Abstract
We have obtained the structure of the bacterial diterpene synthase, tuberculosinol/iso-tuberculosinol synthase (Rv3378c) from Mycobacterium tuberculosis, a target for anti-infective therapies that block virulence factor formation. This phosphatase adopts the same fold as found in the Z- or cis-prenyltransferases. We also obtained structures containing the tuberculosinyl diphosphate substrate together with one bisphosphonate inhibitor-bound structure. These structures together with the results of site-directed mutagenesis suggest an unusual mechanism of action involving two Tyr residues. Given the similarity in local and global structure between Rv3378c and the M. tuberculosis cis-decaprenyl diphosphate synthase (DPPS; Rv2361c), the possibility exists for the development of inhibitors that target not only virulence but also cell wall biosynthesis, based in part on the structures reported here.
Introduction
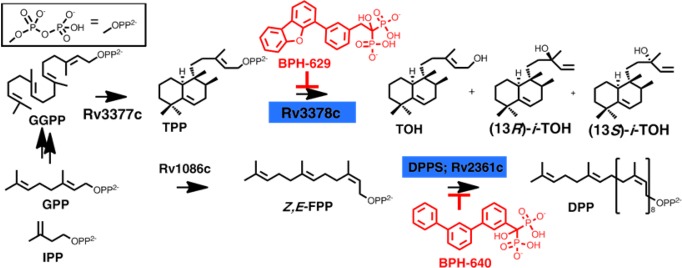
Tuberculosis, caused by the bacterium Mycobacterium tuberculosis, is a major source of morbidity and mortality worldwide with almost two million deaths annually, and the rise in multidrug resistant and extensively drug resistant strains is of great concern.1−3 There is thus considerable interest in the development of new drugs and new leads such as SQ109 and TMC-207,4 which target cell wall biosynthesis and ATP formation, respectively, and also in the development of new therapeutic approaches that target virulence factor (VF) formation. VFs are by definition not essential for bacterial growth outside of host cells but are involved in processes such as invasion, persistence, lysis, and evasion of innate immune system responses, as found for example with staphyloxanthin in Staphylococcus aureus.5,6 This carotenoid VF provides resistance to host-based reactive oxygen species-based killing, and inhibiting staphyloxanthin biosynthesis is a novel route to anti-infective therapy.6 In M. tuberculosis, one class of VFs are the tuberculosinols:7−9 tuberculosinol (TOH) and the (±)-iso-tuberculosinols (iso-TOH), Scheme 1. The latter were first isolated from a marine sponge from the Nosy Be island, Madagascar, and are thus also known as (±)-nosyberkols10 (Scheme 1). The original structures proposed7−10 have been confirmed by total synthesis.11,12 The biosynthesis of the tuberculosinols is catalyzed by two enzymes: Rv3377c, tuberculosinyl (halama-5,13-dien-15-yl) diphosphate synthase, and Rv3378c, tuberculosinol/(±)-iso-tuberculosinol synthase (Scheme 1). Both proteins are essential for bacterial survival inside macrophages13 with the tuberculosinols inhibiting phagolysosome maturation as well as macrophage phagocytosis.8,9,14 Thus, the three-dimensional (3D) structures of both Rv3377c and Rv3378c are of considerable interest since they are thought to represent novel antivirulence therapeutic targets for tuberculosis.15 Rv3377c is a diterpene cyclase that converts geranylgeranyl diphosphate (GGPP) into tuberculosinyl diphosphate (TPP), while Rv3378c converts TPP into TOH and the iso-TOHs, acting as a phosphatase/isomerase, as shown in Scheme 1. Here, we report the structure of Rv3378c both in its apo form as well as bound to its TPP substrate, and to a bisphosphonate inhibitor, BPH-629 (Scheme 1). We also report the structure of the M. tuberculosis cis-decaprenyl diphosphate synthase (Rv2361c) with another bisphosphonate inhibitor, BPH-640 (Scheme 1), which has a similar structure and inhibitor-binding mode as those found with Rv3378c, the phosphatase.
Scheme 1. Biosynthesis of Mycobacterium tuberculosis virulence factors (tuberculosinols) and decaprenyl diphosphate, which is essential for cell wall biosynthesis.
Materials and Methods
Full details are given in the Supporting Information (SI) online.
Result and Discussion
We cloned, expressed, purified, crystallized, and solved the Rv3378c protein structure by using multiple isomorphous replacement in a tetragonal crystal and then by using molecular replacement in a monoclinic crystal, Table 1 and Table S1 in the SI. Residues 46–50 could not be modeled in one crystal, or 82–90 in another, due to lattice packing and disorder. Nevertheless, the two structures superimposed well, and a continuous protein dimer model could be constructed (Figure 1a and Figure S1a in the SI), consistent with the observation that Rv3378c also exists as a dimer in solution.9 The overall fold shows close structural homology to that seen in M. tuberculosis cis-farnesyl diphosphate synthase (Rv1086; 2.33 Å Cα rmsd over 201 residues; PDB ID code 2VFW), M. tuberculosis decaprenyl diphosphate synthase (Rv2361; 2.23 Å over 195 residues; PDB ID code 2VG4), and E. coli undecaprenyl diphosphate synthase (UPPS; 2.44 Å over 203 residues; PDB ID code 1JP3), although there is only ∼12–14% sequence identity with these proteins.
Table 1. Data collection and refinement statistics for the Rv3378c crystals. Values in parentheses are for the outermost resolution shells.
| Rv3378c native 1 | Rv3378c native 2 | Rv3378c Y51F with TPP | Rv3378c Y51F/Y90F with TPP | Rv3378c with 629 | Rv2361c with 640 | |
|---|---|---|---|---|---|---|
| Data Collection | ||||||
| resolution (Å) | 25.0–2.3 (2.4–2.3) | 25.0–2.1 (2.2–2.1) | 25.0–2.7 (2.8–2.7) | 25.0–2.4 (2.5–2.4) | 25.0–2.1 (2.2–2.1) | 50–1.8 (1.9–1.8) |
| space group | P43212 | C2 | P43212 | P43212 | P43212 | P212121 |
| unit cell | ||||||
| a (Å) | 105.9 | 208.4 | 106.5 | 106.3 | 105.4 | 77.8 |
| b (Å) | 105.9 | 56.5 | 106.5 | 106.3 | 105.4 | 89.2 |
| c (Å) | 66.6 | 114.4 | 66.4 | 66.8 | 66.0 | 93.9 |
| β (deg) | 90.3 | 90.3 | 90.3 | 90.3 | 90.3 | 90.0 |
| no. of unique reflections | 18334 (1794) | 77301 (6681) | 11190 (1049) | 15419 (1505) | 21574 (2082) | 58370 (2875) |
| redundancy | 7.9 (8.1) | 3.9 (2.7) | 7.2 (7.7) | 10.3 (10.6) | 10.2 (10.2) | 14.8 (14.8) |
| completeness (%) | 99.9 (100.0) | 98.1 (85.6) | 99.3 (96.8) | 99.7 (100.0) | 99.3 (97.6) | 100.0 (100.0) |
| mean I/σ(I) | 43.6 (6.5) | 19.2 (2.3) | 47.1 (3.6) | 37.2 (4.3) | 52.4 (6.4) | 41.6 (3.4) |
| Rmerge (%) | 5.0 (43.3) | 6.5 (41.6) | 5.6 (39.1) | 6.5 (52.1) | 5.3 (39.7) | 7.8 (69.4) |
| Refinement | ||||||
| no. of chain(s) | 1 | 4 | 1 | 1 | 1 | 2 |
| no. of reflections | 17676 (1634) | 65562 (2237) | 10756 (946) | 14914 (1326) | 20950 (1961) | 55357 (3783) |
| Rwork (95% of data) | 0.212 (0.250) | 0.198 (0.222) | 0.240 (0.342) | 0.231 (0.293) | 0.197 (0.242) | 0.157 (0.227) |
| Rfree (5% of data) | 0.239 (0.308) | 0.239 (0.302) | 0.261 (0.365) | 0.260 (0.363) | 0.237 (0.243) | 0.198 (0.294) |
| rmsd bonds (Å) | 0.007 | 0.006 | 0.006 | 0.008 | 0.008 | 0.025 |
| rmsd angles (deg) | 1.3 | 1.2 | 1.3 | 1.2 | 1.4 | 2.4 |
| Ramachandran Plot (%) | ||||||
| most favored (%) | 95.1 | 96.3 | 94.4 | 94.7 | 96.9 | 94.0 |
| allowed (%) | 4.9 | 3.7 | 5.6 | 5.3 | 3.1 | 5.8 |
| disallowed (%) | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.2 |
| mean B (Å2) /atoms | 55.2/2611 | 28.6/10274 | 89.0/2520 | 55.7/2444 | 46.8/2757 | 26.9/4510 |
| PDB ID code | 3WQK | 3WQL | 3WQN | 4KT8 | 3WQM | 4ONC |
Figure 1.
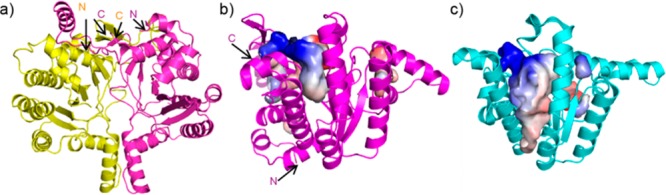
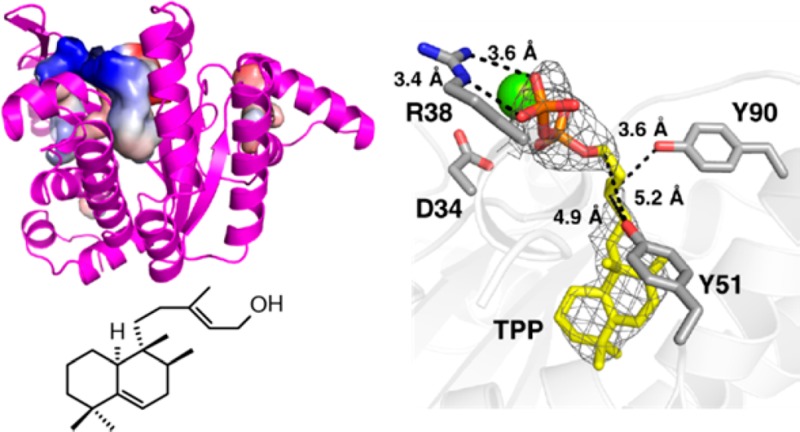
Structures of tuberculosinol/(13R,S)-iso-tuberculosinol synthase (Rv3378c) and undecaprenyl diphosphate synthase (UPPS). (a) Rv3378c dimer. Both N- and C-termini of both monomers locate at the top of the dimer structure. (b) Predicted Rv3378c ligand binding site. (c) Escherichia coli UPPS ligand binding sites (PDB ID code 2E98). Electrostatic surfaces are shown for Rv3378c and UPPS cavities, colored blue in positive region, red in negative region, calculated using PyMOL.
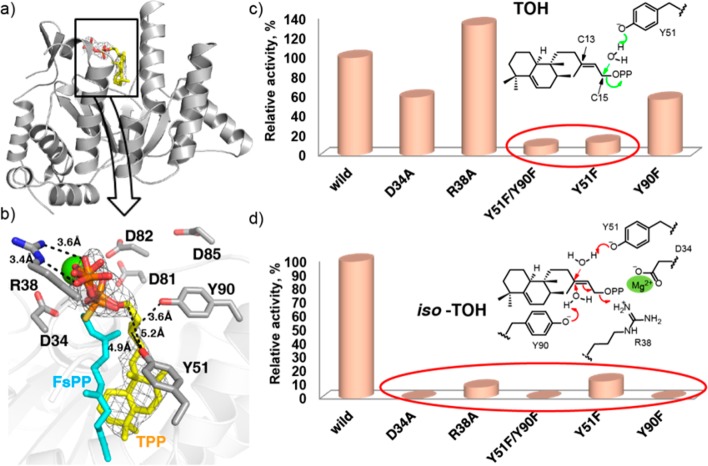
There is a potential substrate-binding site at the “top” of the structure (Figure 1b and Figure S1b in the SI), but this cavity16 is smaller than that seen in UPPS, which makes the C55 diphosphate used in bacterial cell wall biosynthesis (∼600 Å3 versus ∼900–1700 Å3, depending on the presence or absence of bound ligands, Figure 1c and Figure S1c in the SI). The Rv3378c protein thus adopts the same fold as seen in cis-head-to-tail prenyl diphosphate synthases, and this structure is unprecedented in enzymes involved in terpene biosynthesis. Attempts to obtain the structures of Rv3378c in the presence of its substrate TPP (by soaking and cocrystallization) were unsuccessful; therefore, we next used computational docking17 to see how TPP might bind to Rv3378c, to guide the design of inactive mutants. The docking results obtained (Figure S2 in the SI) indicated that the fused rings bound to a large hydrophobic pocket while the allyl side chain docked close to Y51 and Y90, suggesting that these residues might be involved in activating water molecules for nucleophilic attack at C13 and/or C15. To test this hypothesis, we made a Y51F mutant and a Y51F/Y90F double mutant, finding ∼13% and ∼5% wild-type activity, respectively, Figure S3 in the SI. We then obtained structures of both mutants in the presence of TPP. In both, TPP occupies the polar-top/nonpolar-bottom cavity shown in Figure 1b (Figure 2a and Figure S4a in the SI), with more complete electron densities found in the Y51F/Y90F double mutant (Figure 2b and Figure S4b in the SI, yellow) than in the Y51F single mutant (Figure S5 in the SI), due presumably to more residual activity in the Y51F single mutant crystal. A similar though not identical hydrophobic pocket is occupied by S-thiolo farnesyl diphosphate (FSPP) when bound to UPPS, as illustrated in the superposition shown in Figure 2b (FSPP in cyan). In UPPS, the two most conserved residues from a SCORECONS18 analysis (which ranks residues in terms of their conserved nature) are D26 and R30 (Table S2 in the SI) which in UPPS are involved in binding to Mg2+ and farnesyl diphosphate (FPP), facilitating diphosphate activation and removal. These two residues correspond to D34 and R38 in Rv3378c (a 2.2 Å Cα rmsd), and based on this homology to UPPS, might therefore be expected to play a role in diphosphate activation and release.
Figure 2.
Structures and activities of Rv3378c. (a) Rv3378c Y51F/Y90F double mutant structure with bound TPP. (b) Rv3378c/TPP (yellow, electron density contoured at 1.5σ) structure superimposed on S-thiolo-FSPP (cyan), bound to UPPS (PDB code 1X06). Mg2+ is from the UPPS structure. Catalytic residues in Rv3378c are shown with gray sticks. Y51, Y90 are based on the F51, F90 coordinates. (c) Activities of wild-type and Rv3378c mutants in TOH production and proposed catalytic mechanism for TOH formation. (d) Activities of wild-type and Rv3378c mutants in iso-TOH production and proposed catalytic mechanism for iso-TOH formation.
To examine the role of these residues in catalysis, we mutated both to Ala. Activity results for these mutants together with Y51F, Y51F/Y90F, Y90F as well as the single DDXXD mutants reported previously9 are shown in c and d of Figure 2 as well as in Figure S3 in the SI. Whereas the WT protein has 41% iso-TOH activity and 59% TOH activity, D34A, R38A and Y90F mutants produced on average ∼1% iso-TOH but on average 49% TOH, meaning that D34, R38 and Y90 are all important for iso-TOH production, a more SN1-like reaction that requires D34 and R38 to facilitate dephosphorylation and carbocation formation, Figure 2d. On the other hand, Y51 is involved in both TOH and iso-TOH formation since production of both is reduced in the Y51F mutant. In addition, the Y51F/Y90F double mutant has essentially no activity. Taken together, these results indicate that D34, R38, Y51 and Y90 are involved in 13R,13S-iso-tuberculosinol formation (Figure 2d), while it is primarily Y51 that is involved in TOH formation, since both Y51F and the Y51F/Y90F mutants produce little or no TOH, as illustrated in Figure 2c. The single DDXXD mutants have activities ranging from 32% to 53%, whereas the double Asp mutants are not active.9 This suggests that the DDXXD motif, although not immediately adjacent to the active site, may also play a role in catalysis, perhaps by helping remove the Mg2+-PPi formed, since 2 Asps would be required for Mg2+ chelation.
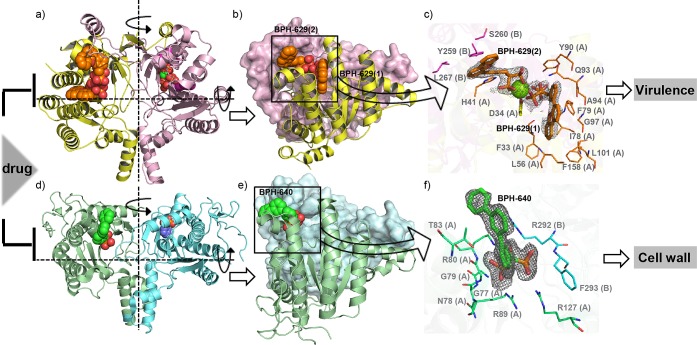
Since Rv3378c is a target for antivirulence therapeutics, we were also interested in discovering Rv3378c inhibitors. We identified BPH-629 as one such inhibitor having a Kd ≈ 560 nM, Figure S6a in the SI. Co-crystallization of Rv3378c and BPH-629 in the presence of Mg2+ yielded a complex structure containing two molecules of BPH-629 and one Mg2+ per Rv3378c monomer. Figure 3a and Figure S7a in the SI show the location of the inhibitor binding sites in the dimer. In each monomer, there are two bound BPH-629 molecules: one (BPH-629 (1)) is buried in the substrate binding site (Figure 3b and Figure S7b in the SI), the other (BPH-629 (2)) is located in the dimer interface, between a helix of monomer A and a loop of monomer B (a and b of Figure 3). In both cases, the bisphosphonate head-groups chelate to a single Mg2+ that is in turn coordinated to the side chain of D34, Figure 3c and Figure S7c in the SI. Moreover, as shown in Figure 3c, the side chain in BPH-629 (1) makes extensive nonpolar interactions with numerous hydrophobic amino acids in the TPP binding site, and BPH-629 (2) is sandwiched between the H41 of monomer A and Y259, S260, and L267 of monomer B.
Figure 3.
Inhibitor-bound structures of Rv3378c and DPPS. (a) Dimeric Rv3378c structure with two bound BPH-629 molecules (shown with spheres) in each monomer (four BPH-629 molecules in the dimer). Because there is only one molecule in an asymmetric unit, the dimer is constructed from the observed monomer structure and its symmetry partners. (b) Front view of Rv3378c dimer. Monomer A and monomer B are shown as a yellow cartoon and a pink surface, respectively. (c) Close-up view of the structure of Rv3378c bound to bisphosphonate inhibitor BPH-629 (thick orange sticks, electron densities contoured at 1.5σ). The BPH-629 molecules both coordinate to the catalytic Mg2+ (green sphere), which in turn coordinates to D34 (yellow sticks) from monomer A. BPH-629 (1) is buried in the substrate binding site in monomer A, formed by hydrophobic residues shown as orange lines. The hydrophobic moiety of BPH-629 (2) is stabilized by Y259, S260, and L267 from monomer B (magenta lines). (d) Dimeric DPPS structure with one bound BPH-640 molecule (shown as spheres) in each monomer. (e) Front view of DPPS dimer. Monomer A and monomer B are shown as a green cartoon and a cyan surface, respectively. (f) Close-up view of the structure of DPPS bound to bisphosphonate inhibitor BPH-640 (thick green sticks, electron densities contoured at 1.5σ). The hydrophobic moiety of BPH-640 is stabilized by residues from monomer A (green lines) and monomer B (cyan lines).
Interestingly, a similar surface binding site is also found in M. tuberculosis DPPS (Rv2361c) in complex with another bisphosphonate, BPH-640, a close analogue of BPH-629, as shown in d, e, and f of Figure 3 and Figure S8a–c in the SI. BPH-640 is a 410 nM inhibitor of DPPS (Figure S6b in the SI) but lacks the O and CH2 groups present in BPH-629. Full crystallographic data acquisition and structure refinement details for this structure are shown in Table 1, and electron density results are shown in Figure 3f. As with BPH-629 (2), BPH-640 also occupies a dimer interface binding site, sandwiched between G77, N78, G79, R80, T83, R89, and R127 of monomer A and R292 and F293 of monomer B (Figure 3f).
The results shown in Figure 3 indicate that the terpene synthase (phosphatase) Rv3378c and the cis-prenyl transferase DPPS both have the same overall fold. They both also exist as dimers in solution9,19 as well as in crystals (a and d of Figure 3), and both are inhibited by structurally similar bisphosphonates, with closely related binding modes (Figure 3b and e) and similar affinities. These observations lead to the idea that in the future it may be possible to design multitarget inhibitors20−22 that target both enzymes in TB bacteria inside macrophages, inhibiting growth (DPPS/cell wall biosynthesis) as well as virulence (Rv3378c/tuberculosinol biosynthesis/invasion).
Summary
We report the first structure of any bacterial diterpene synthase, tuberculosinol/iso-tuberculosinol synthase (Rv3378c) from M. tuberculosis, a target for anti-infective therapies that block virulence factor formation. This phosphatase adopts the same fold as found in the Z- or cis-prenyltransferases. We also obtained structures in the presence of TPP and with a bisphosphonate inhibitor where one molecule occupied the TPP substrate binding site; the second is located at the dimer interface site. An unusual surface site was also found with a related bisphosphonate inhibitor bound to the M. tuberculosis DPPS (Rv2361c). In addition, we used the X-ray structures of Rv3378c together with the results of site-directed mutagenesis to propose mechanisms of action for formation of tuberculosinol and the iso-tuberculosinols in which two Tyr residues play an important role. Given the similarity in local and global structures of Rv3378c and Rv2361c, the possibility exists that in the future it might be possible to develop multitarget inhibitors that target not only virulence, but also cell wall biosynthesis, in part based on the structures reported here.
Acknowledgments
This work was supported by the National Basic Research Program of China (2011CB710800 and 2011CBA00805), the Tianjin Municipal Science and Technology Commission (12ZCZDSY12500), and the United States Public Health Service (National Institutes of Health Grant GM065307). X.F. was supported by a Predoctoral Fellowship from the American Heart Association, Midwest Affiliate (13PRE14510056). We thank the National Synchrotron Radiation Research Center of Taiwan for beamtime allocation and data collection assistance.
Supporting Information Available
Experimental materials, methods, supporting tables and figures. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
¶ H.C.C., X.F., and T.P.K. contributed equally.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Ernst J. D. Nat. Rev. Immunol. 2012, 12, 581. [DOI] [PubMed] [Google Scholar]
- Lienhardt C.; Glaziou P.; Uplekar M.; Lonnroth K.; Getahun H.; Raviglione M. Nat. Rev. Microbiol. 2012, 10, 407. [DOI] [PubMed] [Google Scholar]
- Zumla A.; Hafner R.; Lienhardt C.; Hoelscher M.; Nunn A. Nat. Rev. Drug. Discovery 2012, 11, 171. [DOI] [PubMed] [Google Scholar]
- Koul A.; Arnoult E.; Lounis N.; Guillemont J.; Andries K. Nature 2011, 469, 483. [DOI] [PubMed] [Google Scholar]
- Liu G. Y.; Essex A.; Buchanan J. T.; Datta V.; Hoffman H. M.; Bastian J. F.; Fierer J.; Nizet V. J. Exp. Med. 2005, 202, 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. I.; Liu G. Y.; Song Y.; Yin F.; Hensler M. E.; Jeng W. Y.; Nizet V.; Wang A. H.; Oldfield E. Science 2008, 319, 1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano C.; Hoshino T. ChemBioChem 2009, 10, 2060. [DOI] [PubMed] [Google Scholar]
- Hoshino T.; Nakano C.; Ootsuka T.; Shinohara Y.; Hara T. Org. Biomol. Chem. 2011, 9, 2156. [DOI] [PubMed] [Google Scholar]
- Nakano C.; Ootsuka T.; Takayama K.; Mitsui T.; Sato T.; Hoshino T. Biosci. Biotechnol. Biochem. 2011, 75, 75. [DOI] [PubMed] [Google Scholar]
- Rudi A.; Aknin M.; Gaydou E.; Kashman Y. J. Nat. Prod. 2004, 67, 1932. [DOI] [PubMed] [Google Scholar]
- Maugel N.; Mann F. M.; Hillwig M. L.; Peters R. J.; Snider B. B. Org. Lett. 2010, 12, 2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangler J. E.; Carson C. A.; Sorensen E. J. Chem. Sci. 2010, 1, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pethe K.; Swenson D. L.; Alonso S.; Anderson J.; Wang C.; Russell D. G. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 13642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann F. M.; Xu M.; Chen X.; Fulton D. B.; Russell D. G.; Peters R. J. J. Am. Chem. Soc. 2009, 131, 17526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann F. M.; Prisic S.; Hu H.; Xu M.; Coates R. M.; Peters R. J. J. Biol. Chem. 2009, 284, 23574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendlich M.; Rippmann F.; Barnickel G. J. Mol. Graph. Modell. 1997, 15, 359. [DOI] [PubMed] [Google Scholar]
- Friesner R. A.; Murphy R. B.; Repasky M. P.; Frye L. L.; Greenwood J. R.; Halgren T. A.; Sanschagrin P. C.; Mainz D. T. J. Med. Chem. 2006, 49, 6177. [DOI] [PubMed] [Google Scholar]
- Valdar W. S. Proteins 2002, 48, 227. [DOI] [PubMed] [Google Scholar]
- Wang W.; Dong C.; McNeil M.; Kaur D.; Mahapatra S.; Crick D. C.; Naismith J. H. J. Mol. Biol. 2008, 381, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Cao R.; Yin F.; Hudock M. P.; Guo R. T.; Krysiak K.; Mukherjee S.; Gao Y. G.; Robinson H.; Song Y.; No J. H.; Bergan K.; Leon A.; Cass L.; Goddard A.; Chang T. K.; Lin F. Y.; Van Beek E.; Papapoulos S.; Wang A. H.; Kubo T.; Ochi M.; Mukkamala D.; Oldfield E. J. Am. Chem. Soc. 2009, 131, 5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morphy J. R.; Harris C. J.. Designing Multi-Target Drugs: Royal Society of Chemistry: London, 2012. [Google Scholar]
- Lin F. Y.; Zhang Y.; Hensler M.; Liu Y. L.; Chow O. A.; Zhu W.; Wang K.; Pang R.; Thienphrapa W.; Nizet V.; Oldfield E. ChemMedChem 2012, 7, 561. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.