Abstract
Context
Some melanomas arising from acral, mucosal, and chronically sun-damaged sites harbor activating mutations and amplification of the type III transmembrane receptor tyrosine kinase KIT. We explored the effects of KIT inhibition using imatinib mesylate in this molecular subset of disease.
Objective
To assess clinical effects of imatinib mesylate in patients with melanoma harboring KIT alterations.
Design, Setting, and Patients
A single-group, open-label, phase 2 trial at 1 community and 5 academic oncology centers in the United States of 295 patients with melanoma screened for the presence of KIT mutations and amplification between April 23, 2007, and April 16, 2010. A total of 51 cases with such alterations were identified and 28 of these patients were treated who had advanced unresectable melanoma arising from acral, mucosal, and chronically sun-damaged sites.
Intervention
Imatinib mesylate, 400 mg orally twice daily.
Main Outcome Measures
Radiographic response, with secondary end points including time to progression, overall survival, and correlation of molecular alterations and clinical response.
Results
Two complete responses lasting 94 (ongoing) and 95 weeks, 2 durable partial responses lasting 53 and 89 (ongoing) weeks, and 2 transient partial responses lasting 12 and 18 weeks among the 25 evaluable patients were observed. The overall durable response rate was 16% (95% confidence interval [CI], 2%–30%), with a median time to progression of 12 weeks (interquartile range [IQR], 6–18 weeks; 95% CI, 11–18 weeks), and a median overall survival of 46.3 weeks (IQR, 28 weeks-not achieved; 95% CI, 28 weeks-not achieved). Response rate was better in cases with mutations affecting recurrent hotspots or with a mutant to wild-type allelic ratio of more than 1 (40% vs 0%, P=.05), indicating positive selection for the mutated allele.
Conclusions
Among patients with advanced melanoma harboring KIT alterations, treatment with imatinib mesylate results in significant clinical responses in a subset of patients. Responses may be limited to tumors harboring KIT alterations of proven functional relevance.
Melanoma causes the greatest morbidity and mortality of all skin cancers.1 An estimated 68 130 invasive melanomas were diagnosed and 8700 deaths due to metastatic disease were recorded in the United States in 2010.2 Dacarbazine,3 interleukin 2,4 and ipilimumab are approved for the treatment of metastatic melanoma by the US Food and Drug Administration. Only ipilimumab has been shown to improve overall survival.3–5
Melanomais composed of several biologically distinct subtypes, each with unique genetic and clinical features,6 and each likely to respond differently to any one therapeutic strategy. The most common melanoma subtype in the United States arises from non−chronically sun-damaged (non-CSD) skin and often harbors activating mutations in BRAF.7 Melanoma arising from mucosal, acral, and CSD sites infrequently have BRAF mutations, but commonly have amplifications or activating mutations of KIT.8,9 KIT is a type III transmembrane receptor tyrosine kinase.10 Binding of its ligand, stem cell factor, results in receptor dimerization, autophosphorylation, and activation of several signaling pathways; thereby, mediating cancer cell growth, proliferation, invasion, metastasis, and inhibition of apoptosis.
The importance of KIT in normal melanocyte development is well established11–13; however, its role as an oncogene and therapeutic target in melanoma has only recently become clear. Although KIT is expressed in some melanomas, loss of expression is observed with progression of disease from superficial to invasive to metastatic stages, suggesting that KIT possesses tumor suppressive functions.14–18 Furthermore, 3 phase 2 studies of metastatic melanoma treated with imatinib mesylate, an orally available ATP-competitive inhibitor of several tyrosine kinases including KIT, did not demonstrate clinical activity.19–21 These trials accrued before the discovery of activating mutations of KIT in melanoma and did not select patients based on the presence of KIT mutations or amplification.
KIT is an established therapeutic target in cancers with activating mutations of KIT, such as gastrointestinal stromal tumors (GIST), and significant benefit is achieved with various small molecule inhibitors of KIT including imatinib mesylate.22 Several melanoma cell lines with KIT mutations are highly sensitive to imatinib mesylate.23–25 Furthermore, several patients with melanoma harboring KIT alterations, including a K642E mutation as well as a 7-codon duplication of exon 11, have been reported to achieve major durable responses to imatinib mesylate.26,27
Given the preclinical and anecdotal clinical activity of imatinib mesylate observed in KIT mutant melanoma, we conducted this study to test the hypothesis that inhibition of KIT in a molecularly selected subgroup of patients with melanomas harboring mutations or amplification of KIT will result in objective regression and disease control. We further explored whether the identification of functionally relevant KIT alterations would allow us to better select patients most likely to respond to KIT inhibition.
METHODS
Patients
Between April 23, 2007, and April 16, 2010, 328 patients were enrolled from 1 community and 5 academic oncology centers in the United States for molecular screening and determination of eligibility. Eligible patients included those patients aged 18 years or older with metastatic melanoma arising from acral, mucosal, and body sites with signs of CSD harboring mutations or amplification of KIT. Patients required an Eastern Cooperative Oncology Group performance status of 0 (able to be fully active and perform all predisease activities without restriction), 1 (unable to perform physically strenuous activity but ambulatory and able to perform work of a light or sedentary nature, such as light housework or office work), or 2 (ambulatory and capable of all self-care, but unable to perform any work activities),28 life expectancy of 3 months or longer, measurable disease according to the Response Evaluation Criteria in Solid Tumors (RECIST),29 and adequate bone marrow, hepatic, and renal function. Prior systemic therapy and treated stable brain metastases were permitted.
All patients were informed of the investigational nature of the study and provided written informed consent in accordance with institutional and federal guidelines. The protocol was approved by each center’s institutional review board. Race/ethnicity was self-reported by the participants.
Molecular Screening
Tumor specimens were tested for KIT mutations and amplification. DNA for mutation analysis was extracted from formalin-fixed, paraffin-embedded specimens as previously published.30 Polymerase chain reaction assays using primers specific for KIT exons 9, 11, 13, 17, and 18; NRAS exons 1 and 2; BRAF exon 15; and GNAQ exon 5 were used, followed by Sanger sequencing. Polymerase chain reaction products were purified using ExoSAP-IT (USB Corporation, Cleveland, Ohio) and directly sequenced in the forward and reverse directions using the Applied Biosystems 3730 capillary DNA analyzer (Applied Biosystems, Foster City, California).
Fluorescence in situ hybridization was performed on formalin-fixed, paraffin-embedded sections as previously described.23 Human BAC clones RP11-722F1 and CTD-3180G20 spanning KIT in 4q12 (Invitrogen, Carlsbad, California), reference clones RP11-365H22 (Wellcome Trust Sanger Institute, Hinxton, Cambridge, England) and RP11-799E21 (BACPAC Resources, Oakland, California) for proximal 4q, and RP11-19F13 (Wellcome Trust Sanger Institute) and RP11-461G20(BACPAC Resources) proximal 4p were used.31 BAC DNA was labeled by nick translation with red dUTP (KIT), orange dUTP (4q reference), or green dUTP (4p reference) from Enzo Life Sciences Inc, Plymouth Meeting, Pennsylvania. Analysis used a Zeiss Axioplan epifluorescence microscope with motorized stage and Isis 5 imaging software (MetaSystems Group, Waltham, Massachusetts). Amplification was defined as a KIT-to-centromere ratio of more than 2.5, with a ratio of more than 10 representing high-level amplification. Polysomy was defined as the presence of discrete red, orange, and green signal groups.
Study Procedures
Patients whose tumors harbored a KIT alteration were eligible to receive imatinib mesylate, 400mgorally twice daily in 6-week cycles until unacceptable toxicity or disease progression. Imatinib mesylate was supplied by the Division of Cancer Treatment and Diagnosis at the National Cancer Institute. Imatinib mesylate was reduced to 400 mg/d in the setting of toxicity, with one further reduction to 300 mg/d permitted. Study participation was discontinued if adverse effects persisted.
Safety evaluations were conducted within 14 days of treatment initiation, every 2 weeks for 18 weeks, and every 6 weeks thereafter. Evaluation included physical examination, complete blood cell count, and clinical chemistry panel, including lactate dehydrogenase. Adverse events were graded according to the National Cancer Institute Common Terminology Criteria for Adverse Events version 3.0 (available at http://www.ctep.info.nih.gov/protocolDevelopment/electronic_applications/docs/ctcaev3.pdf).
Radiographic imaging and tumor measurements were performed within 4 weeks of treatment initiation, every 6weeks for 18 weeks, and every 12 weeks subsequently. Response was judged according to RECIST version 1.0.29 All RECIST responses were confirmed by a central radiologist (J.T.).To be considered durable, changes in tumor measurements were confirmed by repeat radiologic assessment performed no less than 6 weeks after the initial response was observed. Stable disease was defined as a less than 30% decreaseorlessthan20% increase in the sum of the longest diameters of the target lesions, taking as reference the baseline tumor measurements. Patients receiving 2 or more weeks of imatinib mesylate were evaluable for response. Patients receiving at least 1week of imatinib mesylate were evaluable for toxicity.
Statistical Analysis
The primary end point of this single-group, open-label, phase 2 study was durable objective response defined as partial response or complete response according to RECIST version 1.0.29 Secondary end points included time to progression, overall survival, and correlation of molecular alterations and clinical response.
A Simon 2-stage mini-max design32 was used to assess the primary end point of response, with the following parameters: 10% not promising response rate, 30% promising response rate, and .10 type I and II error rates. If 1 or more durable responses were observed in the first 16 evaluable patients, accrual continued until 25 evaluable patients were identified. If 5 or more durable responses were observed, imatinib mesylate would be considered worthy of further testing in this patient population.
Survival time was defined as the time from initiation of imatinib mesylate to the date of death or last follow-up. Time to progression was defined as the time from initiation of imatinib mesylate to the date of progression or last follow-up. Survival distributions were estimated using the Kaplan-Meier method.33 Comparisons were made by log-rank test. Baseline patient characteristics and adverse events were reported using summary statistics and frequency tables. Adverse events were reported as the most severe manifestation of each event category during any cycle of treatment. Fisher exact test was used to compare molecular alteration and response. Statistical analysis was conducted using SAS version 9.2 (SAS Institute Inc, Cary, North Carolina). All significance tests were 2-sided and used a 5% level of significance.
RESULTS
Molecular Screening
Three hundred twenty-eight patients consented to molecular screening. The flow of patients and tumor testing is shown in eFigure 1 (available at http://www.jama.com). Mutation status for KIT, BRAF, NRAS, and GNAQ, and KIT amplification status was determined in 295 cases, including 85 acral melanomas, 93 mucosal melanomas, and 87 melanomas clinically determined to arise from CSD sites. Fifteen melanomas arising from skin without clinical signs of CSD, 1 primary meningeal melanoma, and 14 melanomas of unknown primary site were inadvertently included for screening. KIT mutations or amplification were found in 23.8% (20 of 84 cases) of acral melanomas, 24.7% (23 of 93 cases) of mucosal melanomas, and 9.2% (8 of 87 cases) of melanomas clinically determined to arise from CSD sites. Histopathological confirmation of grade 2 or higher solar elastosis defining CSD34 was performed in 39 of 87 melanomas (44.8%) with clinical signs of CSD. Clinical and histopathological assessments of CSD were concordant in 32 cases (82%). The proportion of histopathologically confirmed CSD melanomas harboring KIT mutations or amplification was 18.8% (6 of 32 cases), twice that observed in melanomas clinically determined to arise from CSD sites (Table 1).NoKIT alterations were identified in the meningeal melanoma, non-CSD melanomas, or melanomas of unknown primary tested.
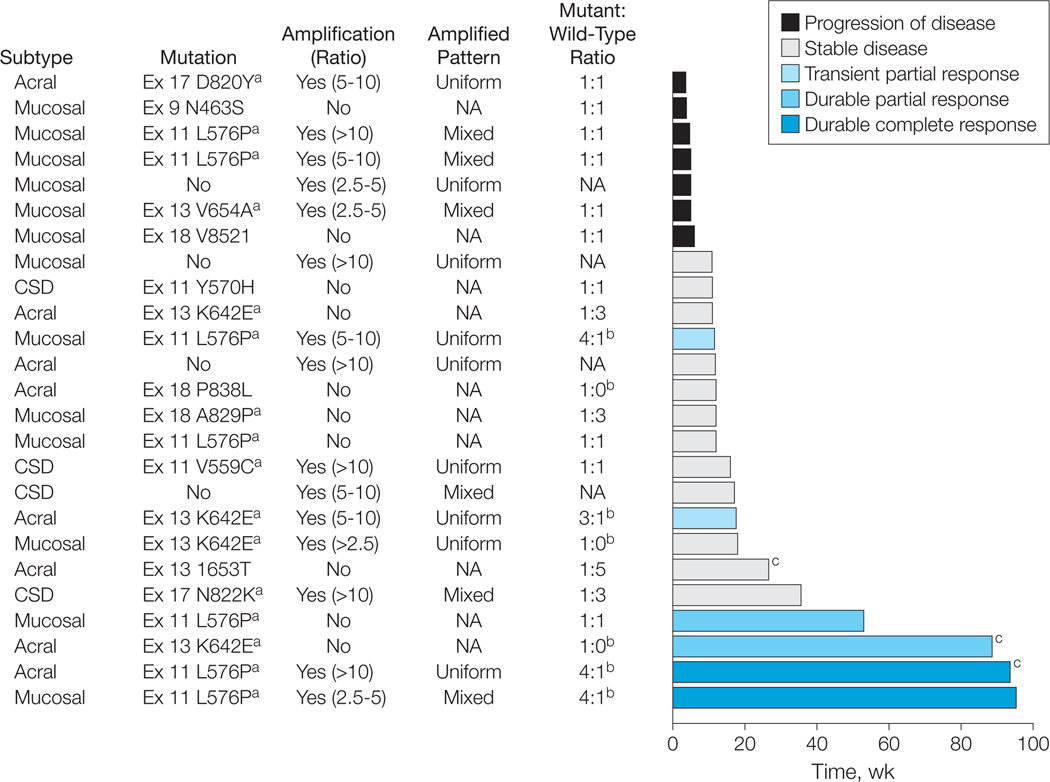
Figure 1. Treatment Response Over Time by Melanoma Subtype and Genetic Alteration of KIT.
CSD indicates melanoma arising from chronically sun-damaged skin; NA, not applicable. The melanoma subtype, KIT mutational and amplification status, the ratio of mutant to wild-type alleles as determined by their respective electropherogram peak heights, and the RECIST (Response Evaluation Criteria in Solid Tumors) response achieved for each patient treated with imatinib mesylate is shown. Amplification was defined as a KIT-to-centromere ratio of 2.5 or more, with the pattern of amplification described as uniform (homogeneous) or mixed (heterogeneous) across the tumor specimen. The best response as determined by RECIST is identified by the color of the bar. The length of the bar represents the duration of time in weeks that each patient remained receiving therapy.
a Mutations previously identified in melanoma8,9 and gastrointestinal stroma tumors.35,36
b Tumors with mutant to wild-type allelic ratios of more than 1:1, which are those hypothesized most likely to respond to therapy with KIT inhibition.
c Patients receiving treatment at the time of this report.
Table 1.
Frequency of Alterations Identified in KIT, BRAF, NRAS, and GNAQ by Melanoma Subtype
| Melanoma | No./Total No. (%) of Patients | ||||||
|---|---|---|---|---|---|---|---|
| KIT |
BRAF Mutation |
NRAS Mutation |
GNAQ Mutation |
||||
| Mutation | Amplification | Both | Either | ||||
| Acral (n = 85) | 18/84 (21.4) | 8/83 (9.6) | 6/82 (7.3) | 20/84 (23.8) | 19/84 (22.6) | 13/84 (15.5) | 1/80 (1.2) |
| Mucosal (n = 93) | 17/93 (18.3) | 14/91 (15.2) | 8/91 (8.8) | 23/93 (24.7) | 8/92 (8.7) | 11/92 (12.0) | 0/92 (0) |
| CSD, centrally reviewed (n = 32)a | 5/32 (15.6) | 3/30 (10.0) | 2/30 (6.7) | 6/32 (18.8) | 7/32 (21.9) | 0/32 (0) | 0/31 (0) |
| Total | 40/209 (19.1) | 25/204 (12.2) | 16/203 (7.9) | 49/209 (23.4) | 34/208 (16.3) | 24/208 (11.5) | 1/203 (0.5%) |
Abbreviation: CSD, chronically sun-damaged.
Among the 87 patients with melanoma arising from skin with clinical evidence of CSD, 8 (9.2%) harbored either a mutation or amplification of KIT. When limiting assessment to the 32 patients with melanoma arising from sun-damaged skin confirmed histopathologically by central review, only 6 cases harbored a KIT mutation or amplification. Overall, 51 cases harboring a mutation or amplification of KIT were identified; however, the subset of cases described in this Table includes only 49 such cases.
Fifty-one cases harbored KIT alterations, including 26 cases (51%) harboring KIT mutations only, 9 (18%) harboring KIT amplification only, and 16 (31%) concurrently harboring both KIT mutations and amplification. The distribution and frequency of mutations identified is shown in eFigure 2. Forty-eight of 51 cases with KIT alterations had no concomitant mutations in BRAF, NRAS, or GNAQ. In total, 162 of 295 samples (54.9%) tested had mutations in BRAF, NRAS, or GNAQ, or a mutation or amplification of KIT (eTable 1).
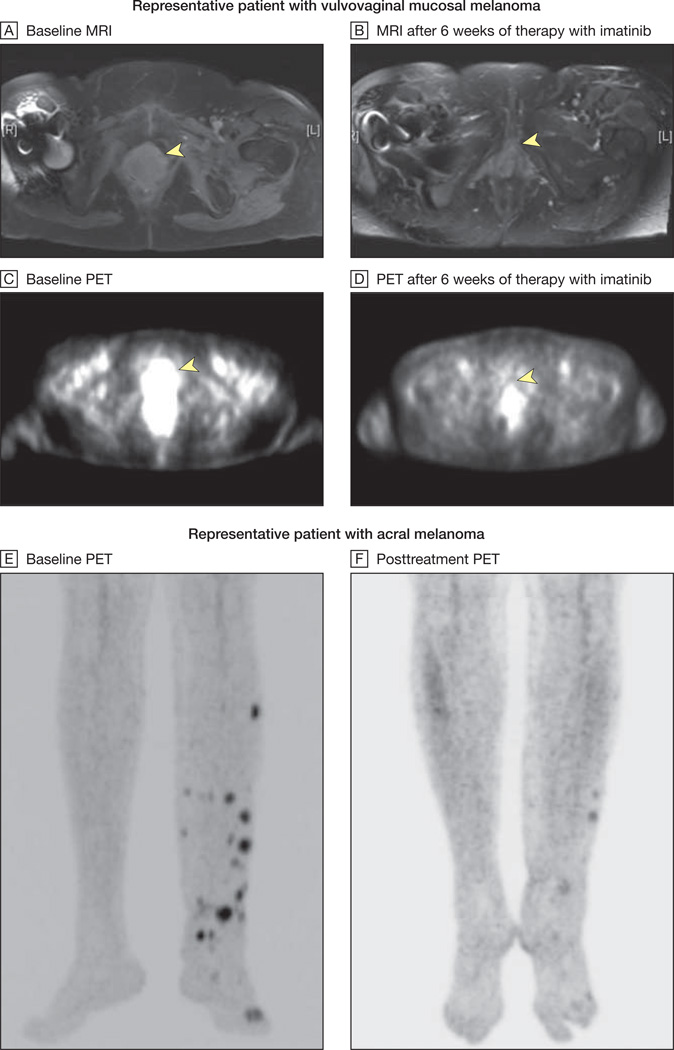
Figure 2. Radiographic Responses to Imatinib Mesylate.
MRI indicates magnetic resonance imaging; PET, positron emission tomography. MRI and PET scans from a representative patient with vulvovaginal mucosal melanoma harboring both an exon 11 L576P mutation and amplification of KIT are shown at baseline (yellow arrows, A and C) and after 6 weeks of therapy with imatinib mesylate (yellow arrows, B and D). The arrowhead in A indicates a heterogeneous multilobulated mass distending the entire vagina and extending inferiorly to the introitus. This lesion demonstrates intense fluorodeoxyglucose uptake on PET imaging (arrowhead in C). After 6 weeks of therapy with imatinib mesylate, there was significant shrinkage in the size of this mass with interval resolution of hypermetabolic activity (arrowheads in B and D indicate baseline locations of tumor). This patient ultimately achieved a complete radiographic response to therapy at her week 12 scan. Baseline and posttreatment PET images of a second representative patient with an acral melanoma harboring an exon 11 576P mutation and amplification are shown in E and F. This patient ultimately achieved a complete radiographic and pathological response to therapy at his week 12 scan.
Treatment
Twenty-eight of 51 patients whose tumors harbored KIT mutations or amplification were treated during the study (13 [46%] with mucosal melanoma, 10 [36%] with acral melanoma, and 5 [18%] with melanoma arising from CSD sites—3 arising from the head and neck and 2 arising from the trunk). The remaining 23 patients were not treated during the study because of no evidence of active disease (n=9), deceased (n=5), receiving other therapy (n=5), ineligible due to brain metastasis (n=1), ineligible due to increased creatinine (n=1), ineligible due to active human immunodeficiency virus infection (n=1), and ineligible due to the inability to wean off methadone, a contraindicated medication per protocolduetoCYP3A4interactions with imatinib mesylate (n=1). None of the patients who were treated had mutationsin BRAF, NRAS, or GNAQ. Baseline patient characteristics are shown in Table 2.Twenty-five patients were evaluable for the primary end point of response. Three were evaluable for toxicity only (2 patients stopped treatment within 2 weeks of starting therapy due to rapid disease progression and 1 stopped treatment due to the development of drug rash with eosinophilia and systemic symptoms syndrome within 10 days of initiating imatinib mesylate).
Table 2.
Demographic and Baseline Characteristics of the Patients
| Characteristic | No. (%) of Patients (n = 28) |
|
|---|---|---|
| Sex | ||
| Female | 15 (54) | |
| Male | 13 (46) | |
| Age, median (range), y | 71 (49–88) | |
| Race/ethnicity | ||
| White | 27 (96) | |
| Black | 1 (4) | |
| Non-Hispanic or non-Latino | 26 (93) | |
| Hispanic or Latino | 2 (7) | |
| Clinical melanoma subtype | ||
| Mucosal | 13 (46) | |
| Acral | 10 (36) | |
| Chronically sun-damaged | 5 (18) | |
| Increased lactate dehydrogenasea | 11 (39) | |
| ECOG performance status | ||
| 0 | 19 (68) | |
| 1 | 8 (28) | |
| 2 | 1 (4) | |
| Prior systemic regimens for metastatic disease | ||
| 0 | 7 (25) | |
| 1 | 14 (50) | |
| ≥2 | 7 (25) | |
Abbreviation: ECOG, Eastern Cooperative Oncology Group.
Indicates lactate dehydrogenase level of more than the upper limit of normal.
The most common adverse events observed are shown in eTable 2. Thirteen of 28 patients (46%) who were treated required a dose reduction to 400 mg/d, with 2 requiring a second dose reduction to 300 mg/d.
Clinical Outcomes
Two patients achieved durable complete responses, 2 achieved durable partial responses, 2 achieved transient partial responses, and 5 achieved stable disease lasting 12 or more weeks. No significant association between clinical melanoma subtype (mucosal: 23%; 95% confidence interval [CI], 8%–50%; acral: 38%; 95% CI, 14%–69%; and CSD: 0%; 95% CI, 0%–49%; P = .36) or lactate dehydrogenase (normal: 38%; 95% CI, 18%–61%; vs increased: 0%; 95%CI, 0%–30%; P = .06) and response was observed. RECIST response, time to progression, and KIT alteration identified for each evaluable patient are shown in Figure 1. Representative radiographic images of patients achieving responses are shown in Figure 2. Because 2 of the 6 responses were transient and not sustained on a follow-up scan performed 6 weeks after the initial response was documented, the predetermined end point of 5 responses of 25 evaluable patients who were treated was not met.
The median time to progression was 12 weeks (interquartile range [IQR], 6–18 weeks; 95% CI, 11–18 weeks). The 4 patients who achieved durable responses maintained disease control for more than 1 year. Durable disease control was observed in patients achieving stable disease, with 2 such patients maintaining stability for more than 6 months. Median survival from the time of initiation of therapy was 46.3 weeks (IQR, 28 weeks-not achieved; 95% CI, 28 weeks-not achieved), with 10 patients (40%) alive at the time of analysis (Figure 3).
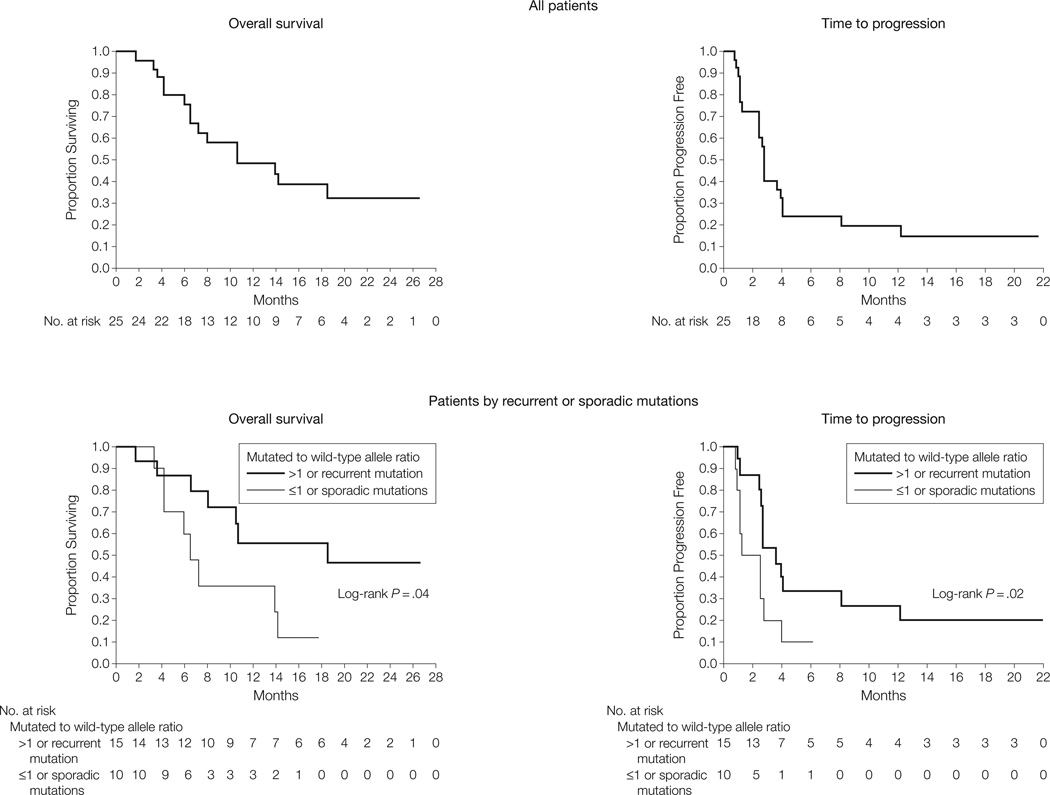
Figure 3. Overall Survival and Time to Progression of All 25 Patients and Those With Recurrent or Sporadic Mutations.
Kaplan-Meier curves for overall survival (15 deaths) and time to progression (22 progression events) of the 25 evaluable patients are shown. The median overall survival was 10.7 months (95% confidence interval [CI], 6.5 months-not achieved) and the median time to progression was 2.8 months (95% CI, 2.5–4.0 months). Mutations repeatedly identified in melanoma or gastrointestinal stromal tumors and those associated with a mutated to wild-type allele ratio of more than 1 are those most likely to be pathogenetically relevant. The subset of patients with tumors harboring such alterations achieved an overall survival and time to progression greater than that achieved in cases without such characteristics.
Five patients underwent optional serial tumor biopsies to determine histopathological and immunohistochemical effects of therapy. One case demonstrating decreased cellularity and reduced tumor proliferation in a clinically responding metastasis compared with both pretreatment sample as well as a clinically progressing lesion is shown in eFigure 3.
Association of Molecular Alterations and Clinical Response
We observed that KIT utations are widely distributed over the coding region as shown in eFigure 2. Many KIT mutations identified in our study have not previously been reported in melanoma8,9 or GIST35,36 and were present only in individual cases, suggesting that some may represent passenger mutations rather than bona fide driver alterations. Specific amino acid changes encountered repeatedly in cancer are likely to be of functional relevance. Interestingly, all 6 responses occurred in tumors with L576P or K642E mutations, the most common mutations in melanoma.8,9 The proportion of responders observed in the 13 cases whose tumors harbored mutations recurrently found in GIST37,38 or melanoma8,9 (V559C, L576P, V642E, and N822K) is 46%, higher than in the group as a whole.
Another method to separate driver from passenger mutations is to identify evidence of positive selection for the mutation. We investigated whether relative abundance of mutant KIT influenced response to imatinib mesylate in 2 ways. First, we assessed whether patients with tumors harboring KIT that was both mutated and amplified were more likely to achieve tumor response than those patients whose tumors harbored either alteration alone. Although a greater likelihood of response was observed in these cases, the difference was not statistically significant (36%; 95% CI, 15%–65%; vs 14%; 95% CI, 4%–40%; P = .35).
We then evaluated whether the ratio of the mutant to wild-type KIT allele influenced response to imatinib mesylate. Mutations associated with a mutant to wild-type allele ratio of more than 1 are likely to be pathogenetically relevant. In a pure population of diploid tumor cells with a heterozygous mutation, this ratio is expected to be 1. As tumor samples typically contain normal cell contamination, the allelic ratio of heterozygous samples is often smaller than 1. Allelic ratios of more than 1 indicate the presence of an independent genetic event, such as amplification of the mutant allele, deletion of the wild-type allele, or loss of heterozygosity, which shifts the ratio in favor of the mutant allele. Such positive selection of the mutated allele suggests that the mutation is a functionally relevant event. Subgroup analysis reveals a higher response rate in cases with an allelic ratio of more than 1 (71% vs 6%; P = .002), with a longer median time to progression (18.0 weeks [IQR, 12.0–95.4 weeks; 95% CI, 11.6–95.4 weeks] vs 11.0 weeks [IQR, 5.0–16.0 weeks; 95% CI, 5.0–12.0 weeks]; P = .01) and extended median survival (median not achieved [IQR, 46.5 weeks-not achieved; 95% CI, 34.8 weeks-not achieved] vs 31.3 weeks [IQR, 18.1–80.5 weeks; 95% CI, 18.1–61.5 weeks]; P = .03).
COMMENT
These results demonstrate that a subset of melanomas with genetic alterations of KIT respond to treatment with imatinib mesylate. Durable responses were observed in 16% (95% CI, 2%–30%) of these patients, with all sustained for more than 1 year. Two additional responses were observed but not sustained on a follow-up scan performed 6 weeks after the initial response was documented. Therefore, this study did not meet its predetermined end point. Nevertheless, the responses achieved in this population preselected based on the presence of KIT mutations or amplification compare favorably to that observed in prior trials of molecularly unselected patients,19–21 in which response was limited to 1 of 62 evaluable participants treated.
This study additionally highlights the challenges in identifying appropriate patients for treatment with KIT inhibition. Seventy-four percent of BRAF mutant melanomas harbor a substitution of glutamic acid for valine at amino acid 600 (the V600E mutation).39 By contrast, mutations in KIT are more widely distributed over the coding region. Treatment of patients with melanoma harboring the oncogenic V600E BRAF mutation with an effective inhibitor of RAF, such as PLX4032 (Plexxikon, RG7204; Roche Pharmaceuticals, Basel, Switzerland), results in tumor response in 81% of cases.40 The more modest results in this study suggest that only select KIT alterations are truly oncogenic and indicative of an effective therapeutic target.
By using more selective molecular criteria, we may better identify patients who will respond to imatinib mesylate. Response to imatinib mesylate is predicted to be dependent on the region of the protein affected by a mutation, with some mutations affecting the binding affinity of imatinib mesylate to KIT as previously demonstrated in in vitro and clinical studies of GIST.35,37,41 Prior observations in GIST demonstrated the sensitivity of K642E and N822K mutations and the resistance of V654A and D820Y mutations to imatinib mesylate.35,41,42 Concordant with these findings, patients with melanoma harboring these resistant mutations progressed, although disease stability and responses were observed in those patients whose tumors harbored K642E and N822K mutations (Figure 1). Imatinib mesylate–resistance in GIST commonly results from the development of secondary KIT mutations, including V654A, D820Y, N822K, and A829P.35,36 These mutations were identified as primary mutations in several melanomas and may also predict a lower probability of response with imatinib mesylate. Indeed, the best response we observed in such cases was stable disease. Importantly, although an in vitro study demonstrated poor sensitivity of a melanoma cell line harboring an L576P mutation to imatinib mesylate,43 we observed dramatic in vivo responses in patients with melanomas harboring this mutation.
To separate bona fide driver from passenger alterations of KIT, we sought evidence for tumor selection of specific mutations. We observed that imatinib mesylate has greater activity in tumors harboring recurrent KIT mutations found in melanoma or GIST, as well as in tumors with a mutant KIT allele in greater abundance than the wild-type allele. When combining those patients whose tumors have an allelic ratio of more than 1 with those whose tumors harbor recurrent primary mutations found in GIST or melanoma, we observed a better response rate, time to progression, and overall survival compared with other cases (Figure 3). These criteria may serve as indicators of genetic events relevant to oncogenesis and should be investigated further as predictive markers of response to KIT inhibition.
In conclusion, our data show that imatinib mesylate therapy in patients with melanoma harboring specific KIT alterations results in clinical responses, consistent with the paradigm of oncogene addiction.44 Prediction of response to KIT inhibition can be improved beyond what we report herein by identifying tumors that harbor KIT alterations of functional relevance.
Acknowledgments
Funding/Support: This study was funded by grant R01FD003445-01 from the US Food and Drug Administration; grants N01CM62206 and P30 CA008748 from the National Institutes of Health; American Society of Clinical Oncology; American Skin Association; and Live4Life Foundation. The Division of Cancer Treatment and Diagnosis at the National Cancer Institute provided the imatinib mesylate for this clinical trial.
Role of the Sponsors: The US Food and Drug Administration, National Institutes of Health, American Society of Clinical Oncology, American Skin Association, and Live4Life Foundation had no role in the design and conduct of the study, in the collection, management, analysis, and interpretation of the data, or in the preparation or approval of the manuscript.
Footnotes
Author Contributions: Dr Carvajal had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Carvajal, Wolchok, Takebe, Bastian, Schwartz.
Acquisition of data: Carvajal, Antonescu, Wolchok, Chapman, Roman, Teitcher, Busam, Chmielowski, Lutzky, Pavlick, Fusco, Cane, Takebe, Vemula, Bastian, Schwartz.
Analysis and interpretation of data: Carvajal, Antonescu, Wolchok, Chapman, Roman, Panageas, Busam, Chmielowski, Lutzky, Vemula, Bouvier, Bastian, Schwartz.
Drafting of the manuscript: Carvajal, Antonescu, Panageas, Bastian, Schwartz.
Critical revision of the manuscript for important intellectual content: Carvajal, Antonescu, Wolchok, Chapman, Roman, Teitcher, Panageas, Busam, Chmielowski, Lutzky, Pavlick, Takebe, Vemula, Bouvier, Bastian, Schwartz.
Statistical analysis: Carvajal, Panageas.
Obtained funding: Carvajal, Wolchok, Bastian, Schwartz.
Administrative, technical, or material support: Carvajal, Chapman, Roman, Busam, Lutzky, Pavlick, Fusco, Cane, Vemula, Bouvier, Schwartz.
Study supervision: Carvajal, Wolchok, Chapman, Takebe, Bastian, Schwartz.
Conflict of Interest Disclosures: All authors have completed and submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Dr Carvajal reported being a consultant for Novartis and receiving research support from the American Society of Clinical Oncology, National Institutes of Health, and the US Food and Drug Administration. Dr Wolchok reported being a consultant for Novartis. Dr Chapman reported being a consultant for and receiving research support for travel, accommodations, and meeting expenses from Roche-Genentech. Dr Chmielowski reported being a consultant for Celgene and Prometheus and receiving payment for lectures from Novartis and Prometheus. Dr Lutzy reported being a consultant for Bristol-Myers Squibb, receiving payment for lectures from Millennium and Bristol-Myers Squibb, and receiving research support from Memorial Sloan-Kettering Cancer Center. Dr Bastian reported being a consultant for Novartis, Abbott, sanofi-aventis, DermTech, and Genomel, having patents filed by the University of California, and receiving research support from the US Food and Drug Administration, National Institutes of Health, Melanoma Research Alliance, Canadian Institutes of Health Research, Canadian Cancer Society, European Commission, Jubilaeumsfonds of the Oesterreichische National Bank, and Deutsche Forchungsgemeinschaft. Dr Schwartz reported being a consultant for Pfizer, Tragara, and Pharmagap, and has received research support from the US Food and Drug Administration. No other authors reported any disclosures.
Previous Presentation: Presented in part at the Molecular Phenotype of Melanoma Subtypes and Patient-Specific Therapy Clinical Science Symposium, 2009 Annual Meeting of the American Society of Clinical Oncology; June 2, 2009; Orlando, Florida.
Online-Only Material: eFigures 1 through 3 and eTables 1 and 2 are available at http://www.jama.com.
Additional Contributions: Margaret Leversha, PhD, and Kalyani Chadalavada (both at Memorial Sloan-Kettering Cancer Center, New York, New York), and the Memorial Sloan-Kettering Cancer Center Molecular Cytogenetics Core provided assistance with the conduct of the fluorescence in situ hybridization studies. Dr Leversha and Ms Chadalavada did not receive any compensation for their work on this study.
REFERENCES
- 1.Geller AC, Miller DR, Annas GD, Demierre MF, Gilchrest BA, Koh HK. Melanoma incidence and mortality among US whites, 1969–1999. JAMA. 2002;288(14):1719–1720. doi: 10.1001/jama.288.14.1719. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 3.Chapman PB, Einhorn LH, Meyers ML, et al. Phase III multicenter randomized trial of the Dartmouth regimen versus dacarbazine in patients with meta-static melanoma. J Clin Oncol. 1999;17(9):2745–2751. doi: 10.1200/JCO.1999.17.9.2745. [DOI] [PubMed] [Google Scholar]
- 4.Atkins MB, Lotze MT, Dutcher JP, et al. High-dose recombinant interleukin 2 therapy for patients with metastatic melanoma: analysis of 270 patients treated between 1985 and 1993. J Clin Oncol. 1999;17(7):2105–2116. doi: 10.1200/JCO.1999.17.7.2105. [DOI] [PubMed] [Google Scholar]
- 5.Hodi FS, O′Day SJ, McDermott DF, et al. Improved survival with ipilimumab in patients with meta-static melanoma. N Engl J Med. 2010;363(8):711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353(20):2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 7.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 8.Curtin JA, Busam K, Pinkel D, Bastian BC. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol. 2006;24(26):4340–4346. doi: 10.1200/JCO.2006.06.2984. [DOI] [PubMed] [Google Scholar]
- 9.Beadling C, Jacobson-Dunlop E, Hodi FS, et al. KIT gene mutations and copy number in melanoma subtypes. Clin Cancer Res. 2008;14(21):6821–6828. doi: 10.1158/1078-0432.CCR-08-0575. [DOI] [PubMed] [Google Scholar]
- 10.Yarden Y, Kuang WJ, Yang-Feng T, et al. Human proto-oncogene c-kit: a new cell surface receptor tyrosine kinase for an unidentified ligand. EMBO J. 1987;6(11):3341–3351. doi: 10.1002/j.1460-2075.1987.tb02655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hou L, Panthier JJ, Arnheiter H. Signaling and tran-scriptional regulation in the neural crest-derived melanocyte lineage: interactions between KIT and MITF. Development. 2000;127(24):5379–5389. doi: 10.1242/dev.127.24.5379. [DOI] [PubMed] [Google Scholar]
- 12.Wehrle-Haller B. The role of Kit-ligand in mela-nocyte development and epidermal homeostasis. Pigment Cell Res. 2003;16(3):287–296. doi: 10.1034/j.1600-0749.2003.00055.x. [DOI] [PubMed] [Google Scholar]
- 13.Alexeev V, Yoon K. Distinctive role of the cKit receptor tyrosine kinase signaling in mammalian melanocytes. J Invest Dermatol. 2006;126(5):1102–1110. doi: 10.1038/sj.jid.5700125. [DOI] [PubMed] [Google Scholar]
- 14.Huang S, Luca M, Gutman M, et al. Enforced c-KIT expression renders highly metastatic human melanoma cells susceptible to stem cell factor-induced apoptosis and inhibits their tumorigenic and metastatic potential. Oncogene. 1996;13(11):2339–2347. [PubMed] [Google Scholar]
- 15.Zakut R, Perlis R, Eliyahu S, et al. KIT ligand (mast cell growth factor) inhibits the growth of KIT-expressing melanoma cells. Oncogene. 1993;8(8):2221–2229. [PubMed] [Google Scholar]
- 16.Lassam N, Bickford S. Loss of c-kit expression in cultured melanoma cells. Oncogene. 1992;7(1):51–56. [PubMed] [Google Scholar]
- 17.Montone KT, van Belle P, Elenitsas R, Elder DE. Proto-oncogene c-kit expression in malignant melanoma: protein loss with tumor progression. Mod Pathol. 1997;10(9):939–944. [PubMed] [Google Scholar]
- 18.Natali PG, Nicotra MR, Winkler AB, Cavaliere R, Bigotti A, Ullrich A. Progression of human cutaneous melanoma is associated with loss of expression of c-kit proto-oncogene receptor. Int J Cancer. 1992;52(2):197–201. doi: 10.1002/ijc.2910520207. [DOI] [PubMed] [Google Scholar]
- 19.Wyman K, Atkins MB, Prieto V, et al. Multi-center phase II trial of high-dose imatinib mesylate in metastatic melanoma: significant toxicity with no clinical efficacy. Cancer. 2006;106(9):2005–2011. doi: 10.1002/cncr.21834. [DOI] [PubMed] [Google Scholar]
- 20.Ugurel S, Hildenbrand R, Zimpfer A, et al. Lack of clinical efficacy of imatinib in metastatic melanoma. Br J Cancer. 2005;92(8):1398–1405. doi: 10.1038/sj.bjc.6602529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim KB, Eton O, Davis DW, et al. Phase II trial of imatinib mesylate in patients with metastatic melanoma. Br J Cancer. 2008;99(5):734–740. doi: 10.1038/sj.bjc.6604482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Demetri GD, von Mehren M, Blanke CD, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med. 2002;347(7):472–480. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- 23.Antonescu CR, Busam KJ, Francone TD, et al. L576P KIT mutation in anal melanomas correlates with KIT protein expression and is sensitive to specific kinase inhibition. Int J Cancer. 2007;121(2):257–264. doi: 10.1002/ijc.22681. [DOI] [PubMed] [Google Scholar]
- 24.Ashida A, Takata M, Murata H, Kido K, Saida T. Pathological activation of KIT in metastatic tumors of acral and mucosal melanomas. Int J Cancer. 2009;124(4):862–868. doi: 10.1002/ijc.24048. [DOI] [PubMed] [Google Scholar]
- 25.Jiang X, Zhou J, Yuen NK, et al. Imatinib targeting of KIT-mutant oncoprotein in melanoma. Clin Cancer Res. 2008;14(23):7726–7732. doi: 10.1158/1078-0432.CCR-08-1144. [DOI] [PubMed] [Google Scholar]
- 26.Hodi FS, Friedlander P, Corless CL, et al. Major response to imatinib mesylate in KIT-mutated melanoma. J Clin Oncol. 2008;26(12):2046–2051. doi: 10.1200/JCO.2007.14.0707. [DOI] [PubMed] [Google Scholar]
- 27.Lutzky J, Bauer J, Bastian BC. Dose-dependent, complete response to imatinib of a metastatic mucosal melanoma with a K642E KIT mutation. Pigment Cell Melanoma Res. 2008;21(4):492–493. doi: 10.1111/j.1755-148X.2008.00475.x. [DOI] [PubMed] [Google Scholar]
- 28.Oken MM, Creech RH, Tormey DC, et al. Toxicity and response criteria of the Eastern Cooperative Oncology Group. Am J Clin Oncol. 1982;5(6):649–655. [PubMed] [Google Scholar]
- 29.Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors: European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92(3):205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 30.Bastian BC, LeBoit PE, Hamm H, Bröcker EB, Pinkel D. Chromosomal gains and losses in primary cutaneous melanomas detected by comparative genomic hybridization. Cancer Res. 1998;58(10):2170–2175. [PubMed] [Google Scholar]
- 31.Fiegler H, Carr P, Douglas EJ, et al. DNA micro-arrays for comparative genomic hybridization based on DOP-PCR amplification of BAC and PAC clones. Genes Chromosomes Cancer. 2003;36(4):361–374. doi: 10.1002/gcc.10155. [DOI] [PubMed] [Google Scholar]
- 32.Simon R. Optimal two-stage designs for phase II clinical trials. Control Clin Trials. 1989;10(1):1–10. doi: 10.1016/0197-2456(89)90015-9. [DOI] [PubMed] [Google Scholar]
- 33.Kaplan E, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 34.Landi MT, Bauer J, Pfeiffer RM, et al. MC1R germ-line variants confer risk for BRAF-mutant melanoma. Science. 2006;313(5786):521–522. doi: 10.1126/science.1127515. [DOI] [PubMed] [Google Scholar]
- 35.Heinrich MC, Corless CL, Blanke CD, et al. Molecular correlates of imatinib resistance in gastrointestinal stromal tumors. J Clin Oncol. 2006;24(29):4764–4774. doi: 10.1200/JCO.2006.06.2265. [DOI] [PubMed] [Google Scholar]
- 36.Antonescu CR, Besmer P, Guo T, et al. Acquired resistance to imatinib in gastrointestinal stromal tumor occurs through secondary gene mutation. Clin Cancer Res. 2005;11(11):4182–4190. doi: 10.1158/1078-0432.CCR-04-2245. [DOI] [PubMed] [Google Scholar]
- 37.Heinrich MC, Corless CL, Demetri GD, et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol. 2003;21(23):4342–4349. doi: 10.1200/JCO.2003.04.190. [DOI] [PubMed] [Google Scholar]
- 38.Debiec-Rychter M, Sciot R, Le Cesne A, et al. KIT mutations and dose selection for imatinib in patients with advanced gastrointestinal stromal tumours. Eur J Cancer. 2006;42(8):1093–1103. doi: 10.1016/j.ejca.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 39.Long GV, Menzies AM, Nagrial AM, et al. Prognostic and clinicopathologic associations of oncogenic BRAF in metastatic melanoma. J Clin Oncol. 2011;29(10):1239–1246. doi: 10.1200/JCO.2010.32.4327. [DOI] [PubMed] [Google Scholar]
- 40.Flaherty KT, Puzanov I, Kim KB, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363(9):809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen LL, Trent JC, Wu EF, et al. A missense mutation in KIT kinase domain 1 correlates with imatinib resistance in gastrointestinal stromal tumors. Cancer Res. 2004;64(17):5913–5919. doi: 10.1158/0008-5472.CAN-04-0085. [DOI] [PubMed] [Google Scholar]
- 42.Chen H, Isozaki K, Kinoshita K, et al. Imatinib inhibits various types of activating mutant kit found in gastrointestinal stromal tumors. Int J Cancer. 2003;105(1):130–135. doi: 10.1002/ijc.11025. [DOI] [PubMed] [Google Scholar]
- 43.Woodman SE, Trent JC, Stemke-Hale K, et al. Activity of dasatinib against L576P KIT mutant melanoma: molecular, cellular, and clinical correlates. Mol Cancer Ther. 2009;8(8):2079–2085. doi: 10.1158/1535-7163.MCT-09-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weinstein IB. Cancer: addiction to oncogenes: the Achilles heal of cancer. Science. 2002;297(8):63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]