Abstract
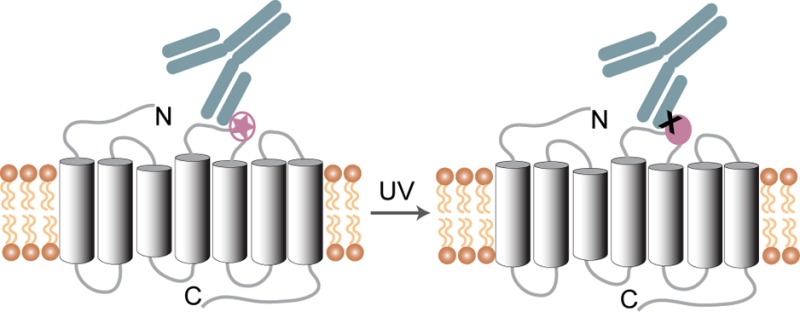
We developed a strategy for creating epitope maps of monoclonal antibodies (mAbs) that bind to G protein-coupled receptors (GPCRs) containing photo-cross-linkers. Using human CXC chemokine receptor 4 (CXCR4) as a model system, we genetically incorporated the photolabile unnatural amino acid p-azido-l-phenylalanine (azF) at various positions within extracellular loop 2 (EC2). We then mapped the interactions of the azF-CXCR4 variants with mAb 12G5 using targeted loss-of-function studies and photo-cross-linking in whole cells in a microplate-based format. We used a novel variation of a whole cell enzyme-linked immunosorbent assay to quantitate cross-linking efficiency. 12G5 cross-linked primarily to residues 184, 178, and 189 in EC2 of CXCR4. Mapping of the data to the crystal structure of CXCR4 showed a distinct mAb epitope footprint with the photo-cross-linked residues clustered around the loss-of-function sites. We also used the targeted photo-cross-linking approach to study the interaction of human CC chemokine receptor 5 (CCR5) with PRO 140, a humanized mAb that inhibits human immunodeficiency virus-1 cellular entry, and 2D7. The mAbs produced distinct cross-linking patterns on EC2 of CCR5. PRO 140 cross-linked primarily to residues 174 and 175 at the amino-terminal end of EC2, and 2D7 cross-linked mainly to residues 170, 176, and 184. These results were mapped to the recent crystal structure of CCR5 in complex with maraviroc, showing cross-linked residues at the tip of the maraviroc binding crevice formed by EC2. As a strategy for mapping mAb epitopes on GPCRs, our targeted photo-cross-linking method is complementary to loss-of-function mutagenesis results and should be especially useful for studying mAbs with discontinuous epitopes.
Monoclonal antibodies (mAbs), which offer stable protein scaffolds with variable domains capable of high target affinity and selectivity, have emerged as an important class of therapeutic biologicals.1−3 Several mAbs to the human immunodeficiency virus-1 (HIV-1) coreceptors CXC chemokine receptor 4 (CXCR4) and CC chemokine receptor 5 (CCR5) have demonstrated antiviral activity and have been advanced for clinical applications in HIV-1 fusion and entry inhibition.4−7 Among them, we decided to study the interaction of CXCR4 with mAb 12G58 and that of CCR5 with two mAbs, 2D79 and PRO 140.10 mAb 12G5 exhibits moderate to strong potency as an HIV-1 entry inhibitor.11 2D7 is a potent HIV-1 entry inhibitor that is awaiting successful humanization.12 PRO 140, a humanized form of the PA14 antibody, showed HIV-1 entry inhibition in several preclinical studies, and it is currently in phase 2b clinical trials (http://clinicaltrials.gov) as a potential therapeutic agent in the treatment of HIV-1 infection.13,14 These antibodies seem to recognize conformation-dependent epitopes in the chemokine receptors formed by amino acid residues either exclusively in extracellular loop 2 (EC2) or along with the N-terminal tail.10−12,15−20 In fact, as conformation-sensitive mAbs, 12G5 and 2D7 are routinely used to detect cell surface expression levels of CXCR4 and CCR5, respectively.
Topological mAb epitope maps for CXCR4 and CCR5 would be useful for designing HIV-1 entry inhibitors.21−23 Traditional site-directed mutagenesis and variations thereof are routinely used for mapping the epitopes of antibodies on target proteins.24 Other methods include but are not limited to shotgun mutagenesis,19 site-directed masking,24 and phage and bacterial surface display.25,26 The highest-resolution epitope maps can be obtained from structural analyses of antigen–antibody co-complexes, which permit direct visualization of contact sites.27 The recently revealed antagonist-bound crystal structures of CXCR4 and CCR5 facilitate identification of antibody accessible sites on the extracellular surface of the receptor.28,29 However, in the absence of antibody-bound cocrystal structures, precise identification of contact sites and binding modes can be challenging. We previously developed an amber codon suppression technology to introduce an unnatural amino acid (UAA) at an engineered amber nonsense codon in expressed GPCRs.30,31 The technology relies on the use of an orthogonal aminoacyl-tRNA synthetase (aa-RS)/suppressor tRNA pair to site-selectively introduce UAAs such as p-benzoyl-l-phenylalanine (BzF) and p-azido-l-phenylalanine (azF) into target proteins.32−34 We optimized the methodology in mammalian cells by using an engineered Tyr-RS,34 and a chimera of human and Bacillus stearothermophilus tRNATyr to heterologously express low-abundance GPCRs.35 We recently exploited the physical and chemical properties of these UAAs in several different studies. AzF, which is also IR active, was used in Fourier transform infrared (FTIR) difference spectroscopy studies to monitor conformational changes associated with rhodopsin activation.36,37 Recently, azF in CCR5 was chemically ligated to triarylphosphine-conjugated FLAG peptides38 to label GPCRs in whole cells.39,40 Photolabile BzF and azF were used to map ligand binding sites on CXCR4 and CCR5 via a method called targeted photo-cross-linking.41−43 A covalent complex of CXCR4 incorporating BzF and its peptide ligand T140 revealed that the benzophenone carbonyl group needs to be ∼3 Å from the nearest atom in the ligand.42 This study was corroborated by later work using azF- and BzF-substituted CCR5 in complex with maraviroc.41 These results agree well with structural studies that indicate that the requirement is in the range of 2–4 Å.44
Here, we describe a microplate-based detection strategy, with potential for high throughput, which is based on targeted loss-of-function mutagenesis and subsequent photo-cross-linking using genetically encoded UAAs to study antibody–receptor complexes. The method relies on a sensitive cell-based enzyme-linked immunosorbent assay (ELISA) to detect fluorimetrically the transiently bound or photo-cross-linked mAb. We used the strategy to map complexes between 12G5 and CXCR4, with a focus on the role of residues in EC2. We also created maps that depict the contribution of residues in EC2 on CCR5 for binding of mAbs 2D7 and PRO 140. In our method, we describe two parallel assays: one that identifies loss-of-binding azF mutants and another that identifies photo-cross-linked residues. In essence, we use the same mutants to identify and confirm the primary “hot spot” of interaction, as well as proximal sites that may be not indispensable for binding yet allow the formation of a stable covalent adduct. This is the first description, to the best of our knowledge, of whole cell detection of photo-cross-linked mAb–GPCR complexes. Our targeted mutagenesis and photo-cross-linking approach should provide a general framework for mapping any GPCR epitope.
Materials and Methods
Materials
Antibodies were obtained from the following sources: 12G5 (eBioscience, catalog no. 14-9999), 2D7 (BD Pharmingen, catalog no. 555990), T21/8 (eBioscience, catalog no. 14-1957), PRO 140 (gift from J. P. Moore at Weill Cornell Medical College), 1D4 (National Cell Culture Center), and horseradish peroxidase (HRP)-labeled goat anti-mouse (KPL, catalog no. 474-1806) and goat anti-human (Jackson Immuno Research, catalog no. 709-036-149). Protein A/G UltraLink was purchased from Pierce (catalog no. 53132), and p-azido-l-phenylalanine (azF) was purchased from Chem-Impex International (catalog no. 06162).
Plasmids
Plasmid pSVBpUC carrying the amber suppressor tRNA gene and plasmid pcDNA3.1(+) carrying the azF aminoacyl-tRNA synthetase gene were described previously.35 Human CXCR4 and human CCR5 genes were in pcDNA3.1(+) and contained a C-terminal 1D4 epitope tag (TETSQVAPA).45 Amber mutations (TAG) were introduced using the QuikChange Lightning Site-Directed Mutagenesis Kit (Stratagene).
Expression of azF Mutants in Mammalian Cells
HEK293T cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, catalog no. 10566) supplemented with 10% fetal bovine serum (FBS) (Gemini) at 37 °C with 5% CO2. The cells were transfected simultaneously with three vectors encoding the wild-type (wt) or amber mutant receptor, suppressor tRNA, and aminoacyl-tRNA synthetase using Lipofectamine 2000 (Invitrogen). The ratio of transfected DNA in micrograms was 1:1:0.1. For transfection of the wt receptor, the microgram amount of DNA was maintained at a 1:10 ratio of the UAA mutants. The total microgram amount of DNA for all transfections was maintained at the same value by supplementing the wt and mock transfections with empty vector pcDNA3.1(+). For example, a transfection in a 10 cm dish uses 3.5 μg of amber receptor DNA, 3.5 μg of suppressor tRNA, and 0.35 μg of aminoacyl-tRNA synthetase together with 19 μL of Lipofectamine 2000. These components are diluted approximately 1:2 or 1:38 for a transfection in a well of a six-well or 96-well plate, respectively. FBS was maintained at 10% in DMEM in the presence or absence of 0.5 mM azF. All assays were performed ∼40 h after transfection.
ELISA-Based Detection
Cells expressing wt or azF mutant receptors were washed two times with PBS containing Ca2+ and Mg2+ (PBSc/m, Gibco, catalog no. 14040) supplemented with 0.5% (w/v) bovine serum albumin, Fraction V (Calbiochem, catalog no. 126575) (Pc/mB). For 12G5 and 1D4 detection, the cells were fixed with 4% (v/v) paraformaldehyde (Thermo Scientific) for 20 min at room temperature (RT). Subsequently, the wells were washed three times with Pc/mB and for 1D4 analysis only, followed by permeabilization with 0.2% Triton X-100 for 20 min at RT. After three washes with Pc/mB, the wells were incubated with the primary antibody (1 μg/mL 1D4, 1.5 μg/mL 12G5, 0.5 μg/mL T21/8, 1.5 μg/mL 2D7, or 9.1 μg/mL PRO 140) for 1.5 h on ice. After three washes with Pc/mB, the wells were incubated with the secondary antibody (HRP-coupled anti-mouse or anti-human) for 1 h at RT. The wells were washed three times with Pc/mB, followed by one wash with PBSc/m. The cells were then treated for 15 min with a detection buffer mixture containing Amplex Red (2.6 mg/mL) (Invitrogen, catalog no. A12222), 20 mM H2O2, and PBS in a 1:10:90 ratio. Fluorescence emission at 590 nm from the wells was detected via excitation at 530 nm in a multiwell fluorescence plate reader instrument (CytoFluor II, PerSeptive Biosystems).
Western Blot Detection
Cells expressing wt or azF mutant receptors from each well were harvested in PBS (Gibco, catalog no. 14200) supplemented with 0.1 mM PMSF and 10 μg/mL aprotinin and spun at 1100g for 3 min. The cell pellets were solubilized for 1 h on a nutator at 4 °C in a buffer containing 1.5% (w/v) n-dodecyl β-d-maltoside (DDM) (Anagrade, Affymetrix), 30 mM Tris-HCl (pH 7.4), 150 mM sodium chloride (NaCl), 10% (v/v) glycerol, and 5 mM EDTA and supplemented with PMSF and a protease inhibitor cocktail (Roche, catalog no. 11836170001). The lysate was centrifuged at 15800g for 10 min at 4 °C, and the supernatant fraction was treated with NuPAGE-LDS gel loading buffer (Invitrogen), supplemented with 100 mM DTT. The samples were then loaded on 4 to 12% Bis-Tris gels (Invitrogen) and electrophoresed in MOPS gel running buffer. After the proteins in the gel had been transferred to a PVDF membrane (Millipore, catalog no. IPVH00010) at 18 V for 45 min using a semidry transfer apparatus (Bio-Rad), the membrane was blocked in TBS-T [10 mM Tris-HCl buffer (pH 7.4), 150 mM NaCl, and 0.05% (v/v) Tween 20] supplemented with 5% (w/v) nonfat dry milk for 1 h at RT. The membranes were then incubated with 0.5 μg/mL 1D4 antibody in PBS supplemented with 0.5% (w/v) BSA (PB buffer) overnight at 4 °C. The next day the membrane was washed extensively in TBS-T, followed by incubation with the HRP-coupled goat anti-mouse antibody diluted 1:20000 in TBS-T supplemented with 5% (w/v) milk for 1 h at RT. Following TBS-T washes as described above, the protein bands were revealed with enhanced chemiluminescence detection reagents (Pierce) and detected with HyBlot CL autoradiography film (Denville Scientific).
ELISA-Based Detection of Photo-Cross-Linked Samples
After primary antibody incubation, the plates were placed on a cold pack and exposed to 365 nm UV light (Spectroline Maxima ML-3500S) for 15 min at 4 °C at a distance of 3 in. from the source. Subsequently, the wells were washed twice, each time with 150 μL of 50 mM glycine-HCl buffer (pH 2.5) supplemented with 500 mM NaCl (G500 buffer). Then the wells were washed once with Pc/mB, followed by secondary antibody incubation as described above.
Western Blot Detection of Photo-Cross-Linked Samples
After the cells in PBS had been harvested in the absence of protease inhibitors, the pellet was resuspended in PB buffer containing the appropriate conformation-dependent antibody. The cell suspension was then incubated while being shaken in a 12-well plate at 4 °C for 1.5 h. The plate was exposed to 365 nm UV light for 15 min at 4 °C. After the cells had been harvested, the cell pellet was washed twice with G500 buffer. This was followed by one wash with PB buffer. After the cell pellets had been solubilized, the supernatant fraction from the clarified lysate was incubated with Protein A/G UltraLink (Pierce, catalog no. 53132) overnight while being shaken at 4 °C. The next day the samples were centrifuged at 1900g for 3 min, and the supernatant fraction was aspirated. The packed resin was incubated with 1.5× LDS sample buffer, supplemented with 200 mM DTT, for 1 h at 37 °C while being shaken. The samples were centrifuged again, followed by separation of the eluted samples from the resin. The eluate was then loaded on gels and analyzed with a Western blot as described above.
Results
Expression of azF-CXCR4 Mutants
We first studied the interaction of mAb 12G58 with CXCR4 as a model system to demonstrate the feasibility of our cell-based microplate photo-cross-linking approach. We used site-directed mutagenesis to generate a panel of 19 CXCR4 mutants incorporating azF at positions 177–195 within EC2 (Figure 1a) and transiently transfected them in HEK293T cells. We confirmed expression of full-length receptors by probing with mAb 1D4, which detects the C-terminal C9 epitope tag on CXCR4. Full-length CXCR4 UAA mutants expressed to detectable levels only in the presence of added azF in the cell growth media, as indicated by Western blot analysis (Figure S1, Supporting Information). We tested the ability of 12G5 to recognize the azF-CXCR4 variants using a whole cell ELISA-based detection strategy.
Figure 1.
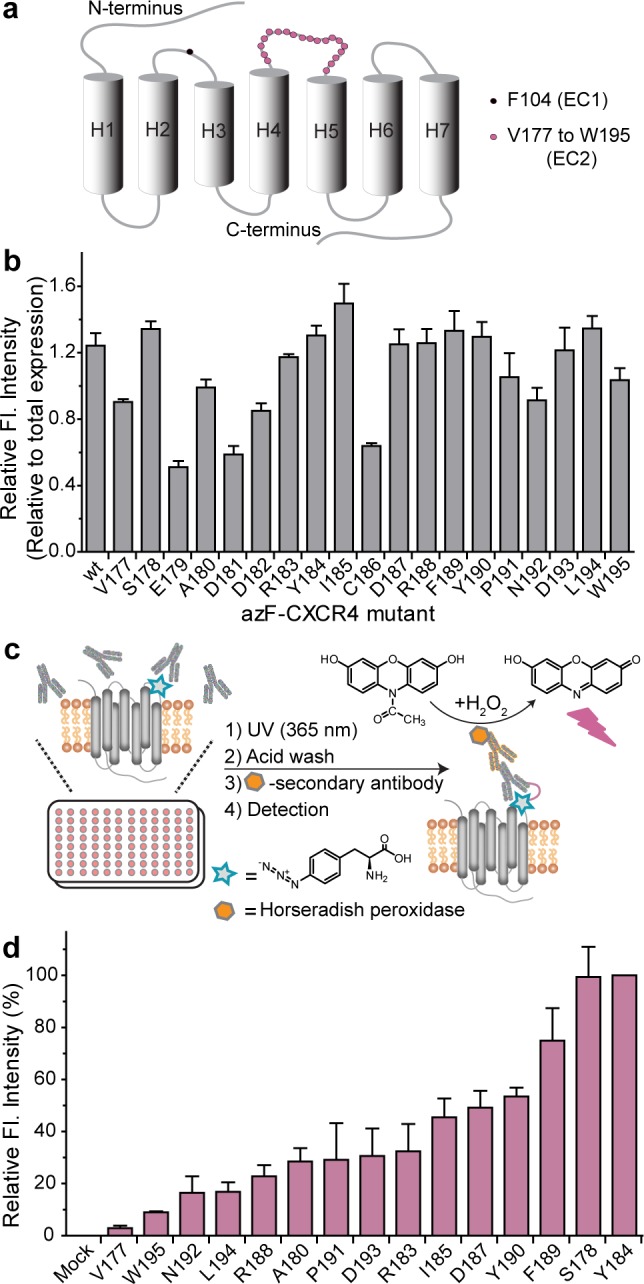
ELISA analysis of binding and photo-cross-linking of azF-CXCR4 mutants to 12G5 mAb. (a) CXCR4 schematic showing sites of azF incorporation. (b) Binding signal normalized to total expression, which is detected in permeabilized HEK293T cells using the 1D4 antibody to a C-terminal epitope. (c) Experimental scheme for photo-cross-linking and detection. HEK293T cells expressing the mutants were incubated with 12G5 and then exposed to UV light. After being washed with a low-pH, high-salt buffer, the cells were incubated with the HRP-coupled anti-mouse secondary antibody. The complexes were detected by HRP-catalyzed formation of a fluorescent product from Amplex Red. (d) Relative photo-cross-linking level obtained by normalizing to the signal from Y184azF-CXCR4. Fluorescence emission at 590 nm was detected with excitation at 530 nm in a multiwell fluorescence plate reader instrument (CytoFluor II, PerSeptive Biosystems). Error bars in panels b and d represent the standard error of the mean from three independent trials, each performed in duplicate or triplicate.
Most of the variants bound appreciably to the conformation-sensitive mAb 12G5, suggesting that azF incorporation did not significantly perturb the epitope structure (Figure 1b). CXCR4 variants incorporating azF at positions 179, 181, 182, and 186 bound poorly to 12G5. These results agree well with literature reports on the observed importance of E179, D181, and D182 for 12G5 binding.11,15−17 Most of the previously reported data were obtained from FACS analyses of alanine scanning mutants in COS, HEK293T, and BHK21 cells. Because C186 and C109 form a disulfide bond in the crystal structure,28 azF substitution at position 186 presumably disrupted the tertiary structure of the receptor, thereby weakening 12G5 binding.11,57
Cell-Based Photo-Cross-Linking of mAb 12G5 to azF-CXCR4 Mutants
To map the 12G5 mAb epitope on CXCR4 with precision, we evaluated the cross-linking efficiency of the UAA mutants to 12G5 in a cell-based ELISA format (Figure 1c). The azF-CXCR4 mutants were expressed in a 96-well plate, incubated with 12G5, and subsequently exposed to UV light. This step was followed by a stringent wash with a low-pH glycine buffer supplemented with salt to remove the transiently bound but non-cross-linked antibody from the photo-cross-linked, covalently associated antibody. Low-pH acid washes have been employed to strip cell surface-bound 12G558,59 and chemokines60,61 in flow cytometric assays. In our assay, this wash step is conducted to circumvent the necessity of solubilizing the cells and analyzing the samples by electrophoresis and Western blotting to separate and quantify noncovalent and covalent complexes after photo-cross-linking. As a result, our photo-cross-linking experiments proceed rapidly to detection, as opposed to the long time scales of gel electrophoresis-based analyses.
The receptor–antibody complexes were then fluorimetrically detected as described above. The results depicted in ascending order of signal strength showed the most intense signals when azF was introduced at positions 184 and 178, followed by position 189 (Figure 1d), under similar total expression levels (Figure S2, Supporting Information). There was no detectable signal above mock when azF was at positions 179, 181, 182, and 186 (data not shown). In comparison, sites in EC1 (F104), TM5 (L210), and IC3 (G231) did not produce corresponding signals under the same conditions (Figure S7, Supporting Information). It was interesting to note that positions 178 and 184 surround the primary 12G5 interaction epitope identified by our azF scan and previous reports of Ala scans, constituting residues 179, 181, and 182 (Figure 4).
Figure 4.
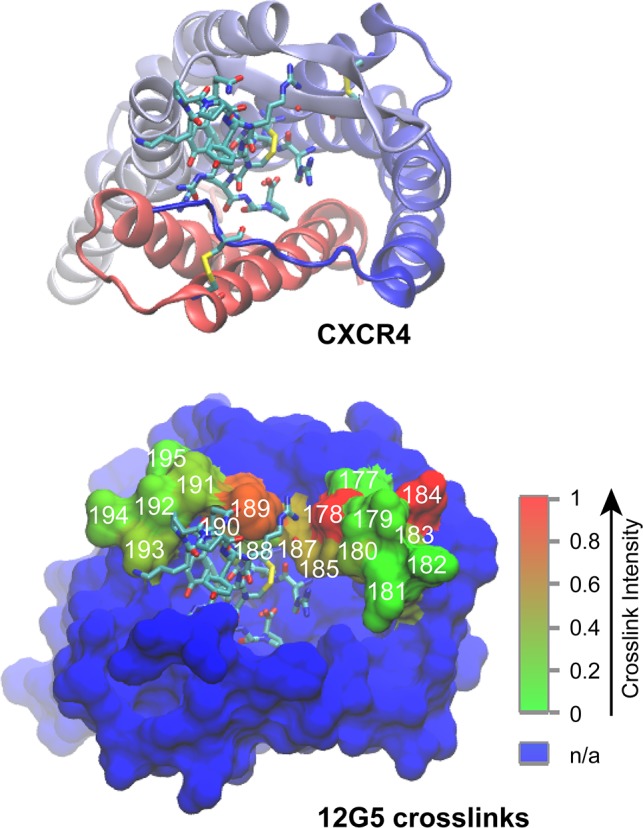
Interpretation of cross-linking data according to the crystal structure of the CXCR4–CVX15 complex (Protein Data Bank entry 3OEU).28 Cartoon representation of CXCR4 with the N-terminus colored blue, fading to red at the C-terminus. The cyclopeptide ligand CVX15 and disulfide-linked Cys residues are shown as sticks (cyan carbons). Surface representation of CXCR4 with residues cross-linked to 12G5 depicted in red fading to green. Residues substituted with azF are numbered in white. The color bar indicates the range of relative cross-linking signals from non-cross-linked sites (green) to cross-linked sites (red). The blue surface corresponds to residues not tested in this study. Molecular graphics were prepared with VMD version 1.9.1.66
We corroborated our results with Western blot analysis, where detection proceeded after immunoprecipitation of the antibody-containing complexes with Protein A/G. We detected UV exposure-dependent protein bands only in the S178-, Y184-, and F189azF-CXCR4 mutants, whereas wt, F104azF-CXCR4 (in EC1), and C186azF-CXCR4 did not produce similar bands under the same conditions (Figure 2 and Figure S8a, Supporting Information). Considering that CXCR4 has a molecular mass of ∼40 kDa and mAb heavy and light chains have molecular masses of 50 and 25 kDa, respectively, we speculate that the observed UV-dependent bands are complexes between CXCR4 dimers and mAb heavy chains.
Figure 2.
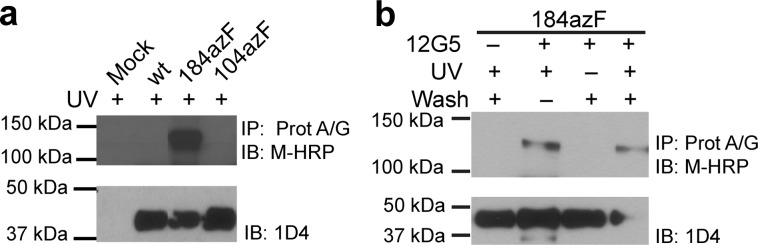
Western blot analysis of photo-cross-linking of CXCR4 variants to 12G5. Detergent-solubilized cells were incubated with Protein A/G UltraLink resin, and immunoprecipitated samples were probed with the HRP-coupled anti-mouse secondary antibody. Receptor expression in crude lysates was detected with the 1D4 antibody to a C-terminal epitope. (a) UV-exposed samples containing CXCR4 incorporating azF at position 104 (EC1) or 184 (EC2) are compared to wt and mock transfections. Specific bands appeared only in the Y184azF-CXCR4 lane. (b) Appearance of unique bands in the Y184azF-CXCR4 sample tested in the presence and absence of 12G5, UV exposure, and subsequent stringent washes. The bands are specific to 12G5 incubation and UV exposure. Blots are representative of at least two independent trials.
Expression of azF-CCR5 Mutants
We were interested in comparing the epitope maps of mAbs 2D79 and PRO 14010 in the CCR5 EC2 loop. Amber mutations were introduced at 25 sites from position 166 to 189 (in EC2) and 263 (in EC3, for comparison), and azF was incorporated (Figure 3a). We detected full-length expressed receptors only in the presence of added azF, as indicated by Western blot analysis (Figure S3, Supporting Information). Next we measured the interactions between 2D7 and the panel of azF-CCR5 mutants. Cell surface expression was monitored with T21/8,46 an antibody targeting an N-terminal epitope of CCR5 (Figure S4b, Supporting Information). Under similar surface expression levels of receptors, 2D7 bound reasonably well to most of the CCR5 mutants, with a loss of function observed with receptors incorporating azF at positions 171–173 and 178 (Figure S4a, Supporting Information).
Figure 3.
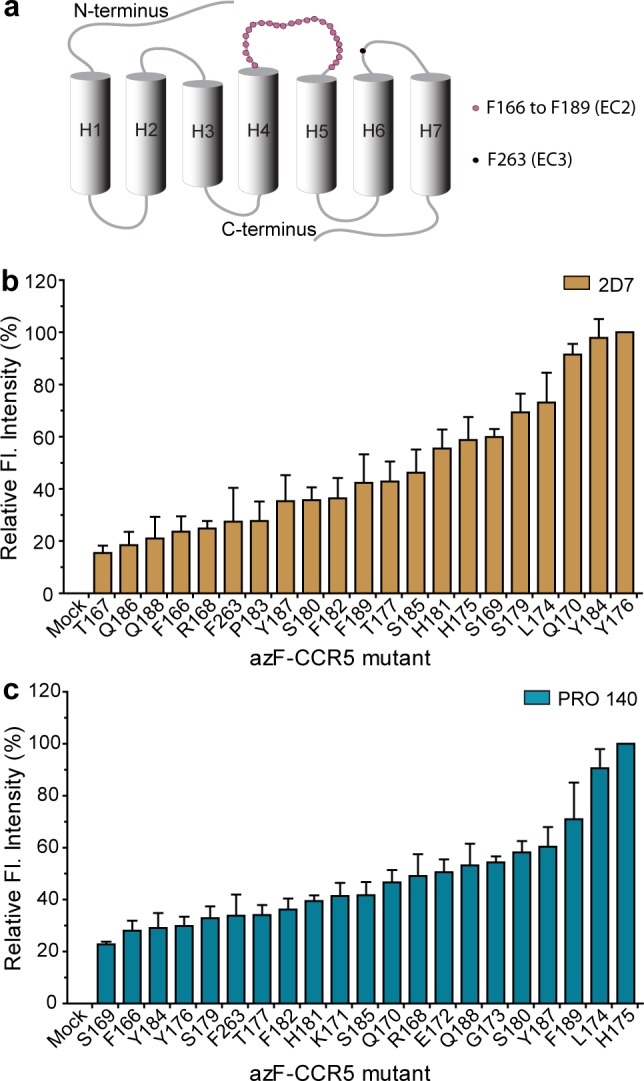
ELISA analysis of binding and photo-cross-linking of azF-CCR5 mutants, expressed in HEK293T cells. (a) CCR5 schematic showing sites of azF incorporaton. (b) Photo-cross-linking to 2D7, relative levels obtained by normalizing to Y176azF-CCR5. (c) Photo-cross-linking to PRO 140, relative levels obtained by normalizing to H175azF-CCR5. Error bars represent the standard error of the mean from three or four independent trials, each performed in duplicate or triplicate.
Cell-Based Photo-Cross-Linking of 2D7 and PRO 140 to azF-CCR5 Mutants
We then extended our photo-cross-linking strategy to study the epitope maps of 2D7 and PRO 140 on CCR5. The azF-CCR5 mutants were expressed in 96-well plates and incubated with T21/8, 2D7, or PRO 140. After UV treatment of the wells containing 2D7 and PRO 140 antibody-bound receptors, the plates were washed and the antibody–receptor complexes were fluorimetrically detected as described for the CXCR4–12G5 photo-cross-linking experiment.
We observed that azF at positions 170, 176, and 184 in CCR5 cross-linked most efficiently to 2D7 (Figure 3b). On the other hand, azF at positions 174 and 175 cross-linked most efficiently to PRO 140 (Figure 3c). For both CXCR4 and CCR5, mutants that did not bind appreciably to these mAbs did not produce detectable signals under cross-linking conditions (Figures S4–S6, Supporting Information).
Discussion
The HIV infection process is initiated by membrane fusion with host cells, whereby the envelope glycoprotein (Env) interacts with the CD4 receptor.21 This binding event triggers conformational changes in Env, followed by interactions of Env with coreceptor CCR5 or CXCR4. Strains of HIV-1 are classified as M-tropic (uses CCR5 for entry), T-tropic (uses CXCR4 for entry), or dual-tropic based on the expression pattern of chemokine receptors on the cell types they infect. Dual-tropic strains use both coreceptors for entry. Current antiretroviral therapy approaches include blocking viral entry with inhibitors that target CCR5 and CXCR4.47 To this end, several small molecule and peptide agonists and antagonists have been developed and characterized for their efficacy as potential HIV therapeutics.48 Additionally, highly specific mAbs have provided an alternative anti-HIV approach.1,49 Moreover, nanobodies, single variable domains of heavy chain camelid antibodies, are being investigated for their potential to target and modulate the function of receptors like CXCR4.50 At least 30 FDA-approved mAb drugs are currently being used to treat cancers, osteoporosis, macular degeneration, and autoimmune diseases.51,52 One challenge of using mAbs to target coreceptors is the possibility of heterologous post-translational modifications, such as tyrosine sulfation.53
Determining the epitope of a mAb is not always straightforward. In some cases, information can be obtained from biochemical assays like alanine scanning mutagenesis that help identify interaction hot spots in the binding interface.54−56 A high-throughput alanine scanning shotgun mutagenesis strategy was used to map the epitopes of several conformation-dependent mAbs on CCR5.19 Studies of antibody–receptor complexes have been complemented with other methodologies like site-directed masking,24 phage and bacterial surface display,25,26 and CLIPS technology.18 In CLIPS, conformational peptide libraries are created for detecting conformational, discontinuous, and complex epitopes on oligomeric proteins. A library of linear peptides is chemically converted into a matrix of conformationally constrained CLIPS peptides, and the potential epitopes are identified by monitoring binding affinity with the antibody of interest.
Here we employed a method for expressing GPCRs with site-specifically introduced azF using an orthogonal aa-RS/suppressor tRNA pair.35,36 The photolabile azF can serve as a chemical reporter of protein interaction sites. Upon UV irradiation, aryl azides form nitrenes that can insert into C–H and N–H bonds or undergo ring expansion to form adducts with amines.33 We sought to identify positions in CXCR4 and CCR5 that when substituted with azF would photo-cross-link to their respective mAbs 12G5, 2D7, and PRO 140 upon UV irradiation.
We first demonstrated the feasibility of a microplate cell-based, photo-cross-linking approach to investigate the interaction between 12G5 and CXCR4 at a single-residue level. The amber codon was introduced successively from position 177 to 195 in EC2 to incorporate azF (Figure 1a). After the expression of full-length receptors had been verified by Western blot analysis, their interaction with 12G5 was interrogated with a whole cell ELISA in a 96-well plate format. We next adapted the ELISA detection strategy to measure whether UV irradiation of the azF CXCR4 variants in the presence of 12G5 produced covalent cross-links between the receptor and mAb at the cell surface. We observed significant cross-links at positions 178, 184, and 189 (Figure 1d). The results are graphically presented on a structural model of CXCR4 (Figure 4).
We then studied the cross-linking maps of 2D7 and PRO 140 on the molecular surface of CCR5. Similar to the 12G5–CXCR4 cross-linking results, we identified positions 170 and 176, which surround the putative primary 2D7 binding epitope constituting K171 and E172, and also found position 184 (Figures 3b and 5). Immunoblot analysis also supported this observation with unique UV-dependent protein bands (Figure S8b, Supporting Information). The sizes of the UV exposure-dependent bands are consistent with complexes between CCR5 monomers and 2D7 heavy chains for Q170 and Y184azF, and a light chain for Y176azF. It is noteworthy that two different positions, 174 and 175, formed the strongest cross-links with PRO 140, a humanized form of the PA14 antibody (Figure 3c). We also note that R168 and Y176 are essential in PA14 recognition, as previously determined by alanine scanning mutagenesis.10 The two positions identified in our photo-cross-linking screen, 174 and 175, are proximal to the key residue Y176 (Figure 5). Interestingly, PA14 exhibits a multidomain epitope, spanning D2 in the N-terminus and residue D95 in EC1, in addition to EC2 residues.
Figure 5.
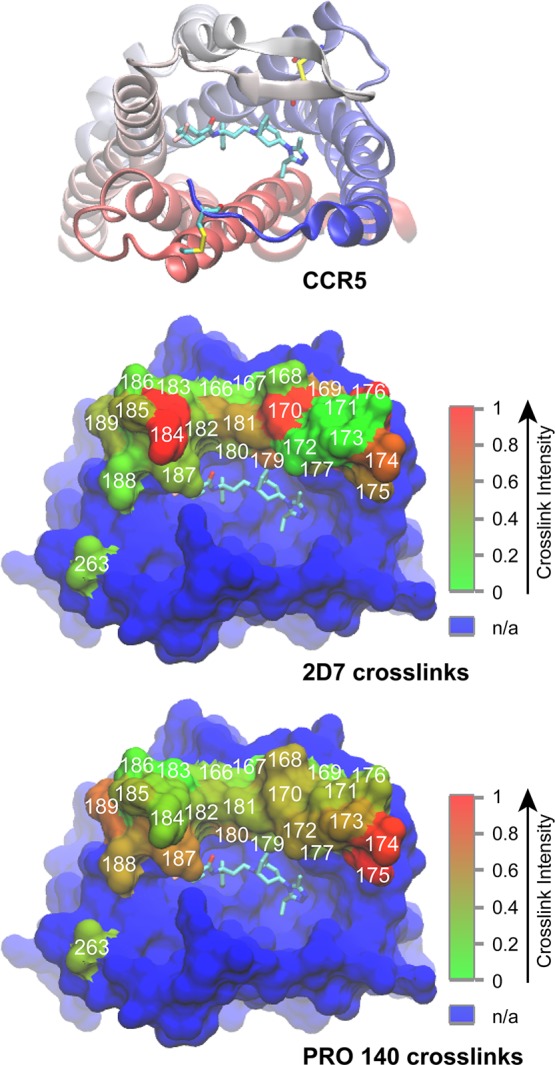
Interpretation of cross-linking data according to the crystal structure of the CCR5–maraviroc complex (Protein Data Bank entry 4MBS).29 Ribbon diagram of CCR5 as viewed from the extracellular surface of the receptor with the N-terminus colored blue, fading to red at the C-terminus. Maraviroc and disulfide-linked Cys residues are shown as sticks (cyan carbons). Surface representation of CCR5 with residues cross-linked to 2D7 (middle) and PRO 140 (bottom) colored red fading to green. Residues substituted with azF are numbered in white. The color bar indicates the range of relative cross-linking signals from non-cross-linked sites (green) to cross-linked sites (red). The blue surface corresponds to residues not tested in this study. Molecular graphics were prepared with VMD version 1.9.1.66
Various studies using alanine point mutants and synthetic peptide scaffolds have collectively reported the importance of amino acid residues Q170, K171, E172, G173, W190, T195, and L196 of CCR5 EC2 in 2D7 recognition.10,12,18−20 The positions common to all of the previous studies, and therefore perhaps the most important, are K171 and E172. Upon evaluating 2D7 recognition of the azF-CCR5 variants, we found reasonable binding of 2D7 to all azF variants, except those in positions 171–173 and 178. The loss-of-function positions were consistent with literature reports on the key role of K171, E172, and G173 for 2D7 recognition and the importance of C178 in forming a necessary disulfide bond.18,62,63
Although 2D7 and PA14/PRO 140 are both potent HIV-inhibitory mAbs,4 it is interesting that there is a lack of synergy between PA14 and 2D7 in inhibiting HIV-1 viral fusion to the host cells.10 We find that the two mAbs have nonoverlapping epitopes in EC2 of CCR5, and PRO 140 additionally recognizes residues in the N-terminus.4 The results of the photo-cross-linking screen with CCR5 described here highlight unique azF-substituted positions upon interrogation with the two different mAbs. Collectively, this information points to alternate binding and viral inhibition modes of these two mAbs. Upon closely examining the EC2 photo-cross-link maps in CCR5, we see that L174azF-CCR5 cross-links to 2D7 with moderate efficiency and is a primary photo-cross-linked position with PRO 140. This observation leads us to postulate that there is a potential overlap in the epitopes surrounding residue 174. In fact, the crystal structure of CCR5 bound to maraviroc highlights the spatial proximity of Y176,29 which is important in PA14 recognition,10 and L174 (Figure 5). Also, L174 is close to the 2D7 binding epitope comprising residues K171, E172, and G173.10,18−20,63 Considering the HIV inhibitory potential of both mAbs, we hypothesize the possibility of designing a synthetic ligand with druglike properties that would span interaction sites in common to 2D7 and PRO 140 recognition.
It is interesting to note for all the mAbs tested that photo-cross-linked residues identified in two chemokine receptors, CXCR4 and CCR5, were in general clustered around known hot spots of interaction. These hot spots have been reported earlier and also identified in two of our azF scans. We previously observed a similar pattern of key binding residues surrounded by efficiently photo-cross-linking residues in studies of the T140–CXCR4 complex.42 Here we find that 12G5 and 2D7 produced similar patterns of cross-linking, with the strongest cross-linked sites spanning two regions, one surrounding the core epitopes and another in the C-terminal halves of the EC2 loop. Taking a closer look at this secondary site, we note that Y190 in CXCR4 is reportedly important for 12G5 recognition. Therefore, it is not surprising that position 189, which is located close to Y190, would produce a cross-link. However, position 184 in the second half of the CCR5 EC2, which cross-linked efficiently with 2D7, does not have any known proximal hot spots of interaction. 2D7 has a predicted split epitope based on an earlier phage display-identified peptide sequence containing residues 170-QKEGL-174 and 195-TL-196 corresponding to two distal regions of the CCR5 EC2 loop, which was shown to bind efficiently to 2D7.12 A clear limitation of phage display experiments is that the mapping of the linear epitope on the phage to the complex protein epitope on the receptor is not unequivocal. In fact, the maraviroc-bound structure of CCR5 shows that residues T195 and L196, which were identified as part of the epitope in the phage display study, are buried in TM5 and perhaps not easily accessible to the mAb. Instead, our experiments suggest that the secondary mAb contact site may be closer to Y184.
We and others have used photo-cross-linking as a tool to form covalent adducts of protein co-complexes. In general, residues that photo-cross-link are likely to be involved in binding interactions that are at the periphery of the protein–protein binding interface and are therefore weaker than those in the core hot spot. Interaction hot spots are characterized by high-affinity interactions that are driven by burial of interacting residues excluding bulk solvent. Computational biologists discuss the presence of an “O-ring” around these hot spots.55,64 The O-ring comprises hydrophobic residues that exclude water and thereby determine the solvation pattern at the binding interface. This underscores their importance as a surrounding seal of contacting residues that are not detected by alanine scans but are still important to the affinity, albeit with a smaller contribution. Comprehensive knowledge of all these contacting and interacting residues from complementary techniques is therefore important for the creation of detailed structural and biochemical maps of protein–protein interactions that can facilitate drug design efforts.
Recent work from our group also highlights the importance of photo-cross-linking in elucidating the binding site of small molecule and peptide ligands on GPCRs.41−43 Specifically, the photo-cross-linking data were evaluated with existing ligand binding models to provide information about spatial orientation in the binding site. Identifying suitable sites in target GPCRs for photo-cross-linking to mAbs can also be applied to structural studies. The crystal structure of the photo-cross-linked complex of the oncoprotein gankyrin with the C-terminal domain of the S6 proteasomal protein has been reported to 2.05 Å resolution.44 This study used BzF as the photolabile residue and highlights structural requirements for forming the cross-link. As another case example, the crystal structure of neurotensin receptor NTSR1 has been reported in an activelike conformation bound to the C-terminal portion of the agonist NTS.65 Extensive alanine scanning mutagenesis was employed to identify stabilizing mutations to facilitate crystallization in the agonist-bound form.
In summary, we have developed a microplate-based photo-cross-linking strategy based on genetic encoding of azF into chemokine receptors to facilitate structural mapping of receptor–mAb interactions. Antibodies 12G5, 2D7, and PRO 140 used in this study are known to interact, at least in part, with residues in EC2 of their respective target receptors. Alanine scanning mutagenesis studies have been used previously to pinpoint loss-of-binding mutations, which correspond to hot spots of interaction between protein partners. Our method of targeted loss-of-function mutagenesis in combination with photo-cross-linking is designed to study the interaction of the antibody with residues that are substituted with photolabile UAAs. We propose using both screens in a complementary fashion to map the binding surface epitope of a mAb on a target cell-surface receptor. In principle, a photo-cross-link is formed upon UV exposure only when the UAA does not disrupt protein interactions and indicates a threshold distance between the two protein partners. On the basis of previous reports, the distance dependence of the photo-cross-linking reaction is predicted to be in the range of 3 Å.41,42,67 In future work, cross-linking information can be further enhanced with mass spectrometric analyses to identify specific cross-linked residues of the antibody. We envision the possibility of a detailed analysis of the binding mode and orientation even with limited availability of co-complex crystal structures, a critical step in the design of high-potency therapeutics. Many transient interactions and low-affinity complexes are difficult to study by alanine scanning or crystallography. “Stapling”’ the GPCR–mAb interaction with the chemical cross-links may facilitate such studies. Our microplate-based format of probing antibody epitopes on receptors can be generalized to any GPCR–antibody interaction of choice and also to map the epitopes of extracellular ligands.
Acknowledgments
We thank Yamina A. Berchiche, Manija A. Kazmi, Saranga Naganathan, and Minyoung Park for helpful discussions and scientific assistance. We also thank John P. Moore for providing us with PRO 140 and Raymond C. Stevens for providing Protein Data Bank coordinates for CCR5 in advance of publication.
Glossary
Abbreviations
- azF
p-azido-l-phenylalanine
- CCR5
CC chemokine receptor 5
- CXCR4
CXC chemokine receptor 4
- EC
extracellular
- ELISA
enzyme-linked immunosorbent assay
- GPCR
G protein-coupled receptor
- HIV
human immunodeficiency virus
- HRP
horseradish peroxidase
- mAb
monoclonal antibody
- UAA
unnatural amino acid
- wt
wild type.
Supporting Information Available
Experimental methods and supplementary figures. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
This paper was published ASAP on February 18, 2014, with an incorrect version of Figure S1 in the Supporting Information. The corrected version was reposted on February 24, 2014.
S.R.-S. was supported in part by National Institutes of Health Training Grant T32 DK-07313. We also thank the Danica Foundation and the Crowley Family Fund.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Herr D. R. (2012) Potential use of G protein-coupled receptor-blocking monoclonal antibodies as therapeutic agents for cancers. Int. Rev. Cell Mol. Biol. 45–81. [DOI] [PubMed] [Google Scholar]
- Nelson A. L.; Dhimolea E.; Reichert J. M. (2010) Development trends for human monoclonal antibody therapeutics. Nat. Rev. Drug Discovery 9, 767–774. [DOI] [PubMed] [Google Scholar]
- Schall T. J.; Proudfoot A. E. I. (2011) Overcoming hurdles in developing successful drugs targeting chemokine receptors. Nat. Rev. Immunol. 11, 355–363. [DOI] [PubMed] [Google Scholar]
- Olson W. C.; Jacobson J. M. (2009) CCR5 monoclonal antibodies for HIV-1 therapy. Curr. Opin. HIV AIDS 4, 104–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strizki J. M.; Turner J. D.; Collman R. G.; Hoxie J.; GonzalezScarano F. (1997) A monoclonal antibody (12G5) directed against CXCR-4 inhibits infection with the dual-tropic human immunodeficiency virus type 1 isolate HIV-1(89.6) but not the T-tropic isolate HIV-1(HxB). J. Virol. 71, 5678–5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight A.; Wilkinson D.; Simmons G.; Talbot S.; Picard L.; Ahuja M.; Marsh M.; Hoxie J. A.; Clapham P. R. (1997) Inhibition of human immunodeficiency virus fusion by a monoclonal antibody to a coreceptor (CXCR4) is both cell type and virus strain dependent. J. Virol. 71, 1692–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji C.; Brandt M.; Dioszegi M.; Jekle A.; Schwoerer S.; Challand S.; Zhang J.; Chen Y.; Zautke L.; Achhammer G.; Baehner M.; Kroetz S.; Heilek-Snyder G.; Schumacher R.; Cammack N.; Sankuratri S. (2007) Novel CCR5 monoclonal antibodies with potent and broad-spectrum anti-HIV activities. Antiviral Res. 74, 125–137. [DOI] [PubMed] [Google Scholar]
- Endres M. J.; Clapham P. R.; Marsh M.; Ahuja M.; Turner J. D.; McKnight A.; Thomas J. F.; StoebenauHaggarty B.; Choe S.; Vance P. J.; Wells T. N. C.; Power C. A.; Sutterwala S. S.; Doms R. W.; Landau N. R.; Hoxie J. A. (1996) CD4-independent infection by HIV-2 is mediated by Fusin/CXCR4. Cell 87, 745–756. [DOI] [PubMed] [Google Scholar]
- Wu L. J.; LaRosa G.; Kassam N.; Gordon C. J.; Heath H.; Ruffing N.; Chen H.; Humblias J.; Samson M.; Parmentier M.; Moore J. P.; Mackay C. R. (1997) Interaction of chemokine receptor CCR5 with its ligands: Multiple domains for HIV-1 gp120 binding and a single domain for chemokine binding. J. Exp. Med. 186, 1373–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson W. C.; Rabut G. E. E.; Nagashima K. A.; Tran D. N. H.; Anselma D. J.; Monard S. P.; Segal J. P.; Thompson D. A. D.; Kajumo F.; Guo Y.; Moore J. P.; Maddon P. J.; Dragic T. (1999) Differential inhibition of human immunodeficiency virus type 1 fusion, gp120 binding, and CC-chemokine activity by monoclonal antibodies to CCR5. J. Virol. 73, 4145–4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnec X.; Quan L.; Olson W. C.; Hazan U.; Dragic T. (2005) Anti-CXCR4 monoclonal antibodies recognizing overlapping epitopes differ significantly in their ability to inhibit entry of human immunodeficiency virus type 1. J. Virol. 79, 1930–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana S.; Kennedy M.; King L. R.; Golding H. (2005) Identification of a linear peptide recognized by monoclonal antibody 2D7 capable of generating CCR5-specific antibodies with human immunodeficiency virus-neutralizing activity. J. Virol. 79, 6791–6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murga J. D.; Franti M.; Pevear D. C.; Maddon P. J.; Olson W. C. (2006) Potent antiviral synergy between monoclonal antibody and small-molecule CCR5 inhibitors of human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 50, 3289–3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A.; Ketas T. J.; Nagashima K. A.; Zhao L.; Cilliers T.; Morris L.; Moore J. P.; Maddon P. J.; Olson W. C. (2001) Potent, broad-spectrum inhibition of human immunodeficiency virus type 1 by the CCR5 monoclonal antibody PRO 140. J. Virol. 75, 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baribaud F.; Edwards T. G.; Sharron M.; Brelot T.; Heveker N.; Price K.; Mortari F.; Alizon M.; Tsang M.; Doms R. W. (2001) Antigenically distinct conformations of CXCR4. J. Virol. 75, 8957–8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brelot A.; Heveker N.; Montes M.; Alizon M. (2000) Identification of residues of CXCR4 critical for human immunodeficiency virus coreceptor and chemokine receptor activities. J. Biol. Chem. 275, 23736–23744. [DOI] [PubMed] [Google Scholar]
- Brelot A.; Heveker N.; Adema K.; Hosie M. J.; Willett B.; Alizon M. (1999) Effect of mutations in the second extracellular loop of CXCR4 on its utilization by human and feline immunodeficiency viruses. J. Virol. 73, 2576–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmerman P.; Puijk W. C.; Meloen R. H. (2009) Functional reconstitution of structurally complex epitopes using CLIPSTM technology. Biopolymers 92, 351–351. [Google Scholar]
- Paes C.; Ingalls J.; Kampani K.; Sulli C.; Kakkar E.; Murray M.; Kotelnikov V.; Greene T. A.; Rucker J. B.; Doranz B. J. (2009) Atomic-Level Mapping of Antibody Epitopes on a GPCR. J. Am. Chem. Soc. 131, 6952–6954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.; Rao E.; Dioszegi M.; Kondru R.; DeRosier A.; Chan E.; Schwoerer S.; Cammack N.; Brandt M.; Sankuratri S.; Ji C. (2007) The second extracellular loop of CCR5 contains the dominant epitopes for highly potent anti-human immunodeficiency virus monoclonal antibodies. Antimicrob. Agents Chemother. 51, 1386–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasse P. J. (2012) The molecular basis of HIV entry. Cell. Microbiol. 14, 1183–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.; Yoder A. (2009) Chemokine coreceptor signaling in HIV-1 infection and pathogenesis. PLoS Pathog. 5, e1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didigu C. A.; Doms R. W. (2012) Novel Approaches to Inhibit HIV Entry. Viruses 4, 309–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ipsen H.; Henmar H.; Bolwig C.; Duffort O.; Barber D.; Polo F.; Larsen J. (2004) Mapping of Der p 2 antibody binding epitopes by site directed mutagenesis. J. Allergy Clin. Immunol. 113, S144. [Google Scholar]
- Rockberg J.; Lofblom J.; Hjelm B.; Uhlen M.; Stahl S. (2008) Epitope mapping of antibodies using bacterial surface display. Nat. Methods 5, 1039–1045. [DOI] [PubMed] [Google Scholar]
- Sidhu S. S. (2000) Phage display in pharmaceutical biotechnology. Curr. Opin. Biotechnol. 11, 610–616. [DOI] [PubMed] [Google Scholar]
- Wyatt R.; Kwong P. D.; Desjardins E.; Sweet R. W.; Robinson J.; Hendrickson W. A.; Sodroski J. G. (1998) The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393, 705–711. [DOI] [PubMed] [Google Scholar]
- Wu B. L.; Chien E. Y. T.; Mol C. D.; Fenalti G.; Liu W.; Katritch V.; Abagyan R.; Brooun A.; Wells P.; Bi F. C.; Hamel D. J.; Kuhn P.; Handel T. M.; Cherezov V.; Stevens R. C. (2010) Structures of the CXCR4 Chemokine GPCR with Small-Molecule and Cyclic Peptide Antagonists. Science 330, 1066–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q.; Zhu Y.; Li J.; Chen Z.; Han G. W.; Kufareva I.; Li T.; Ma L.; Fenalti G.; Li J.; Zhang W.; Xie X.; Yang H.; Jiang H.; Cherezov V.; Liu H.; Stevens R. C.; Zhao Q.; Wu B. (2013) Structure of the CCR5 Chemokine Receptor-HIV Entry Inhibitor Maraviroc Complex. Science 341, 1387–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.; Parrish A. R.; Wang L. (2009) Expanding the Genetic Code for Biological Studies. Chem. Biol. 16, 323–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daggett K. A.; Sakmar T. P. (2011) Site-specific in vitro and in vivo incorporation of molecular probes to study G-protein-coupled receptors. Curr. Opin. Chem. Biol. 15, 392–398. [DOI] [PubMed] [Google Scholar]
- Chin J. W.; Martin A. B.; King D. S.; Wang L.; Schultz P. G. (2002) Addition of a photo-cross-linking amino acid to the genetic code of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 99, 11020–11024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin J. W.; Santoro S. W.; Martin A. B.; King D. S.; Wang L.; Schultz P. G. (2002) Addition of p-azido-l-phenylaianine to the genetic code of Escherichia coli. J. Am. Chem. Soc. 124, 9026–9027. [DOI] [PubMed] [Google Scholar]
- Chin J. W.; Cropp T. A.; Anderson J. C.; Mukherji M.; Zhang Z. W.; Schultz P. G. (2003) An expanded eukaryotic genetic code. Science 301, 964–967. [DOI] [PubMed] [Google Scholar]
- Ye S.; Kohrer C.; Huber T.; Kazmi M.; Sachdev P.; Yan E. C. Y.; Bhagat A.; RajBhandary U. L.; Sakmar T. P. (2008) Site-specific incorporation of keto amino acids into functional G protein-coupled receptors using unnatural amino acid mutagenesis. J. Biol. Chem. 283, 1525–1533. [DOI] [PubMed] [Google Scholar]
- Ye S. X.; Huber T.; Vogel R.; Sakmar T. P. (2009) FTIR analysis of GPCR activation using azido probes. Nat. Chem. Biol. 5, 397–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S. X.; Zaitseva E.; Caltabiano G.; Schertler G. F. X.; Sakmar T. P.; Deupi X.; Vogel R. (2010) Tracking G-protein-coupled receptor activation using genetically encoded infrared probes. Nature 464, 1386–1390. [DOI] [PubMed] [Google Scholar]
- Sletten E. M.; Bertozzi C. R. (2009) Bioorthogonal Chemistry: Fishing for Selectivity in a Sea of Functionality. Angew. Chem., Int. Ed. 48, 6974–6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naganathan S.; Ye S.; Sakmar T. P.; Huber T. (2013) Site-Specific Epitope Tagging of G Protein-Coupled Receptors by Bioorthogonal Modification of a Genetically Encoded Unnatural Amino Acid. Biochemistry 52, 1028–1036. [DOI] [PubMed] [Google Scholar]
- Huber T.; Naganathan S.; Tian H.; Ye S.; Sakmar T. P. (2013) Unnatural Amino Acid Mutagenesis of GPCRs Using Amber Codon Suppression and Bioorthogonal Labeling. Methods Enzymol. 520, 281–305. [DOI] [PubMed] [Google Scholar]
- Grunbeck A.; Huber T.; Abrol R.; Trzaskowski B.; Goddard W. A. III; Sakmar T. P. (2012) Genetically Encoded Photo-cross-linkers Map the Binding Site of an Allosteric Drug on a G Protein-Coupled Receptor. ACS Chem. Biol. 7, 967–972. [DOI] [PubMed] [Google Scholar]
- Grunbeck A.; Huber T.; Sachdev P.; Sakmar T. P. (2011) Mapping the Ligand-Binding Site on a G Protein-Coupled Receptor (GPCR) Using Genetically Encoded Photocross-linkers. Biochemistry 50, 3411–3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunbeck A.; Huber T.; Sakmar T. P. (2013) Mapping a Ligand Binding Site Using Genetically Encoded Photoactivatable Cross-linkers. Methods Enzymol. 520, 307–322. [DOI] [PubMed] [Google Scholar]
- Sato S.; Mimasu S.; Sato A.; Hino N.; Sakamoto K.; Umehara T.; Yokoyama S. (2011) Crystallographic Study of a Site-Specifically Cross-Linked Protein Complex with a Genetically Incorporated Photoreactive Amino Acid. Biochemistry 50, 250–257. [DOI] [PubMed] [Google Scholar]
- Franke R. R.; Sakmar T. P.; Oprian D. D.; Khorana H. G. (1988) A single amino acid substitution in rhodopsin (lysine-248-leucine) prevents activation of transducin. J. Biol. Chem. 263, 2119–2122. [PubMed] [Google Scholar]
- Pollok-Kopp B.; Schwarze K.; Baradari V. K.; Oppermann M. (2003) Analysis of ligand-stimulated CC chemokine receptor 5 (CCR5) phosphorylation in intact cells using phosphosite-specific antibodies. J. Biol. Chem. 278, 2190–2198. [DOI] [PubMed] [Google Scholar]
- Ray N.; Dorns R. W. (2006) HIV-1 coreceptors and their inhibitors. Curr. Top. Microbiol. Immunol. 303, 97–120. [DOI] [PubMed] [Google Scholar]
- Seibert C.; Sakmar T. P. (2004) Small-molecule antagonists of CCR5 and CXCR4: A promising new class of anti-HIV-1 drugs. Curr. Pharm. Des. 10, 2041–2062. [DOI] [PubMed] [Google Scholar]
- Webb D. R.; Handel T. M.; Kretz-Rommel A.; Stevens R. C. (2013) Opportunities for functional selectivity in GPCR antibodies. Biochem. Pharmacol. 85, 147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahnichen S.; Blanchetot C.; Maussang D.; Gonzalez-Pajuelo M.; Chow K. Y.; Bosch L.; De Vrieze S.; Serruys B.; Ulrichts H.; Vandevelde W.; Saunders M.; De Haard H. J.; Schols D.; Leurs R.; Vanlandschoot P.; Verrips T.; Smit M. J. (2010) CXCR4 nanobodies (VHH-based single variable domains) potently inhibit chemotaxis and HIV-1 replication and mobilize stem cells. Proc. Natl. Acad. Sci. U.S.A. 107, 20565–20570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scolnik P. A. (2009) mAbs: A business perspective. MAbs 1, 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliwkowski M. X.; Mellman I. (2013) Antibody Therapeutics in Cancer. Science 341, 1192–1198. [DOI] [PubMed] [Google Scholar]
- Seibert C.; Sakmar T. P. (2008) Toward a framework for sulfoproteomics: Synthesis and characterization of sulfotyrosine-containing peptides. Biopolymers 90, 459–477. [DOI] [PubMed] [Google Scholar]
- Cunningham B. C.; Wells J. A. (1989) High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science 244, 1081–1085. [DOI] [PubMed] [Google Scholar]
- Moreira I. S.; Fernandes P. A.; Ramos M. J. (2007) Hot spots: A review of the protein–protein interface determinant amino-acid residues. Proteins: Struct., Funct., Bioinf. 68, 803–812. [DOI] [PubMed] [Google Scholar]
- Kortemme T.; Baker D. (2002) A simple physical model for binding energy hot spots in protein-protein complexes. Proc. Natl. Acad. Sci. U.S.A. 99, 14116–14121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabot D. J.; Zhang P. F.; Quinnan G. V.; Broder C. C. (1999) Mutagenesis of CXCR4 identifies important domains for human immunodeficiency virus type 1 X4 isolate envelope-mediated membrane fusion and virus entry and reveals cryptic coreceptor activity for R5 isolates. J. Virol. 73, 6598–6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signoret N.; Rosenkilde M. M.; Klasse P. J.; Schwartz T. W.; Malim M. H.; Hoxie J. A.; Marsh M. (1998) Differential regulation of CXCR4 and CCR5 endocytosis. J. Cell Sci. 111, 2819–2830. [DOI] [PubMed] [Google Scholar]
- Signoret N.; Oldridge J.; PelchenMatthews A.; Klasse P. J.; Tran T.; Brass L. F.; Rosenkilde M. M.; Schwartz T. W.; Holmes W.; Dallas W.; Luther M. A.; Wells T. N. C.; Hoxie J. A.; Marsh M. (1997) Phorbol esters and SDF-1 induce rapid endocytosis and down modulation of the chemokine receptor CXCR4. J. Cell Biol. 139, 651–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N. M.; Luo Z. W.; Luo J. S.; Liut D. X.; Hall J. W.; Pomerantz R. J.; Huang Z. W. (2001) Structural and functional characterization of human CXCR4 as a chemokine receptor and HIV-1 co-receptor by mutagenesis and molecular modeling studies. J. Biol. Chem. 276, 42826–42833. [DOI] [PubMed] [Google Scholar]
- Amara A.; LeGall S.; Schwartz O.; Salamero J.; Montes M.; Loetscher P.; Baggiolini M.; Virelizier J. L.; ArenzanaSeisdedos F. (1997) HIV coreceptor downregulation as antiviral principle: SDF-1α-dependent internalization of the chemokine receptor CXCR4 contributes to inhibition of HIV replication. J. Exp. Med. 186, 139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C.; Lee B.; Vakili J.; Doranz B. J.; Govaerts C.; Migeotte I.; Sharron M.; Dupriez V.; Vassart G.; Doms R. W.; Parmentier M. (1999) Extracellular cysteines of CCR5 are required for chemokine binding, but dispensable for HIV-1 coreceptor activity. J. Biol. Chem. 274, 18902–18908. [DOI] [PubMed] [Google Scholar]
- Lee B.; Sharron M.; Blanpain C.; Doranz B. J.; Vakili J.; Setoh P.; Berg E.; Liu G.; Guy H. R.; Durell S. R.; Parmentier M.; Chang C. N.; Price K.; Tsang M.; Doms R. W. (1999) Epitope mapping of CCR5 reveals multiple conformational states and distinct but overlapping structures involved in chemokine and coreceptor function. J. Biol. Chem. 274, 9617–9626. [DOI] [PubMed] [Google Scholar]
- Bogan A. A.; Thorn K. S. (1998) Anatomy of hot spots in protein interfaces. J. Mol. Biol. 280, 1–9. [DOI] [PubMed] [Google Scholar]
- White J. F.; Noinaj N.; Shibata Y.; Love J.; Kloss B.; Xu F.; Gvozdenovic-Jeremic J.; Shah P.; Shiloach J.; Tate C. G.; Grisshammer R. (2012) Structure of the agonist-bound neurotensin receptor. Nature 490, 508–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W.; Dalke A.; Schulten K. (1996) VMD: Visual molecular dynamics. J. Mol. Graphics Modell. 14, 33–38. [DOI] [PubMed] [Google Scholar]
- Grunbeck A.; Sakmar T. P. (2013) Probing G Protein-coupled Receptor-ligand Interactions with Targeted Photoactivatable Cross-linkers. Biochemistry 52, 8625–8632. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


