Abstract
Objective
Enamelysin (MMP20) and kallikrein 4 (KLK4) are believed to be necessary to clear proteins from the enamel matrix of developing teeth. MMP20 is expressed by secretory stage ameloblasts, while KLK4 is expressed from the transition stage throughout the maturation stage. The aim of this study is to investigate the activation of KLK4 by MMP20 and the inactivation of MMP20 by KLK4.
Design
Native pig MMP20 (pMMP20) and KLK4 (pKLK4) were isolated directly from enamel scrapings from developing molars. Recombinant human proKLK4 (rh-proKLK4) was activated by incubation with pMMP20 or recombinant human MMP20 (rhMMP20), and the resulting KLK4 activity was detected by zymography. Reaction products were isolated by reverse-phase high performance liquid chromatography (RP-HPLC), and their N-termini characterized by Edman degradation. The pMMP20 was incubated with pKLK4 under mildly acidic or under physiologic conditions, and enzyme activity was analyzed by zymography. The catalytic domain of rhMMP20 was incubated with pKLK4 or recombinant human KLK4 (rhKLK4) and the digestion products were characterized by zymography and Edman degradation.
Results
Both pMMP20 and rhMMP20 activated rh-proKLK4 by cleaving at the propeptide-enzyme junction used in vivo. The pMMP20 was inactivated by pKLK4 under physiologic conditions, but not under mildly acidic conditions. Both pKLK4 and rhKLK4 cleaved MMP20 principally at two sites in the catalytic domain of MMP20.
Conclusions
MMP20 activates proKLK4 and KLK4 inactivates MMP20 in vitro, and these actions are likely to occur during enamel formation in vivo.
Keywords: Enamel, MMP20, KLK4, Proteases, Tooth
1. Introduction
Enamel formation progresses in two stages. During the secretory stage enamel mineral is initially deposited as thin ribbons that lengthen along a mineralization front at the enamel surface.1,2 These ribbons crystallize into hydroxyapatite and mature slowly, by growing in width and thickness. Most crystal maturation occurs after the enamel layer has reached its final thickness, during the maturation stage. Enamel proteins are abundant during the secretory and early maturation stages of amelogenesis, but are virtually absent from the matrix by the late maturation stage.3 The disappearance of enamel proteins is associated with two secreted proteases: enamelysin (MMP20)4 and kallikrein 4 (KLK4).5
MMP20 has been shown to be expressed by secretory stage ameloblasts by in situ hybridization6–8 and immunohistochemistry.9 Enamel proteins are processed by MMP20 to provide space within the deeper enamel matrix that allows the enamel crystallites to grow in width and thickness. In addition to the role of processing enamel proteins, junctional complexes present on ameloblasts may also be cleaved by MMP20 to foster cell movement, which is necessary for the formation of the decussating enamel rod pattern.10,11 Another proposed function of MMP20 is to activate KLK4.12
KLK4 is specifically expressed by transition and maturation stage ameloblasts.13 Enamel proteins are progressively degraded by KLK4 within the enamel matrix so the enamel crystallites can grow in width and thickness,14–16 and the digestion products are reabsorbed into ameloblasts during the maturation stage.13 In the absence of KLK4 expression, there is substantial retention of enamel proteins within the enamel matrix layer.17,18 Although there is no MMP20 activity in enamel from first molars of day 15 wild-type mice corresponding to the maturation stage, MMP20 activity was retained in enamel of day 15 KLK4 null mice,18 suggesting that MMP20 might be inactivated by KLK4.
Other proteases may play a role in enamel formation. Signal-peptide-peptidase-like 2a (Sppl2a) knockout mice show maturation stage enamel defects, but SPPL2A resides in lysosomes/late endosomes and is not active in the extracellular matrix.19 MMP9 has been proposed to cleave amelogenin during the secretory stage of enamel formation,20 but these findings are inconsistent with the observations that MMP20 catalyzes the cleavages that generate all of the accumulated amelogenin amelogenin products in secretory stage pig enamel,21,22 and MMP9 mutations cause Metaphyseal Anadysplasia, which is not associated with enamel defects.23 Chymostrypsin C is associated with enamel formation, but the levels are marginal compared with the pancreas.24 There is no evidence of enamel defects associated with CTRC mutations, which increase the risk of pancreatitis by diminishing its protective trypsin-degrading activity.25
In this study we provide in vitro evidence for the activation of proKLK4 by MMP20 and the inactivation of MMP20 by specific KLK4 cleavages.
2. Materials and methods
2.1. Preparation and extraction of soft and hard enamel
Tooth germs of permanent molars were surgically extracted from the mandibles of deceased 5-month-old pigs from the Meat Market of Metropolitan Central Wholesale Market (Shinagawa, Tokyo). The enamel organ epithelia (EOE) and dental pulp tissues were removed using tissue forceps. The soft, cheese-like enamel was separated from the crowns using a spatula. Early maturation-stage enamel samples, containing KLK4, were obtained by scraping the remaining hard, chalky enamel. Both soft and hard enamel shavings were homogenized in Sörensen buffer (pH 7.4), made by mixing Na2HPO4 and KH2PO4 to achieve a final phosphate concentration of 50 mM and a pH of 7.4. Soluble fraction (N extract) was collected by centrifugation. Insoluble material was further homogenized in 50 mM carbonate-bicarbonate buffer (pH 10.8) and soluble fraction (AL extract) was collected by centrifugation.
2.2. Isolation of porcine enamelysin (pMMP20)
The AL extract obtained from soft enamel was buffer-changed to 50 mM Tris–HCl and 6 M urea buffer (pH 7.4) with YM-3 membrane (Millipore Corporation, Billerica, MA, USA) and fractionated on a Heparin Sepharose 6 Fast Flow column (1.6 cm × 20 cm, GE Healthcare, Uppsala, Sweden) with buffer A: 50 mM Tris–HCl and 6 M urea (pH 7.4). Proteins were eluted with a step gradient of NaCl (0, 0.05, 0.1, 0.2 and 1 M) in buffer A at a flow rate of 0.2 mL/min at 4 °C while monitoring the absorbance at 280 nm. The MMP20 fraction eluted in the second peak, was concentrated and buffer exchanged to 50 mM Tris–HCl (pH 7.4) using an Amicon Ultra-4 Centrifugal Filter (Millipore Corporation). The protein content of the MMP20 fraction was measured using the Pierce 660 nm Protein Assay kit (Thermo Scientific, Rockford, IL, USA) and the sample was stored at −80 °C.
2.3. Isolation of porcine kallikrein 4 (pKLK4)
The N extract obtained from hard enamel was raised to 40% saturation by the addition of ammonium sulfate and the precipitate was removed by centrifugation. The supernatant was raised to 65% saturation and the precipitate (containing KLK4) was pelleted by centrifugation. The 40–65% saturation pellet was resupended in 5 mL of 0.5 M acetic acid and the precipitate was removed by centrifugation. The supernatant was fractionated by reversed-phase high-performance liquid chromatography (RP-HPLC) using a Discovery C18 column (4.6 mm × 25 cm, Sigma–Aldrich/Supelco, Bellefonte, PA, USA) and eluted with a linear gradient (20 to 80% B in 60 min) at a flow rate of 1.0 mL/min. Buffer A was 0.05% trifluoroacetic acid (TFA); buffer B was 0.1% TFA in 80% aqueous acetonitrile. Protein was detected by absorbance at 220 nm. The KLK4 fraction eluted between 57 and 61 min, and was concentrated and buffer-exchanged to 50 mM Tris–HCl (pH 7.4) using an Amicon Ultra-4 Centrifugal Filter (Millipore Corporation). The protein content of the KLK4 fraction was measured using a Pierce 660 nm Protein Assay kit (Thermo Scientific) and the sample was stored at −80 °C.
2.4. Activation of recombinant proKLK4 by porcine and recombinant MMP20
Recombinant human MMP20 (rhMMP20) purchased from Enzo Life Sciences (Farmingdale, NY, USA) and recombinant human proKLK4 (rh-proKLK4) purchased from R&D Systems (Minneapolis, MN, USA) were separately dissolved in 150 µL of 50 mM Tris–HCl/10 mM CaCl2 buffer containing 5 mM benzamidine and 1 mM phenylmethanesulfonylfluoride (PMSF, pH 7.4). The rh-proKLK4 (10 µg each) was activated with 30 µg of pMMP20 or 1 µg of rhMMP20 for 5 h at 37 °C. The activation was stopped by adding 1,10-phenanthroline to a final concentration of 10 mM. Both rh-proKLK4 and activated KLK4 were analyzed by Western Blotting. KLK4 activity was detected by casein zymography. The reaction aliquot (approximately 7 µg) was characterized by RP-HPLC using a Discovery C18 column (4.6 mm × 25 cm, Sigma–Aldrich/Supelco). The column was eluted with a linear gradient (0–100% B in 55 min) at a flow rate of 1.0 mL/min. Buffer A was 0.05% TFA; buffer B was 0.1% TFA in 80% aqueous acetonitrile. Protein was detected by absorbance at 220 nm. The N-terminus of activated KLK4 was analyzed by Edman degradation.
2.5. Digestion of porcine MMP20 by porcine KLK4
The pMMP20 (30 µg) was dissolved with 200 µL of 50 mM Tris–HC1 and 10 mM ethylenediaminetetraacetic acid (EDTA, pH 7.4) or 50 mM sodium acetate and 10 mM EDTA (pH 5.5) and digested with pKLK4 (10 µg) at 37 °C. Reaction aliquots at 0, 5, 10 and 20 h were analyzed by casein gel zymography.
2.6. Cleavage experiment of recombinant MMP20 by porcine KLK4
The rhMMP20 (10 µg) was dissolved with 200 µL of 50 mM Tris–HCl/10 mM EDTA (pH 7.4) and was incubated with pKLK4 (10 µg) or recombinant human KLK4 (1 µg), which was activated by rhMMP20, at 37 °C for 20 h. Reaction aliquots at 0 and 20 h were analyzed by casein gel zymography and the digestion products at 20 h were characterized by RP-HPLC using a Discovery C18 column (4.6 mm × 25 cm, Sigma– Aldrich/Supelco). The column was eluted with a linear gradient (0–100% B in 55 min) at a flow rate of 1.0 mL/min. Buffer A was 0.05% TFA; buffer B was 0.1% TFA in 80% aqueous acetonitrile. The RP-HPLC elution was monitored at 220 nm and the N-terminus of the generated peaks were analyzed by Edman degradation.
2.7. SDS-polyacrylamide gel electrophoresis (SDS-PAGE)
SDS-PAGE was performed using NuPAGE 4–12% Bis-Tris gel (Invitrogen, Carlsbad, CA, USA). Samples were dissolved in NuPAGE LDS sample buffer (Invitrogen), and electrophoresis was carried out at 200 V for 35 min with NuPAGE MES SDS running buffer (Invitrogen). The gel was stained with Simply Blue Safe Stain (Invitrogen) or Stains-All (Sigma–Aldrich, St. Louis, MO, USA). The apparent molecular weights of the protein bands were estimated by comparison with SeeBlue Plus2 Pre-Stained Standard (Invitrogen).
2.8. Enzymograms
Zymography was carried out using Novex 12% Zymogram Casein Gel and Novex 10% Zymogram Gelatin Gel (Invitrogen). Samples were dissolved in NuPAGE LDS sample buffer (Invitrogen), and electrophoresis was carried out at 30 mA for about 1 h with Novex Tris Glycine SDS running buffer (Invitrogen). The gel was shaken gently in 2.5% Triton X-100 solution for 1 h at room temperature with one buffer change and then incubated overnight with 10 mM CaCl2 or 10 mM EDTA in 50 mM Tris–HCl (pH 7.4) or with 10 mM CaCl2 in 50 mM sodium acetate buffers (pH 5.5). Proteinase activities were visualized as unstained bands after the gel was stained with Coomassie Brilliant Blue (CBB).
2.9. Densitometry
The active bands of MMP20 and KLK4 on zymograms after the digestion were quantified using ImageJ densitometry software.
2.10. Western blot analysis
Three commercial antibodies with specificities for the centre region of human KLK4 (#ab71234), the amino-terminal end of human MMP20 (#ab39038), and the hemopexin domain of human MMP20 (#ab84737) were purchased from Abcam (Abcam, Cambridge, MA, USA). Samples fractionated by SDS-PAGE were electrotransferred onto a Hybond-ECL membrane (GE Healthcare). The three antibodies were incubated overnight at 1:1,000 dilutions. The secondary antibody was diluted 1:10,000 and incubated for 3 h. Immunopositive bands were visualized by chemiluminescent detection using the ECL Advance Western Blotting Detection Kit (GE Healthcare).
2.11. Automated Edman degradation, amino acid analysis and mass analysis
Automated Edman degradation was performed with an Applied Biosystems Procise 494 HT protein sequencing system at APRO life Science Institute Inc. (Naruto, Tokushima, Japan) and with an Applied Biosystems Procise 494 cLC protein sequencing system at Japan Bio Services Co., LTD (Asaka, Saitama, Japan).
3. Results
3.1. Isolation of porcine MMP20 and KLK4
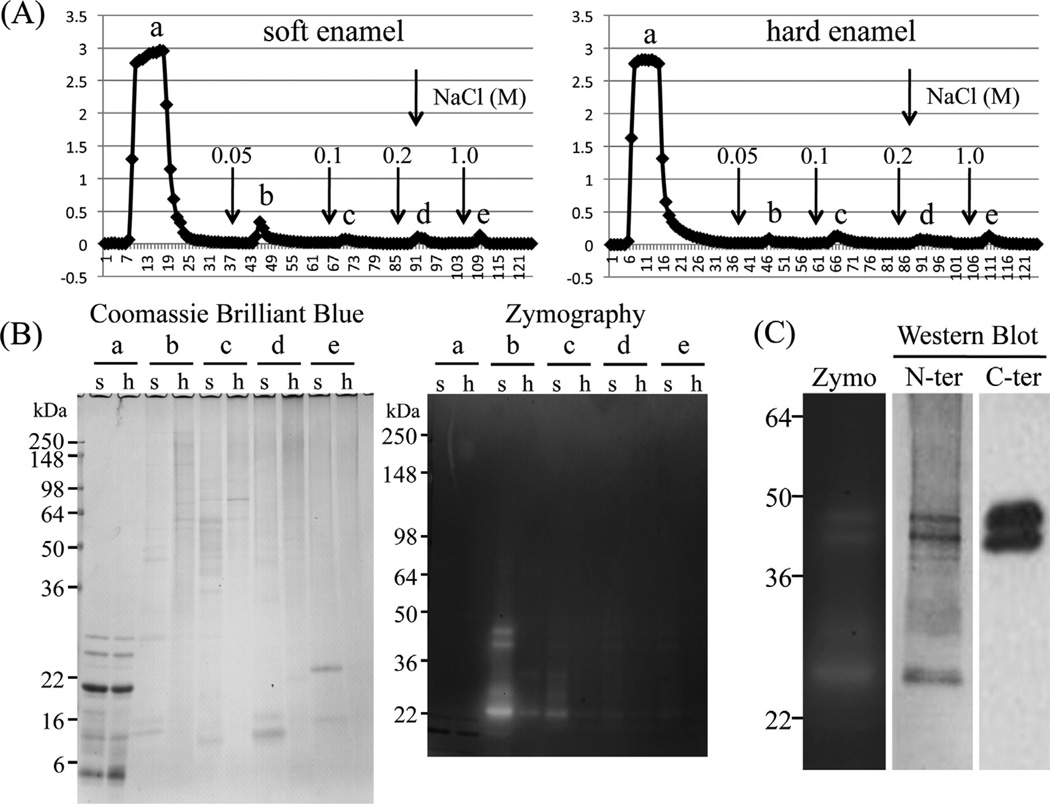
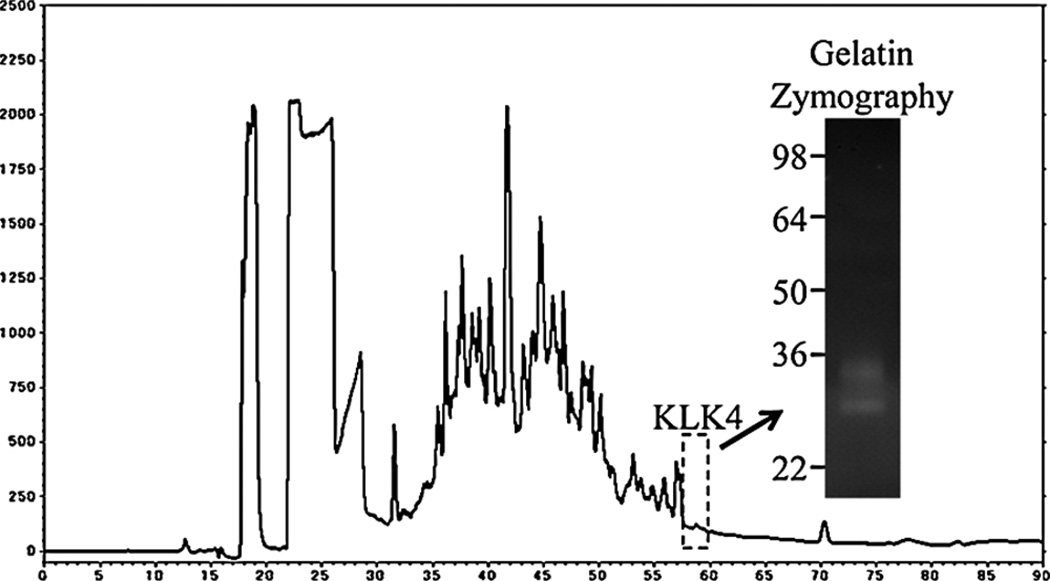
Porcine MMP20 was purified from the AL extracts of soft or hard enamel. Soft enamel was scraped from developing pig teeth for affinity chromatography using a heparin Sepharose column, which fractionated the extracts into five peaks (Fig. 1A). Porcine MMP20 mainly eluted in the second peak from soft enamel, and casein zymography confirmed the presence of MMP20 activity. Little MMP20 was detected in each fraction from hard enamel (Fig. 1B). Western blot analysis with both N-terminal and C-terminal antibodies detected the familiar MMP20 doublet migrating at 41 and 46 kDa. The MMP20 catalytic domain, having an apparent molecular weight of 25–27 kDa, was also observed by casein zymography and Western blotting using the MMP20 amino-terminal antibody (Fig. 1C). Porcine KLK4 isolated from hard enamel was evident as two bands migrating at 34 and 30 kDa on gelatin zymograms (Fig. 2).
Fig. 1.
Isolation of porcine MMP20. (A) Heparin Sepharose chromatograms showing absorbance at 280 nm for AL extracts from soft enamel (290 mg) and hard enamel (150 mg). Downward pointing arrows are starting point of the step gradient with 0.05, 0.1, 0.2 and 1 M NaCl. (B) SDS-PAGE and casein zymogram showing each fractions a, b, c, d and e on Heparin chromatograms from soft (s) and hard (h) enamel. (C) Casein zymography (Zymo) and Western blot using MMP20 amino-terminal end- (N-ter) and hemopexin domain- (C-ter) specific antibodies showing final prepared MMP20 used for this study.
Fig. 2.
Isolation of porcine KLK4. C18-chromatogram of N extract, 40–65% ammonium sulfate precipitating and the gelatin zymogram of KLK4 fraction collected at from 57.5 to 60 min (dotted square).
3.2. Activation of recombinant proKLK4 by porcine and recombinant MMP20
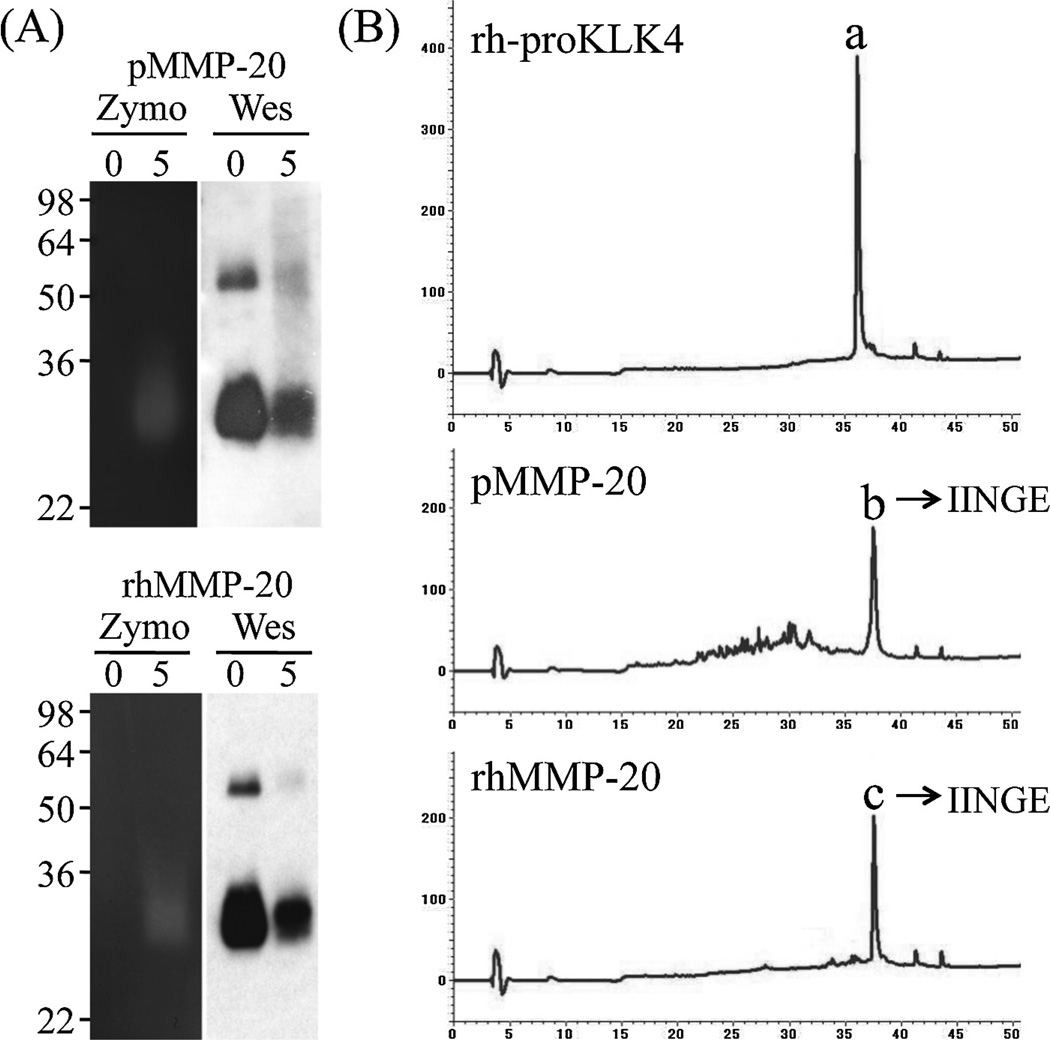
Incubation of rh-proKLK4 (substrate) with pMMP20 or rhMMP20 generated active smear bands migrating at 29 kDa on casein gel zymograms, which were identified as KLK4 by Western blot analyses using KLK4 antibodies raised against rh-proKLK4 or activated KLK4 (Fig. 3A). The original rh-proKLK4 displayed a single chromatographic peak on RP-HPLC with a retention time at 36 min (Fig. 3B). A generated peak from rh-proKLK4 by activation with both pMMP20 and rhMMP20 eluted at 37.5 min and Edman sequencing of both peaks gave the same I-I-N-G-E sequence (Fig. 3B).
Fig. 3.
Activation of recombinant human proKLK4 (substrate) by MMP20. (A) Both native porcine MMP20 (pMMP20) and recombinant human MMP20 (rhMMP20) were used to activate recombinant human proKLK4 (rh-proKLK4) for 0 and 5 h. Both MMP20s activated rh-proKLK4 and the purified product of the digestion exhibited at a smear band above 30 kDa on a casein zymogram (Zymo), and on Western blots (Wes), demonstrating that the product was both active and KLK4. (B) C-18 RP-HPLC chromatograms showing rh-proKLK4 original material (a) with a retention time of 36 min; the generation of peaks (b) and (c) with retention times of 37.5 min after activation with pMMP20 and rhMMP20. The protein sequence of each activated KLK4 peaks (b) and (c) gave IINGE, which corresponds to the N-terminus of active KLK4.
3.3. Digestion of porcine MMP20 by porcine KLK4
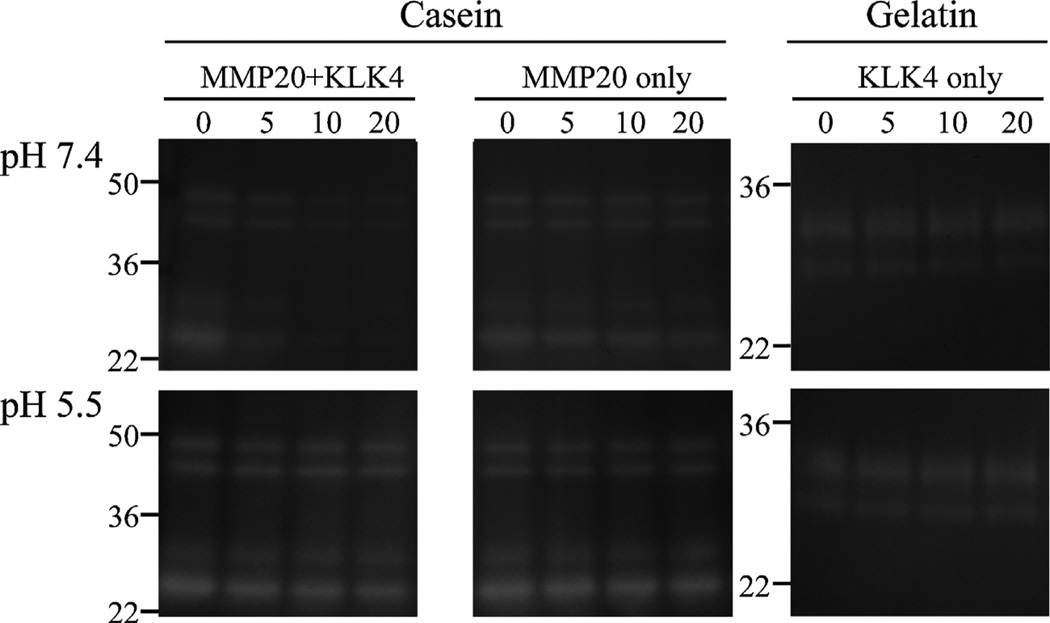
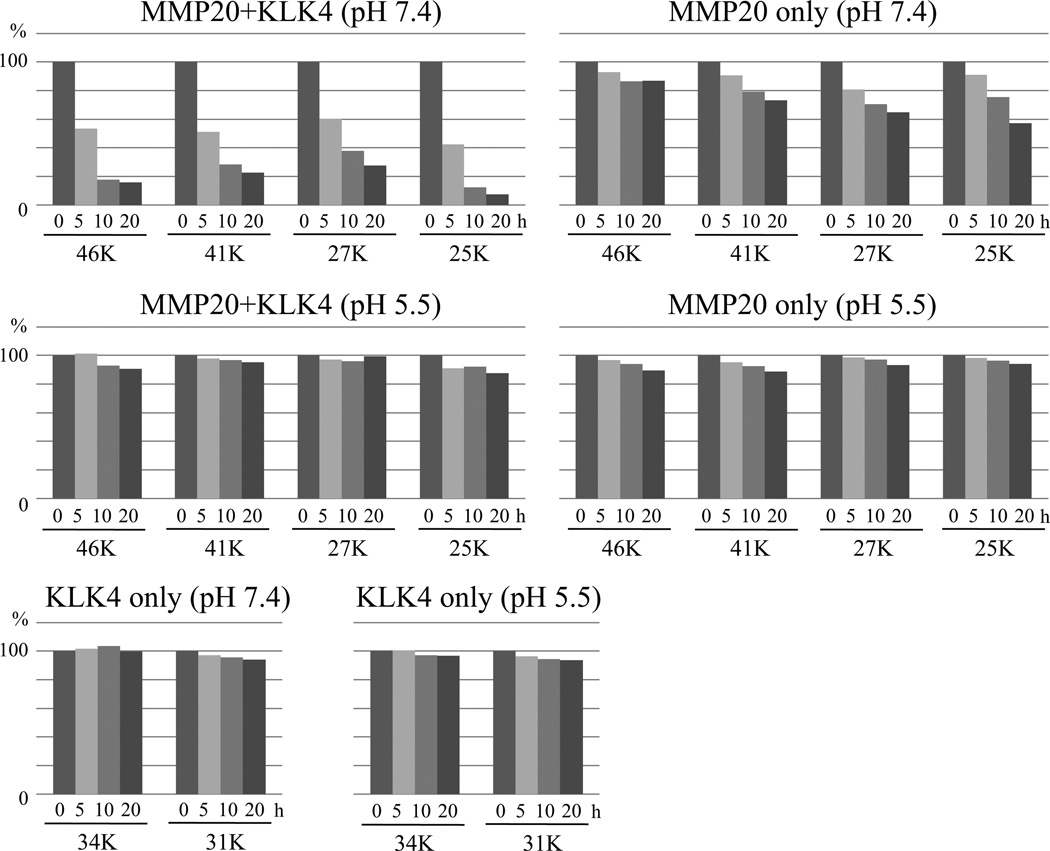
The pMMP20 was digested by pKLK4 under two pH conditions (Fig. 4). At pH 7.4, pKLK4 degraded pMMP20 within 20 h, at which time the residual 46, 41, 27 and 25 kDa active pMMP20 bands had dropped to approximately 20% of pre-incubation levels (Fig. 5). In contrast, in the active pMMP20-only control, the residual active pMMP20 bands decreased only modestly, to 60–80% of pre-incubation levels, apparently due to autolysis (Fig. 5). At pH 5.5, active pMMP20 bands were stable when incubated with or without pKLK4. The pKLK4-only samples also were stable under both pH conditions. These results demonstrate that KLK4 degrades MMP20 in vitro.
Fig. 4.
Digestion of porcine MMP20 (pMMP20) by KLK4 under physiologic (pH 7.4) or mildly acidic (pH 5.5) conditions. Purified native porcine KLK4 (pKLK4) was used to digest pMMP20 for 0, 5, 10 and 20 h. Both pMMP20 and pKLK4 alone were run as controls. The pKLK4 catalyzed pMMP20 under pH 7.4, but not under pH 5.5.
Fig. 5.
Quantification analyses of active pMMP20 and pKLK4 bands on zymograms following 5, 10, 15, and 20 h incubations together or separately at pH 7.4 or 5.5. The area of each active band on the zymogram gel was determined using ImageJ densitometry software and normalized to the density of 0 h band.
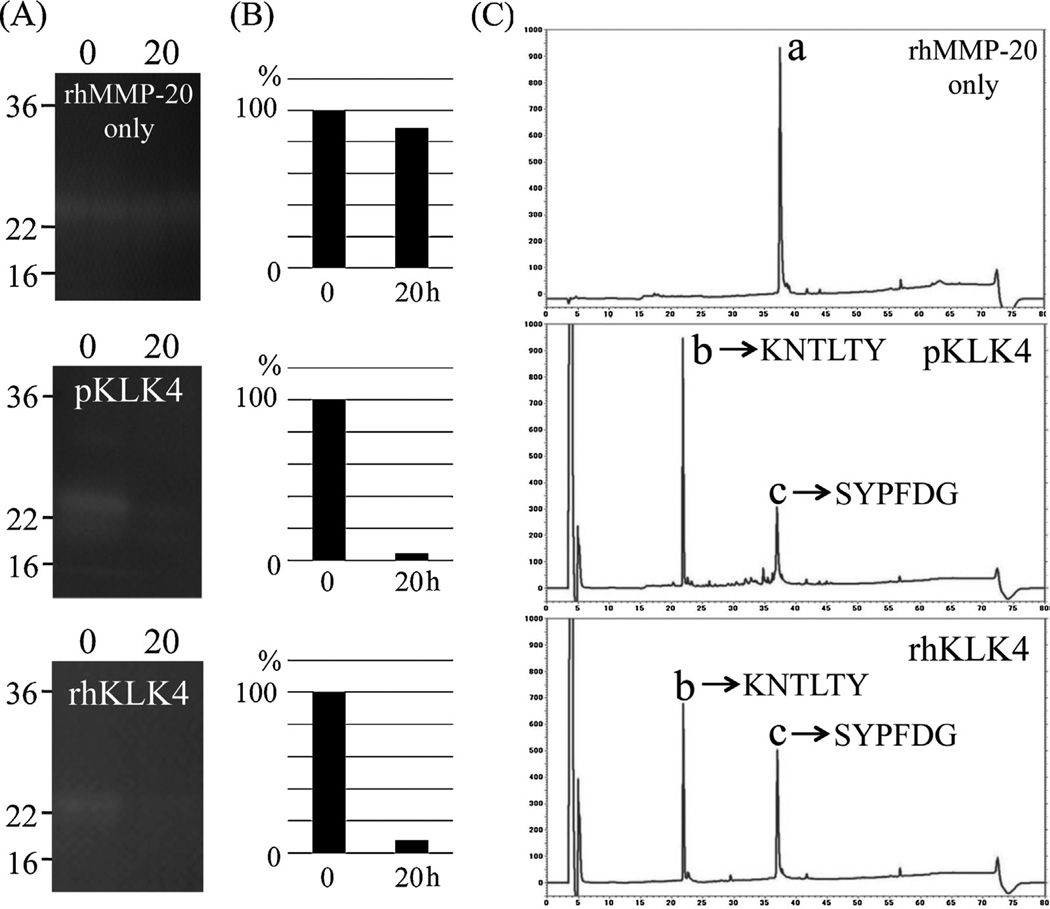
3.4. Cleavage experiment of recombinant MMP20 by porcine KLK4
Since KLK4 was able to degrade MMP20, we characterized the locations of the pMMP20 cleavage sites catalyzed by KLK4. We used commercial rhMMP20, which comprises the catalytic domain, and further purified it using RP-HPLC prior to KLK4 digestion (data not shown). The rhMMP20 was digested by pKLK4 and rhKLK4 which was activated by rhMMP20 under the condition of pH 7.4. Both KLK4 proteins reduced the activity of rhMMP20 (Fig. 6A). The original rhMMP20 displayed a single chromatographic peak on RP-HPLC with a retention time at 37.5 min (Fig. 6B). The digestion of rhMMP20 by pKLK4 and rhKLK4 generated two major chromatographic peaks at 22 and 37 min and Edman sequencing of those peaks gave K-N-T-L-T-Y and S-Y-P-F-D-G, respectively (Fig. 6B).
Fig. 6.
Inactivation of recombinant human MMP20 (rhMMP20) (substrate) by KLK4. (A) Both native porcine KLK4 (pKLK4) and recombinant human KLK4 (rhKLK4) activated by rhMMP20 were used to digest rhMMP20 for 0 or 20 h. The rhMMP20 alone was incubated for 20 h as a control and no degradation occurred before and after the incubation (top). Both pKLK4 (centre) and rhKLK4 (bottom) cleaved rhMMP20 and exhibited almost no activity on a casein zymogram after 20 h. (B) Quantification analysis of active bands before and after incubation. The active bands were normalized to the density of the 0 h band. (C) The digestion of (rhMMP20) was clearer on the chromatographic peaks of intact MMP20 and its digestion products after incubation with KLK4. C-18 RP-HPLC chromatograms after the digestion for 20 h showing rhMMP20 original material (a) with a retention time of 37.5 min; the generation of peaks (b) and (c) with retention times of 22 and 37 min after digestion with pKLK4 and rhKLK4. The protein sequences for the generated MMP20 peaks (b) and (c) are provided on the chromatograms.
4. Discussion
4.1. Isolation of MMP20 and KLK4
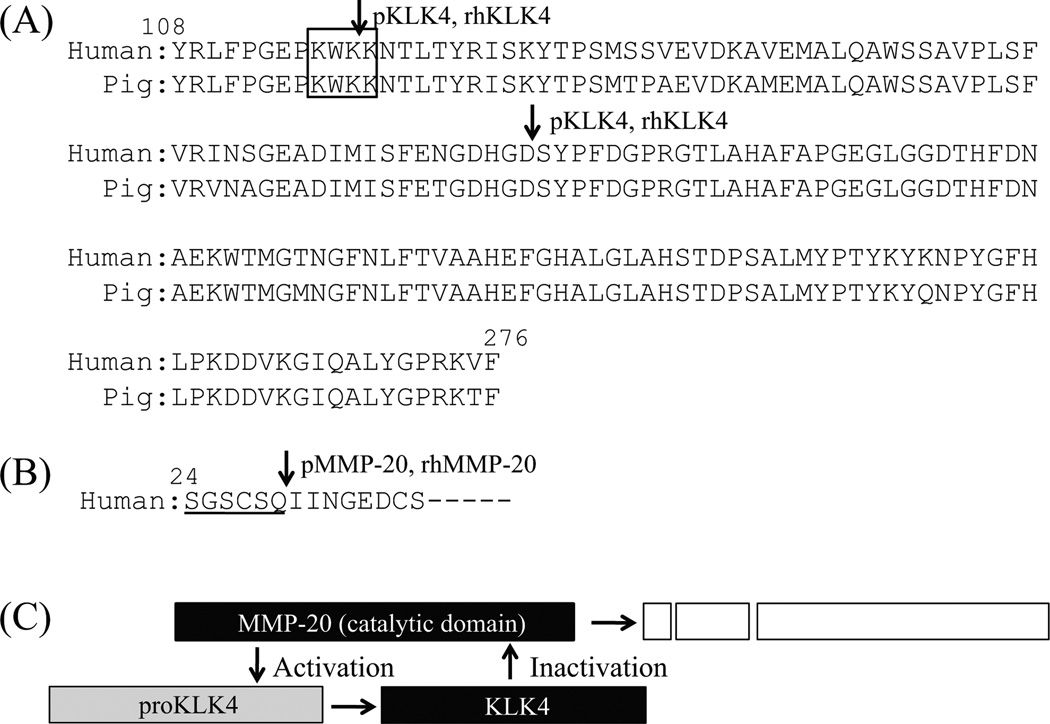
Our previous purification protocol for MMP20 started with anion exchange chromatography.26,27 In this study, we improved the isolation protocol for MMP20. Certain consensus sequences have been proposed for heparin-binding proteins, such as BBXB or BXBB, where B denotes a positively charged amino acid residue.28 We found that MMP20 possesses one heparin-binding site (K116–K119) in the catalytic domain (Fig. 7A), which is conserved in human and pig. Based upon this finding, we were able to isolate porcine MMP20 by applying approximately 300 mg of soft enamel AL extract on a Heparin affinity column.
Fig. 7.
Summary of the activation and inactivation of MMP20 and KLK4. (A) Pairwise sequence alignment of catalytic domain in human and pig MMP20. The square indicates a consensus amino acid sequence for heparin-binding. Downward pointing arrows are inactivation sites catalyzed by pKLK4 and/or rhKLK4 (after Lys118 and Asp178) as identified by Edman degradation analysis. All inactivation sites are conserved in human and pig MMP20. (B) Sequence of propeptide-enzyme junction of human KLK4. Downward pointing arrow marks an activation site (after Gln29) by both pMMP20 and rhMMP20 as identified by Edman degradation analysis. Underline indicates a propeptide region of human KLK4. (C) Diagram showing the activation and inactivation by MMP20-KLK4 interaction. The MMP20 activates proKLK4 to KLK4 by cleaving at appropriate activation site of propeptide-enzyme junction. The KLK4 inactives MMP20 by cleaving two major sites in the catalytic domain.
4.2. Activation of KLK4 by MMP20
In situ hybridization studies have demonstrated that MMP20 is expressed by secretory stage ameloblasts, while KLK4 is expressed from the transition stage throughout the maturation stage.6–8,29 In general, kallikrein related proteases are secreted as inactive pro-forms (proKLKs) or zymogens that are activated extracellularly by proteolytic release of their N-terminal propeptide, and this is a key step in their functional regulation. MMP20 has been shown to be a broad activator of proKLKs.30 Previously we demonstrated that recombinant pig proKLK4 can be activated by recombinant pig MMP20 (rpMMP20).12 The rpMMP20 also cleaved a fluorescent peptide analogue of the KLK4 propeptide-enzyme junction. Here we demonstrated that both pMMP20 and rhMMP20 activate rh-proKLK4 by cleaving precisely at the propeptide-enzyme junction at Gln29-Ile30 used in vivo (Fig. 7B). Recently we demonstrated that KLK4 is active in the MMP20 null mice, suggesting that KLK4 can be activated by protease(s) other than MMP20.18 An in vitro study demonstrated that dipeptidyl peptidase I (DPPI) activates proKLK4.31 These findings support the conclusion that KLK4 is activated by not only MMP20 but also other proteases.
4.3. Inactivation of MMP20 by KLK4
In KLK4 null mice MMP20 activity persists longer than in wild-type mice, suggesting KLK4 may inactivate MMP20 in vivo.18 We demonstrated that KLK4, which was activated by MMP20, cleaves the catalytic domain of MMP20 under neutral pH conditions. Maturation stage ameloblasts modulate between ruffle-ended and smooth-ended phases.32 The enamel beneath ruffle-ended ameloblasts is mildly acidic. Bicarbonate ion neutralize the matrix under smooth-ended ameloblasts.33 Because degradation of MMP20 by KLK4 occurs only at neutral and not at acidic pH, our findings suggest that the cleavage of MMP20 by KLK4 mainly progresses at physiologic pH under smooth-ended ameloblasts.
The catalytic domain of MMP20 shares high sequence similarity with other MMPs and use the same ligands as other MMPs to bind two zinc and two calcium ions: H176, D178, H191, H204, H226, H230 and H236 for zinc ions, and D166, D183, G184, R186, T188, G198, G200, D202, D206 and E209 for calcium ions.34,35 We demonstrated that both pKLK4 and rhKLK4 were able to catalyze specific cleavages after Lys118-Lys119 and Asp178-Ser179 within the catalytic domain of rhMMP20 (Fig. 7A). The latter cleavage site may affect the residues coordinating the zinc and calcium ions and thereby cause the inactivation of MMP20.
4.4. MMP20-KLK4 activation and inactivation interactions
MMP20 is expressed by secretory stage ameloblasts7 and it slowly processes enamel proteins into a group of cleavage products that accumulate in the enamel.22,36,37 Furthermore, enamel proteins are progressively degraded by KLK4, which is expressed by transition and maturation stage ameloblasts.13 In addition to the processing and degradation for enamel proteins, we conclude that the MMP20 is able to activate pro-KLK4, and the activated KLK4 is able to inactivate MMP20 remaining in enamel during the transition and early maturation stage (Fig. 7C).
Acknowledgments
The authors thank Mr. Yasuaki Fukuta of APRO life Science Institute Inc. (Naruto, Tokushima, Japan) and Mr. Minoru Yasui of Japan Bio Services Co., LTD (Asaka, Saitama, Japan) for their help in amino acid sequence analysis.
Funding
This study was supported by Grant-in-Aid for Research Activity Start-up Number 24890265 (YY), and National Institutes of Health Grant DE 019775 (JPS) from NIDCR.
Footnotes
Competing interests
There are no conflicts of interest to declare from the authors.
Ethical approval
All experimental procedures involving the use of animals were reviewed and approved by the Ethics Committee of the Institute of Tsurumi University, Yokohama, Japan.
Contributor Information
James P. Simmer, Email: jsimmer@umich.edu.
John D. Bartlett, Email: jbartlett@forsyth.org.
Takeo Karakida, Email: karakida-t@tsurumi-u.ac.jp.
Shinichiro Oida, Email: oida-s@tsurumi-u.ac.jp.
REFERENCES
- 1.Ronnholm E. The amelogenesis of human teeth as revealed by electron mircoscopy II. The development of the enamel crystallites. J Ultrastructure Res. 1962;6:249–303. doi: 10.1016/s0022-5320(62)80036-7. [DOI] [PubMed] [Google Scholar]
- 2.Boyde A. The development of enamel structure. Proc R Soc Med. 1967;60:923–928. [PMC free article] [PubMed] [Google Scholar]
- 3.Smith CE, Pompura JR, Borenstein S, Fazel A, Nanci A. Degradation and loss of matrix proteins from developing enamel. Anat Rec. 1989;224:292–316. doi: 10.1002/ar.1092240219. [DOI] [PubMed] [Google Scholar]
- 4.Bartlett JD, Simmer JP, Xue J, Margolis HC, Moreno EC. Molecular cloning and mRNA tissue distribution of a novel matrix metalloproteinase isolated from porcine enamel organ. Gene. 1996;183:123–128. doi: 10.1016/s0378-1119(96)00525-2. [DOI] [PubMed] [Google Scholar]
- 5.Cloning and characterization of a cDNA encoding human EMSP1. In: Simmer JP, Ryu OH, Qian Q, Zhang C, Cao X, Sun X, et al., editors. Chemistry and biology of mineralized tissues: proceedings of the sixth international conference; 1998. [Google Scholar]
- 6.Hu JC, Sun X, Zhang C, Liu S, Bartlett JD, Simmer JP. Enamelysin and kallikrein-4 mRNA expression in developing mouse molars. Eur J Oral Sci. 2002;110:307–315. doi: 10.1034/j.1600-0722.2002.21301.x. [DOI] [PubMed] [Google Scholar]
- 7.Simmer JP, Sun X, Yamada Y, Zhang CH, Bartlett JD, Hu JC-C. Enamelysin and kallikrein-4 expression in the mouse incisor. In: Kobayashi I, Ozawa H, editors. Biomineralization: formation, diversity, evolution and application. Proceedings of the 8th international symposium on biomineralization. Hadano, Japan: Tokai University Press; 2004. pp. 348–352. [Google Scholar]
- 8.Begue-Kirn C, Krebsbach PH, Bartlett JD, Butler WT. Dentin sialoprotein, dentin phosphoprotein, enamelysin and ameloblastin: tooth-specific molecules that are distinctively expressed during murine dental differentiation. Eur J Oral Sci. 1998;106:963–970. doi: 10.1046/j.0909-8836.1998.eos106510.x. [DOI] [PubMed] [Google Scholar]
- 9.Fukae M, Tanabe T, Uchida T, Lee SK, Ryu OH, Murakami C, et al. Enamelysin (matrix metalloproteinase-20): localization in the developing tooth and effects of pH and calcium on amelogenin hydrolysis. J Dent Res. 1998;77:1580–1588. doi: 10.1177/00220345980770080501. [DOI] [PubMed] [Google Scholar]
- 10.Bartlett JD, Yamakoshi Y, Simmer JP, Nanci A, Smith CE. MMP20 cleaves E-cadherin and influences ameloblast development. Cells Tissues Organs. 2011;194:222–226. doi: 10.1159/000324205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartlett JD, Smith CE. Modulation of cell-cell junctional complexes by matrix metalloproteinases. J Dent Res. 2013;92:10–17. doi: 10.1177/0022034512463397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ryu O, Hu JC, Yamakoshi Y, Villemain JL, Cao X, Zhang C, et al. Porcine kallikrein-4 activation, glycosylation, activity, and expression in prokaryotic and eukaryotic hosts. Eur J Oral Sci. 2002;110:358–365. doi: 10.1034/j.1600-0722.2002.21349.x. [DOI] [PubMed] [Google Scholar]
- 13.Simmer J, Hu Y, Richardson A, Bartlett J, Hu JC-C. Why does enamel in Klk4 null mice break above the dentino-enamel junction? Cells Tissues Organs. 2011;194:211–215. doi: 10.1159/000324260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith CE. Cellular and chemical events during enamel maturation. Crit Rev Oral Biol Med. 1998;9:128–161. doi: 10.1177/10454411980090020101. [DOI] [PubMed] [Google Scholar]
- 15.Fukae M, Yamamoto R, Karakida T, Shimoda S, Tanabe T. Micelle structure of amelogenin in porcine secretory enamel. J Dent Res. 2007;86:758–763. doi: 10.1177/154405910708600814. [DOI] [PubMed] [Google Scholar]
- 16.Lu Y, Papagerakis P, Yamakoshi Y, Hu JC, Bartlett JD, Simmer JP. Functions of KLK4 and MMP-20 in dental enamel formation. Biol Chem. 2008;389:695–700. doi: 10.1515/BC.2008.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith CE, Richardson AS, Hu Y, Bartlett JD, Hu JC, Simmer JP. Effect of Kallikrein 4 loss on enamel mineralization: comparison with mice lacking matrix metalloproteinase 20. J Biol Chem. 2011;286:18149–18160. doi: 10.1074/jbc.M110.194258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamakoshi Y, Richardson AS, Nunez SM, Yamakoshi F, Milkovich RN, Hu JC, et al. Enamel proteins and proteases in Mmp20 and Klk4 null and double-null mice. Eur J Oral Sci. 2011;119(Suppl 1):206–216. doi: 10.1111/j.1600-0722.2011.00866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bronckers AL, Gueneli N, Lullmann-Rauch R, Schneppenheim J, Moraru AP, Himmerkus N, et al. The intramembrane protease SPPL2A is critical for tooth enamel formation. J Bone Miner Res. 2013;28:1622–1630. doi: 10.1002/jbmr.1895. [DOI] [PubMed] [Google Scholar]
- 20.Feng J, McDaniel JS, Chuang HH, Huang O, Rakian A, Xu X, et al. Binding of amelogenin to MMP-9 and their co-expression in developing mouse teeth. J Mol Histol. 2012;43:473–485. doi: 10.1007/s10735-012-9423-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryu OH, Fincham AG, Hu CC, Zhang C, Qian Q, Bartlett JD, et al. Characterization of recombinant pig enamelysin activity and cleavage of recombinant pig and mouse amelogenins. J Dent Res. 1999;78:743–750. doi: 10.1177/00220345990780030601. [DOI] [PubMed] [Google Scholar]
- 22.Nagano T, Kakegawa A, Yamakoshi Y, Tsuchiya S, Hu JC, Gomi K, et al. Mmp-20 and Klk4 cleavage site preferences for amelogenin sequences. J Dent Res. 2009;88:823–828. doi: 10.1177/0022034509342694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lausch E, Keppler R, Hilbert K, Cormier-Daire V, Nikkei S, Nishimura G, et al. Mutations in MMP9 and MMP13 determine the mode of inheritance and the clinical spectrum of metaphyseal anadysplasia. Am J Hum Genet. 2009;85:168–178. doi: 10.1016/j.ajhg.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lacruz RS, Smith CE, Smith SM, Hu P, Bringas P, Jr, Sahin-Toth M, et al. Chymotrypsin C (caldecrin) is associated with enamel development. J Dent Res. 2011;90:1228–1233. doi: 10.1177/0022034511418231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou J, Sahin-Toth M. Chymotrypsin C mutations in chronic pancreatitis. J Gastroenterol Hepatol. 2011;26:1238–1246. doi: 10.1111/j.1440-1746.2011.06791.x. http://dx.doi.org/10.1111/j.l440-1746.2011.06791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamakoshi Y, Hu JC-C, Ryu OH, Tanabe T, Oida S, Fukae M, et al. A comprehensive strategy for purifying pig enamel proteins. In: Kobayashi I, Ozawa H, editors. Biomineralization: formation, diversity, evolution and application. Proceedings of the 8th international symposium on biomineralization. Hadano, Japan: Tokai University Press; 2003. pp. 326–332. [Google Scholar]
- 27.Yamada Y, Yamakoshi Y, Gerlach R, Hu C, Matsumoto K, Fukae M, et al. Purification and characterization of enamelysin from secretory stage pig enamel. Arch Comp Biol Tooth Enam. 2003;8:21–25. [Google Scholar]
- 28.Shimazaki K, Uji K, Tazume T, Kumura H, Shimo-Oka T. Approach to identification and comparison of the heparin-interacting sites of lactoferrin using synthetic peptides. Excerpta Medica Int Congr Ser. 2000;1195:37–46. [Google Scholar]
- 29.Hu JC, Ryu OH, Chen JJ, Uchida T, Wakida K, Murakami C, et al. Localization of EMSP1 expression during tooth formation and cloning of mouse cDNA. J Dent Res. 2000;79:70–76. doi: 10.1177/00220345000790011301. [DOI] [PubMed] [Google Scholar]
- 30.Yoon H, Blaber SI, Li W, Scarisbrick IA, Blaber M. Activation profiles of human kallikrein-related peptidases by matrix metalloproteinases. Biol Chem. 2013;394:137–147. doi: 10.1515/hsz-2012-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tye CE, Pham CT, Simmer JP, Bartlett JD. DPPI may activate KLK4 during enamel formation. J Dent Res. 2009;88:323–327. doi: 10.1177/0022034509334240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Josephsen K, Fejerskov O. Ameloblast modulation in the maturation zone of the rat incisor enamel organ A light and electron microscopic study. J Anat. 1977;124:45–70. [PMC free article] [PubMed] [Google Scholar]
- 33.Sasaki S, Takagi T, Suzuki M. Cyclical changes in pH in bovine developing enamel as sequential bands. Arch Oral Biol. 1991;36:227–231. doi: 10.1016/0003-9969(91)90090-h. [DOI] [PubMed] [Google Scholar]
- 34.Andreini C, Banci L, Bertini I, Luchinat C, Rosato A. Bioinformatic comparison of structures and homology-models of matrix metalloproteinases. J Proteome Res. 2004;3:21–31. doi: 10.1021/pr0340476. [DOI] [PubMed] [Google Scholar]
- 35.Arendt Y, Banci L, Bertini I, Cantini F, Cozzi R, Del Conte R, et al. Catalytic domain of MMP20 (Enamelysin)—the NMR structure of a new matrix metalloproteinase. FEBS Lett. 2007;581:4723–4726. doi: 10.1016/j.febslet.2007.08.069. [DOI] [PubMed] [Google Scholar]
- 36.Iwata T, Yamakoshi Y, Hu JC, Ishikawa I, Bartlett JD, Krebsbach PH, et al. Processing of ameloblastin by MMP-20. J Dent Res. 2007;86:153–157. doi: 10.1177/154405910708600209. [DOI] [PubMed] [Google Scholar]
- 37.Chun YH, Yamakoshi Y, Yamakoshi F, Fukae M, Hu JC, Bartlett JD, et al. Cleavage site specificity of MMP-20 for secretory-stage ameloblastin. J Dent Res. 2010;89:785–790. doi: 10.1177/0022034510366903. [DOI] [PMC free article] [PubMed] [Google Scholar]