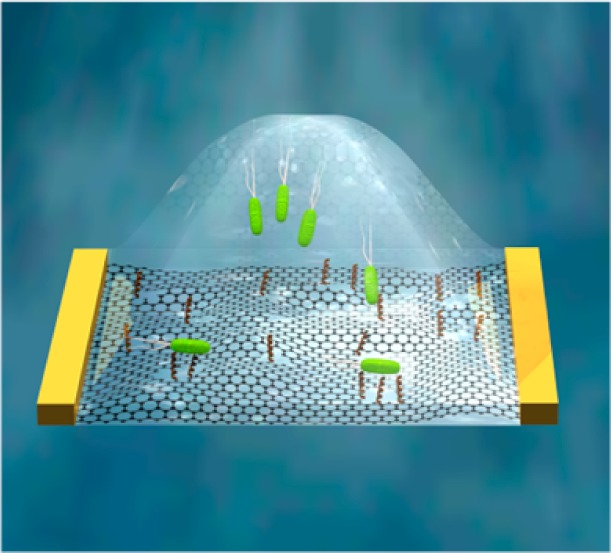
Herein, we utilized a recently developed graphene derivative, holey reduced graphene oxide (hRGO; Figure 1a),36 which demonstrates p-type semiconductor transfer characteristics and is endowed with an abundance of oxygen-containing groups (especially on the edges of holes),37 as the transducer element in FET devices as a proof-of-principle for the creation of assay sensors. For this purpose, an AMP, Magainin I (GIGKFLHSAGKFGKAFVGEIMKS),38 was covalently functionalized to hRGO, yielding a gram-negative specific biosensor that operates by taking advantage of the electrostatic interaction between positively charged Magainin I and anionic lipopolysaccharides.39 AMPs, which are inherent to many organisms’ immune systems, recognize target pathogens by interacting with surface components of microbial cells.38,40,41 While the exact mechanism for their antimicrobial activities remains undetermined, the microbicidal or microbiostatic activity is generally postulated to occur via membrane disruption.42 To date, AMPs have been successfully employed for the detection of pathogens utilizing impedance sensors43 and fluorescence assays38,41 and are therefore attractive candidates for recognition elements in new sensing platforms or materials.
Figure 1.
(a) TEM image of hRGO. (b) Schematic illustration of an AMP-functionalized hRGO FET for the selective detection of gram-negative bacteria cells.
Figure 1b represents a schematic illustration of an AMP-functionalized hRGO-based FET device for E. coli O157:H7 detection. hRGO (Figure 1a) was first synthesized via enzymatic oxidation,36 in which a sample of graphene oxide was subjected to 8 days of HRP/H2O2 oxidation in a phosphate buffer (0.1 M, pH 7.0) to produce holey graphene oxide. Next, to reduce the oxidized carbon nanomaterial, a mixture containing 5.0 mL of 0.125 wt % holey graphene oxide, 4.8 mL of nanopure water, 200 μL of hydrazine hydrate (50 wt %), and 35 μL of NH4OH (28 wt %) was stirred for 5 min and heated at 95 °C for 1 h. The suspension containing hRGO was subsequently dialyzed against distilled water with 0.5% NH4OH to remove the hydrazine. The resulting product consists of graphene flakes with holes of 26.7 ± 12.8 nm diameter and neck widths of 8.9 ± 6.9 nm.36 Next, the as-synthesized product was diluted in water to 0.01 mg/mL and deposited between interdigitated electrodes (Au/Ti, 100 nm/30 nm, 10 μm spacing) using an alternating-current dielectrophoresis method with a bias voltage of 10 Vpp at 300 kHz for 60 s.44 The transfer characteristics of bare hRGO devices were then measured in a 1 mM phosphate-buffered saline (PBS) solution after drying for several hours at 120 °C in air.
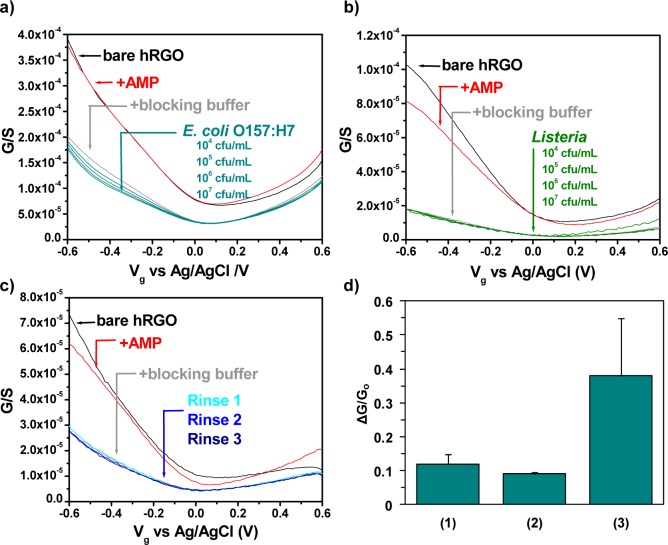
Functionalization was accomplished by activating carboxylic groups on bare hRGO devices with 1-ethyl-3-[3-(dimethylamino)propyl]carbodiimide/N-hydroxysuccinimide [EDC/NHS; 100 and 25 nM, respectively, in a 50 mM 2-(N-morpholino)ethanesulfonic acid buffer] for 30 min.45,46 After a thorough rinsing with PBS, activated devices were incubated overnight with AMP (1 μM in PBS), resulting in the formation of amide bonds between Magainin I and activated hRGO, observable as a decrease in the FET device conductance in the p-type region (Figure 2a,b). After incubation with a blocking buffer (BB; 0.1% Tween 20 in PBS) for 1 h, a further decrease in the conductance was detected as a result of Tween occupying nonspecific binding sites on hRGO. Utilization of this BB is important to ensure a good device performance, with the lack of a BB leading to both the allowance of nonspecific interactions and low response, possibly related to the conformation of AMP on the device surface (Figure S1 in the Supporting Information, SI). Upon subsequent exposure to heat-killed E. coli O157:H7 (104–107 cfu/mL in PBS for 1 h at each concentration, a time sufficient to ensure ample time for cell capture, as evidenced in Figure S2 in the SI), the devices demonstrated a further response in the p-type region that was attributed to the specific interaction between the attached AMP and bacterial cells in solution. Presumably, this interaction induces electron transfer with hRGO, which decreases the conductivity of the device by depleting the main carriers (i.e., holes); however, electrostatic gating may also contribute to the observed response, and so the overall mechanism is complex. The limit of detection for this sensor was calculated to be 803 cfu/mL (Figure S3 in the SI). As a control experiment, functionalized devices were exposed to gram-positive Listeria cells, which do not interact with Magainin I. After incubation with Listeria (104–107 cfu/mL in PBS; Figure 2b), the transfer characteristics changed negligibly, comparable to a device treated equally with PBS (Figure 2c and the procedure in the SI), thereby indicating minimal to no binding between AMP and the control bacterial cells. Small changes in the local pH and differences in the ionic strength do not have much of an effect on the sensor at −0.5 Vg (Figure S4 in the SI). Additional experiments involving specific targets (E. coli O157:H7 and Salmonella, also a gram-negative bacterium specific to Magainin I), and nonspecific targets (bovine serum albumin and Listeria), agreed with these results and are summarized in Figure S5 in the SI. Upon exposure of bare hRGO devices to 107 cfu/mL E. coli or Listeria, a significant, nonspecific response was observed (Figure S6 in the SI), providing evidence that the prior response (i.e., Figure 2a) resulted from the specific interaction of AMP with bacteria. Unfortunately, because of the strong interaction of AMP with lipopolysaccharides at pH 7.4, the sensors cannot be refreshed and therefore are not reusable.
Figure 2.
Electronic detection of bacteria–AMP interactions. Conductance (G) versus gate voltage (Vg) of bare hRGO FET devices, after functionalization with AMP, after incubation with a BB, and after incubation with either (a) 104–107 cfu/mL E. coli O157:H7 or (b) 104–107 cfu/mL Listeria. The limit of detection was calculated to be 803 cfu/mL. (c) Effect of rinsing on AMP and Tween 20 attachment. The response to rinsing was approximately equivalent to the response from Listeria. (d) Relative response at Vg = −0.5 V to E. coli of unfunctionalized (1), noncovalently functionalized (2), and covalently functionalized (3) hrGO devices blocked with a BB. Averaged from four devices; error bars indicate 1 standard deviation.
Additional control experiments were implemented to examine the effect of covalent attachment (Figures 2d and S7 in the SI). In the first control, bare hRGO was incubated with a BB and subsequently exposed to 107 cfu/mL E. coli O157:H7 (1). In the second control, hRGO devices were first incubated with an AMP solution without EDC/NHS activation, successively incubated with a BB, and finally exposed to 107 cfu/mL E. coli (2). The results of these experiments are summarized in Figure 2d, which indicates that devices functionalized covalently with AMP (3) demonstrate a much larger response to E. coli. These results suggest that covalent attachment of AMP to hRGO device surfaces is integral for achieving a superior sensor performance versus nonspecific binding and noncovalent functionalization of AMP, respectively.
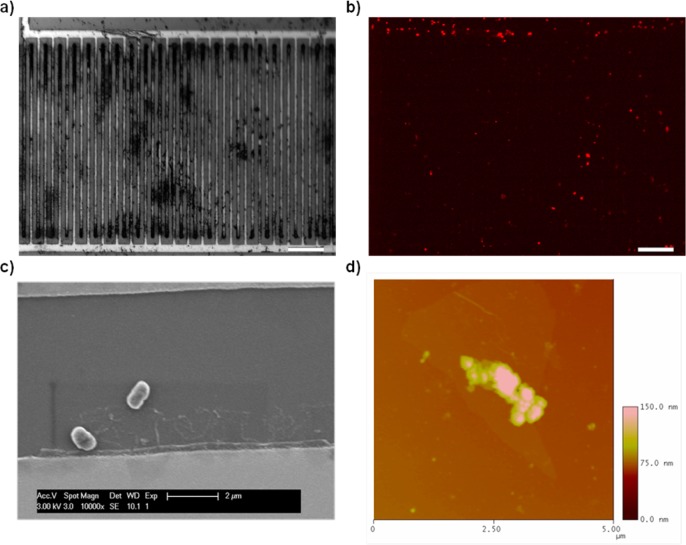
To visualize the attachment of bacterial cells to the surface of functionalized FET devices after exposure to E. coli, fluorescence microscopy, scanning electron microscopy (SEM), and atomic force microscopy (AFM) were performed (imaging details in the SI). Bright-field and fluorescence micrographs of propidium iodide (PI)-stained cells provided visual proof that bacterial cells are bound to the device (Figure 3a,b). For SEM imaging, palladium was first sputtered onto the surface of the device to increase contrast. Micrographs (Figure 3c) revealed that rodlike E. coli cells were attached to the surface of the functionalized hRGO surface, thereby supplying the measured FET device response. Additionally, the attachment of E. coli cells to the surface of AMP-functionalized hRGO was confirmed by AFM (Figure 3d).
Figure 3.
(a) Optical micrograph of a single functionalized device after exposure to 107 cfu/mL E. coli. (b) Same device under a red fluorescent protein filter, showing PI-stained cell fluorescing. Scale bar for parts a and b is 50 μm. (c) SEM image of the functionalized device surface after incubation with E. coli O157:H7. The scale bar 2 μm (d) AFM image over an area of 5 μm2 depicting the attachment of bacteria to the surface of hRGO.
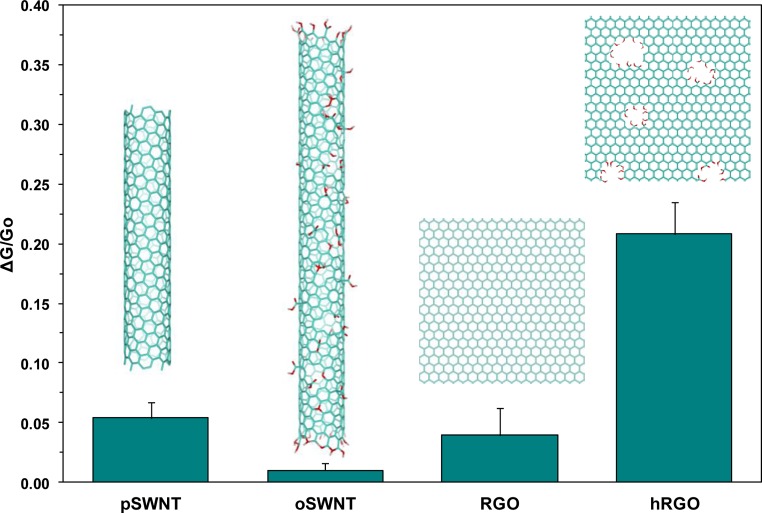
In order to examine the efficiency of hRGO versus other carbon nanomaterials, reduced graphene oxide (RGO), commercially available pristine SWNTs (pSWNTs), and oxidized SWCNTs (oSWNTs) were also employed as the transducer element in FET devices to detect bacteria. Similar to hRGO-based FETs, RGO, pSWNT, and oSWNT FET devices were covalently functionalized with Magainin I. After EDC/NHS coupling, the conductance decreased as before (Figure S6 in the SI). Next, incubation with a BB induced a further decrease in the conductance, and after incubation with 107 cfu/mL E. coli, there was an additional decrease in the conductance in response to bacterial cells. When compared with hRGO FET devices, however, the mean relative changes in the conductance upon exposure to E. coli were significantly lower (Figure 4), despite the higher concentration of SWNTs on the device surface, as seen previously47. For RGO and pSWNTs, this low response may be attributed to the amount of oxygen functionality (i.e., carboxyl groups) available for coupling. At low concentrations of this functional group, minimal AMP would be bound, which would result in insufficient binding of the bacteria. The low efficiency of oSWNTs, which underperformed all other samples, can be attributed to the availability of the oxygen content as well as the loss of the electronic efficiency from oxidization. According to the manufacturer, oSWNTs contain 1.1 ± 0.1 mol % carboxylic acid groups, as determined by acid–base titration, while pSWNTs did not yield detectable results.48 X-ray photoelectron spectroscopy and energy-dispersive X-ray analysis of RGO and hRGO samples determined the oxygen content between 4.3–4.5 and 20.5–25.2 atom %, respectively (Figures S7 and S8 in the SI), which confirmed that the abundance of oxygen functionalities coupled with the preservation of the electronic properties is directly correlated to the larger response of hRGO to gram-negative bacteria after covalent functionalization with AMP.
Figure 4.
Comparison of the mean normalized responses (Vg = −0.5 V) of four Magainin I functionalized carbon nanomaterials to 107 cfu/mL E. coli. Averaged from four devices; the error bars represent 1 standard deviation.
Conclusion
hRGO affords a rich chemistry that facilitates the functionalization of highly sensitive sensors while retaining the useful electronic properties of similar analogues. By exploiting interactions between Magainin I and gram-negative bacteria and transducing those interactions into conductance changes using hRGO-based FET devices, we have demonstrated a simple and selective methodology for the detection of gram-negative bacteria that outperformed other tested carbon nanomaterials. Because of the preservation of good electronic properties despite the high content of oxygen moieties on hRGO and the relative ease of functionalization, this approach could feasibly be targeted to a broad variety of bacterial species using a library of AMPs in order to elucidate the content of complex biological media through the use of sensor arrays, and future work will be directed toward this application.
Acknowledgments
Y.C. and Y.Z. received financial support through graduate student fellowships from Bayer Material Science. G.P.K. received financial support through EPA STAR Graduate Fellowship FP-91713801. This work was supported by NIEHS Award R01ES019304 and NSF CAREER Award 0954345.
Supporting Information Available
Experimental details, imaging information, and elemental analysis. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Geim A. K.; Novoselov K. S. The Rise of Graphene. Nat. Mater. 2007, 6, 183–191. [DOI] [PubMed] [Google Scholar]
- Zhu Y.; Murali S.; Cai W.; Li X.; Suk J. W.; Potts J. R.; Ruoff R. S. Graphene and Graphene Oxide: Synthesis, Properties, and Applications. Adv. Mater. 2010, 22, 3906–3924. [DOI] [PubMed] [Google Scholar]
- Castro Neto A. H.; Guinea F.; Peres N. M. R.; Novoselov K. S.; Geim A. K. The Electronic Properties of Graphene. Rev. Mod. Phys. 2009, 81, 109–162. [Google Scholar]
- Lee C.; Wei X.; Kysar J. W.; Hone J. Measurement of the Elastic Properties and Intrinsic Strength of Monolayer Graphene. Science 2008, 321, 385–388. [DOI] [PubMed] [Google Scholar]
- Geim A. K. Graphene: Status and Prospects. Science 2009, 324, 1530–1534. [DOI] [PubMed] [Google Scholar]
- Allen M. J.; Tung V. C.; Kaner R. B. Honeycomb Carbon: A Review of Graphene. Chem. Rev. 2010, 110, 132–145. [DOI] [PubMed] [Google Scholar]
- Kauffman D. R.; Star A. Graphene versus Carbon Nanotubes for Chemical Sensor and Fuel Cell Applications. Analyst 2010, 135, 2790–2797. [DOI] [PubMed] [Google Scholar]
- Novoselov K. S.; Falko V. I.; Colombo L.; Gellert P. R.; Schwab M. G.; Kim K. A Roadmap for Graphene. Nature 2012, 490, 192–200. [DOI] [PubMed] [Google Scholar]
- Han M. Y.; Özyilmaz B.; Zhang Y.; Kim P. Energy Band-Gap Engineering of Graphene Nanoribbons. Phys. Rev. Lett. 2007, 98, 206805. [DOI] [PubMed] [Google Scholar]
- Yang X.; Dou X.; Rouhanipour A.; Zhi L.; Rader H. J.; Mullen K. Two-Dimensional Graphene Nanoribbons. J. Am. Chem. Soc. 2008, 130, 4216–4217. [DOI] [PubMed] [Google Scholar]
- Nakada K.; Fujita M.; Dresselhaus G.; Dresselhaus M. S. Edge State in Graphene Ribbons: Nanometer Size Effect and Edge Shape Dependence. Phys. Rev. B: Condens. Matter Mater. Phys. 1996, 54, 17954–17961. [DOI] [PubMed] [Google Scholar]
- Son Y.-W.; Cohen M. L.; Louie S. G. Energy Gaps in Graphene Nanoribbons. Phys. Rev. Lett. 2006, 97, 216803. [DOI] [PubMed] [Google Scholar]
- Barone V.; Hod O.; Scuseria G. E. Electronic Structure and Stability of Semiconducting Graphene Nanoribbons. Nano Lett. 2006, 6, 2748–2754. [DOI] [PubMed] [Google Scholar]
- Bai J.; Zhong X.; Jiang S.; Huang Y.; Duan X. Graphene Nanomesh. Nat. Nanotechnol. 2010, 5, 190–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.; Safron N. S.; Han E.; Arnold M. S.; Gopalan P. Fabrication and Characterization of Large-Area, Semiconducting Nanoperforated Graphene Materials. Nano Lett. 2010, 10, 1125–1131. [DOI] [PubMed] [Google Scholar]
- Liu H.; Ryu S.; Chen Z.; Steigerwald M. L.; Nuckolls C.; Brus L. E. Photochemical Reactivity of Graphene. J. Am. Chem. Soc. 2009, 131, 17099–17101. [DOI] [PubMed] [Google Scholar]
- Huang Y.; Dong X.; Liu Y.; Li L.-J.; Chen P. Graphene-Based Biosensors for Detection of Bacteria and their Metabolic Activities. J. Mater. Chem. 2011, 21, 12358–12362. [Google Scholar]
- Huang J.; Qi L.; Li J. In Situ Imaging of Layer-by-Layer Sublimation of Suspended Graphene. Nano Res. 2010, 3, 43–50. [Google Scholar]
- Liu L.; Ryu S.; Tomasik M. R.; Stolyarova E.; Jung N.; Hybertsen M. S.; Steigerwald M. L.; Brus L. E.; Flynn G. W. Graphene Oxidation: Thickness-Dependent Etching and Strong Chemical Doping. Nano Lett. 2008, 8, 1965–1970. [DOI] [PubMed] [Google Scholar]
- Lin Y.; Watson K. A.; Kim J.-W.; Baggett D. W.; Working D. C.; Connell J. W. Bulk Preparation of Holey Graphene via Controlled Catalytic Oxidation. Nanoscale 2013, 5, 7814–7824. [DOI] [PubMed] [Google Scholar]
- Bieri M.; Treier M.; Cai J.; Ait-Mansour K.; Ruffieux P.; Groning O.; Groning P.; Kastler M.; Rieger R.; Feng X.; Mullen K.; Fasel R. Porous Graphenes: Two-Dimensional Polymer Synthesis with Atomic Precision. Chem. Commun. 2009, 6919–6921. [DOI] [PubMed] [Google Scholar]
- Zhao X.; Hayner C. M.; Kung M. C.; Kung H. H. Flexible Holey Graphene Paper Electrodes with Enhanced Rate Capability for Energy Storage Applications. ACS Nano 2011, 5, 8739–8749. [DOI] [PubMed] [Google Scholar]
- Zhao X.; Hayner C. M.; Kung M. C.; Kung H. H. In-Plane Vacancy-Enabled High-Power Si–Graphene Composite Electrode for Lithium-Ion Batteries. Adv. Energy Mater. 2011, 1, 1079–1084. [Google Scholar]
- Radich J. G.; Kamat P. V. Making Graphene Holey. Gold-Nanoparticle-Mediated Hydroxyl Radical Attack on Reduced Graphene Oxide. ACS Nano 2013, 7, 5546–5557. [DOI] [PubMed] [Google Scholar]
- Zhou X.; Zhang Y.; Wang C.; Wu X.; Yang Y.; Zheng B.; Wu H.; Guo S.; Zhang J. Photo-Fenton Reaction of Graphene Oxide: A New Strategy to Prepare Graphene Quantum Dots for DNA Cleavage. ACS Nano 2012, 6, 6592–6599. [DOI] [PubMed] [Google Scholar]
- Kim S. N.; Rusling J. F.; Papadimitrakopoulos F. Carbon Nanotubes for Electronic and Electrochemical Detection of Biomolecules. Adv. Mater. 2007, 19, 3214–3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen B. L.; Kichambare P. D.; Star A. Carbon Nanotube Field-Effect-Transistor-Based Biosensors. Adv. Mater. 2007, 19, 1439–1451. [Google Scholar]
- Shen H.; Zhang L.; Liu M.; Zhang Z. Biomedical Applications of Graphene. Theranostics 2012, 2, 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Dong X.; Chen P. Biological and Chemical Sensors Based on Graphene Materials. Chem. Soc. Rev. 2012, 41, 2283–2307. [DOI] [PubMed] [Google Scholar]
- Münzer A. M.; Michael Z. P.; Star A. Carbon nanotubes for the label-free detection of biomarkers. ACS Nano 2013, 7, 7448–7453. [DOI] [PubMed] [Google Scholar]
- Villamizar R. A.; Maroto A.; Rius F. X.; Inza I.; Figueras M. J. Fast Detection of Salmonella Infantis with Carbon Nanotube Field Effect Transistors. Biosens. Bioelectron. 2008, 24, 279–283. [DOI] [PubMed] [Google Scholar]
- So H.-M.; Park D.-W.; Jeon E.-K.; Kim Y.-H.; Kim B. S.; Lee C.-K.; Choi S. Y.; Kim S. C.; Chang H.; Lee J.-O. Detection and Titer Estimation of Escherichia coli Using Aptamer-Functionalized Single-Walled Carbon-Nanotube Field-Effect Transistors. Small 2008, 4, 197–201. [DOI] [PubMed] [Google Scholar]
- Zelada-Guillén G. A.; Riu J.; Düzgün A.; Rius F. X. Immediate Detection of Living Bacteria at Ultralow Concentrations Using a Carbon Nanotube Based Potentiometric Aptasensor. Angew. Chem., Int. Ed. 2009, 48, 7334–7337. [DOI] [PubMed] [Google Scholar]
- Hernández R.; Vallés C.; Benito A. M.; Maser W. K.; Xavier Rius F.; Riu J. Graphene-Based Potentiometric Biosensor for the Immediate Detection of Living Bacteria. Biosens. Bioelectron. 2014, 54, 553–557. [DOI] [PubMed] [Google Scholar]
- Mannoor M. S.; Tao H.; Clayton J. D.; Sengupta A.; Kaplan D. L.; Naik R. R.; Verma N.; Omenetto F. G.; McAlpine M. C. Graphene-Based Wireless Bacteria Detection on Tooth Enamel. Nat. Commun. 2012, 3, 763–763. [DOI] [PubMed] [Google Scholar]
- Kotchey G. P.; Allen B. L.; Vedala H.; Yanamala N.; Kapralov A. A.; Tyurina Y. Y.; Klein-Seetharaman J.; Kagan V. E.; Star A. The Enzymatic Oxidation of Graphene Oxide. ACS Nano 2011, 5, 2098–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedala H.; Sorescu D. C.; Kotchey G. P.; Star A. Chemical Sensitivity of Graphene Edges Decorated with Metal Nanoparticles. Nano Lett. 2011, 11, 2342–2347. [DOI] [PubMed] [Google Scholar]
- Kulagina N. V.; Shaffer K. M.; Anderson G. P.; Ligler F. S.; Taitt C. R. Antimicrobial Peptide-Based Array for Escherichia coli and Salmonella Screening. Anal. Chim. Acta 2006, 575, 9–15. [DOI] [PubMed] [Google Scholar]
- Lee I. H.; Cho Y.; Lehrer R. I. Effects of pH and Salinity on the Antimicrobial Properties of Clavanins. Infect. Immun. 1997, 65, 2898–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasloff M. Antimicrobial Peptides of Multicellular Organisms. Nature 2002, 415, 389–395. [DOI] [PubMed] [Google Scholar]
- Kulagina N. V.; Lassman M. E.; Ligler F. S.; Taitt C. R. Antimicrobial Peptides for Detection of Bacteria in Biosensor Assays. Anal. Chem. 2005, 77, 6504–6508. [DOI] [PubMed] [Google Scholar]
- Strauss J.; Kadilak A.; Cronin C.; Mello C. M.; Camesano T. A. Binding, Inactivation, and Adhesion Forces between Antimicrobial Peptide Cecropin P1 and Pathogenic E. coli. Colloids Surf., B 2010, 75, 156–164. [DOI] [PubMed] [Google Scholar]
- Mannoor M. S.; Zhang S.; Link A. J.; McAlpine M. C. Electrical Detection of Pathogenic Bacteria via Immobilized Antimicrobial Peptides. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 19207–19212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z. B.; Liu X. J.; Campbell E. E. B.; Zhang S. L. Alternating Current Dielectrophoresis of Carbon Nanotubes. J. Appl. Phys. 2005, 98, 056103. [Google Scholar]
- Singh K. V.; Pandey R. R.; Wang X.; Lake R.; Ozkan C. S.; Wang K.; Ozkan M. Covalent Functionalization of Single Walled Carbon Nanotubes with Peptide Nucleic Acid: Nanocomponents for Molecular Level Electronics. Carbon 2006, 44, 1730–1739. [Google Scholar]
- Jung D.-H.; Kim B. H.; Ko Y. K.; Jung M. S.; Jung S.; Lee S. Y.; Jung H.-T. Covalent Attachment and Hybridization of DNA Oligonucleotides on Patterned Single-Walled Carbon Nanotube Films. Langmuir 2004, 20, 8886–8891. [DOI] [PubMed] [Google Scholar]
- Chen Y.; Vedala H.; Kotchey G. P.; Audfray A.; Cecioni S.; Imberty A.; Vidal S.; Star A. Electronic Detection of Lectins Using Carbohydrate-Functionalized Nanostructures: Graphene versus Carbon Nanotubes. ACS Nano 2011, 6, 760–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H.; Bhowmik P.; Zhao B.; Hamon M. A.; Itkis M. E.; Haddon R. C. Determination of the Acidic Sites of Purified Single-Walled Carbon Nanotubes by Acid–Base Titration. Chem. Phys. Lett. 2001, 345, 25–28. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.