Abstract
Bacterial chemoreceptors cluster in highly ordered, cooperative, extended arrays with a conserved architecture, but the principles that govern array assembly remain unclear. Here we show images of cellular arrays as well as selected chemoreceptor complexes reconstituted in vitro that reveal new principles of array structure and assembly. First, in every case, receptors clustered in a trimers-of-dimers configuration, suggesting this is a highly favored fundamental building block. Second, these trimers-of-receptor dimers exhibited great versatility in the kinds of contacts they formed with each other and with other components of the signaling pathway, although only one architectural type occurred in native arrays. Third, the membrane, while it likely accelerates the formation of arrays, was neither necessary nor sufficient for lattice formation. Molecular crowding substituted for the stabilizing effect of the membrane and allowed cytoplasmic receptor fragments to form sandwiched lattices that strongly resemble the cytoplasmic chemoreceptor arrays found in some bacterial species. Finally, the effective determinant of array structure seemed to be CheA and CheW, which formed a “superlattice” of alternating CheA-filled and CheA-empty rings that linked receptor trimers-of-dimer units into their native hexagonal lattice. While concomitant overexpression of receptors, CheA, and CheW yielded arrays with native spacing, the CheA occupancy was lower and less ordered, suggesting that temporal and spatial coordination of gene expression driven by a single transcription factor may be vital for full order, or that array overgrowth may trigger a disassembly process. The results described here provide new insights into the assembly intermediates and assembly mechanism of this massive macromolecular complex.
Motile bacteria sense and respond to their environment through a networked system of chemoreceptors.1 These receptors are best understood in Escherichia coli, where an extended array of methyl-accepting chemotaxis proteins (MCPs) is found at the pole of each cell. Dimeric MCPs are anchored in the inner cell membrane and sense stimuli through their periplasmic ligand-binding domains. The resulting signal, either attractive or repulsive, is transferred down the length of the MCPs through conformational changes in various domains, culminating in a change in the activity of a histidine kinase, CheA, bound to the cytoplasmic tip of the MCP dimer. CheA also functions as a homodimer, performing trans-autophosphorylation and the subsequent transfer of a phosphoryl group to one of two response regulators. One of these, CheY, binds to the flagellar motor when phosphorylated, triggering a switch in the predominant direction of rotation, and thus effecting “tumbles” that interrupt linear “runs” and change the search direction. The other response regulator, CheB, is a methylesterase whose activity is stimulated by phosphorylation. The balance of the activities of CheB and the methyltransferase CheR dictates the methylation state of specific glutamate residues in the MCPs that are responsible for adaptation.
The polar chemoreceptor array has a highly regular structure: trimers of MCP dimers are linked in extended hexagonal lattices, with 12 nm spacing between the centers of adjacent hexagons. Associated molecules of CheA and CheW, a coupling protein, form rings linking trimers-of-receptor dimers into hexagons and neighboring hexagons into the extended lattice.2,3 This arrangement and spacing is highly conserved among different bacterial species and between different signaling states in E. coli.4−6 The structure of the extended lattice is important because it gives rise to one of the most striking aspects of the chemotaxis system, its high degree of cooperativity. Signal amplification in vivo can lead to apparent Hill coefficients (nH) ranging from 10 to 27 depending on the type of receptor and its modification state, indicative of a highly cooperative system.7,8 It remains unclear, however, how this extended, regular lattice forms.
To study the structure and function of the chemoreceptor array, a variety of protocols have been explored to reconstitute complexes in vitro. Such samples have been used to study phosphotransfer,9−12 cooperativity,13−15 stability,16,17 and protein–protein interactions.18−21 Here, we apply electron cryotomography (ECT) to image native chemoreceptor arrays as well as selected chemoreceptor complexes reconstituted in vitro. We find that the stoichiometry, mixing order, and the presence of membranes and crowding agents all effect higher-order structure. Our results point to an assembly process in which coordinated production of receptors and CheA and CheW in the presence of stabilizing membranes and the dense cytoplasmic environment all contribute to the formation of fully ordered, extended lattices.
Materials and Methods
Strains and Growth Conditions
Strains used in this study are listed in Table S1 of the Supporting Information. pCO3 was derived from pJC322 by polymerase chain reaction-based site-directed mutagenesis to generate tsrA413T. Strains were grown to midexponential phase in Tryptone Broth (TB) at 30 °C, with appropriate antibiotics. Expression of Tsr from pCO3 was induced with 250 μM isopropyl β-d-1-thiogalactopyranoside (IPTG). Expression of CheA and CheW was induced from pPM25 with 100 μM sodium salicylate. Strain UU2619 was lysed by incubation with 300 ng/mL pencillin for 1 h at 30 °C. Strain CO4 was lysed by treatment with 2 mg/mL lysozyme for 45 min at 37 °C, followed by treatment with 1 mg/mL DNase for 30 min at 37 °C. Samples were kept on ice until they were frozen for ECT.
Electron Cryotomography (ECT)
A 20 μL cell culture was mixed with a pelleted 100 μL colloidal gold solution, BSA treated to avoid aggregation.23 Three microliters of this cell/gold mixture was then applied to R2/2 copper Quantifoil grids (Quantifoil Micro Tools). After excess liquid had been blotted away using a Vitrobot (FEI), the sample was plunge-frozen in a liquid ethane/propane mixture.23,24 Images were collected using either an FEI Polara G2 (FEI Co., Hillsboro, OR) 300 kV field emission gun electron microscope at California Institute of Technology, an FEI TITAN Krios (FEI Co.) 300 kV field emission gun at the University of California (Los Angeles, CA), or an FEI TITAN Krios (FEI Co.) 300 kV field emission gun with an image corrector for lens aberration correction at Janelia Farms. All microscopes were equipped with Gatan (Pleasanton, CA) image filters. California Institute of Technology and Janelia Farms microscopes were outfitted with a K2 Summit counting electron detector camera (Gatan), and the University of California microscope was outfitted with a 4 megapixel CCD (Gatan). Data were collected using UCSFtomo25 or BatchTomo (FEI Co.) using cumulative electron doses of approximately ≤160 e/A2 for each individual tilt series. The images were CTF corrected, aligned, and reconstructed using weighted back projection using the IMOD software package.26 SIRT reconstructions were calculated using TOMO3D.27 Subvolume averaging and symmetrizing were conducted using PEET.28
Classification by Missing Wedge Effect-Corrected Principle Component Analysis (WMD-corrected PCA) Using PEET
WMD-corrected PCA, which attempts to compensate for the missing wedge effect in the electron cryotomogram, and k-means clustering was performed using PEET.28 Subvolumes were chosen from a single array patch and contained one to six receptor hexagons, and associated density above and below. Varying the cube size of the subvolume did not affect the results. Classes with fewer than 10 particles were discarded, as they likely contained misaligned or false particles, and the resulting subvolume averages were too noisy to interpret. The results of WMD-corrected PCA, including variances and information criteria, are summarized in Table S2 of the Supporting Information.
Purification of Signaling Components and Assembly in Vitro
Two different types of in vitro reconstitutions were tested employing full length Tsr, CheA, and CheW.
In method A, Tsr-containing inner membranes were prepared essentially as described previously,21,29 with some modifications. Briefly, Tsr expression was induced from plasmid pJC322 with 1 mM IPTG for 4 h in HCB326, an E. coli strain lacking native chemotaxis proteins. Cells were collected, resuspended in lysis buffer [50 mM KH2PO4 (pH 7.5), 5 mM DTT, 10 mM EDTA, 1 mM 1,10-phenanthroline, 10% glycerol, and 1 mM phenylmethanesulfonyl fluoride (PMSF)], and lysed with a Constant Cell Disruption System (Constant Systems, Kennesaw, GA). Cell debris was removed by centrifugation and the supernatant equilibrated with 10 mM aqueous iodoacetamide. Membranes were isolated by ultracentrifugation and passage over a sucrose gradient and resuspended in Tsr reaction buffer [50 mM HEPES (pH 7.5), 50 mM KCl, and 5 mM MgCl2] with 1 mM PMSF. Membrane suspensions were stored at −80 °C until they were used. The membrane protein content was determined by a modified BCA assay (Pierce Biotechnology, Rockford, IL). Membranes typically contained 20% Tsr, determined by densitometry of Coomassie-stained sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) gels.
To reconstitute signaling complexes, native membranes containing Tsr were combined with purified His6-CheW and His6-CheA (prepared as described in refs (21) and (29)). The following ternary complex components were combined: 12 μM Tsr, 6 μM His6-CheW, and 2 μM His6-CheA in Tsr reaction buffer. Samples were incubated at room temperature for 15 min, extruded through a 27 gauge needle, and incubated again for 30 min at room temperature before being washed. The resulting sample was immediately placed on ice until they were imaged.
Method B was a modification of several previous protocols.16,17,19,20,29 The E. coli serine receptor (Tsr) was overexpressed in gutted E. coli strain UU1581, which lacks all chemotaxis proteins, including receptors and adaptation enzymes, using plasmid pJC3.22 Inside-out, inner bacterial membrane vesicles containing Tsr were isolated as previously described.11,18 The total protein concentration in the membranes was determined by the BCA assay, and the fraction of total protein represented by receptors (typically ∼20%) was determined by ImageJ densitometry of SDS–PAGE gels.
Signaling complexes were reconstituted by combining 6.7 μM Tsr receptor, 5 μM CheA kinase, and 10 μM CheW adaptor protein in activity buffer [160 mM NaCl, 6 mM MgCl2, 50 mM Tris, and 3 mM EDTA (pH 7.5)] for 45 min at 22 °C in the presence of 0.5 mg/mL BSA, 2 mM TCEP, and 2 mM PMSF. Samples were centrifuged at 21000g for 7 min, and pellets were washed twice to remove unbound CheA and CheW by resuspension in a 10-fold excess of activity buffer (without BSA, TCEP, and PMSF) and repelleting. After the final wash, pellets were resuspended in the original volume of activity buffer, resulting in functional, stable complexes.16,17,19,20 The resulting sample was immediately placed on ice, shipped overnight, and then cryo-frozen and prepared for ECT.
Ordered Assembly of Array Components in Vitro
Cytoplasmic fragments of the Tar receptor without the transmembrane and HAMP domains (amino acids 1–256) and with full methylation (CF4Q) were generated. CF4Q, CheW, CheA, and CheY were expressed and purified as previously described.30 Protein purity was assessed via SDS–PAGE analysis, and protein concentrations were determined using a BCA assay (Thermo Fisher Scientific). All lipids were purchased from Avanti Polar Lipids, and large unilamellar vesicles (LUVs) were prepared as previously described.31 PEG 8000 (Fluka) and d-(+)-trehalose (Sigma-Aldrich) were prepared as 40% (w/v) stock solutions in deionized water and passed through a 0.22 μm syringe filter prior to being used. A modified kinase buffer [50 mM potassium phosphate, 50 mM KCl, and 5 mM MgCl2 (pH 7.5)] was used for sample preparation.
Formation and characterization of kinase-active ternary complexes followed published methods,30,31 further specified as follows. Vesicle-mediated CF4Q ternary complexes were prepared by incubating 30 μM CF, 12 μM CheW, and 6 μM CheA with 580 μM total LUVs (1:1 DOPC:DOGS-NTA-Ni2+ ratio), while PEG-mediated CF4Q complexes were prepared by incubating 50 μM CF, 20 μM CheW, and 12 μM CheA with final concentrations of 7.5% (w/v) PEG 8000 and 4% (w/v) trehalose. For both PEG- and vesicle-mediated complexes, CF was added last to minimize CF-promoted aggregation,32 and samples were incubated overnight at 25 °C in a circulating water bath and subjected to an enzyme-coupled assay and gel-based cosedimentation assay to check for phosphorylation activity and ternary complex formation.
Results
WT E. coli Chemoreceptor Arrays Are Superlattices of Alternating CheA-Filled and CheA-Empty Rings
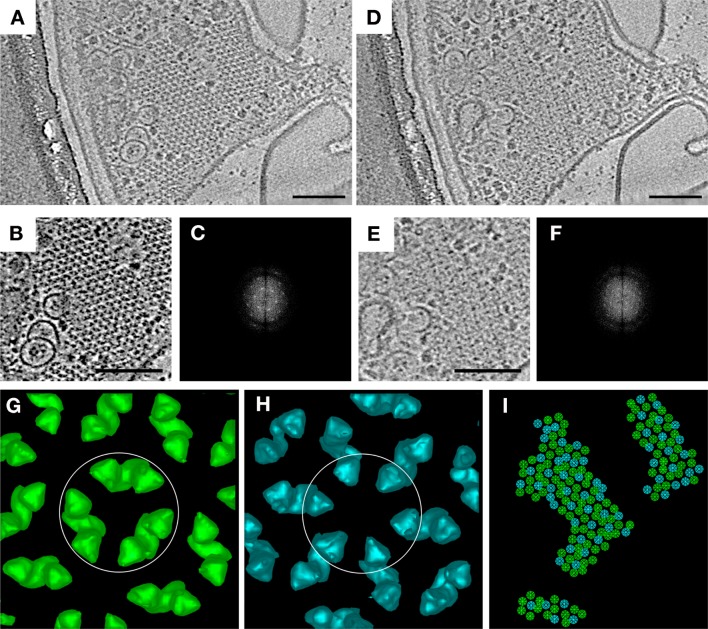
In E. coli polar chemoreceptor arrays, dimers of CheA link adjacent trimers of MCP dimers. On the basis of the crystal structures of these components and their complexes, it is apparent that in the extended hexagonal lattice, not all hexagons can be occupied by three CheA dimers. Rather, a regular pattern was predicted in which CheA-filled hexagons alternate with CheA-empty hexagons.3 While this hypothesis was strongly supported by the demonstration that there are two different kinds of array hexagons in a tomogram (“filled” and “empty”), the arrangement of these units was not reported.2
Because of further advances in sample preparation (receptors locked in a single state), data collection (thinner sample, direct detector), and processing (contrast transfer function correction), we are now often able to visualize CheA dimers individually in tomograms within intact arrays. This allowed us to confirm the order of the “superlattice” of CheA-filled and CheA-empty hexagons directly. To do this, we analyzed tomograms of wild-type (WT) cells expressing serine-sensing receptors in the demethylated state (Tsr-EEEE). This modification state of the receptors promotes stable packing of the P1 and P2 domains of CheA,6 leading to a strong keel-like density that facilitates identification of CheA dimers in tomograms. We observed a layer of CheA below the MCP hexagons in tomograms, which appeared to be highly ordered, as confirmed by Fourier transform (Figure 1). Individual CheW molecules were not identifiable, likely because of their smaller size. We then used principal component analysis (PCA) to identify classes of hexagons in a tomographic slice on the basis of CheA occupancy. Only two classes of receptor hexagons were observed: one in which each pair of Tsr trimers is linked by a CheA dimer and one in which none of them is (Figure 1G,H). When we forced more classes to exist, only additional filled and empty classes resulted, confirming that there are very few if any partially filled hexagons. These two classes were present in a roughly 2:1 ratio (117 filled rings and 64 empty rings). By mapping the classes back onto the tomographic slice, we found a strictly alternating pattern (Figure 1I), confirming that native arrays are a superlattice. The trimers-of-receptor dimers lie at the vertices of a hexagonal lattice with 12 nm spacing. Connected to the cytoplasmic tips of the receptors, the associated CheA (together with CheW) forms another lattice. Here, the three CheA dimers linking one receptor hexagon lie at the vertices of a larger hexagonal lattice with a spacing of 21 nm. This results in a hexagonal array of receptors linked to a lattice of alternating CheA-filled and CheA-empty hexagons. This pattern is reflected in the power spectrum shown in Figure 1F. We also classified arrays from cells expressing Tsr in the methylated state (Tsr-QQQQ). While the classification did not result in a high degree of confidence, it did separate the hexagons into the same two classes: one in which each pair of Tsr trimers is linked by a CheA dimer and one in which none of them is. The resulting CheA localization pattern resembled that seen in Tsr-EEEE, but with some errors in the distribution pattern, likely because of a higher number of misclassified subvolumes because of the smaller keel density of CheA (Figure S1 of the Supporting Information).
Figure 1.
Classification of E. coli chemoreceptor array hexagons reveals ordered CheA occupancy. (A) Tomographic slice through an array patch at the level of the chemoreceptors. (B) Cutout of the patch, and corresponding power spectrum (C), revealing hexagonal lattice. (D) Tomographic slice of the same array patch below the level of the chemoreceptors, showing CheA. (E) Cutout of the patch, and corresponding power spectrum (F), revealing ordered occupancy by CheA. (G–I) Classification by principal components analysis and k-means clustering of hexagons in the same array patch results in two distinct classes: hexagons linked by three CheA dimers (green symbols, subvolume average circled in panel G) and hexagons lacking CheA dimers (turquoise symbols, subvolume average circled in panel H). The organization of class averages is shown in panel I. Scale bars are 100 nm, and power spectra are not to scale.
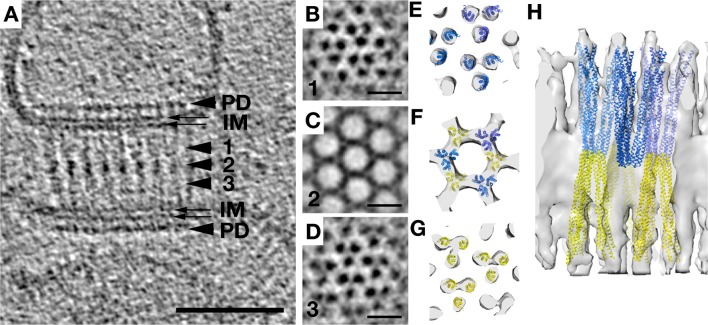
Overexpressed Chemoreceptors, in the Absence of CheA and CheW, Form Zippers
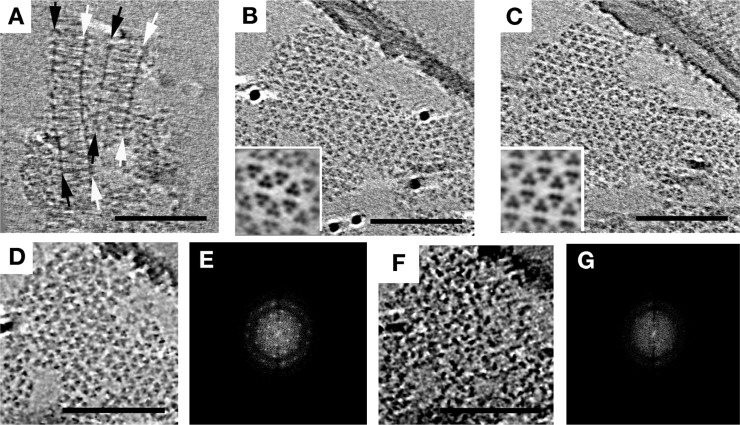
As different investigators have explored different protocols to characterize array structure and function, one of the earliest strategies was to simply overexpress receptors, often in the absence of CheA and CheW. Strongly overexpressed Tsr chemoreceptors are known to form non-native ordered arrays termed “zippers” in which two receptor layers interact with one another at their normally CheA–CheW-binding, cytoplasmic tips, creating characteristic membrane invaginations.33−36 We investigated the structure of these zippers at higher resolution using a preparation of E. coli Tsr in purified inner membranes. Interestingly, we found that zippers survived cell lysis and membrane purification, indicating that the interactions between the kinase-binding domains of the MCPs at their membrane-distal tips are highly stable. Importantly, the fundamental building block in zippers was seen to be trimers of dimers, just as in native arrays, but when viewed from the top, zippers exhibited tighter packing, with triangular lattices at the top and bottom and a hexagonal pattern at midsection (Figure 2). This complicated pattern was the result of two layers of receptors linked in a hexagonal lattice with a center-to-center spacing of 9 nm at the midsection, with alternating trimers facing opposite directions (Figure 2 and Movie S1 of the Supporting Information).
Figure 2.
Overexpression of Tsr without sufficient CheA and CheW results in zippers. (A) A side view of a receptor zipper reveals two layers of membrane-bound receptors interacting at their membrane-distal tips. PD denotes periplasmic domains and IM inner membrane leaflets. The scale bar is 50 nm. Arrows indicate relative positions of subvolume averages shown at the right in panels B–D. Scale bars are 10 nm. (E–H) Model of receptor density from the subvolume average and manually fitted Tsr crystal structure from ref (46) in top view (E–G, levels roughly corresponding to B–D, respectively) and side view (H), showing the arrangement of receptors. Blue and yellow colors indicate receptors of opposing orientation.
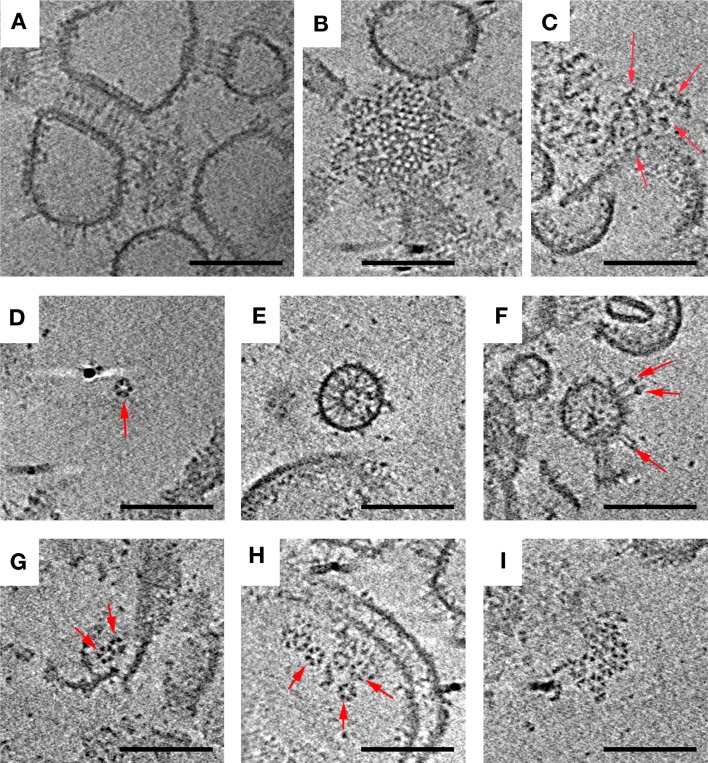
In Vitro Assembly of Array Components Results in Functional Complexes, Hexagons, and Small Arrays
Another strategy that has been used to reconstitute chemoreceptor systems in vitro is to add excess purified CheA and CheW to overexpressed receptors purified within their native E. coli membranes. Different variations of this basic strategy have been explored. One important variable appears to be the length of time CheA and CheW are allowed to interact with the receptors, as the largest Hill coefficients have been measured after the longest incubation times (exceeding 4 h13). Here we isolated inner membrane vesicles containing Tsr and then combined them with purified CheA and CheW for 15–45 min.16,37 This type of preparation is known to generate functional receptor–CheA–CheW units in which receptors bind attractant serine and regulate CheA kinase activity in the normal way. ECT revealed Tsr zippers similar to those observed in Tsr inner membranes prior to addition of CheA and CheW (see above), as well as large, loosely associated aggregates (Figure 3; overview in Figure S2 of the Supporting Information). The zippers likely formed within the cell and remained associated throughout lysis and addition of CheA and CheW. Inner membrane preparations are known to yield an ∼80:20 molar ratio of inside-out to right-side-out receptors.17 Both receptor orientations are observed in the images, and the outward-pointing cytoplasmic tips dominate as expected (Figure 3 and Figure S2 of the Supporting Information).
Figure 3.
In vitro reconstitution of signaling complexes produces a variety of structures. Arrangements observed included receptor zippers with 9 nm center-to-center hexagonal spacing (side view, A; top view, B), loosely ordered aggregates (C), individual hexagons of six trimers of dimers (D), receptors oriented inward (E) and outward (F) from vesicles, linked hexagons (G), multiple unlinked hexagons (H), and the largest 12 nm hexagonal array patch observed (I). Arrows indicate structures of interest. Scale bars are 100 nm.
Besides the expected zippers, a range of assembly intermediates was observed, providing insight into the mechanism of array assembly. We also found partial or full Tsr hexagons, double hexagons, and small arrays formed from multiple hexagons. Individual receptor hexagons were fully occupied by three CheA dimers (Figure S3 of the Supporting Information). No large, nativelike arrays were detected, however, consistent with the low Hill coefficient observed for this type of preparation.16 We have not observed small assembly intermediates in native cells but would likely to be unable to resolve them because of the relative thickness of the sample.
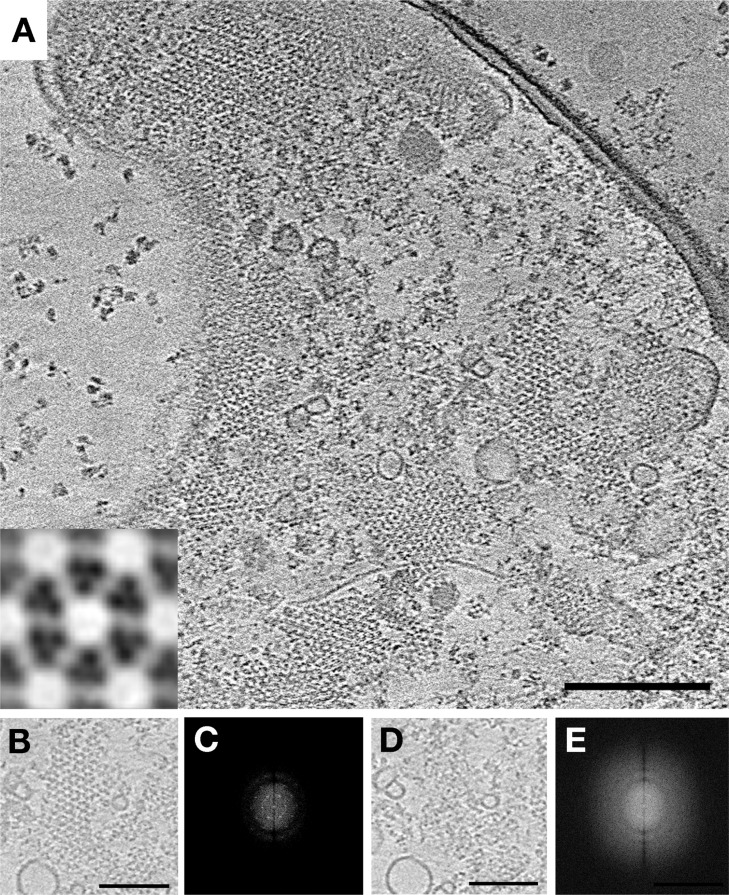
Co-overexpression of Chemoreceptors, CheA, and CheW Yields Nativelike Hexagonal Arrays, but CheA Occupancy Is Diminished
Suspecting that optimal array formation may require simultaneous production of receptors, CheA, CheW, some investigators have tried co-overexpression. Concomitant overexpression of CheA and CheW has in fact already been shown to produce large arrays without membrane invaginations.38 In cells overexpressing Tsr from one plasmid and CheA and CheW from another, we observed arrays with the native 12 nm hexagonal spacing (Figure 4). We again used a Tsr variant (Tsr-A413T) that locks the P1 and P2 domains of CheA into an identifiable “keel” to investigate CheA occupancy.6 By immunoblotting, we determined that Tsr and CheA had been overexpressed to similar extents [25.5 and 26 times their WT levels, respectively (Figure S4 of the Supporting Information)], but classification of hexagons in tomograms revealed significantly lower and less ordered occupancy of CheA than in native arrays (Figure S5 of the Supporting Information). Two major classes were observed with zero or two CheA dimers per ring. Direct observation of CheA dimers in the tomograms confirmed the lack of any superlattice order of CheA-filled or CheA-empty rings.
Figure 4.
Co-overexpression of Tsr, CheA, and CheW restores WT array structure. (A) A tomographic slice of a lysed E. coli cell overexpressing the chemotaxis proteins Tsr-A413T, CheA, and CheW reveals extended well-ordered hexagonal arrays with 12 nm center-to-center spacing. The inset shows a higher-magnification subvolume average showing the top view of a single hexagon. (B) Array patch at the level of the receptors and the corresponding power spectrum (C). (D) Same array patch at the level of CheA and the corresponding power spectrum (E) showing a lack of order in the CheA arrangement. Scale bars are 100 nm, and power spectra are not to scale.
Ordered, Vesicle-Mediated Assembly of Receptor Fragments, CheA, and CheW in Vitro Leads to Large Arrays
The overexpression experiments described above suggest that in addition to proper ratios of receptors, CheA, and CheW, assembly of native arrays depends on proper temporal and perhaps spatial ordering of the process. To expore this possibility, we used a system in which soluble MCPs lacking their transmembrane domains could be added to mixtures of Ni2+-NTA-conjugated lipids and CheA and CheW in different orders.31,32 His-tagged cytoplasmic fragments of the aspartate-sensing Tar chemoreceptor lacking their periplasmic ligand-binding domains, transmembrane domains, and HAMP domains (Tar-CF) were purified. We found that by first adding purified CheA and CheW to Ni2+-NTA-tagged lipids and then adding the soluble receptor fragments, we could form vesicle-associated arrays containing at least 20 hexagons with a 12 nm center-to-center spacing (Figure 5). The ordered patches may have been even larger, but unfortunately, the high degree of curvature of the vesicles precluded accurate estimation of the total hexagon number, as well as visualization of the organization of CheA dimers. Zippers and loose aggregates of MCPs were not observed.
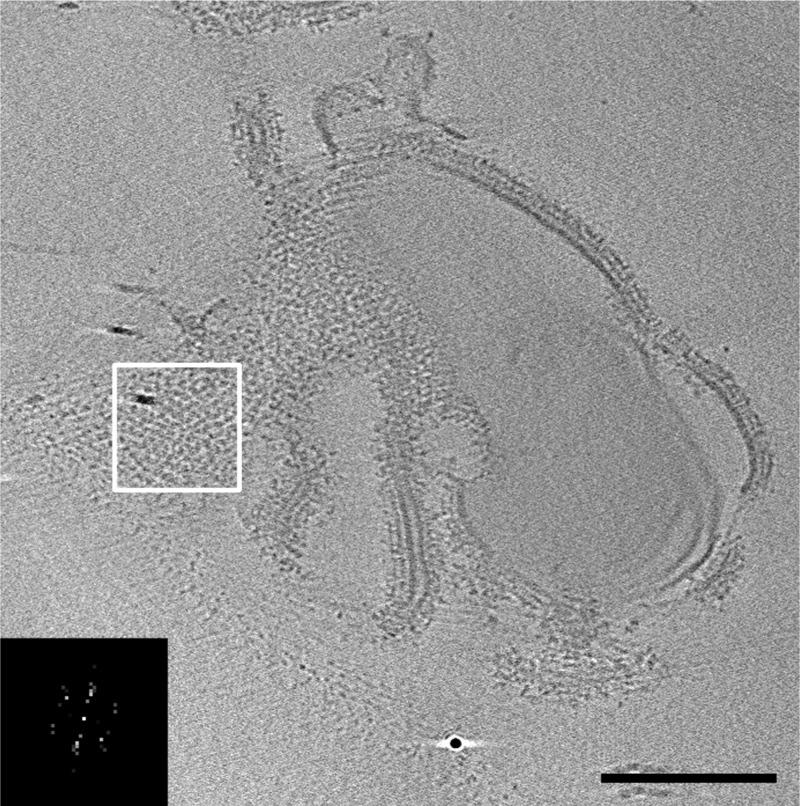
Figure 5.
Addition of MCPs after CheA and CheW produces extended 12 nm arrays. Vesicle-mediated assembly of Tar-CF, CheA, and CheW leads to extended arrays, shown in a tomographic slice. The inset shows a power spectrum (not to scale) of the white-boxed region that shows the hexagonal order of the array, with a 12 nm center-to-center spacing. The scale bar is 200 nm.
The Membrane Is Not Essential for Array Formation and Can Be Replaced by Crowding Agents
Membrane interactions are essential for transmembrane chemoreceptor array formation and function.39−41 However, many bacteria contain soluble cytoplasmic chemoreceptor arrays that do not associate with any membrane42,43 [e.g., Vibrio cholerae (Figure S6 of the Supporting Information)]. If cytoplasmic receptors can form extended arrays without an organizing membrane, can normally polar chemoreceptors do so as well? To test this, we again purified soluble cytoplasmic fragments of the Tar receptor, as well as CheA and CheW, from E. coli. Again we assembled complexes in vitro using these components, with CheA and CheW present in stoichiometric excess, but this time in the absence of lipid vesicles. To mimic cellular conditions, we included the molecular crowding agents PEG-8000 and trehalose in the assembly reaction mixture.30 Interestingly, extended arrays formed with an architecture identical to those of the cytoplasmic clusters in Rhodobacter sphaeroides and V. cholerae in vivo (Figure S6 of the Supporting Information). Two CheA and CheW base plates, 31 nm apart, flanked two hexagonal lattices of chemoreceptor trimers with a 12 nm center-to-center spacing to form a “sandwich” (Figure 6). In contrast to zippers, in which receptors interact at the kinase-binding tip, in this case the two receptor layers interacted at their membrane-proximal tips. As observed for polar chemoreceptors,3,5,44 the kinase-binding regions near the CheA and CheW base plates were well ordered, with a decreasing level of order toward the midsection of the sandwich (Figure S7 of the Supporting Information). To assess CheA occupancy, we classified an array patch and observed hexagon classes with zero, one, two, and three CheA dimers, indicating less order than in native membrane-bound arrays (Figure S8 of the Supporting Information), consistent with direct observation (Figure 6F,G). Interestingly, we were able to assemble functional complexes in the absence of membranes only from Tar-CF in the methylated (QQQQ) adaptation state, not in the demethylated (EEEE) state, even in the presence of higher concentrations of molecular crowding agents, suggesting that the methylated receptor state may form more stable complexes.
Figure 6.
E. coli Tar chemoreceptors lacking transmembrane regions form extended arrays in the presence of CheA, CheW, and molecular crowding agents. Tomographic slices showing extended arrays. (A) A side view reveals two parallel CheA and CheW base plates (arrows) spaced 31 nm apart. Top views of the chemoreceptors close to either base plate (B and C, corresponding to white and black arrows in A, respectively) reveal a well-ordered, hexagonal arrangement with a center-to-center spacing of 12 nm. Insets show enlarged subvolume averages. (D) Array patch at the level of the receptors and the corresponding power spectrum (E). (F) Same array patch at the level of CheA and the corresponding power spectrum (G), showing the lack of order. Scale bars are 100 nm, and power spectra are not to scale.
Discussion
Here we explored the structure and assembly of chemoreceptor arrays by imaging both native arrays and selected in vitro preparations that yield functional receptor–CheA–CheW complexes. We found that native arrays are not only hexagonally ordered, but a superlattice of alternating CheA-filled and CheA-empty rings exists. When Tsr receptors are overexpressed in the absence of CheA and CheW, stable double-layer zippers form as previously observed, and this study reveals that the receptors are still arranged as trimers-of-receptor dimers, though packed in a non-native lattice. When receptor-containing membranes are incubated with purified CheA and CheW, isolated “functional units” (pairs of trimers-of-receptor dimers linked by CheA dimers) and “rings” (six trimers-of-receptor dimers linked by three CheA dimers) were found, as were clusters of rings forming small arrays, but no large, nativelike arrays were observed (as expected given the low Hill coefficient reported for this type of preparation16). Instead, the observed small complexes and arrays are proposed to be early intermediates in the assembly of native arrays. Larger, more extended 12 nm hexagonal arrays are produced by co-overexpression of the receptor, CheA, and CheW, or by reconstituting receptor cytoplasmic domains with CheA and CheW on Ni-NTA lipid vesicles. The same receptor cytoplasmic domains form sandwiched arrays upon being incubated with CheA and CheW in the absence of membranes, but in the presence of crowding agents.
One of the principles that emerges from these observations and others already in the literature is that with the exception of some crystal structures, for example the Thermatoga receptors, where the receptors were arranged in “hedgerows” of dimers,45 receptors always form trimers of dimers linked together tightly at their kinase-binding tips but splaying outward toward their ligand-binding tips. However, the degree of splaying observed by ECT is less than that seen in the crystal structure.3,6,46 This same building block is seen in native arrays, zippers, reconstituted mixtures, and cytoplasmic arrays. This structure probably forms rapidly within cells and is highly stable, remaining intact through cell lysis and/or diverse purification–reconstitution procedures.
The second principle that emerges is that the trimers-of-receptor dimers unit exhibits striking versatility in the kinds of contacts it can form with other trimers-of-receptor dimers and other components of the signaling pathway. In native arrays, trimers of receptor dimers form extended hexagonal arrays anchored in the membrane at their ligand-binding tips and associating laterally through networked CheA and CheW. In the absence of CheA and CheW, trimers bind each other at their kinase-binding tips in antiparallel fashion to form superstable zippers. A more recent crystal structure reveals two potential CheA- and CheW-binding sites along each dimer.555 In the absence of membrane binding, either through deletion of transmembrane domains or in endogenous cytoplasmic chemoreceptors, MCPs again form bilayers, but in these cases through interactions at the ends of the receptors distal to their CheA-binding tips. Because there are so many possible structures that can form, while the architecture seen in cells is universally conserved,4 the assembly process within a cell must be tightly regulated.
For transmembrane receptors, the membrane likely accelerates array formation by holding newly synthesized receptors in a plane and then stabilizes arrays after formation.39−41 These findings reveal, however, another assembly principle: the membrane is neither necessary for proper array organization (evidenced by endogenous cytoplasmic clusters in many organisms and in vitro assembly of cytoplasmic fragments of normally polar receptors) nor sufficient (evidenced by zippering of receptors in the absence of coupling proteins). Active signaling complexes of cytoplasmic receptor fragments can be generated via association with lipid vesicles47 and can also be generated in the absence of membrane binding, by increasing the extent of molecular crowding to mimic the cellular environment.30 Our new results show that both of these preparations contain extended arrays with a 12 nm spacing equivalent to that of intact receptor arrays. In addition, our results also show that extended arrays are always stabilized on both faces, by either membranes or CheA and CheW base plates (arrays stabilized on only one side have not been observed). Molecular crowding agents increase the apparent local concentration of components by excluding volume, shifting the equilibria of biomolecular interactions in favor of associated states. The ability of molecular crowding agents to promote arrays of CheA and CheW and cytoplasmic receptor domains underscores the dramatic role that the dense cellular environment can play in assembly.48−50
As a final assembly principle, the effective determinant of array structure seems to be CheA and CheW, for both membrane-bound and cytoplasmic arrays. In native arrays, we find nearly complete, and highly ordered, CheA occupancy. Filled hexagons containing three CheA dimers surround an empty hexagon containing none. This leads to a slightly higher receptor:CheA ratio in arrays (6:1) compared to the total concentration ratio in cells (3.4:1).51 It may be that cells contain excess CheA and CheW to promote correct assembly. When Tsr, CheA, and CheW are decoupled and overexpressed, the resulting arrays exhibit the same 12 nm hexagonal packing as native arrays, but we observe less-than-native CheA incorporation, suggesting that the assembly process may become defective when the precise stoichiometry and temporal control provided by native transcription is disrupted. Alternatively, overexpression could activate a putative disassembly mechanism responsible for removing CheA and destabilizing the array, thereby preventing excess array growth, as previously suggested.17 Intriguingly, Bacillus subtilis has been reported to have a much higher ratio of MCP to CheA than E. coli, approximately 23 receptor dimers to one CheA dimer52 versus 3.4:1 for E. coli.51 Thus, at least in B. subtilis, it appears that relatively little CheA is required to nucleate a morphologically correct array with respect to receptor packing.
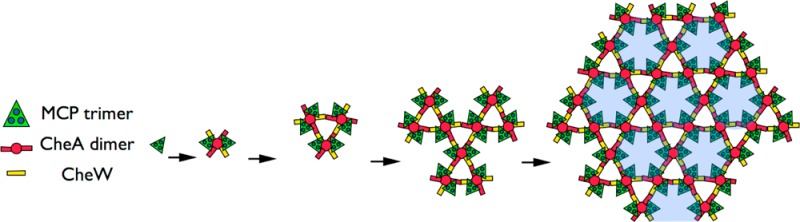
We imagine then that in cells, receptors are inserted into the membrane as they are produced and quickly dimerize. Next, receptor dimers trimerize, and then CheA dimers capture trimers-of-receptor dimers into six-receptor functional units before any off-pathway complexes form. These functional units either unite through CheW with existing arrays or link together to form CheA-filled rings, which then unite to existing arrays. Given the known interactions of CheA and CheW, both processes lead directly to the highly cooperative superlattice of alternating CheA-filled and CheA-empty rings (Figure 7 and Movie S1 of the Supporting Information).
Figure 7.

Model of array assembly. Schematic showing sequential assembly of the core functional unit (two trimers-of-receptor dimers, one CheA dimer, and two CheW monomers) forming from individual trimers-of-receptor dimers, and subsequently coalescing into individual hexagons, which in turn assemble into the extended superlattice. Empty hexagons without associated CheA are colored blue.
The special conditions that exist within cells that allow and guide this assembly process may, however, be challenging to mimic in vitro. Given that, the smallest functional unit that displays full regulation of kinase activity is the six-receptor-dimer, one-CheA-dimer, two-CheW unit that also seems to be the basic building block of array assembly, and all the reconstitution protocols explored here produce such functional units. Their biochemical and biophysical properties can therefore be studied effectively as long as care is taken not to include the signal from non-native structures that may also be present. This can be done through CheA- or CheW-readout methods (rather than simply monitoring receptors); for instance, monitoring the effects of mutagenesis, cross-linking, or protein modification on kinase activity measures only the effects within functional complexes.18−21,53−55 Notably, certain reconstitutions have also already exhibited Hill coefficients close to those observed for cellular arrays,13 suggesting that nativelike higher-order structures must be present. We hope the interplay between EM and in vitro reconstitution methods, together with the application of the new assembly principles revealed here, will eventually allow the production of even larger-than-cellular arrays with the fully native structure for enhanced structural, biophysical, and biochemical characterization of array properties.
Acknowledgments
We thank Drs. Zhiheng Yu and Jason de la Cruz for microscopy support at Howard Hughes Medical Institute Janelia Farms and Dr. Dan Toso and Associate Director Ivo Atanasov for support using the University of California TITAN Krios microscope. We thank Drs. Gongpu Zhao and Peijun Zhang for initial electron micrographs of the PEG-mediated complexes of CF, CheA, and CheW. We thank Dr. Sandy Parkinson for the gift of α-Tsr and α-CheA antibodies, strains, and plasmids, as well as for sharing results prior to publication. We thank Dr. John Heumann for assistance with PEET software.
Supporting Information Available
Two supporting tables, eight supporting figures, and a movie summarizing the main findings of this paper. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Present Address
# M.L.W.: Novartis Institutes for Biomedical Research, Cambridge, MA 02139.
This work was supported by National Institutes of Health Grant R01-GM085288 to L.K.T., National Institute of General Medical Sciences Grant GM101425 to G.J.J., National Institutes of Health Grant R01-GM040731 to J.J.F., National Institutes of Health Grant RO1-GM055984 to L.L.K., and National Institutes of Health CBI training grant T32 GM008505 to H.L.H.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Hazelbauer G. L.; Falke J. J.; Parkinson J. S. (2008) Bacterial chemoreceptors: High-performance signaling in networked arrays. Trends Biochem. Sci. 33, 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Hu B.; Morado D. R.; Jani S.; Manson M. D.; Margolin W. (2012) Molecular architecture of chemoreceptor arrays revealed by cryoelectron tomography of Escherichia coli minicells. Proc. Natl. Acad. Sci. U.S.A. 109, E1481–E1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briegel A.; Li X.; Bilwes A. M.; Hughes K. T.; Jensen G. J.; Crane B. R. (2012) Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proc. Natl. Acad. Sci. U.S.A. 109, 3766–3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briegel A.; Ortega D. R.; Tocheva E. I.; Wuichet K.; Li Z.; Chen S.; Müller A.; Iancu C. V.; Murphy G. E.; Dobro M. J.; Zhulin I. B.; Jensen G. J. (2009) Universal architecture of bacterial chemoreceptor arrays. Proc. Natl. Acad. Sci. U.S.A. 106, 17181–17186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briegel A.; Beeby M.; Thanbichler M.; Jensen G. J. (2011) Activated chemoreceptor arrays remain intact and hexagonally packed. Mol. Microbiol. 82, 748–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briegel A.; Ames P.; Gumbart J.; Oikonomou C. M.; Parkinson J. S.; Jensen G. J. (2013) The mobility of two kinase domains in the Escherichia coli chemoreceptor array varies with signaling state. Mol. Microbiol. 89, 831–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X.-S.; Parkinson J. S. (2014) Unorthodox sensory adaptation site in the Escherichia coli serine chemoreceptor. J. Bacteriol. 196, 641–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourjik V.; Berg H. C. (2004) Functional interactions between receptors in bacterial chemotaxis. Nature 428, 437–441. [DOI] [PubMed] [Google Scholar]
- Schuster S. C.; Swanson R. V.; Alex L. A.; Bourret R. B.; Simon M. I. (1993) Assembly and function of a quaternary signal transduction complex by surface plasmon resonance. Nature 365, 343–347. [DOI] [PubMed] [Google Scholar]
- Ninfa E. G.; Stock A.; Mowbray S.; Stock J. B. (1991) Reconstitution of the bacterial chemotaxis signal transduction system from purified components. J. Biol. Chem. 266, 9764–9770. [PubMed] [Google Scholar]
- Gegner J. A.; Graham D. R.; Roth A. F.; Dahlquist F. W. (1992) Assembly of an MCP receptor, CheW, and kinase CheA complex in the bacterial chemotaxis signal transduction pathway. Cell 70, 975–982. [DOI] [PubMed] [Google Scholar]
- Borkovich K. A.; Kaplan N.; Hess J. F.; Simon M. I. (1989) Transmembrane signal transduction in bacterial chemotaxis involves ligand-dependent activation of phosphate group transfer. Proc. Natl. Acad. Sci. U.S.A. 86, 1208–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G.; Weis R. M. (2000) Covalent modification regulates ligand binding to receptor complexes in the chemosensory system of Escherichia coli. Cell 100, 357–365. [DOI] [PubMed] [Google Scholar]
- Lai R.-Z.; Manson J. M. B.; Bormans A. F.; Draheim R. R.; Nguyen N. T.; Manson M. D. (2005) Cooperative signaling among bacterial chemoreceptors. Biochemistry 44, 14298–14307. [DOI] [PubMed] [Google Scholar]
- Bornhorst J. A.; Falke J. J. (2000) Attractant regulation of the aspartate receptor–kinase complex: Limited cooperative interactions between receptors and effects of the receptor modification state. Biochemistry 39, 9486–9493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slivka P. F.; Falke J. J. (2012) Isolated bacterial chemosensory array possesses quasi- and ultrastable components: Functional links between array stability, cooperativity, and order. Biochemistry 51, 10218–10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbse A. H.; Falke J. J. (2009) The core signaling proteins of bacterial chemotaxis assemble to form an ultrastable complex. Biochemistry 48, 6975–6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A. S.; Kohout S. C.; Gilman K. A.; Falke J. J. (2006) CheA kinase of bacterial chemotaxis: Chemical mapping of four essential docking sites. Biochemistry 45, 8699–8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natale A. M.; Duplantis J. L.; Piasta K. N.; Falke J. J. (2013) Structure, Function, and On–Off Switching of a Core Unit Contact between CheA Kinase and CheW Adaptor Protein in the Bacterial Chemosensory Array: A Disulfide Mapping and Mutagenesis Study. Biochemistry 52, 7753–7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piasta K. N.; Ulliman C. J.; Slivka P. F.; Crane B. R.; Falke J. J. (2013) Defining a key receptor-CheA kinase contact and elucidating its function in the membrane-bound bacterial chemosensory array: A disulfide mapping and TAM-IDS study. Biochemistry 52, 3866–3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underbakke E. S.; Zhu Y. M.; Kiessling L. L. (2011) Protein footprinting in a complex milieu: Identifying the interaction surfaces of the chemotaxis adaptor protein CheW. J. Mol. Biol. 409, 483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames P.; Studdert C. A.; Reiser R. H.; Parkinson J. S. (2002) Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 99, 7060–7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iancu C. V.; Tivol W. F.; Schooler J. B.; Dias D. P.; Henderson G. P.; Murphy G. E.; Wright E. R.; Li Z.; Yu Z.; Briegel A.; Gan L.; He Y.; Jensen G. J. (2007) Electron cryotomography sample preparation using the Vitrobot. Nat. Protoc. 1, 2813–2819. [DOI] [PubMed] [Google Scholar]
- Tivol W.; Briegel A.; Jensen G. J. (2008) An Improved Cryogen for Plunge Freezing. Microscopy and Micoanalysis 14, 375–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q. S.; Keszthelyi B.; Branlund E.; Lyle J. M.; Braunfeld M. B.; Sedat J. W.; Agard D. A. (2007) UCSF tomography: An integrated software suite for real-time electron microscopic tomographic data collection, alignment and reconstruction. J. Struct. Biol. 157, 138–147. [DOI] [PubMed] [Google Scholar]
- Kremer J. R.; Mastronarde D. N.; McIntosh J. R. (1996) Computer visualization of three-dimensional data using Imod. J. Struct. Biol. 116, 71–76. [DOI] [PubMed] [Google Scholar]
- Agulleiro J. I.; Fernandez J. J. (2011) Fast tomographic reconstruction on multicore computers. Bioinformatics 27, 582–583. [DOI] [PubMed] [Google Scholar]
- Nicastro D.; Schwartz C. L.; Pierson J.; Gaudette R.; Porter M. E.; McIntosh J. R. (2006) The molecular architecture of axonemes revealed by cryoelectron tomography. Science 313, 944–948. [DOI] [PubMed] [Google Scholar]
- Borkovich K. A.; Simon M. I. (1991) Coupling of receptor function to phosphate-transfer reactions in bacterial chemotaxis. Methods Enzymol. 200, 205–214. [DOI] [PubMed] [Google Scholar]
- Fowler D. J.; Weis R. M.; Thompson L. K. (2010) Kinase-active signaling complexes of bacterial chemoreceptors do not contain proposed receptor-receptor contacts observed in crystal structures. Biochemistry 49, 1425–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunanayake Mudiyanselage A. P. K. K.; Yang M.; Accomando L. A.-R.; Thompson L. K.; Weis R. M. (2013) Membrane association of a protein increases rate, extent, and specificity of chemical cross-linking. Biochemistry 52, 6127–6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefusco D. J.; Shrout A. L.; Besschetnova T. Y.; Weis R. M. (2007) Formation and activity of template-assembled receptor signaling complexes. Langmuir 23, 3280–3289. [DOI] [PubMed] [Google Scholar]
- Zhang P.; Bos W.; Heymann J.; Gnaegi H.; Kessel M.; Peters P. J.; Subramaniam S. (2004) Direct visualization of receptor arrays in frozen-hydrated sections and plunge-frozen specimens of E. coli engineered to overproduce the chemotaxis receptor Tsr. J. Microsc. 216, 76–83. [DOI] [PubMed] [Google Scholar]
- Weis R. M.; Hirai T.; Chalah A.; Kessel M.; Peters P. J.; Subramaniam S. (2003) Electron microscopic analysis of membrane assemblies formed by the bacterial chemotaxis receptor Tsr. J. Bacteriol. 185, 3636–3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefman J.; Zhang P.; Hirai T.; Weis R. M.; Juliani J.; Bliss D.; Bos E.; Peters P. J.; Subramaniam S. (2004) Three-dimensional electron microscopic imaging of membrane invaginations in Escherichia coli overproducing the chemotaxis receptor Tsr. J. Bacteriol. 186, 5052–5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khursigara C. M.; Wu X.; Zhang P.; Lefman J.; Subramaniam S. (2008) Role of HAMP domains in chemotaxis signaling by bacterial chemoreceptors. Proc. Natl. Acad. Sci. U.S.A. 105, 16555–16560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underbakke E. S.; Zhu Y. M.; Kiessling L. L. (2008) Isotope-coded affinity tags with tunable reactivities for protein footprinting. Angew. Chem. 120, 98323–99826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P.; Khursigara C. M.; Hartnell L. M.; Subramaniam S. (2007) Direct visualization of Escherichia coli chemotaxis receptor arrays using cryo-electron microscopy. Proc. Natl. Acad. Sci. U.S.A. 104, 3777–3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A. S.; Falke J. J. (2004) Side chains at the membrane-water interface modulate the signaling state of a transembrane receptor. Biochemistry 43, 1763–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draheim R. R.; Bormans A. F.; Lai R.-Z.; Manson M. D. (2006) Tuning a bacterial chemoreceptor with protein-membrane interactions. Biochemistry 45, 14655–14664. [DOI] [PubMed] [Google Scholar]
- Amin D. N.; Hazelbauer G. L. (2012) Influence of membrane lipid composition on a transmembrane bacterial chemoreceptor. J. Biol. Chem. 287, 41697–41705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuichet K.; Zhulin I. B. (2010) Origins and diversification of a complex signal transduction system in prokaryotes. Sci. Signaling 3, ra. 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich L. E.; Zhulin I. B. (2010) The MiST2 database: A comprehensive genomics resource on microbial signal transduction. Nucleic Acids Res. 38, D401–D407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briegel A.; Ding H. J.; Li Z.; Werner J.; Gitai Z.; Dias D. P.; Jensen R. B.; Jensen G. J. (2008) Location and architecture of the Caulobacter crescentus chemoreceptor array. Mol. Microbiol. 69, 30–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-Y.; Borbat P. P.; Gonzalez-Bonet G.; Bhatnagar J.; Pollard A. M.; Freed J. H.; Bilwes A. M.; Crane B. R. (2006) Reconstruction of the chemotaxis receptor-kinase assembly. Nat. Struct. Mol. Biol. 13, 400–407. [DOI] [PubMed] [Google Scholar]
- Kim K. K.; Yokota H.; Kim S. H. (1999) Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature 400, 787–792. [DOI] [PubMed] [Google Scholar]
- Li X.; Fleetwood A. D.; Bayas C.; Bilwes A. M.; Ortega D. R.; Zhulin I. B.; Crane B. R. (2013) The 3.2 Å Resolution Structure of a Receptor:CheA:CheW Signaling Complex Defines Overlapping Binding Sites and Key Residue Interactions within Bacterial Chemosensory Arrays. Biochemistry 52, 3852–3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrout A. L.; Montefusco D. J.; Weis R. M. (2003) Template-directed assembly of receptor signalling complexes. Biochemistry 42, 13379–13385. [DOI] [PubMed] [Google Scholar]
- Zhou H.-X.; Rivas G.; Minton A. P. (2008) Macromolecular crowding and confinement: Biochemical, biophysical, and potential physiological consequences. Annu. Rev. Biophys. 37, 375–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H.-X. (2013) Influence of crowded cellular environments on protein folding, binding, and oligomerization: Biological consequences and potentials of atomistic modeling. FEBS Lett. 587, 1053–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis R. J. (2001) Macromolecular crowding: Obvious but underappreciated. Trends Biochem. Sci. 26, 597–604. [DOI] [PubMed] [Google Scholar]
- Li M.; Hazelbauer G. L. (2004) Cellular stoichiometry of the components of the chemotaxis signalling complex. J. Bacteriol. 186, 3687–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannistraro V. J.; Glekas G. D.; Rao C. V.; Ordal G. W. (2011) Cellular stoichiometry of the chemotaxis proteins in Bacillus subtilis. J. Bacteriol. 193, 3220–3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chervitz S. A.; Falke J. J. (1995) Lock on/off disulfides identify the transmembrane signaling helix of the aspartate receptor. J. Biol. Chem. 270, 24043–24053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chervitz S. A.; Lin C.; Falke J. J. (1995) Transmembrane signaling by the aspartate receptor: Engineered disulfides reveal static regions of the subunit interface. Biochemistry 34, 9722–9733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sferdean F. C.; Weis R. M.; Thompson L. K. (2012) Ligand Affinity and Kinase Activity Are Independent of Bacterial Chemotaxis Receptor Concentration: Insight into Signaling Mechanisms. Biochemistry 51, 6920–6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.