Abstract
G protein-coupled receptors (GPCRs) are versatile signaling proteins that mediate complex cellular responses to hormones and neurotransmitters. Recent advances in GPCR crystallography have provided inactive and active state structures for rhodopsin and the β2 adrenergic receptor (β2AR). Although these structures suggest a two-state ‘on-off’ mechanism of receptor activation, other biophysical studies and observed signaling versatility suggest that GPCRs are highly dynamic and exist in a multitude of functionally distinct conformations. To fully understand how GPCRs work, we must characterize these conformations and determine how ligands affect their energetics and rates of interconversion. This brief review will compare and contrast the dynamic properties of rhodopsin and β2AR that shed light on the role of structural dynamics in their distinct signaling behaviors.
Introduction
G protein-coupled receptors exhibit a complex profile of signaling and regulatory behavior upon activation by endogenous or synthetic agonists. For most GPCRs, binding of the endogenous hormone or neurotransmitter leads to conformational changes at the cytoplasmic ends of the transmembrane (TM) segments that provide an interaction interface for cytosolic proteins including heterotrimeric G proteins, G protein-coupled receptor kinases (GRKs) and arrestins. In addition, GPCRs have been shown to localize to specific signaling compartments at the plasma membrane through interactions between specific sequences in their carboxyl termini or third intracellular loops and scaffolding proteins [1–4]. More recent evidence suggests that some GPCRs may signal from intracellular compartments such as endosomes [5,6].
Many GPCRs can signal in absence of endogenous agonists, a phenomenon termed basal activity. GPCR ligands can induce a broad range of signaling responses. At saturating concentrations, ligands can induce the maximal G protein signaling response (full agonists), induce submaximal signaling (partial agonists) or decrease basal levels of signaling (inverse agonists). Furthermore, some ligands can act as agonists of one signaling pathway while acting as inverse agonists of an alternative pathway (biased agonists).
There is a growing body of evidence that this functional versatility is due to structural plasticity. GPCRs can no longer be described as simple bimodal switches, but rather exist as ensembles of discrete conformations with energetics that can be influenced by ligands, cytosolic signaling and regulatory proteins, lipids, pH, ions and possibly transmembrane voltage gradients [7–9]. This structural plasticity may contribute in part to current challenges in GPCR drug discovery. Further complicating our understanding of GPCR signaling is the role of homo- and heterooligomers. While oligomerization of GPCRs has been extensively studied, [10,11], the structure and dynamics of dimers/oligomers and their roles in receptor function and physiology are not fully understood [12].
This review will focus on the very narrow topic of protein dynamics of individual GPCR protomers. Proteins are often conceptualized as the rigid entities we observe in crystal structures. However all proteins exhibit dynamic character at several levels, from femtosecond bond vibrations, to side-chain motions that occur on the picosecond to nanosecond timescales, to larger domain motions that happen over microseconds to seconds [13]. Here we will review the possible role that protein dynamics plays in the functional differences for the two most extensively studied GPCR model systems: rhodopsin and the β2 adrenergic receptor (β2AR). As will be discussed below, these receptors have very similar structures in their inactive and active states, but differ in signaling efficiency, complexity and kinetics.
Rhodopsin is a highly efficient photoreceptor
Rhodopsin remains the best-characterized GPCR by biophysical methods to date. This can be attributed in part to its physiologic importance, its natural abundance, its biochemical stability, the ability to monitor its functional state by the spectroscopic properties of its covalent ligand retinal, and the ability to precisely time its activation by light. Although rhodopsin has long served as a prototypical GPCR, its function as a light sensor is uniquely specialized for both sensitivity and fidelity. In the inactive state, rhodopsin is covalently bound to 11-cis-retinal which acts as a highly efficacious inverse agonist to suppress basal activity. The virtual absence of basal activity ensures high signal fidelity in the visual system. Illumination by light induces isomerization of 11-cis-retinal to all-trans-retinal, which acts as a highly efficacious agonist for activation of the specialized visual system G protein transducin (Gt). In the absence of light, retinal isomerization has a very high energy barrier (~45 kcal/mol) that prevents basal signaling [14]. Upon retinal isomerization, rhodopsin activation follows a series of short-lived intermediates before reaching an equilibrium between several metarhodopsin (Meta) states [15]. In physiological settings, the Meta I state progresses to the Meta II states, which are fully active and capable to coupling to Gt, within milliseconds. As seen below, virtually every light activated rhodopsin proceeds to the active Meta II state even in the absence of transducin. It has been estimated that a single active rhodopsin molecule can activate hundreds of transducin molecules [16]. Subsequent hydrolysis of the covalent link between retinal and rhodopsin yields the apoprotein opsin. The remarkable signal amplification in response to a photon is due in part to the efficiency of rhodopsin as well as the highly organized structure of the rod outer segments of the retina [17].
The β2AR is a versatile but less efficient signaling machine
The switch-like behavior of rhodopsin from completely inactive to fully active in milliseconds is likely unique to this GPCR and essential for its role in vision. In contrast, like many other GPCRs the β2AR exhibits varying levels of basal activity and couples to two different G proteins (Gs and Gi) under physiologic conditions. In addition, the β2AR has been shown to signal through arrestins in a G protein independent manner [18,19]. The β2AR has been a pharmaceutical target for the treatment of asthma and chronic lung disease. Consequently, there is a rich diversity of ligands that span the efficacy spectrum. It has been observed that the efficacy profile for ligands differs for different signaling pathways [20,21], supporting the existence of multiple ligand-specific signaling conformations.
Unlike the highly efficient activation of rhodopsin by light-induced isomerization of retinal, the activation of the β2AR by its natural agonist adrenaline is much less efficient. Comparing the energetics of retinal isomerization to adrenaline binding offers some insight into this difference. Retinal isomerization provides approximately 35 kcal/mol in the transition of rhodopsin to bathorhodopsin [22], which is the first photo-activated intermediate. This energy is then transferred in multiple steps to achieve the Meta II conformation. In contrast, adrenaline binds with relatively low affinity to the β2AR (Ki of ~1 μM) in the absence of Gs, and this binding event generates only ~8.2 kcal/mol of energy for β2AR activation. Furthermore, binding of adrenaline to the β2AR is dependent on the concentration of the hormone. Rapid activation of the receptor, therefore, requires a high concentration of adrenaline delivered at a synapse or via the circulation. Due to the relatively low affinity and rapid dissociation rate of adrenaline, it is possible that not every binding event leads to a signaling event. As will be shown below, even when bound to a high affinity agonist at saturating concentrations, only a fraction of the β2AR achieves a fully active conformation.
Methodological challenges in studying dynamics of GPCRs
Before proceeding, it is important to acknowledge the technical challenges and limitations in studying the dynamic properties of GPCRs and other membrane proteins. Methods used to study protein structure and dynamics include crystallography, nuclear magnetic resonance (NMR) spectroscopy, fluorescence spectroscopy and electron paramagnetic resonance (EPR) spectroscopy. To obtain information about the dynamics of specific structural domains such as a TM segment, it is often necessary to chemically modify the TM at specific sites (such as a single reactive cysteine) with small probes (typically < 500 Da) that are sensitive to the local molecular environment and can be detected by fluorescence, EPR or NMR spectroscopy. These methods generally require access to pure, functional protein necessitating extraction of the GPCR from a native lipid bilayer using detergents. The process of purifying proteins in detergent leads to loss of interactions with native lipids. Thus studies are often carried out in detergent solutions or in artificial lipid bilayers. The amounts of protein required for these studies vary from 10–100 micrograms for fluorescence studies to 1–10 milligrams for crystallographic trials and NMR experiments.
Another concern in understanding protein dynamics is the functional state of the protein. A significant fraction of GPCRs purified from expression systems such as insect cells, yeast and bacteria (particularly from inclusion bodies) may be nonfunctional due to improper folding during biosynthesis or denaturation during purification. Accurate interpretation of biophysical studies requires that virtually all of the protein be functional. Therefore it is essential to include ligand affinity chromatography in the purification protocol and validate the fraction of functional receptor during biophysical experiments.
An exception to the use of purified protein to study protein dynamics can be found in the elegant studies using fluorescent protein reporters and FlAsH tags as described by Lohse et al. in this issue. These approaches allow the study of protein kinetics in native cell membranes; however, due to the nature and size of the fluorescent probes, this approach is limited in its ability to monitor the structure and dynamics of specific structural domains. A technology of promise for future studies is the use of unnatural amino acids incorporated using suppressor tRNA methods [23]. This approach may allow site-specific labeling of GPCRs with small probes in living cells.
Given these experimental caveats, we have limited our comparisons of the dynamic properties of the β2AR and rhodopsin to purified protein studied in similar environments, most often the long alkyl chain maltoside detergent dodecylmaltoside. Thus, differences in dynamics can be attributed to the intrinsic differences in these proteins rather than differences in the experimental environment.
Models of β2AR and rhodopsin activation
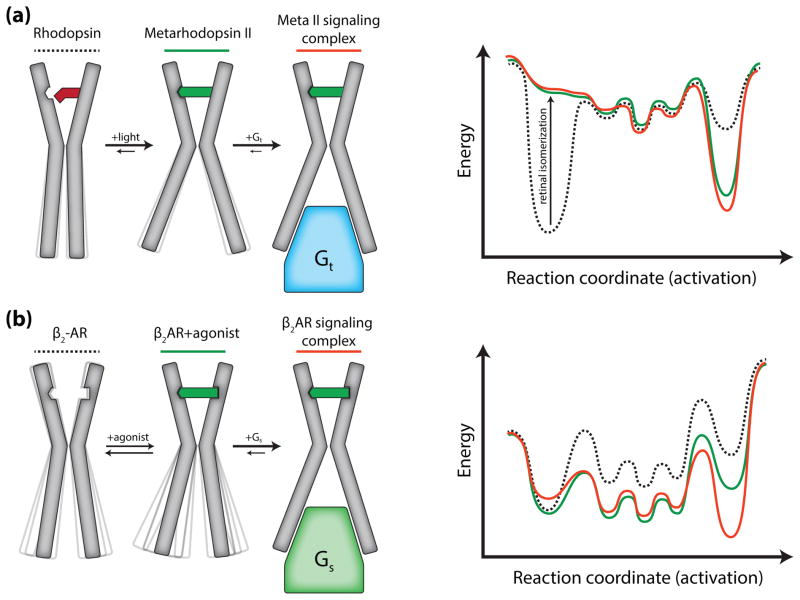
The results of crystallography experiments and other biophysical studies discussed below provide support for two different models of activation for rhodopsin and β2AR (Fig. 1). Rhodopsin exists predominantly in two distinct conformations: inactive (bound to 11-cis-retinal) or active (all-trans-retinal-bound Meta II). While there are other well-characterized intermediates, their lifetimes are too short to have direct roles in interactions with signaling proteins [15]. This is illustrated in the cartoon and simple energy landscapes shown in Fig. 1a. The light-activated state of rhodopsin is relatively stable such that the cytoplasmic surface of nearly all Meta II molecules is in the open conformation, even in the absence of transducin. Transducin contributes relatively little to the stability of this open conformation. In contrast, functional and biophysical studies suggest that the β2AR exists in an ensemble of low energy conformations with different functional properties (Fig. 1b). Even when bound to a nearly irreversible agonist with a dissociation half-life of 30 hours, the active state is not fully stabilized and the receptor becomes more heterogeneous. Only in the presence of the G protein is the active conformation fully stabilized.
Figure 1.
Differing models for activation of rhodopsin and the β2AR. (a) 11-cis-retinal bound rhodopsin is inactive with a cytoplasmic domain incapable of coupling to transducin (Gt). As the inactive conformation is the lowest energy state, dark rhodopsin displays minimal conformational heterogeneity. Light induced isomerization of the ligand to all-trans-retinal increases the energy of the inactive conformation resulting in a transition to the activated Meta II state and an opening of the cytoplasmic domain. The C-terminus of transducin interacts with the Meta II state to form the signaling complex. (b) Unliganded β2AR is conformationally dynamic as a result of smaller energetic differences between inactive, intermediate, and active states. Agonist binding increases β2AR dynamics by decreasing the energy of intermediate and active states. However, agonists do not fully stabilize the active state, and agonist bound β2AR primarily exists in inactive and intermediate conformations. Gs further stabilizes the active conformation and formation of the signaling complex is required for the receptor to completely transition to the active state.
Insights into dynamics from inactive and active state crystal structures
Recent advances in protein engineering, in meso crystallography, and micro-focus X-ray diffraction data collection have enabled the structural characterization of many GPCRs. Among these, three have been crystallized in inactive and active conformations, including rhodopsin [24–28], the β2AR [29–31], and the M2 muscarinic receptor [32,33]. While only the β2AR has been crystalized in complex with a G protein and a G protein mimetic nanobody (Nb80), rhodopsin has been crystallized in complex with the carboxyl terminal peptide of transducin (Gαt-CT) as a surrogate for the intact G protein. Comparison of inactive and active structures indicates a conserved set of changes required to engage a G protein. The most dramatic conformational change associated with receptor activation is a 7–14 angstrom displacement of transmembrane 6 (TM6) accompanied by more subtle rearrangements of TM5 and TM7.
Although these crystal structures provide high-resolution insights into GPCR activation, they represent two endpoints of a complex conformational ensemble. Crystallogenesis usually traps low energy receptor conformations, and as a result, crystal structures usually provide only limited insights into protein dynamics. Nevertheless, heterogeneity in conformation of the same protein among multiple crystal structures can provide some clues into protein dynamics. In cases where there are two or more molecules in the asymmetric unit, one may observe distinct conformations. Several inactive-state structures of rhodopsin have two molecules in the asymmetric unit. Comparison of these two molecules reveals a root mean squared deviation (RMSD) of 0.5 Å, and a single conformation of TM6 with an interaction between the Glu134, Arg135 and Glu247. This interaction has been described as the “ionic lock” responsible for stabilizing the inactive state of rhodopsin. While all of the inactive-state structures of the β2AR to date have only one molecule in the asymmetric unit, there are several structures of the highly homologous β1AR having two molecules in the asymmetric unit. Among the various inactive β1AR monomers, TM6 exists in two different conformations depending on the state of the ionic lock. While the overall RMSD between these β1AR conformations is low (1.3 Å), the cytoplasmic end of TM6 is displaced outward by 7 Å in the conformation with a broken ionic lock [34].
Differences in protein dynamics might also be deduced by comparing the methods required to obtain active state structures of rhodopsin and the β2AR. Active state structures of the β2AR have only been obtained in complex with a G protein [31] or G protein mimetic nanobody (Nb80)[30] to stabilize the open conformation of TM6. In the absence of such a stabilizing interaction at the cytoplasmic domain of the receptor, the β2AR either fails to form crystals or crystallizes in an inactive conformation, even when bound to a covalent agonist [35]. These results suggest that the active state of the β2AR is a relatively high-energy state in the absence of its G protein, and even a high affinity agonist with virtually complete occupancy at the receptor binding pocket cannot fully stabilize the active state. Long time-scale molecular dynamics simulations further support this hypothesis. When starting with the active state structure bound to an agonist, the cytoplasmic domains collapse to the inactive state within 11 microseconds of simulation [35].
In contrast, there are several active state structures of rhodopsin including ligand-free opsin and Meta II, crystallized both with and without Gαt-CT (Park, Scheerer et al. 2008, Scheerer, Park et al. 2008, Choe, Kim et al. 2011, Standfuss, Edwards et al. 2011). All of these structures are remarkably similar (RMSD < 0.5 Å) suggesting that, at least under the conditions used for crystallography, the active conformation of rhodopsin is a low energy state even in the absence of an agonist and transducin.
Insights into structural dynamics from biophysical studies
Numerous studies have applied biophysical methods to characterize structural changes and dynamics of rhodopsin and the β2AR. These have been reviewed more extensively elsewhere [7,15], and include more recent applications of NMR [36–39], and time-dependent derivatization with chemical probes [40]. Additionally, molecular dynamics simulations have provided insight into the activation of the β2AR [41]. However, for the purpose of this review, we will focus on a few studies that highlight a difference in the dynamic behavior of rhodopsin and the β2AR that may be responsible for functional differences.
Support for differences in protein dynamics between the β2AR and rhodopsin comes from a few studies that characterize steady-state distributions of receptor conformations in detergent-solubilized purified protein. As noted above, detergents are necessary for extraction and purification of GPCRs from native membranes. They maintain the protein in a uniform molecular environment and facilitate studies by EPR and NMR spectroscopy where relatively high concentrations of protein are required. Detergents are amphipathic molecules that act as a surrogate for a membrane environment, with the hydrophobic component binding to the transmembrane segments and hydrophilic end facing outward towards the aqueous environment. There is a broad spectrum of structurally different detergents, and the sensitivity to membrane proteins to different detergents is related to the overall stability of the membrane protein. Relatively few detergents are able to extract GPCRs in a functional state. Consistent with the greater overall stability of rhodopsin, it is more tolerant to detergents having short alkyl chains or charged head-groups than the β2AR and most other GPCRs. For the studies discussed below, the detergent dodecylmaltoside is used to extract, purify and study both rhodopsin and the β2AR. The β2AR in dodecylmaltoside is able to bind both agonists and antagonists with the same rank order of affinity as in a lipid environment. Both the β2AR and rhodopsin can couple to their respective G proteins in dodecylmaltoside solutions. Thus, while not a native environment, these biochemical preparations provide a suitable surrogate for comparing the structural dynamics of these receptors that are relevant to their functional differences.
Rhodopsin dynamics
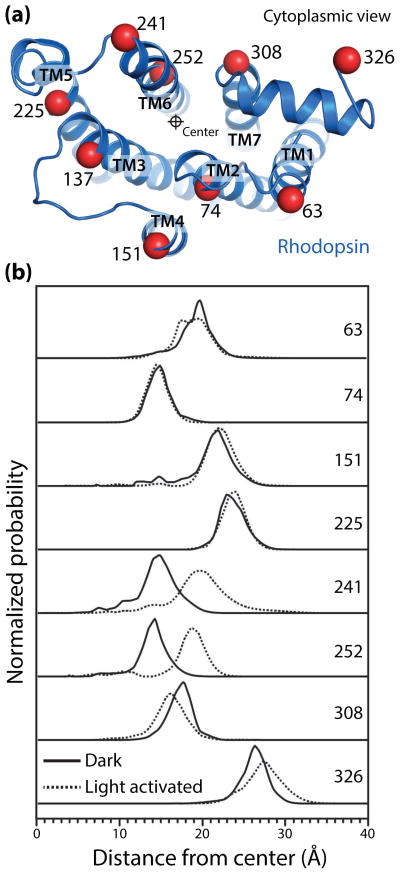
To directly examine structural changes in rhodopsin, Altenbach et al. utilized double electron-electron resonance (DEER) spectroscopy to map the conformational changes in the cytoplasmic domain of rhodopsin upon receptor activation [42]. In an intensive set of experiments, rhodopsin was site specifically labeled with a nitroxide probe at nine sites in a pairwise manner (Fig. 2a), yielding a total of 17 distance constraints for inactive and light-activated receptor. A global analysis of these distance restraints established that, upon light activation of rhodopsin, TM6 is displaced outward by 5 Å (Fig. 2b). This change is accompanied with smaller conformational changes in TM7 and helix 8. The conformation for light activated rhodopsin observed by these DEER experiments was subsequently shown to be highly similar to the crystal structure of opsin and Meta II. While crystal structures of Meta II required low pH or constitutively activated mutants, the DEER spectroscopy experiments were performed at more physiological pH conditions. DEER spectroscopy requires cryogenic temperatures that may affect receptor conformation. Light induced conformational changes have also been examined by room temperature 19F fluorine NMR spectroscopy of trifluoroethanethiol labeled rhodopsin [43]. Like the DEER spectroscopy results, 19F-NMR spectra also show a complete transition to the active conformation upon illumination by light. Multiple lines of spectroscopic evidence, therefore, indicate that isomerization of 11-cis-retinal by light induces a complete transition to Meta II, with virtually no receptor in inactive or intermediate conformations. These studies strongly indicated that the active Meta II conformation of rhodopsin is the lowest energy state in the presence of all-trans-retinal and in the absence of transducin.
Figure 2.
Double electron-electron resonance (DEER) spectroscopy of rhodopsin activation (adapted from Altenbach et al. [42]). (a) View of the cytoplasmic surface of inactive rhodopsin. Nine sites, depicted with Cα carbons as red spheres, were site-specifically labeled pairwise with nitroxide spin probes. Distance distributions between these sites were determined for inactive and light-activated rhodopsin. (b) The displacement of each transmembrane helix from a central reference point is shown. Light activation of rhodopsin results in a 5 Å outward displacement of TM6, with smaller changes in TM1 and TM7. Notably, these data show that light activation results in a complete shift to the activated Meta II conformation in the absence of transducin, with almost no receptor in inactive or intermediate conformations.
β2AR dynamics
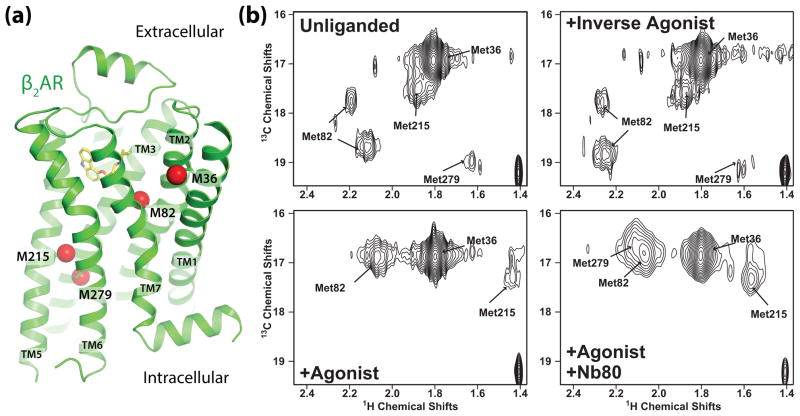
While DEER studies have not been reported for the β2AR, recent NMR studies reveal dynamic behavior supporting the model in Fig. 1b. In these studies, a modified β2AR having only four native methionines was biosynthetically labeled with 13CH3ε-methionine (Fig. 3a), purified in dodecylmaltoside, and HSQC spectra were obtained in the absence of ligand, the presence of an inverse agonist (carazolol), and the presence of a high affinity agonist (BI-167107) with and without the G protein mimetic nanobody 80 (Nb80) [44]. The four methionines, which could be resolved and assigned by mutagenesis (Fig. 3b), allow simultaneous monitoring of conformational changes around the binding pocket (M82) and the cytoplasmic ends of TM5 (M215) and TM6 (M279). In the absence of ligand or when bound to the inverse agonist carazolol, the β2AR exhibited conformational heterogeneity as evidenced by two well-resolved peaks representing the single M82. These two peaks represent two distinct conformations having different chemical environments around M82 that exchange on a slow timescale (seconds). When bound to the agonist alone, the two M82 peaks shifted upfield and merged into a single more intense peak. In contrast, upon agonist binding the peak originating from M215 in TM5 became notably weaker and shifted upfield, and the peak from M279 in TM6 was no longer visible. The weakening or loss of intensity of peaks representing M215 and M279 suggests that, when bound to a high affinity agonist, the cytoplasmic ends of TM5 and TM6 exist in several conformational states that exchange on an intermediate (millisecond) timescale (Fig. 1b). In the presence of agonist and the G protein mimetic Nb80, a new peak representing M279 appears and the peak representing M215 strengthens and shifts downfield, consistent with a more stable, uniform conformation. Notably, the peaks for M215 and M279 observed in the presence of Nb80 are not detected for β2AR bound to agonist alone, suggesting that the active conformation exists at very low levels for agonist bound receptor. Taken together, these results are consistent with the model in Fig. 1b and suggest that when bound to agonist alone the β2AR is more conformationally heterogeneous than in the unliganded or inverse agonist bound conformation, and that the G protein mimetic Nb80 is required to stabilize the active state. These results are in agreement with earlier studies using fluorescence lifetime spectroscopy showing that agonist binding leads to conformational heterogeneity, and that agonists and partial agonists stabilize distinct conformational states [45]. While the experiments above were performed on detergent solubilized receptor, the results are supported by experiments in which fluorophore-labeled, purified β2AR was reconstituted into synthetic lipid bilayers in the presence and absence of the G protein Gs. Agonist alone did not fully stabilize the conformation observed with agonist and G protein together [46].
Figure 3.
13CH3ε-methionine NMR spectroscopy of β2AR (adapted from Nygaard et al. [44]). (a) View of the β2AR transmembrane helices showing 13CH3ε-methionine labeled carbons as red spheres. NMR peaks in HSQC spectra originate from four distinct sites. (b) HSQC spectra of β2AR in four states: unliganded, bound to inverse agonist carazolol, to agonist BI-167107, and to BI-167107 with the G protein mimetic nanobody Nb80.
Conclusions and future directions
We are only beginning to appreciate the role of protein dynamics in GPCR signaling. By necessity, studies that provide the greatest structural insights involve the use of purified protein in non-native environments. These experiments provided evidence for fundamental differences between the β2AR and rhodopsin. The very efficient coupling of retinal isomers to the cytoplasmic surface of rhodopsin is essential for its highly efficient response to light. By contrast, the relatively inefficient coupling of the ligand-binding site to the cytoplasmic surface in the β2AR may play a role in its more complex functional repertoire and the diverse responses to different synthetic ligands. A more complete understanding of the role of dynamics in GPCR function will require measurement of the timescales of receptor dynamics from milliseconds to seconds and the ability to monitor conformational changes in single receptor molecules as they function in a native cell membrane.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Insel PA, Head BP, Ostrom RS, Patel HH, Swaney JS, Tang CM, Roth DM. Caveolae and lipid rafts: G protein-coupled receptor signaling microdomains in cardiac myocytes. Ann N Y Acad Sci. 2005;1047:166–172. doi: 10.1196/annals.1341.015. [DOI] [PubMed] [Google Scholar]
- 2.Shcherbakova OG, Hurt CM, Xiang Y, Dell’acqua ML, Zhang Q, Tsien RW, Kobilka BK. Organization of {beta}-adrenoceptor signaling compartments by sympathetic innervation of cardiac myocytes. J Cell Biol. 2007;176(4):521–533. doi: 10.1083/jcb.200604167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kovacs JJ, Hara MR, Davenport CL, Kim J, Lefkowitz RJ. Arrestin development: Emerging roles for beta-arrestins in developmental signaling pathways. Dev Cell. 2009;17(4):443–458. doi: 10.1016/j.devcel.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiang Y, Kobilka B. The pdz-binding motif of the beta2-adrenoceptor is essential for physiologic signaling and trafficking in cardiac myocytes. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(19):10776–10781. doi: 10.1073/pnas.1831718100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Irannejad R, Tomshine JC, Tomshine JR, Chevalier M, Mahoney JP, Steyaert J, Rasmussen SG, Sunahara RK, El-Samad H, Huang B, von Zastrow M. Conformational biosensors reveal gpcr signalling from endosomes. Nature. 2013;495(7442):534–538. doi: 10.1038/nature12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calebiro D, Nikolaev VO, Lohse MJ. Imaging of persistent camp signaling by internalized g protein-coupled receptors. J Mol Endocrinol. 2010;45 (1):1–8. doi: 10.1677/JME-10-0014. [DOI] [PubMed] [Google Scholar]
- 7.Deupi X, Kobilka BK. Energy landscapes as a tool to integrate gpcr structure, dynamics, and function. Physiology (Bethesda) 2010;25 (5):293–303. doi: 10.1152/physiol.00002.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhattacharya S, Vaidehi N. Computational mapping of the conformational transitions in agonist selective pathways of a g-protein coupled receptor. J Am Chem Soc. 2010;132(14):5205–5214. doi: 10.1021/ja910700y. [DOI] [PubMed] [Google Scholar]
- 9.Ben-Chaim Y, Chanda B, Dascal N, Bezanilla F, Parnas I, Parnas H. Movement of ‘gating charge’ is coupled to ligand binding in a g-protein-coupled receptor. Nature. 2006;444(7115):106–109. doi: 10.1038/nature05259. [DOI] [PubMed] [Google Scholar]
- 10.Rivero-Muller A, Jonas KC, Hanyaloglu AC, Huhtaniemi I. Di/oligomerization of gpcrs-mechanisms and functional significance. Progress in molecular biology and translational science. 2013;117:163–185. doi: 10.1016/B978-0-12-386931-9.00007-6. [DOI] [PubMed] [Google Scholar]
- 11.Terrillon S, Bouvier M. Roles of g-protein-coupled receptor dimerization. EMBO Rep. 2004;5(1):30–34. doi: 10.1038/sj.embor.7400052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lambert NA. Gpcr dimers fall apart. Sci Signal. 2010;3(115):pe12. doi: 10.1126/scisignal.3115pe12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450(7172):964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 14.Baylor DA. Photoreceptor signals and vision. Proctor lecture Investigative ophthalmology & visual science. 1987;28(1):34–49. [PubMed] [Google Scholar]
- 15.Hofmann KP, Scheerer P, Hildebrand PW, Choe HW, Park JH, Heck M, Ernst OP. A g protein-coupled receptor at work: The rhodopsin model. Trends Biochem Sci. 2009;34(11):540–552. doi: 10.1016/j.tibs.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 16.Stryer L. Visual excitation and recovery. J Biol Chem. 1991;266(17):10711–10714. [PubMed] [Google Scholar]
- 17.Palczewski K. G protein-coupled receptor rhodopsin. Annu Rev Biochem. 2006;75:743–767. doi: 10.1146/annurev.biochem.75.103004.142743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Azzi M, Charest PG, Angers S, Rousseau G, Kohout T, Bouvier M, Pineyro G. Beta-arrestin-mediated activation of mapk by inverse agonists reveals distinct active conformations for g protein-coupled receptors. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(20):11406–11411. doi: 10.1073/pnas.1936664100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shenoy SK, Drake MT, Nelson CD, Houtz DA, Xiao K, Madabushi S, Reiter E, Premont RT, Lichtarge O, Lefkowitz RJ. Beta-arrestin-dependent, g protein-independent erk1/2 activation by the beta2 adrenergic receptor. J Biol Chem. 2006;281(2):1261–1273. doi: 10.1074/jbc.M506576200. [DOI] [PubMed] [Google Scholar]
- 20.Violin JD, Lefkowitz RJ. Beta-arrestin-biased ligands at seven-transmembrane receptors. Trends Pharmacol Sci. 2007;28(8):416–422. doi: 10.1016/j.tips.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 21.Galandrin S, Bouvier M. Distinct signaling profiles of beta1 and beta2 adrenergic receptor ligands toward adenylyl cyclase and mitogen-activated protein kinase reveals the pluridimensionality of efficacy. Mol Pharmacol. 2006;70(5):1575–1584. doi: 10.1124/mol.106.026716. [DOI] [PubMed] [Google Scholar]
- 22.Cooper A. Energy uptake in the first step of visual excitation. Nature. 1979;282(5738):531–533. doi: 10.1038/282531a0. [DOI] [PubMed] [Google Scholar]
- •23.Naganathan S, Ye S, Sakmar TP, Huber T. Site-specific epitope tagging of g protein-coupled receptors by bioorthogonal modification of a genetically encoded unnatural amino acid. Biochemistry. 2013;52 (6):1028–1036. doi: 10.1021/bi301292h. This paper describes a promising approach for labeling GPCRs in cells for study in their native contexts. [DOI] [PubMed] [Google Scholar]
- 24.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, et al. Crystal structure of rhodopsin: A g protein-coupled receptor. Science. 2000;289(5480):739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 25.Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Crystal structure of the ligand-free g-protein-coupled receptor opsin. Nature. 2008;454(7201):183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 26.Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, Choe HW, Hofmann KP, Ernst OP. Crystal structure of opsin in its g-protein-interacting conformation. Nature. 2008;455(7212):497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- ••27.Choe HW, Kim YJ, Park JH, Morizumi T, Pai EF, Krauss N, Hofmann KP, Scheerer P, Ernst OP. Crystal structure of metarhodopsin ii. Nature. 2011;471(7340):651–655. doi: 10.1038/nature09789. The structure of metarhodopsin II with and without the Gαt-CT is presented in this paper. [DOI] [PubMed] [Google Scholar]
- •28.Standfuss J, Edwards PC, D’Antona A, Fransen M, Xie G, Oprian DD, Schertler GF. The structural basis of agonist-induced activation in constitutively active rhodopsin. Nature. 2011;471(7340):656–660. doi: 10.1038/nature09795. The structure of metarhodopsin II and its comparison with DEER spectroscopy data from [42] is presented here. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Yao XJ, Weis WI, Stevens RC, Kobilka BK. Gpcr engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318(5854):1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- ••30.Rasmussen SG, Choi HJ, Fung JJ, Pardon E, Casarosa P, Chae PS, Devree BT, Rosenbaum DM, Thian FS, Kobilka TS, Schnapp A, et al. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature. 2011;469(7329):175–180. doi: 10.1038/nature09648. To stabilize the active state of the β2AR for crystallographic study, the authors utilized camelid antibody fragments (nanobodies) that specifically bind to the β2AR in the active state. The resulting G protein mimetic nanobody Nb80 has been used to characterize the β2AR active state both by crystallographic and biophysical experiments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••31.Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, Mathiesen JM, et al. Crystal structure of the beta2 adrenergic receptor-gs protein complex. Nature. 2011;477(7366):549–555. doi: 10.1038/nature10361. The structure of the only GPCR-G protein complex solved to date is described in this paper. The structure of the β2AR-Gs complex yielded key insight into the conformation of a fully active receptor engaging its cognate G protein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haga K, Kruse AC, Asada H, Yurugi-Kobayashi T, Shiroishi M, Zhang C, Weis WI, Okada T, Kobilka BK, Haga T, Kobayashi T. Structure of the human m2 muscarinic acetylcholine receptor bound to an antagonist. Nature. 2012;482(7386):547–551. doi: 10.1038/nature10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kruse AC, Ring AM, Manglik A, Hu J, Hu K, Eitel K, Hubner H, Pardon E, Valant C, Sexton PM, Christopoulos A, et al. Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature. 2013;504(7478):101–106. doi: 10.1038/nature12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •34.Moukhametzianov R, Warne T, Edwards PC, Serrano-Vega MJ, Leslie AG, Tate CG, Schertler GF. Two distinct conformations of helix 6 observed in antagonist-bound structures of a beta1-adrenergic receptor. Proc Natl Acad Sci U S A. 2011;108(20):8228–8232. doi: 10.1073/pnas.1100185108. This paper highlights structural heterogeneity in TM6 of inactive, thermostabilized β1AR as revealed by multiple crystal structures. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenbaum DM, Zhang C, Lyons JA, Holl R, Aragao D, Arlow DH, Rasmussen SG, Choi HJ, Devree BT, Sunahara RK, Chae PS, et al. Structure and function of an irreversible agonist-beta(2) adrenoceptor complex. Nature. 2011;469(7329):236–240. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •36.Kofuku Y, Ueda T, Okude J, Shiraishi Y, Kondo K, Maeda M, Tsujishita H, Shimada I. Efficacy of the beta(2)-adrenergic receptor is determined by conformational equilibrium in the transmembrane region. Nat Commun. 2012;3:1045. doi: 10.1038/ncomms2046. Along with [44], this paper utilized 13CH3ε-methionine NMR spectroscopy to examine how ligands alter the conformational ensemble of the β2AR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bokoch MP, Zou Y, Rasmussen SG, Liu CW, Nygaard R, Rosenbaum DM, Fung JJ, Choi HJ, Thian FS, Kobilka TS, Puglisi JD, et al. Ligand-specific regulation of the extracellular surface of a g-protein-coupled receptor. Nature. 2010;463(7277):108–112. doi: 10.1038/nature08650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu JJ, Horst R, Katritch V, Stevens RC, Wuthrich K. Biased signaling pathways in beta2-adrenergic receptor characterized by 19f-nmr. Science. 2012;335(6072):1106–1110. doi: 10.1126/science.1215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim TH, Chung KY, Manglik A, Hansen AL, Dror RO, Mildorf TJ, Shaw DE, Kobilka BK, Prosser RS. The role of ligands on the equilibria between functional states of a g protein-coupled receptor. J Am Chem Soc. 2013;135(25):9465–9474. doi: 10.1021/ja404305k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kahsai AW, Xiao K, Rajagopal S, Ahn S, Shukla AK, Sun J, Oas TG, Lefkowitz RJ. Multiple ligand-specific conformations of the beta2-adrenergic receptor. Nat Chem Biol. 2011;7(10):692–700. doi: 10.1038/nchembio.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •41.Dror RO, Arlow DH, Maragakis P, Mildorf TJ, Pan AC, Xu H, Borhani DW, Shaw DE. Activation mechanism of the beta2-adrenergic receptor. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(46):18684–18689. doi: 10.1073/pnas.1110499108. The use of long timescale molecular dynamics to understand GPCR function is illustrated by this article. Simulation of the β2AR suggests a loose allosteric coupling between ligand binding and receptor conformational change, which is in agreement with experimental data derived from spectroscopic experiments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••42.Altenbach C, Kusnetzow AK, Ernst OP, Hofmann KP, Hubbell WL. High-resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(21):7439–7444. doi: 10.1073/pnas.0802515105. This article utilized DEER spectroscopy to directly examine the conformation of the cytoplasmic domain of rhodopsin in both dark and light activated states. In the absence of crystallographic constraints and at physiological pH, the authors show movements in TMs 1, 6, and 7 associated with rhodopsin activation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klein-Seetharaman J, Getmanova EV, Loewen MC, Reeves PJ, Khorana HG. Nmr spectroscopy in studies of light-induced structural changes in mammalian rhodopsin: Applicability of solution (19)f nmr. Proceedings of the National Academy of Sciences of the United States of America. 1999;96 (24):13744–13749. doi: 10.1073/pnas.96.24.13744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••44.Nygaard R, Zou Y, Dror RO, Mildorf TJ, Arlow DH, Manglik A, Pan AC, Liu CW, Fung JJ, Bokoch MP, Thian FS, et al. The dynamic process of beta(2)-adrenergic receptor activation. Cell. 2013;152(3):532–542. doi: 10.1016/j.cell.2013.01.008. This paper examines the conformational dynamics of the β2AR in the transmembrane region by solution state NMR spectroscopy. The studies show that agonists do not fully stabilize the active state of β2AR, and that the active conformation is only achieved in the presence of a G protein mimetic nanobody, Nb80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghanouni P, Gryczynski Z, Steenhuis JJ, Lee TW, Farrens DL, Lakowicz JR, Kobilka BK. Functionally different agonists induce distinct conformations in the g protein coupling domain of the beta 2 adrenergic receptor. J Biol Chem. 2001;276(27):24433–24436. doi: 10.1074/jbc.C100162200. [DOI] [PubMed] [Google Scholar]
- 46.Yao XJ, Velez Ruiz G, Whorton MR, Rasmussen SG, DeVree BT, Deupi X, Sunahara RK, Kobilka B. The effect of ligand efficacy on the formation and stability of a gpcr-g protein complex. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(23):9501–9506. doi: 10.1073/pnas.0811437106. [DOI] [PMC free article] [PubMed] [Google Scholar]