Abstract
Background
Liver dysfunction and cirrhosis affect vasculature in several organ systems and cause impairment of organ functions, thereby increasing morbidity and mortality. Establishment of a mouse model of hepatopulmonary syndrome (HPS) would provide greater insights into the genetic basis of the disease. Our objectives were to establish a mouse model of lung injury after common bile duct ligation (CBDL) and to investigate pulmonary pathogenesis for application in future therapeutic approaches.
Methods
Eight-week-old Balb/c mice were subjected to CBDL. Immunohistochemical analyses and real-time quantitative reverse transcriptional polymerase chain reaction were performed on pulmonary tissues. The presence of HPS markers was detected by western blot and microarray analyses.
Results
We observed extensive proliferation of CD31-positive pulmonary vascular endothelial cells at 2 weeks after CBDL and identified 10 upregulated and 9 down-regulated proteins that were associated with angiogenesis. TNF-α and MMP-9 were highly expressed at 3 weeks after CBDL and were less expressed in the lungs of the control group.
Conclusions
We constructed a mouse lung injury model by using CBDL. Contrary to our expectation, lung pathology in our mouse model exhibited differences from that of rat models, and the mechanisms responsible for these differences are unknown. This phenomenon may be explained by contrasting processes related to TNF induction of angiogenic signaling pathways in the inflammatory phase. Thus, we suggest that our mouse model can be applied to pulmonary pathological analyses in the inflammatory phase, i.e., to systemic inflammatory response syndrome, acute lung injury, and multiple organ dysfunction syndrome.
Introduction
Liver dysfunction and cirrhosis affect vasculature in several organ systems, resulting in impairment of organ function that leads to increased morbidity and mortality.[1], [2] Hepatopulmonary syndrome (HPS) is a pulmonary vascular disorder associated with liver cirrhosis and is seen in 5–32% of cirrhotic patients in the setting of pulmonary microvascular dilatation-induced defects in arterial oxygenation.[1], [2] Although HPS occurs more frequently in patients with severe liver disease, patients with severe hepatic dysfunction do not always have HPS. This syndrome can also be seen in children with biliary atresia, and the same disease pattern is seen in the non-cirrhosis setting in congenital heart disease patients with a post-operative status of bidirectional cavopulmonary shunt that excludes hepatic venous return through the pulmonary vasculature.[3]–[5] Experimental rat models of HPS have enabled researchers to investigate the pathogenesis of this disease. Fallon et al. demonstrated that endothelial nitric oxide synthase (eNOS) and inducible nitric oxide synthase (iNOS) in the lungs play an important role in HPS pathogenesis.[5]–[9] Fallon et al. also reported that angiogenesis plays a role in the pathophysiology of HPS and explained the mechanisms underlying the activation of vascular endothelial growth factor A (VEGF-A), which is produced by inflammatory cells such as intravascular monocytes and induces angiogenesis.[10]
Establishment of a mouse model of HPS would help obtain greater insights into the genetic basis of the disease. Although some results have been reported for murine models of hepatic disease, mortality rates in mice are higher than those in rats, and limited data are available on lung pathology after common bile duct ligation (CBDL) in mice.[11]–[14] Our objectives were to establish a mouse model of lung injury by using CBDL and investigate its pulmonary pathogenesis for application in future therapeutic approaches.
Materials and Methods
Animal model
All animals were treated in accordance with standards of the Ehime University Animal Care Committee using approved animal protocols including animal ethics (Permit Number: 05R02-2). The Ehime University Animal Care Committee approved our animal protocols including animal ethics. All animals were treated in accordance with standards of the committee using the approved animal protocols. Eight-week-old Balb/c mice (n = 112) were obtained from Clea Japan Inc. (Tokyo) and were housed under barrier conditions. A standard sterilized laboratory diet and water were available ad libitum. All surgical procedures were performed utilizing clean techniques. All animals were anesthetized with ketamine (0.1 mg/g) and xylazine (0.01 mg/g) administered by intraperitoneal injection. After induction of anesthesia, a median abdominal incision was made and the common bile duct was identified. The duct was dissected carefully under a microscope and doubly ligated with 7–0 Prolene (Ethicon, Somerville, NJ) and transected. In the sham operation (control) group, the duct was dissected without CBDL. The abdominal incision was closed in two layers. After abdominal closure, 1.0 ml of 0.9% saline was injected subcutaneously into each mouse, and the mice were kept warm until recovery was confirmed. We continued to inject 1.0 ml of 0.9% saline subcutaneously every 2 d for 6 d; this procedure led to improved survival rates after surgery.
Mice were sacrificed between 1 and 4 weeks after surgery. Mice were anesthetized with ketamine (0.1 mg/g) and xylazine (0.01 mg/g). After we confirmed that mice were well sedated, they were given 50 µl of heparin (50 mg/ml) subcutaneously.[15] The trachea was surgically exposed and cannulated with a 20G Terumo Surflo Catheter (Terumo Corporation, Tokyo, Japan). Right cardiac ventricles were perfused with cold PBS (10 ml) under physiological pressure with an exit through the severed right atrium. The lungs were inflated and fixed at 23 cm pressure with 4% paraformaldehyde in PBS. The lungs, liver, heart, and spleen were carefully harvested with dissection of excess surrounding tissues.
A method for intrapulmonary shunt-fraction analysis in mice was established by Miniati et al.[15] Briefly, fluorescent microspheres (1.25×105; diameter, 10 µm) (Molecular Probes, Eugene, OR) in PBS were injected into the inferior vena cava and all blood was aspirated from the transected right carotid artery. Aspirated microspheres were then counted manually under a fluorescence microscope (Olympus, Tokyo, Japan).
Immunohistochemistry and immunofluorescence
In brief, deparaffinization and standard antigen retrieval by microwave irradiation were performed on 8-µm sections of 4% paraformaldehyde paraffin-fixed tissues. The sections were then blocked with 3% bovine serum albumin and incubated with primary antibodies against CD31 (BD Biosciences, San Diego, CA), von Willebrand factor (vWF; Chemicon International, Temecula, CA), eNOS, iNOS (BD Biosciences, San Diego, CA), Ly6G, CD68, F4/80, MMP-8, MMP-9, TIMP-1, and TIMP-4 (Abcam, Cambridge, MA). Next, secondary antibodies were applied and the samples were incubated. Sections were developed with 3,3′-diaminobenzidine (DAB; Sigma-Aldrich, St. Louis, MO) and counter-stained with hematoxylin.
Sections were immunostained using antibodies to vWF (Chemicon International, Temecula, CA) and tumor necrosis factor alpha (TNF-α; R&D Systems, Minneapolis, MN), and Alexa Fluor anti-mouse IgG for secondary fluorescent antibodies (Molecular Probes, Eugene, OR). For nuclear staining, mounting medium with DAPI (Vector Laboratories, Burlingame, CA) was used. Pictures were taken using a fluorescence microscope (Olympus, Tokyo, Japan).
Microvascular density and quantitation were assessed as previously described by Zhang et al.[10], [16] This analysis was performed in a blinded manner. Dissected lung sections that were immunostained for CD31 and vWF were used (n = 5), and pulmonary vessels with a diameter of >100 µm were excluded from the analysis. Positive-stained cells were counted at five representative locations under high magnification (40×) using ImageJ software (National Institutes of Health) and results were averaged.
Assembly of CD31-positive cells/magnetic beads complex
Homogenized whole lungs were treated using a solution of type-1 collagenase (400 U/ml) (Worthington, Lakewood, NJ). CD31-positive cells, including pulmonary vascular endothelial cells, were collected with magnetic beads (Dynabeads; Life Technologies, Grand Island, NY) coated with anti-CD31 antibodies (BD Biosciences, San Diego, CA) as follows.[17] The cellular digest was filtered through a sterile 40-µm mesh and washed three times in 10% FBS-DMEM (Wako Pure Chemical Industries, Osaka, Japan). One milliliter of the cell suspension was placed in a tube with the magnetic beads/CD31-antibody complex and rotated for 20 min at 4°C. The bead-bound cells were then washed five times with 10% FBS-DMEM and the isolated cells were used in the following procedures.
Real-time quantitative reverse transcriptional polymerase chain reaction (qRT-PCR)
Whole pulmonary cells and CD31-positive cells were selected as described above. Total RNA was extracted from each cell in the CBDL and sham mouse groups using Isogen II (Nippon Gene, Toyama, Japan). The RNA samples were then treated with an RNase-free DNase (Ambion, Carlsbad, CA) to remove genomic DNA contaminants. Messenger RNA was quantified using SYBR Green PCR master mix (Roche, Branford, CT) with an ABI Prism 7300 Fast Real-Time PCR system. The mRNA expression levels of endothelin-1 (ET-1), endothelial nitric oxide synthase (eNOS), endothelin type A receptor (ET-A), endothelin type B receptor (ET-B), kinase insert domain receptor (KDR), CC chemokine receptor type 1 (CCR1), chemokine CXC motif ligand 3 (CXCL3), CXC chemokine receptor 2 (CXCR2), chemokine CC motif ligand 9 (CCL9), matrix metallopeptidase 9 (MMP-9), interleukin-1 beta (IL-1b), and TNF-α were normalized to the levels of of β-actin. The primer sequences used for the amplification are shown in Table 1. The experiments were performed in triplicate and independently repeated a minimum of three times.
Table 1. Primer sequences for quantitative RT-PCR.
| Target gene | Primer sequence 5'-3' | |
| ET-1 | forward | GTCAACACTCCCGACCACGTT |
| reverse | CTGGTTTGTCTTAGGTGTTCCTC | |
| eNOS | forward | CTGTGGTCTGGTGCTGGTC |
| reverse | TGGGCAACTTGAAGAGTGTG | |
| ET-A | forward | ACCACAGTCCATGCCATCAC |
| reverse | TCAACATCTCACAAGTCATGAG | |
| ET-B | forward | TTGGAGCTGAGATGTGTAAGC |
| reverse | CAGTGAAGCCATGTTGATACC | |
| KDR | forward | GGGATGG TCCTTGCATCAGAA |
| reverse | ACTGGTAGCCACTGGTCTGGTTG | |
| CCR1 | forward | TTAGCTTCCATGCCTGCCTTATA |
| reverse | TCCACTGCTTCAGGCTCTTGT | |
| CXCL3 | forward | GGAGCACCAACTGACAGGAGAGAA |
| reverse | ACCACCCTGCAGGAAGTGTCAA | |
| CXCR2 | forward | GCTCTGACTACCACCCAACCTTGA |
| reverse | AGAAGAGCAGCTGTGACCTGCTGT | |
| CCL9 | forward | CCAGTGGTGGGTGTACCAG |
| reverse | CTCCGATCACTGGGGTTG | |
| MMP9 | forward | CGAACTTCGACACTGACAAGAAGT |
| reverse | GCACGCTGGAATGATCTAAGC | |
| IL-1b | forward | CAACCAACAAGTGATATTCTCCATG |
| reverse | GATCCACACTCTCCAGCTGCA | |
| TNF-α | forward | CTGTAGCCCACGTCGTAGC |
| reverse | TTGAGATCCATGCCGTTG | |
| β-actin | forward | CGGTTCCGATGCCCTGAGGCTCTT |
| reverse | CGTCACACTTCATGATGGAATTGA |
Western blot analysis
CD31-positive cells were isolated as described above and lysed in RIPA buffer (50 mM Tris-HCl containing 0.15 m NaCl, 0.1% SDS, 1% sodium cholate, 1% Nonidet P-40, 1 mm EDTA, and 0.5 mm phenylmethylsulfonyl fluoride, pH 7.4). Proteins from lung tissues were also extracted with RIPA buffer. Equal concentrations of protein from pulmonary CD31-positive cells and whole lungs were fractionated on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to Immun-Blot PVDF Membranes (Bio-Rad Laboratories, Hercules, CA). Samples were incubated with primary antibodies against MMP-9, F4/80, CD31, eNOS, induced nitric oxide synthase (iNOS), and β-actin (BD Biosciences, San Diego, CA). The proteins were detected using an ECL Prime Western Blotting Detection System (GE Healthcare, Buckinghamshire, UK) and imaged on an LAS-4000 luminescent image analyzer (Fujifilm Life Science USA, Stamford, CT).
Microarray analysis
CD31-positive cells were assembled from three mice in each group. Total RNA was extracted from pulmonary CD31-positive cells and whole pulmonary cells with Isogen II and collected as one sample in each group. We used 500 ng of total RNA to generate double-stranded cDNA. The cDNA was transcribed with DIG-labeled nucleotides (Roche Diagnostics, Basel, Switzerland), fragmented, and hybridized to a Gene Chip Mouse Gene 1.0 ST Array (Affymetrix, Santa Clara, CA) according to the manufacturer's instructions. These results were analyzed using the Gene Spring GX 12.1 (Agilent Technologies, Santa Clara, CA) and Ingenuity Pathway Analysis (IPA; Ingenuity Systems, Redwood, CA) software. Functional analysis by IPA identified the biological functions that were most significant to the data set. Fischer's exact test was used to calculate a p-value indicating the probability that each biological function assigned to that data set was due to chance alone. The microarray data are deposited in Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.-gov/geo, Accession number: GSE50088) according to Minimum Information About Microarray Experiment (MIAME) guidelines.
Protein array analysis of serum
Serum from CBDL and sham operated mice was prepared and subjected to mouse angiogenesis array (R&D Systems ARY015, Minneapolis, MN) analysis according to the manufacturer's instructions.
Flow cytometry
Two weeks and 3 weeks after CBDL, bronchoalveolar lavage (BAL) fluid was collected from CBDL and sham operated mice to analyze the neutrophil and macrophage populations. To obtain BAL fluid, the trachea was cannulated with a 20 G angiocatheter and the lungs were lavaged five times with 1 ml of 1%FCS in PBS. The fluid was collected into one FACS tube and centrifuged at 500×g for 5 min at 4°C. The cell pellet was washed in FACS buffer, stained with CD11b, Ly6G, and F4/80 antibodies (BD Biosciences, San Diego, CA), and incubated on ice for 30 minutes. After cells were washed in FACS buffer, they were resuspended in 500 µl of FACS buffer and analyzed with FACS Calibur flowcytometer (Becton Dickinson, San Jose, CA).
Statistical analysis
All measurement values in this study are expressed as means ± SE. All statistical analyses were performed with JMP 8.0 and analyzed using, where appropriate, either paired t test or Wilcoxon signed rank test. All tests were two sided. Results with p-values <0.05 were considered statistically significant.
Results
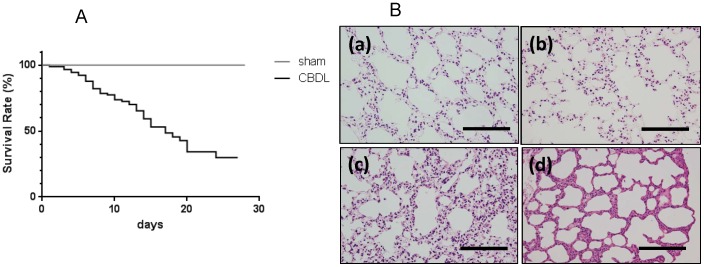
The frequency of mortality was 82.2%, 59.2%, and 34.2% at 1, 2, and 3 weeks, respectively, after CBDL (Fig. 1A). No mortality was observed in the sham group. Significant weight loss occurred beginning at 2 weeks after surgery (Table 2), and appetite loss and low activity were observed prior to death. No bile leakage was observed at autopsy in the abdominal cavity of mice that died before sacrifice. To confirm the level of liver damage resulting from CBDL surgery, serum levels of liver-function markers were measured (n = 5); all of these markers were significantly elevated from 1 week after CBDL (Table 2). The development of ascites, a parameter of liver dysfunction,[14] was observed from 1 week after surgery. To assess lung injury after surgery, arterial blood was drained from the right carotid artery for blood gas analysis. Significantly lower PaO2 was found in the CBDL group at 2 and 3 weeks after surgery (70.0%±2.87% and 68.4%±1.3% respectively) compared with the sham group (87.8%±1.6%) (n = 6). To confirm and compare our results with a well-established experimental hepatopulmonary syndrome model (a rat cirrhotic model of pulmonary microcirculation),[10] we applied the intrapulmonary shunt fraction calculation to our model. The resulting value for the sham group was 2.6%±1.3%. For the CBDL group, the resulting values were 4.1%±0.9% at 2 weeks after CBDL; and 4.9%±2.0% at 3 weeks after CBDL. Differences between sham and CBDL mice were not significant (P = 0.4 at 2 weeks, P = 0.3 at 3 weeks, n = 7). Histologic changes in hematoxylin-eosin staining and pulmonary vascular proliferation in immunohistochemical analysis of the lungs are shown in Figures 1B and 2A. These changes were consistent with findings of a murine CBDL model in which pulmonary angiogenic processes were confirmed by immunostaining imagery.[12]
Figure 1. Survival curves and histological changes in common bile duct ligated (CBDL) mice and sham operated mice.
(A) Kaplan-Meier survival curve in common bile duct ligated (CBDL) mice (n = 44) and sham operated (control) mice (n = 41). (B) HE-stained lung sections of sham and CBDL mice: (a) sham; (b) 1 week after CBDL; (c) 2 weeks after CBDL; (d) 3 weeks after CBDL (×20). Scale bar = 100 µm.
Table 2. Laboratory data after common bile duct ligation in mice.
| sham | CBDL, week | |||
| 1 | 2 | 3 | ||
| AST, IU/l | 64±5.0 | 528±72* | 651±99* | 647±106* |
| ALT, IU/l | 30±2.0 | 520±46* | 492±31* | 618±107* |
| T. Bil, mg/dl | 0.6±0.1 | 12±1.3* | 18±2.6* | 19±2.6* |
| ALP | 315±35 | 1485±243* | 2764±281* | 2709±386* |
| Liver weight, g/10 g BW | 0.05±0.01 | 0.06±0.01 | 0.09±0.01* | 0.09±0.01* |
Values given are means ± SE.
CBDL, common bile duct ligation; AST, aspartate aminotransferase; ALT, alanine aminotransferase; T. Bil, total bilirubin; ALP, alkaline phosphatase; BW, body weight. *p<0.05 vs. sham.
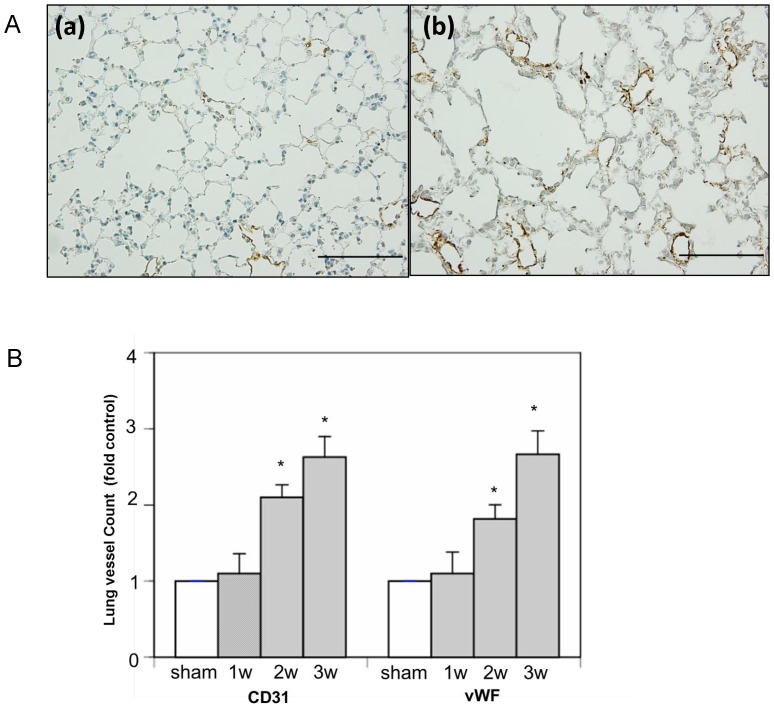
Figure 2. Immunohistochemical analysis of lung tissues in common bile duct ligated (CBDL) and sham-operated (control) mice.
(A) Immunohistochemistry of von Willebrand factor (vWF)-stained lung section of sham and CBDL mice: (a) sham; (b) 3 weeks after CBDL. Scale bar = 100 µm. (B) Graphical summary of lung microvessel counts by immunohistochemical analysis using CD31 and vWF antibodies in mice at 1–3 weeks after CBDL and in sham operated mice. *P<0.05 (CBDL vs. sham).
To investigate angiogenesis in the lung after CBDL by using methods consistent with an established rat model, we performed lung microvessel counts in which we quantified increases in microvessels at 2 and 3 weeks after CBDL in comparison with the sham group (Fig. 2A and B).[10] A significant increase in average lung microvessel count was revealed by both CD31 and vWF staining 2 weeks after CBDL, relative to the results for the sham group. Immunostaining of lungs using eNOS and iNOS antibodies, which were reported to be highly expressed after CBDL in a rat model,[6] revealed no significant differences in eNOS and iNOS expression between the CBDL and sham groups (data not shown). Elevated ET-1, ET-A, ET-B, KDR, and eNOS mRNA expression levels have also been reported after CBDL in rats.[6]–[9], [18] However, our qRT-PCR analyses revealed no significant differences in mRNA expression between the CBDL and sham groups (data not shown). We thus hypothesized that lung pathogenesis in this mouse CBDL model could be different from that in the other models, and we proceeded to apply global analysis of the gene expression profile associated with this lung pathogenesis using a microarray technique.
Identification of genes in mouse CD31-positive pulmonary cells by microarray analysis
Based on our finding that proliferation of CD31-positive pulmonary vascular endothelial cells was more extensive after 2 weeks, we assumed that analysis of genes related to CD31-positive cells would be the key to investigating pulmonary pathogenesis after CBDL. CD31-positive pulmonary cells were selectively collected as described in the Methods, and the purity of the cell fractions was confirmed by qRT-PCR and western blot analyses.
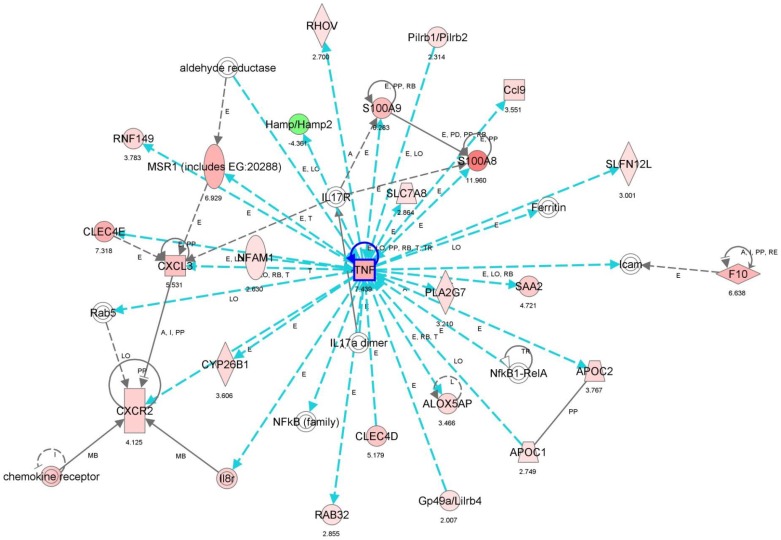
We determined the global gene expression profiles of CD31-positive pulmonary cells in mice 2 and 3 weeks after CBDL, compared to those of sham mice, and found 79 genes that were commonly up-regulated by more than 2-fold in mice that had undergone CBDL (Table 3). This information was used to visualize the network of gene expression patterns, and 34 genes were identified as being connected directly or indirectly to TNF-α, located at the center of the network (Fig. 3). To confirm the localization of TNF-α, immunofluorescence analysis was used (Fig 4). The results indicated that TNF-α was highly expressed in pulmonary vascular endothelial cells in the CBDL mice, when compared with the sham operated mice.
Table 3. Genes up- or down-regulated >2.0 fold in pulmonary cells of mice at 2 and 3 weeks after CBDL, assembled by a magnetic bead/CD31-antibody complex, in comparison with levels in sham operated mice, as determined by the Gene Chip Mouse Gene 1.0 ST Array.
| Gene ID | Symbol | Gene Name Fold change | 2w | 3w |
| 17394 | MMP8 | matrix metallopeptidase 8 (neutrophil collagenase) | 18.597 | 20.121 |
| 74145 | F13A1 | coagulation factor XIII, A1 polypeptide | 12.429 | 7.512 |
| 20201 | S100A8 | S100 calcium binding protein A8 | 11.960 | 13.878 |
| 21926 | TNF | tumor necrosis factor | 7.439 | 2.874 |
| 56619 | CLEC4E | C-type lectin domain family 4, member E | 7.318 | 3.959 |
| 12475 | CD14 | CD14 molecule | 7.262 | 4.187 |
| 13003 | VCAN | versican | 7.093 | 3.964 |
| 100034251 | Gm11428 | predicted gene 11428 | 7.012 | 9.548 |
| 104183|12655 | Chi3l3/Chi3l4 | chitinase 3-like 3 | 6.984 | 3.645 |
| 20288 | MSR1 | macrophage scavenger receptor 1 | 6.929 | 4.557 |
| 14058 | F10 | coagulation factor X | 6.638 | 3.471 |
| 12772 | CCR2 | chemokine (C-C motif) receptor 2 | 6.389 | 4.536 |
| 20202 | S100A9 | S100 calcium binding protein A9 | 6.283 | 7.824 |
| 58217 | TREM1 | triggering receptor expressed on myeloid cells 1 | 5.96 | 5.005 |
| 216799 | NLRP3 | NLR family, pyrin domain containing 3 | 5.68 | 3.363 |
| 20310 | CXCL3 | chemokine (C-X-C motif) ligand 3 | 5.531 | 3.265 |
| 20716 | SERPINA3 | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 | 4.557 | 5.632 |
| 12768 | CCR1 | chemokine (C-C motif) receptor 1 | 4.501 | 4.698 |
| 12489 | Cd33 | CD33 antigen | 4.368 | 4.975 |
| 23845 | CLEC5A | C-type lectin domain family 5, member A | 4.148 | 4.047 |
| 12765 | CXCR2 | chemokine (C-X-C motif) receptor 2 | 4.125 | 5.535 |
| 100038947 | Sirpb1a | signal-regulatory protein beta 1A | 3.944 | 3.915 |
| 83382 | SIGLEC8 | sialic acid binding Ig-like lectin 8 | 3.921 | 3.208 |
| 12273 | C5AR1 | complement component 5a receptor 1 | 3.893 | 2.917 |
| 16819 | LCN2 | lipocalin 2 | 3.862 | 3.671 |
| 16948 | LOX | lysyl oxidase | 3.856 | 3.168 |
| 12986 | CSF3R | colony stimulating factor 3 receptor (granulocyte) | 3.854 | 4.490 |
| 16409 | ITGAM | integrin, alpha M (complement component 3 receptor 3 subunit) | 3.791 | 3.748 |
| 17395 | MMP9 | matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92 kDa type IV collagenase) | 3.576 | 4.147 |
| 20308 | CCL9 | chemokine (C-C motif) ligand 9 | 3.551 | 2.835 |
| 246256 | FCGR3A | Fc fragment of IgG, low affinity IIIa, receptor (CD16a) | 3.511 | 3.572 |
| 74442 | SGMS2 | sphingomyelin synthase 2 | 3.491 | 2.318 |
| 11690 | ALOX5AP | arachidonate 5-lipoxygenase-activating protein | 3.466 | 3.410 |
| 170733 | Klra17 | killer cell lectin-like receptor, subfamily A, member 17 | 3.466 | 3.997 |
| 20568 | SLPI | secretory leukocyte peptidase inhibitor | 3.433 | 2.644 |
| 18173 | SLC11A1 | solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1 | 3.212 | 2.344 |
| 56221 | CCL24 | chemokine (C-C motif) ligand 24 | 3.211 | 2.598 |
| 27226 | PLA2G7 | phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) | 3.21 | 3.009 |
| 18733 | LILRB3 | leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 | 3.173 | 3.046 |
| 13058 | CYBB | cytochrome b-245, beta polypeptide | 3.147 | 2.017 |
| 16176 | IL1B | interleukin 1, beta | 3.147 | 2.196 |
| 170743 | TLR7 | toll-like receptor 7 | 3.136 | 2.434 |
| 278180 | VSIG4 | V-set and immunoglobulin domain containing 4 | 3.131 | 9.911 |
| 12978 | CSF1R | colony stimulating factor 1 receptor | 3.076 | 2.409 |
| 19204 | PTAFR | platelet-activating factor receptor | 3.049 | 2.502 |
| 21810 | TGFBI | transforming growth factor, beta-induced, 68kDa | 3.004 | 2.992 |
| 20556 | SLFN12L | schlafen family member 12-like | 3.001 | 2.139 |
| 12182 | BST1 | bone marrow stromal cell antigen 1 | 2.925 | 2.327 |
| 15368 | HMOX1 | heme oxygenase (decycling) 1 | 2.844 | 2.563 |
| 14293 | FPR1 | formyl peptide receptor 1 | 2.748 | 2.280 |
| 14191 | FGR | Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog | 2.675 | 2.150 |
| 57248 | Ly6a | lymphocyte antigen 6 complex, locus A | 2.673 | 3.938 |
| 20351 | SEMA4A | sema domain, immunoglobulin domain, transmembrane domain and short cytoplasmic domain, 4A | 2.654 | 2.315 |
| 74039 | NFAM1 | NFAT activating protein with ITAM motif 1 | 2.63 | 2.750 |
| 16854 | LGALS3 | lectin, galactoside-binding, soluble, 3 | 2.627 | 2.037 |
| 14131 | FCGR2A | Fc fragment of IgG, low affinity IIa, receptor (CD32) | 2.569 | 2.921 |
| 12267 | C3AR1 | complement component 3a receptor 1 | 2.546 | 3.573 |
| 14127 | FCER1G | Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide | 2.529 | 2.279 |
| 80885 | HCAR2 | hydroxycarboxylic acid receptor 2 | 2.508 | 2.019 |
| 380924 | OLFM4 | olfactomedin 4 | 2.497 | 2.717 |
| 11801 | CD5L | CD5 molecule-like | 2.492 | 5.669 |
| 12983 | CSF2RB | colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) | 2.444 | 2.262 |
| 16658 | MAFB | v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian) | 2.428 | 2.510 |
| 22177 | TYROBP | TYRO protein tyrosine kinase binding protein | 2.331 | 2.255 |
| 11501 | ADAM8 | ADAM metallopeptidase domain 8 | 2.327 | 2.410 |
| 170744 | TLR8 | toll-like receptor 8 | 2.177 | 2.221 |
| 378460 | PRAM1 | PML-RARA regulated adaptor molecule 1 | 2.153 | 2.739 |
| 20375 | SPI1 | spleen focus forming virus (SFFV) proviral integration oncogene spi1 | 2.142 | 2.175 |
| 14728 | Gp49a/Lilrb4 | leukocyte immunoglobulin-like receptor, subfamily B, member 4 | 2.007 | 2.302 |
| 18507 | PAX5 | paired box 5 | −2.068 | −2.522 |
| 14128 | FCER2 | Fc fragment of IgE, low affinity II, receptor for (CD23) | −2.08 | −3.552 |
| 12482 | MS4A1 | membrane-spanning 4-domains, subfamily A, member 1 | −2.130 | −2.302 |
| 12478 | CD19 | CD19 molecule | −2.183 | −2.643 |
| 19695 | Reg3g | regenerating islet-derived 3 gamma | −2.365 | −2.158 |
| 15510 | HSPD1 | heat shock 60 kDa protein 1 (chaperonin) | −2.529 | −2.360 |
| 12517 | CD72 | CD72 molecule | −2.539 | −2.522 |
| 17888 | MYH6 | myosin, heavy chain 6, cardiac muscle, alpha | −3.71 | −2.284 |
| 230899 | NPPA | natriuretic peptide A | −5.536 | −5.525 |
| 15505 | HSPH1 | heat shock 105 kDa/110 kDa protein 1 | −5.641 | −3.651 |
Figure 3. Relevance network of gene signaling pathways identified by Ingenuity Pathway Analysis.
Arrow charts represent all expression values (fold changes). Color-coded gene symbols: red represents up-regulation, green represents down-regulation. Tumor necrosis factor-alpha (TNF-α) plays a central role in pulmonary pathogenesis in common bile duct ligated mice.
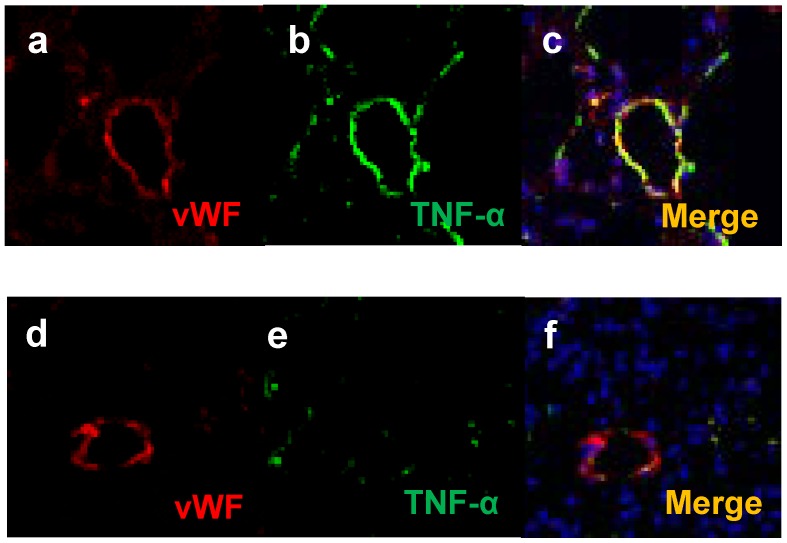
Figure 4. Immunofluorescent localization of von Willebrand factor (vWF) and tumor necrosis factor alpha (TNF-α).
Representative images of lung tissues isolated 3 weeks after the CBDL or sham procedures showing vWF (red), TNF-α (green), and DAPI, nuclear staining marker (blue). (a–c) Staining at 3 weeks after the CBDL and (d–f) sham procedures. TNF-α was more highly expressed in pulmonary endothelial vascular cells at 3 weeks after CBDL than those of the sham operated group (original magnification, ×20).
qRT-PCR verification of microarray data
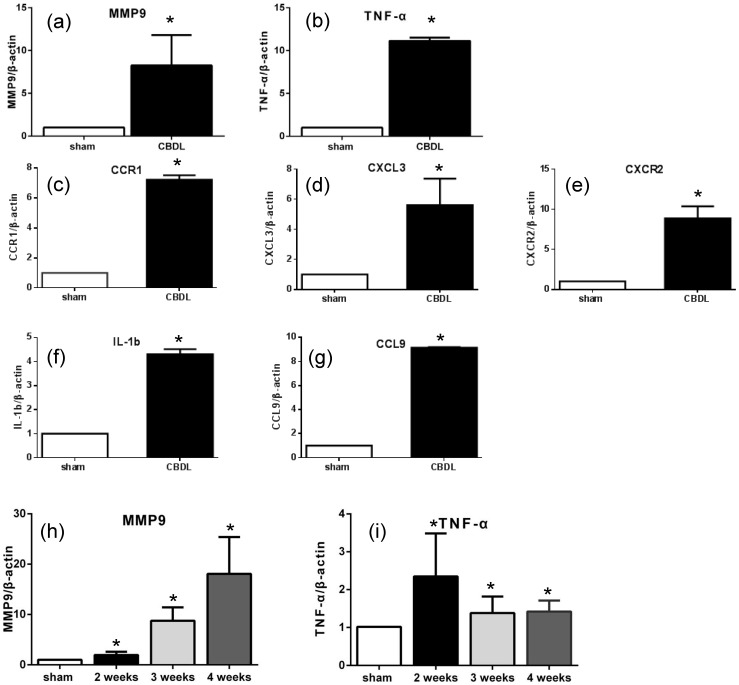
Seven genes associated with angiogenesis were selected for validation, and the directional fold change of each gene was confirmed using qRT-PCR analysis (Fig. 5a-i). Significant differences in mRNA expression of each selected gene were observed when compared with the sham operated group. Peak elevation of mRNA expression in MMP9 was observed at 4 weeks after CBDL, while that in TNF-α was observed 2 weeks after CBDL (Fig. 5h, i).
Figure 5. Real-time PCR analysis of representative genes identified with microarray analysis (a–g) was performed in pulmonary cells assembled by magnetic beads/CD31-antibody complexes in mice 3 weeks after CBDL (CBDL) and in sham operated controls (sham).
(a) MMP9 and (b) TNF-α. (c) CCR1, (d) CXCL3, (e) CXCR2, (f) IL-1b, and (g) CCL9. (h) and (i) were performed in whole pulmonary cells of sham and 1–4 weeks after CBDL groups. (h) MMP9 and (i) TNF-α. Values are expressed as mean ± standard error (n = 3 in each group). Y-axis abbreviations: CCL9, chemokine CC motif ligand 9; MMP9, matrix metallopeptidase 9; TNF-α, tumor necrosis factor alpha; CCR1, CC chemokine type-1 receptor; CXCL3, chemokine CXC motif ligand 3; CXCR2, CXC chemokine receptor; IL-1b, interleukin-1 beta. *P<0.05 (CBDL vs. sham).
Identification of angiogenic protein in serum by protein array analysis
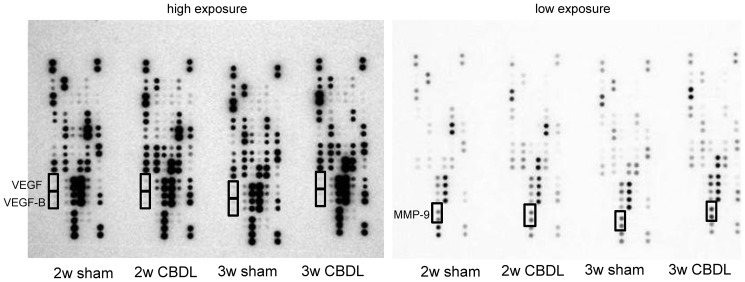
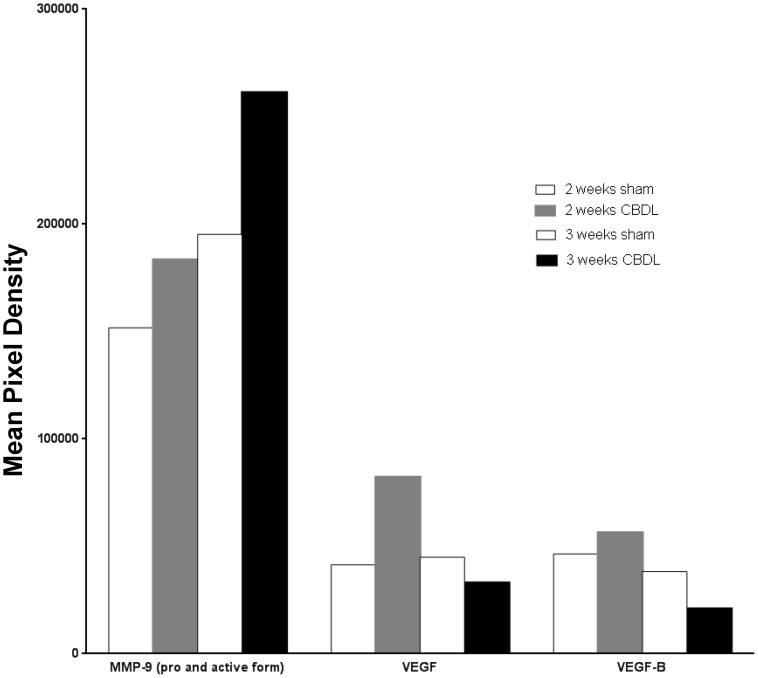
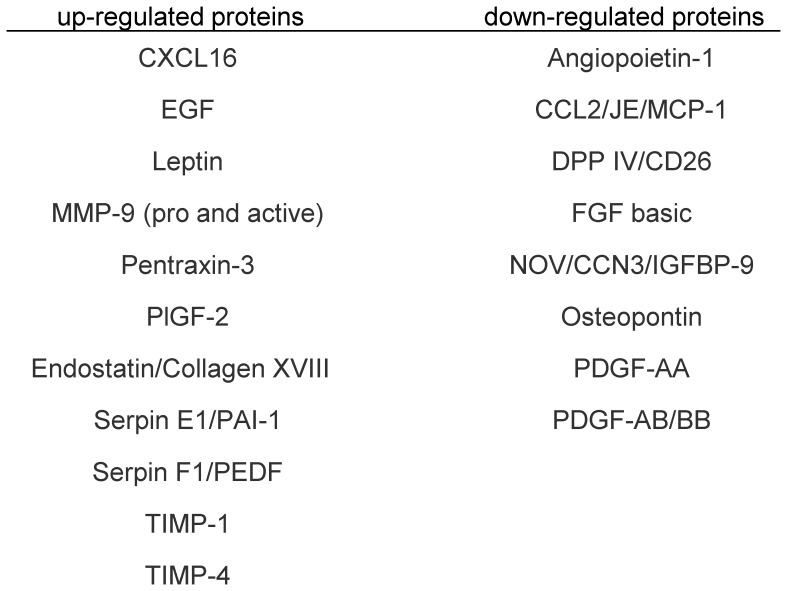
To identify the proteins that were important to angiogenic processes, we analyzed mouse serum 2 and 3 weeks after CBDL in comparison to that of sham mice using protein array analysis focused on angiogenesis (Fig. 6 and 7, Fig. S1). This analysis indicated 10 up-regulated and 9 down-regulated proteins that were associated with angiogenesis (Fig. 8). The up-regulated expression pattern of the MMP-9 gene was also supported by protein analysis of mouse serum (Fig. 6, 7). However, no significant difference was noted between the VEGF and VEGF-B protein levels in CBDL and sham mouse serum, which was different from the data obtained in rat models (Fig. 6, 7).[10], [19]
Figure 6. Protein array analysis of serum from common bile duct ligated (CBDL) and sham operated mice.
Representative proteins identified using protein array analysis in serum isolated from CBDL mice and sham operated (sham) mice.
Figure 7. Protein array analysis of serum from common bile duct ligated (CBDL) and sham operated mice.
Mean pixel density of representative proteins analyzed by protein array in serum isolated from CBDL and sham operated mice.
Figure 8. Protein array analysis of serum from common bile duct ligated (CBDL) and sham operated mice.
Representative list of up-regulated and down-regulated proteins in the serum of common bile duct ligated mice, in comparison with the levels measured in sham operated mice by using protein array analysis. CXCL16, chemokine (CXC motif ligand 16); DLL4, delta-like ligand 4; DPPIV, dipeptidyl peptidase IV; EGF, epidermal growth factor; MMP-9, matrix metallopeptidase 9; FGF, fibroblast growth factor; MCP-1, monocyte chemotactic protein-1; TIMP-1, tissue inhibitor of metalloproteinase-1; CCL2/JE/MCP-1, chemokine (CC motif) ligand 2; NOV/CCN3/IGFBP-9, nephroblastoma overexpressed; VEGF, vascular endothelial growth factor; PDGF, platelet-derived growth factor.
MMPs and TIMPs protein level and localization in impaired lungs after CBDL
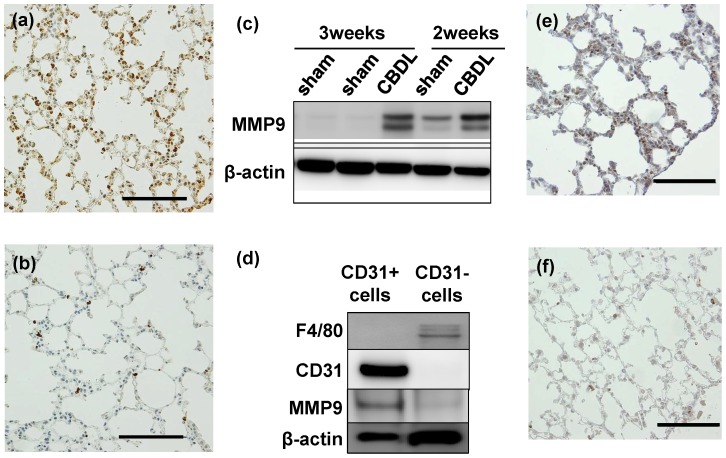
The next step in investigating the mechanism of lung injury was to focus on MMP-9, which we expected to act as a key protein in angiogenesis after CBDL. Immunohistochemistry analyses revealed that MMP-9 was highly expressed in vascular endothelial cells in CBDL mice (3 weeks), contrary to the finding that MMP-9 was less expressed in lungs of the sham group (Fig. 9a and b). Western blot analysis of lung whole cell lysates isolated from impaired lungs also showed high expression of MMP-9 in CBDL mice compared to sham mice (Fig. 9c). In addition, CD31-positive pulmonary vascular cell lysates showed high expression of MMP9, 3 weeks after CBDL in comparison with the CD31-negative pulmonary cells. The CD31-positive pulmonary cells also had lower F4/80 expression than CD31-negative pulmonary cells (Fig. 9d). In light of the results of the microarray analysis, in which MMP8 mRNA was highly expressed in pulmonary endothelial cells, we investigated the expression of MMP8 using immunohistochemistry. We found that MMP8 was highly expressed in the lungs of the mice 3 weeks after CBDL compared with those from sham operated mice. (Fig.9e: lungs from mice 3 weeks after CBDL, Fig.9f: lungs from sham operated mice). We also examined the expression of the MMP inhibitors, TIMP-1 and TIMP-4, in lungs of CBDL and sham operated mice. A significant difference was not found among the CBDL and sham operated mice in either TIMP-1 or TIMP-4 expression in lungs. (data not shown) In addition to these data, we investigated the presence of fibrosis in lungs after CBDL using Masson trichrome staining, since we assumed that our model had the possibility of causing pulmonary fibrosis due to the high level of MMP expression. [20]–[22] However, the results showed that our model did not induce the development of pulmonary fibrosis at either 2 or 3 weeks after CBDL compared with sham operated mice (data not shown).
Figure 9. MMP8 and MMP-9 expression in mice 3 weeks after common bile duct ligation (CBDL).
(a) Immunoreactivity of MMP-9 (matrix metallopeptidase) is stronger than that in sham-operated mice; (b) Immunohistochemical analysis. Scale bar = 100 µm. (c) Immunoblotting of MMP-9 in the whole pulmonary tissue of the sham and of the 2 and 3 weeks after CBDL groups revealed that MMP-9 is highly expressed in the 2 and 3 weeks after CBDL groups, compared with the expression in the sham groups. (d) Immunoblotting of F4/80, CD31, and MMP9 in the CD31-positive pulmonary cell lysates obtained 3 weeks after CBDL showed higher levels of MMP9 expression in pulmonary vascular endothelial cells compared with those of CD31-negative pulmonary cells. (e) Immunoreactivity of MMP-8 is stronger than that in sham-operated mice; (f) Immunohistochemical analysis. Scale bar = 100 µm.
Neutrophils increase in the lungs of mice after CBDL
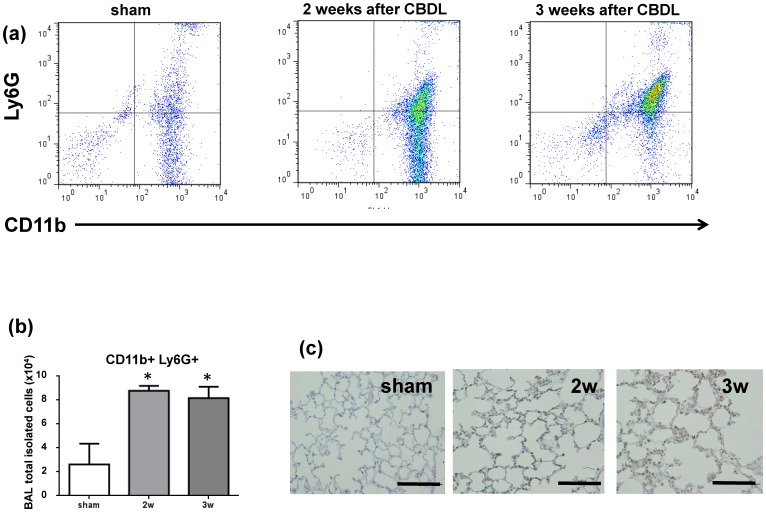
To investigate whether inflammatory cells contribute to the pulmonary pathogenesis in mice after CBDL, cells were collected from BAL fluid obtained from CBDL and sham operated mice and analyzed by flow cytometry. We found that CD11b positive and Ly6G positive cells, which are defined as neutrophils, were significantly increased 2 weeks after CBDL treatment. (Fig.10 a and b) Moreover, immunohistochemistry showed that Ly6G was highly expressed in lungs both at 2 and 3 weeks after CBDL, when compared with the lungs from sham operated mice. We also investigated whether the number of macrophages, i.e., those identified as CD11b-positive and F4/80-positive cells using flow cytometry analysis, were increased in the BAL fluid of mice after CBDL. Surprisingly, we found no significant increase in macrophages contrary to findings from the CBDL rat model (data not shown). [10], [19] In addition, immunohistochemistry did not show expression of either CD68 or F4/80 in the lungs of mice that had CBDL.
Figure 10. CBDL increases the number of neutrophils in bonchoalveolar lavage fluid.
Neutrophils (CD11b-positive and Ly6G-positive cells) identified by flow cytometry were increased 2 and 3 weeks after CBDL. (a) Quantitation of total neutrophils in the BAL fluid showed a significant increase 2 and 3 weeks after CBDL compared with sham operated mice. (b) Ly6G expression is higher in the lungs of mice 2 and 3 weeks after CBDL compared with sham operated mice using immunohistochemical analysis. (Fig. 10c)
Discussion
Cirrhosis results in disorders such as HPS, which is characterized by hypoxemia, high cardiac output, and extrahepatic lesions, all of which increase morbidity and mortality.[1] The same pattern of lung lesions, the pathological mechanism of which has not yet been clarified, is seen in patients with bi-directional cavopulmonary shunts.[4] Models of these diseases have been established in rats,[6], [23], [24] but to the best of our knowledge, only two studies have reported mouse models of pulmonary disease.[11], [14] We hypothesized that if a mouse model were established for this lung pathology, we could use a more extensive approach for resolving lung lesions by using transgenic or knockout mice. Therefore, we established a lung injury model by using CBDL, which incidentally had a higher survival rate than previous mouse models. [11], [14] Contrary to our expectation, lung pathology in our mouse model exhibited differences from that of rat models.[6], [10] The mechanisms responsible for the differences in pulmonary pathogenesis between mice and rats are unknown. In studies of humans with acute obstructive cholangitis (AOC) or other sepsis related to bile duct obstruction, the onset of multiple organ dysfunction (MOD) involving lungs is a complication associated with high morbidity and mortality.[1], [25] Hence, we suggest that our mouse model can be applied to pulmonary pathological analyses in the inflammatory phase, i.e., to systemic inflammatory response syndrome, acute lung injury, and MOD syndrome.[11], [26], [27]
Our findings showed that TNF-α plays an important role in development of pulmonary pathology including angiogenesis not associated with release of VEGF. On the other hand, in the rat model of CBDL, TNF-α plays an important role in pulmonary angiogenesis associated with VEGF-A-mediated angiogenic pathways.[10] This phenomenon may be explained by contrasting processes related to TNF-α induction of angiogenic signaling pathways in the inflammatory phase. TNF-α is secreted by macrophages, monocytes, and endothelial cells during inflammation. Activated neutrophils work to defend against inflammation, while overstimulated neutrophils injure tissues.[28], [29] TNF-α also induces expression of proangiogenic factors and primes vascular endothelial cells for angiogenic sprouting, while simultaneously blocking VEGF-induced proliferation in vitro.[30] TNF-α has also been reported to up-regulate VEGFR-2 expression and promote angiogenesis in vitro.[31], [32] Consequently, these two types of processes may help explain differences between mice and rats in angiogenic pathways after CBDL.
MMPs play an important role in the pulmonary pathogenesis of inflammatory lung diseases in human, which was reported by Kong et al.[33], [34] MMP-8 (neutrophil collagenase), which was elevated in both serum protein and pulmonary endothelial cells in this study, are released by activated neutrophils, and contributes to the formation and progression of acute lung injury and lung fibrosis.[34], [35] This pulmonary pathogenesis was confirmed by the absence of MMP-8 in mice has an antifibrotic effect to bleomycin induced lung fibrosis.[35] MMP-9 (type IV collagenase) has the ability to degrade gelatins and basement membrane collagen.[36] MMP-9 is known to exist in small quantities in healthy human lungs and to be produced at high levels by inflammatory cells (i.e., macrophages, polymorphonuclear neutrophils, and monocytes) in several lung diseases.[37] For a rodent ventilator-induced lung injury model, Kim et al. reported that lungs impaired by high tidal volume had higher expression of MMP-9 than lungs ventilated by low tidal volume and related this finding to neutrophilic inflammation.[38] Keck et al. reported that MMP-9 was a valid marker for predicting the development of lung damage and was required for migration of neutrophils into lung tissue in the rodent acute pancreatitis model.[27], [39] Johnson et al. reported that MMP-9 was required for angiogenesis in ischemic limbs and suggested that macrophages act as the source of MMP-9.[40]
In the present study, we found a high level of MMP9 production in CBDL mice serum on performing protein array analysis (Fig. 6). Strong gene expression and protein production of MMP9 were detected in CD31-positive cells (Fig. 5h). Moreover, invasive and proliferative endothelial cells were mostly observed in CBDL mice by immunohistochemical and HE staining (Fig. 1–2). Taken together, these findings suggest that MMP-9, mainly derived from endothelial cells, may play an important role in regulating angiogenesis in lung injury after CBDL. The next step will be to use MMP-9 knockout mice in analyses of treatment for cirrhosis-induced lung injury. In our model, the reason that lung fibrosis, which was assessed with Masson trichrome staining, was not strongly induced might be explained as follows; firstly, MMP-9, which is secreted from increased neutrophils during inflammation plays a role in suppression of fibrosis in the tissue.[20] Secondly, unbalanced MMP and TIMP expression during the time course we used was reported to contribute to development of experimental pulmonary fibrosis: lung MMP-2 and MMP-9 m-RNA levels have peak levels and less pulmonary fibrosis is present 1 week after induction, while MMPs inhibitors, such as TIMPs, have a different peak and are present at higher levels 3 weeks after pulmonary fibrosis induction [21], [22] Lastly, the mice strain used, which was Balb/c, might have affected the development or severity of pulmonary fibrosis, as Walkin et al. described that this strain is resistant to pulmonary fibrosis, but susceptible to hepatic fibrosis. [41]
Data from our CBDL model in mice did not mimic what is observed in rats particularly in regards to HPS pathophysiology, contrary to our expectations. The TNF-α inhibitor pentoxifylline has a beneficial effect on HPS development in rats model; however, no efficient improvement of arterial oxygenation could be observed in a pilot study of pentoxifylline administration against human HPS patients. [42] These findings suggest that HPS pathophysiology is complex and imply that a number of details regarding the mechanism of HPS are still unclear. However, we believe that our findings contribute to our understanding of HPS pathophysiology in humans and provide insights that may help in the development of efficacious therapies against this disease. [43]
In conclusion, we have shown that pulmonary pathogenesis occurs after CBDL in a mouse model and demonstrated differences between this model and other experimental animal models of hepatopulmonary syndrome. This model, which is similar to systemic inflammatory response syndrome, can be used to assess pulmonary pathology in the activated inflammation phase.
Supporting Information
List of proteins in serum of common bile duct ligated (CBDL) mice and sham operated (control) mice in protein array analysis.
(TIF)
Acknowledgments
We thank Tomomi Shimizu (Department of Cardiovascular Surgery, Ehime University), Yumiko Fukuda (Department of Oral and Maxillofacial Surgery, Ehime University), Yusuke Fujioka (Department of Biochemistry and Molecular Genetics, Ehime University), and Kenji Kameda (Integrated Center for Sciences (INCS) Shigenobu Station, Ehime University) for their professional assistance.
Funding Statement
This work was supported in part by JSPS KAKENHI grant number 11008958 (http://www.jsps.go.jp/english/index.html). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. No additional external funding received for this study.
References
- 1. Zhang J, Fallon MB (2012) Hepatopulmonary syndrome: update on pathogenesis and clinical features. Nat Rev Gastroenterol Hepatol 9: 539–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Machicao VI, Fallon MB (2012) Hepatopulmonary syndrome. Semin Respir Crit Care Med 33: 11–16. [DOI] [PubMed] [Google Scholar]
- 3. Sari S, Oguz D, Sucak T, Dalgic B, Atasever T (2012) Hepatopulmonary syndrome in children with cirrhotic and non-cirrhotic portal hypertension: a single-center experience. Dig Dis Sci 57: 175–181. [DOI] [PubMed] [Google Scholar]
- 4.Schranz D, Michel-Behnke I (2004) Arteriovenous malformations and hepatopulmonary syndrome. Lancet 364: 26–27; author reply 27. [DOI] [PubMed]
- 5.Kochar R, Fallon MB (2011) Pulmonary Manifestations of Liver Disease. Schiff's Diseases of the Liver, Eleventh Edition: 381–391.
- 6. Fallon MB, Abrams GA, Luo B, Hou Z, Dai J, et al. (1997) The role of endothelial nitric oxide synthase in the pathogenesis of a rat model of hepatopulmonary syndrome. Gastroenterology 113: 606–614. [DOI] [PubMed] [Google Scholar]
- 7. Ling Y, Zhang J, Luo B, Song D, Liu L, et al. (2004) The role of endothelin-1 and the endothelin B receptor in the pathogenesis of hepatopulmonary syndrome in the rat. Hepatology 39: 1593–1602. [DOI] [PubMed] [Google Scholar]
- 8. Liu L, Zhang M, Luo B, Abrams GA, Fallon MB (2001) Biliary cyst fluid from common bile duct-ligated rats stimulates endothelial nitric oxide synthase in pulmonary artery endothelial cells: a potential role in hepatopulmonary syndrome. Hepatology 33: 722–727. [DOI] [PubMed] [Google Scholar]
- 9. Luo B, Liu L, Tang L, Zhang J, Ling Y, et al. (2004) ET-1 and TNF-alpha in HPS: analysis in prehepatic portal hypertension and biliary and nonbiliary cirrhosis in rats. Am J Physiol Gastrointest Liver Physiol 286: G294–303. [DOI] [PubMed] [Google Scholar]
- 10. Zhang J, Luo B, Tang L, Wang Y, Stockard CR, et al. (2009) Pulmonary angiogenesis in a rat model of hepatopulmonary syndrome. Gastroenterology 136: 1070–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Beierle EA, Vauthey JN, Moldawer LL, Copeland EM 3rd (1996) Hepatic tumor necrosis factor-alpha production and distant organ dysfunction in a murine model of obstructive jaundice. Am J Surg 171: 202–206. [DOI] [PubMed] [Google Scholar]
- 12. Van Steenkiste C, Trachet B, Casteleyn C, van Loo D, Van Hoorebeke L, et al. (2010) Vascular corrosion casting: analyzing wall shear stress in the portal vein and vascular abnormalities in portal hypertensive and cirrhotic rodents. Lab Invest 90: 1558–1572. [DOI] [PubMed] [Google Scholar]
- 13. Kirkland JG, Godfrey CB, Garrett R, Kakar S, Yeh BM, et al. (2010) Reversible surgical model of biliary inflammation and obstructive jaundice in mice. J Surg Res 164: 221–227. [DOI] [PubMed] [Google Scholar]
- 14. Alaish SM, Torres M, Ferlito M, Sun CC, De Maio A (2005) The severity of cholestatic injury is modulated by the genetic background. Shock 24: 412–416. [DOI] [PubMed] [Google Scholar]
- 15. Miniati D, Jelin EB, Ng J, Wu J, Carlson TR, et al. (2010) Constitutively active endothelial Notch4 causes lung arteriovenous shunts in mice. Am J Physiol Lung Cell Mol Physiol 298: L169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kato T, Kimura T, Ishii N, Fujii A, Yamamoto K, et al. (1999) The methodology of quantitation of microvessel density and prognostic value of neovascularization associated with long-term survival in Japanese patients with breast cancer. Breast Cancer Res Treat 53: 19–31. [DOI] [PubMed] [Google Scholar]
- 17. Dong QG, Bernasconi S, Lostaglio S, De Calmanovici RW, Martin-Padura I, et al. (1997) A general strategy for isolation of endothelial cells from murine tissues. Characterization of two endothelial cell lines from the murine lung and subcutaneous sponge implants. Arterioscler Thromb Vasc Biol 17: 1599–1604. [DOI] [PubMed] [Google Scholar]
- 18. Tang L, Luo B, Patel RP, Ling Y, Zhang J, et al. (2007) Modulation of pulmonary endothelial endothelin B receptor expression and signaling: implications for experimental hepatopulmonary syndrome. Am J Physiol Lung Cell Mol Physiol 292: L1467–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thenappan T, Goel A, Marsboom G, Fang YH, Toth PT, et al. (2011) A central role for CD68(+) macrophages in hepatopulmonary syndrome. Reversal by macrophage depletion. Am J Respir Crit Care Med 183: 1080–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chakrabarti S, Patel KD (2005) Matrix metalloproteinase-2 (MMP-2) and MMP-9 in pulmonary pathology. Exp Lung Res 31: 599–621. [DOI] [PubMed] [Google Scholar]
- 21. Oggionni T, Morbini P, Inghilleri S, Palladini G, Tozzi R, et al. (2006) Time course of matrix metalloproteases and tissue inhibitors in bleomycin-induced pulmonary fibrosis. Eur J Histochem 50: 317–325. [PubMed] [Google Scholar]
- 22. Wang BL, Tu YY, Fu JF, Zhong YX, Fu GQ, et al. (2011) Unbalanced MMP/TIMP-1 expression during the development of experimental pulmonary fibrosis with acute paraquat poisoning. Mol Med Rep 4: 243–248. [DOI] [PubMed] [Google Scholar]
- 23. Tipps RS, Mumtaz M, Leahy P, Duncan BW (2008) Gene array analysis of a rat model of pulmonary arteriovenous malformations after superior cavopulmonary anastomosis. J Thorac Cardiovasc Surg 136: 283–289. [DOI] [PubMed] [Google Scholar]
- 24. Chang SW, Ohara N (1992) Pulmonary circulatory dysfunction in rats with biliary cirrhosis. An animal model of the hepatopulmonary syndrome. Am Rev Respir Dis 145: 798–805. [DOI] [PubMed] [Google Scholar]
- 25. Sugiyama M, Atomi Y (1997) Treatment of acute cholangitis due to choledocholithiasis in elderly and younger patients. Arch Surg 132: 1129–1133. [DOI] [PubMed] [Google Scholar]
- 26. Gonzalez-Lopez A, Albaiceta GM (2012) Repair after acute lung injury: molecular mechanisms and therapeutic opportunities. Crit Care 16: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Keck T, Balcom JHt, Fernandez-del Castillo C, Antoniu BA, Warshaw AL (2002) Matrix metalloproteinase-9 promotes neutrophil migration and alveolar capillary leakage in pancreatitis-associated lung injury in the rat. Gastroenterology 122: 188–201. [DOI] [PubMed] [Google Scholar]
- 28. Chakrabarti S, Zee JM, Patel KD (2006) Regulation of matrix metalloproteinase-9 (MMP-9) in TNF-stimulated neutrophils: novel pathways for tertiary granule release. Journal of Leukocyte Biology 79: 214–222. [DOI] [PubMed] [Google Scholar]
- 29. Phillipson M, Kubes P (2011) The neutrophil in vascular inflammation. Nat Med 17: 1381–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sainson RC, Johnston DA, Chu HC, Holderfield MT, Nakatsu MN, et al. (2008) TNF primes endothelial cells for angiogenic sprouting by inducing a tip cell phenotype. Blood 111: 4997–5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang R, Xu Y, Ekman N, Wu Z, Wu J, et al. (2003) Etk/Bmx transactivates vascular endothelial growth factor 2 and recruits phosphatidylinositol 3-kinase to mediate the tumor necrosis factor-induced angiogenic pathway. J Biol Chem 278: 51267–51276. [DOI] [PubMed] [Google Scholar]
- 32. Giraudo E, Primo L, Audero E, Gerber HP, Koolwijk P, et al. (1998) Tumor necrosis factor-alpha regulates expression of vascular endothelial growth factor receptor-2 and of its co-receptor neuropilin-1 in human vascular endothelial cells. J Biol Chem 273: 22128–22135. [DOI] [PubMed] [Google Scholar]
- 33. Kong MY, Gaggar A, Li Y, Winkler M, Blalock JE, et al. (2009) Matrix metalloproteinase activity in pediatric acute lung injury. Int J Med Sci 6: 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kong MY, Li Y, Oster R, Gaggar A, Clancy JP (2011) Early elevation of matrix metalloproteinase-8 and -9 in pediatric ARDS is associated with an increased risk of prolonged mechanical ventilation. PLoS One 6: e22596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Garcia-Prieto E, Gonzalez-Lopez A, Cabrera S, Astudillo A, Gutierrez-Fernandez A, et al. (2010) Resistance to bleomycin-induced lung fibrosis in MMP-8 deficient mice is mediated by interleukin-10. PLoS One 5: e13242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bendeck MP (2004) Macrophage matrix metalloproteinase-9 regulates angiogenesis in ischemic muscle. Circ Res 94: 138–139. [DOI] [PubMed] [Google Scholar]
- 37. Atkinson JJ, Senior RM (2003) Matrix metalloproteinase-9 in lung remodeling. Am J Respir Cell Mol Biol 28: 12–24. [DOI] [PubMed] [Google Scholar]
- 38. Kim JH, Suk MH, Yoon DW, Lee SH, Hur GY, et al. (2006) Inhibition of matrix metalloproteinase-9 prevents neutrophilic inflammation in ventilator-induced lung injury. Am J Physiol Lung Cell Mol Physiol 291: L580–587. [DOI] [PubMed] [Google Scholar]
- 39. Keck T, Jargon D, Klunsch A, Thomusch O, Richter S, et al. (2006) MMP-9 in serum correlates with the development of pulmonary complications in experimental acute pancreatitis. Pancreatology 6: 316–322. [DOI] [PubMed] [Google Scholar]
- 40. Johnson C, Sung HJ, Lessner SM, Fini ME, Galis ZS (2004) Matrix metalloproteinase-9 is required for adequate angiogenic revascularization of ischemic tissues: potential role in capillary branching. Circ Res 94: 262–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Walkin L, Herrick SE, Summers A, Brenchley PE, Hoff CM, et al. (2013) The role of mouse strain differences in the susceptibility to fibrosis: a systematic review. Fibrogenesis Tissue Repair 6: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tanikella R, Philips GM, Faulk DK, Kawut SM, Fallon MB (2008) Pilot study of pentoxifylline in hepatopulmonary syndrome. Liver Transpl 14: 1199–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roberts KE, Kawut SM, Krowka MJ, Brown RS Jr, Trotter JF, et al.. (2010) Genetic risk factors for hepatopulmonary syndrome in patients with advanced liver disease. Gastroenterology 139: 130–139 e124. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of proteins in serum of common bile duct ligated (CBDL) mice and sham operated (control) mice in protein array analysis.
(TIF)