Abstract
Candida albicans is a ubiquitous fungus, which can cause very serious and sometimes life-threatening infections in susceptible patients. We used Caenorhabditis elegans as a model host to screen a library of C. albicans mutants for decreased virulence and identified SPT20 as important for virulence. The transcription co-activator SPT20 was identified originally as a suppressor of Ty and solo δ insertion mutations, which can cause transcription defects in Saccharomyces cerevisiae. It is resistant to the toxicity caused by overexpression of GAL4-VP16. We constructed a C. albicans spt20Δ/Δ mutant and found the spt20Δ/Δ strain was significantly less virulent than the wild-type strain SC5314 in C. elegans (p < 0.0001), Galleria mellonella (p < 0.01) and mice (p < 0.001). Morphologically, spt20Δ/Δ mutant cells demonstrated a “snow-flake” shape and clustered together; prolonged culture times resulted in increased size of the cluster. The clustered morphology was associated with defects in nuclei distribution, as the nuclei were not observed in many cellular compartments. In addition, the C. albicans spt20Δ/Δ mutant resulted in defects in hyphae and biofilm formation (compared to the wild-type strain, p < 0.05), and sensitivity to cell wall and osmotic stressors, and to antifungal agents. Thus our study demonstrated a role of C. albicans SPT20 in overall morphology and distribution of nuclear material, which may cause the defects in filamentation and biofilm formation directly when this gene is deleted.
Introduction
Candida albicans is part of the human flora that can be isolated from the gastrointestinal tract, vagina, and mouth in healthy individuals. Nevertheless, when the anatomical barrier is damaged or when host immunity declines then an invasive fungal infection can follow [1]. C. albicans is the fourth most common pathogen isolated from blood cultures and candidemia is associated with mortality rates as high as 35% [2] or even higher [3], [4]. A characteristic of C. albicans is that it can make the switch between several morphological forms: budding yeast, pseudohyphae, and true hyphae, according to the environmental conditions [5], [6]. The pathogenic ability of C. albicans is closely related to the change between these morphological forms [7], [8]. Additionally, the formation of hyphae [9] and biofilms [10], the integrity of the cell wall [11], the rapid adaptive capacity to external environmental condition [12] all contribute to virulence and are all related to morphogenesis.
Here we report SPT20 is involved in C. albicans virulence and is essential for hyphal and biofilm formation. The transcription co-activator SPT20 encodes a 604 amino acid nuclear protein that is rich in glutamine and asparagines [13] and was identified originally as a suppressor of Ty and solo δ insertion mutations, which can cause transcription defects in Saccharomyces cerevisiae [14]. There is ample evidence that S. cerevisiae SPT20 mutants have some broader transcriptional defects when compared with the other SPT genes [13]–[15]. Genome-wide expression analysis showed that SPT20 controls about 10% of normal transcription in S. cerevisiae [16]. Of note is that SPT20 has also been identified by another name, ADA5, as it is a member of the ADA group of genes, and it is resistant to the toxicity caused by overexpression of GAL4-VP16 which blocks the transcription factor combined with transcription complexes, thus killing the yeast cells [13]. Interestingly, we found that in C. albicans, SPT20 is associated with cell morphology; spt20Δ/Δ cells cluster together and demonstrate a “snow-flake” shape. Deformity in cell morphology is accompanied by defects in nuclear localization.
Materials and Methods
Strains and growth conditions
C. albicans strains were grown in YPD (1% yeast extract, 2% peptone, and 2% dextrose) at 30°C. Nourseothricin-sensitive clones were identified by their small colony size and confirmed by re-streaking on YPD agar plates containing 200 µg/mL nourseothricin (Werner Bioagents, Jena, Germany) as described previously [17].
For the nematode studies, the C. elegans glp-4; sek-1 strain was propagated on nematode growth medium on lawns of Escherichia coli OP50 using standard methods[18].
Generation of a homozygous mutant and re-integrated strain
The flipper cassette from pSFS2 containing the dominant nourseothricin resistance marker CaSAT1 was used to generate the spt20Δ/Δ mutant [17]. This SAT1 flipping strategy was used to avoid any interference from the auxotrophic selective marker genes in our virulence studies. The DNA fragment for homologous recombination was generated as follows: PCR was used to amplify a 924 bp ApaI-XhoI DNA fragment containing an upstream region and the 5′ end of SPT20 gene with primers SPT20-up-FWD and SPT20-up-RV (Table S1). The fragment was then cloned into the pSFS2 vector upstream of the MAL2p-CaFLP-CaSAT1 cassette, yielding plasmid pSFS2-SPT20up. A 726 bp SacII-SacI fragment containing a down-stream region and the 3′ end end of the SPT20 gene was amplified by PCR with primers SPT20-down-FWD and SPT20-down-RV (Table S1) and cloned into the pSFS2-SPT20up vector downstream of the MAL2p-CaFLP-CaSAT1 cassette using the SacII and SacI restriction sites, yielding the final plasmid pSFS2-SPT20updwn. The pSFS2-SPT20updwn plasmid was digested with ApaI and SacI, and the 5856 bp fragment was incorporated into C. albicans SC5314 using the lithium acetate method [19]. Nourseothricin-resistant transformants were evaluated for the appropriate insert location by PCR (Table S2), and the correct transformants were used for further work. After excising the nourseothricin resistance marker (caSAT1), the same method was used to delete the remaining functional allele of SPT20. Another round of integration/excision yielded SPT20 homozygous mutant spt20Δ/Δ. The primers used to identify all the strains are listed in Table S2.
To re-integrate SPT20 into the SPT20 null mutant, spt20Δ/Δ, a 3640 bp ApaI-XhoI fragment containing the complete open reading frame as well as 790 bp of upstream and 594 bp of downstream flanking sequences was amplified with the primers SPT20-FWD and SPT20-RV (Table S1) and substituted for upstream and 5′ end of SPT20 gene sequence in pSFS2-SPT20updwn at the ApaI and XhoI restriction site to generate the plasmid pSFS2-SPT20Rec. The pSFS2-SPT20Rec plasmid was digested with ApaI and SacI, and the 7848 bp fragment was used to reintroduce the SPT20 sequence into the C. albicans mutant spt20Δ/Δ. Nourseothricin-sensitive derivatives were obtained in the following process, yielding the SPT20 reconstituted strain spt20Δ/SPT20 for further investigations. The primers used to identify the strains are listed in Table S2. We were unable to produce a strain that re-integrated the SPT20 sequence at the second allele site. However, testing of the single loci re-integrated strain exhibited morphology and virulence comparable to the wild-type strain.
In vitro hyphal formation and biofilm growth assays
For hyphal formation, C. albicans strains were incubated with 1 mL serum for 60 min at 37°C with agitation in the dark, then collected by centrifugation and washed twice with phosphate buffered saline (PBS). Cells were suspended in Spider media and incubated at 37°C for 20 h [18], then photographed using an Olympus Bx51 series upright microscope. For biofilm growth evaluation [20], strains of C. albicans were grown in YPD medium overnight at 30°C, diluted to an OD600 of 0.5 in 2 mL Spider medium, and added to a sterile 12-well plate containing a prepared silicone square measuring 1.5 × 1.5 cm (cut from silicone sheets Bentec Medical Inc., Woodland, CA) that had been pretreated overnight with bovine serum (Sigma-Aldrich). The inoculated 12-well plate was incubated with gentle agitation (150 rpm) for 90 min at 37°C for adhesion to occur. The squares were washed with 2 mL of PBS to remove any un-adhered cells, and moved to a fresh 12-well plate containing 2 mL of fresh Spider medium. This plate was incubated at 37°C for an additional 60 h with agitation (150 rpm) to allow for biofilm formation. The silicone platform and attached biofilm were removed from the wells, dried overnight, and weighed the following day. The total biomass of each biofilm was calculated by subtracting the weight of the silicone platform material prior to biofilm growth from the weight after the drying period and adjusting for the weight of a control pad exposed to no cells. The average total biomass for each strain was calculated from four independent samples. Statistical significance was determined by the analyses of variance (ANOVA). For comparison between two C. albicans isolates, the Student t test was used. A p-value of less than 0.05 was considered significant.
Sensitivity assays
The sensitivity to cell stressors was evaluated using the cell wall stress agent sodium dodecyl sulfate (SDS) and osmotic stress agent sodium chloride (NaCl). C. albicans strains were grown overnight and then suspended in YPD at 2×106 cells/mL. Ten fold serial dilutions from 104 to 101 of the wild-type, spt20Δ/Δ and spt20Δ/SPT20 re-integrated strains were spotted in a volume of 5 µL on YPD agar plates with the indicated chemical agent added. Cells were grown under 25°C, 30°C, and 37°C conditions until colonies appeared.
Virulence assay using the C. elegans infection model
The assay was carried out using an established protocol [18]. In brief, freshly grown C. albicans cells were inoculated into 2 mL of YPD broth and allowed to grow overnight at 30°C. The following day, 100 µL of yeast was spread into a square lawn on a 10-cm plate containing brain heart infusion (BHI) agar and kanamycin (45 µg/mL), followed by incubation for approximately 20 h at 30°C. Synchronized adult C. elegans glp-4; sek-1 nematodes grown at 25°C were carefully washed from plates containing their normal food source (E. coli OP50 strain) using sterile M9 buffer. Approximately 400 to 500 washed worms were then added to the center of the C. albicans lawns. The plates were incubated at 25°C for 4 h. Worms were then carefully moved into a 15 mL conical tube and washed four times with sterile M9. Sixty worms were then transferred into a single well of a six-well tissue culture plate (Corning, Inc.) containing 2 mL of liquid medium (80% M9, 20% BHI) and kanamycin (45 µg/mL). Worms were scored daily and dead worms were removed from the assay. Microscopy of nematodes was performed by using Nomarski optics on a Zeiss AxioImager microscope.
To screen for mutations that affect C. albicans virulence, we evaluated a miscellaneous homozygous mutant library from the Fungal Genetics Stock Center (Kansas City, MO), deposited by Dr. Aaron Mitchell. Worms were infected as described above and scored for visibility of fungal filaments protruding through the worms cuticle and survival at 72 h post infection. Survival was determined by the ability to respond to touch with a platinum wire. The control group was infected with strain DAY286, the parent of the mutant strains in the library collection.
Virulence assay using the Galleria mellonella infection model
Inocula were prepared by growing C. albicans strains at 30°C overnight with agitation. Cells were collected with centrifugation and washed 3 times with PBS. Yeast cells were counted using a hemocytometer. G. mellonella larvae (Vanderhorst Wholesale Inc., St. Marys, OH) at the final instar stage were inoculated with 106 cfu of C. albicans suspended in PBS. Each infection group contained 16 randomly chosen larvae of the appropriate weight (330 ± 25 mg). The inoculum was injected in a 10 µL volume directly to the last left, pro-leg using a Hamilton syringe [21]. After inoculation, larvae were incubated at 37°C. A mock inoculation with PBS was included as a control for each experiment to observe for killing due to physical trauma or infection by pathogenic contaminants. The number of dead larvae was scored daily.
Virulence assay using a murine candidiasis model
A disseminated murine candidiasis model [22] was selected for the assay. Strains SC5314, spt20Δ/Δ, spt20Δ/SPT20 were used to infect mice. Cultures were grown overnight then washed 3 times with PBS. CD-1, six-week old, female mice were infected with 1.5 × 106 CFUs suspended in PBS via a tail vein injection in a 100 µl volume. Twelve mice were inoculated per strain tested and observed daily. Animal survival was evaluated by using the Kaplan-Meier method and differences were determined by using the log-rank test (STATA 6; STATA, College Station, TX). A p value of < 0.05 was considered statistically significant.
Fungal burden in the kidneys was assessed using four mice per infection strain [23]. We harvested the kidneys from mice aseptically 2 days post infection. Tissues were weighed and homogenized in sterile PBS by use of a Tissue Tearor (model 398; Biospec Products Inc., Racine, WI). Serial dilutions were plated on YPD agar plates containing 100 µg/mL ampicillin, 100 µg/mL streptomycin, and 45 µg/mL kanamycin. The cfu/g kidney were counted after growth at 30°C for 48 h. Statistical analyses were performed using ANOVA and post hoc (Bonferroni and Student–Newman–Keuls) tests.
Fungal cells staining with periodic acid Schiff (PAS)
Histopathological analysis was performed to assess kidney infestation. The murine candidiasis model was prepared as described above. Two days after inoculation, kidneys were removed aseptically from each mouse before being fixed in 10% neutral buffered formalin. Kidneys were embedded in paraffin and sections were stained with Periodic Acid Schiff (PAS) to reveal the hyphal structure of the fungal pathogens [23].
Staining of nuclei
Cells were stained for nuclei according to the protocol by Kopecka et al. and Fuchs et al. [24], [25]. The strains were grown in YPD overnight at 30°C. Nuclei were identified by staining with 0.1 µg/mL DAPI for 15 min. The stained cells were collected then washed twice with 1% BSA in PBS for 5 min followed by a five minute rinse in 0.1% BSA in PBS. Cells were suspended in Vectashield (Vector Laboratories, Inc.) and visualized with an Olympus microscope. All observations were confirmed with 3 independent cell cultures.
Fixation and staining of Calcofluor White
Cells were fixed by adding 40% Formaldehyde (50 µL) to 450 µL cell cultures for 30 min and washed 2 times with PBS. Subsequently, cells were re-suspended in Calcofluor solution (Sigma) for 5 min at room temperature and washed 5 times with PBS. Then the stained cells were re-suspended in 10 µL mounting medium and observed using an Olympus confocal microscope.
Drug susceptibility tests
For the susceptibility assays, we grew cultures in YPD at 30°C with agitation. Cells were diluted in PBS to a density of 2.0×106 cells/mL. The strains were plated as ten fold serial dilutions, spotting 5 µL of each dilution onto YPD agar plates supplemented with fluconazole (FLC; 2.5 µg/mL), amphotericin B (AMB; 1 µg/mL), 5-fluorocytosine (5FC; 4 µg/mL) or caspofungin (CAS; 0.075 µg/mL) and onto drug-free YPD agar plates as a control. Plates were incubated for 48 h at 35°C.
Ethics statement
All experiments were approved by Massachusetts General Hospital Research Animal Care Subcommittee and as outlined in the Guide for the Care and Use of Laboratory Animals of National Institutes of Health.
Results
Using a C. elegans infection model, we screened a C. albicans mutant library that consisted of 86 strains, for mutants that exhibited decreased virulence compared to strain DAY286. Through the course of our screening assay, we identified the SPT20 mutant strain as causing significantly less death in the C. elegans worms than the control strain DAY286. More specifically, the control infection produced an average of 47.6% dead worms with visible filaments protruding through the worm cuticle with a SD of 10.3%.However, the worms infected with the spt20 mutant strain did not produce any dead worms with protruding filaments. SPT20 deletion strains were developed for further analysis in order to confirm the role of this gene and the contributions of SPT20 in C. albicans virulence.
The spt20Δ/Δ mutant has reduced virulence in the C. elegans and the G. mellonella infection models
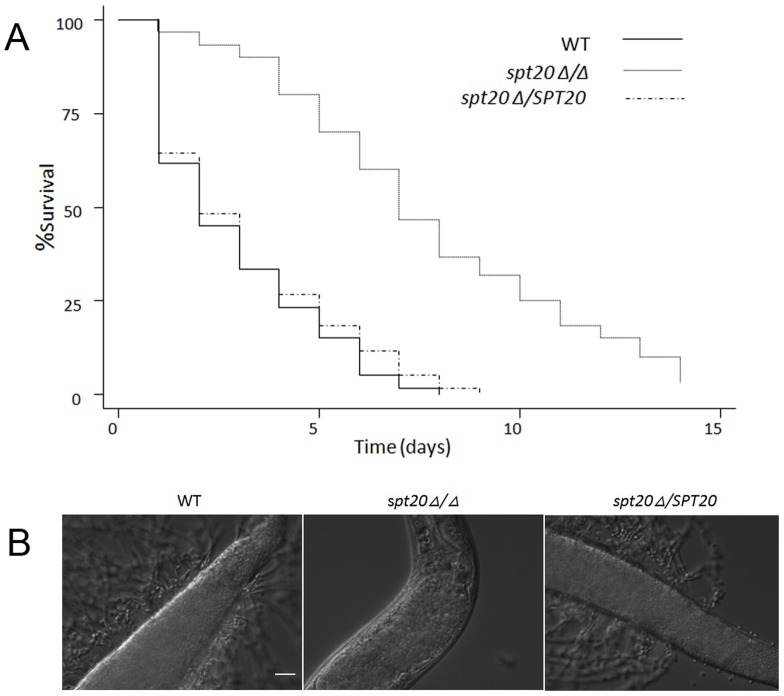
We confirmed the impact of SPT20 deletion in virulence using the C. elegans infection model ( Figure 1 ). In this series of experiments, we performed the nematode assays in more detail with daily scoring of dead/alive nematodes. We found that more than half of the worms infected with the wild-type SC5314 (or the re-integrated strain spt20Δ/SPT20) died within the first 48 h after infection ( Figure 1A ) and almost half the dead worms had visible hyphae piercing the cuticle ( Figure 1B ). At 120 h, >80% of the worms infected with the wild-type strain SC5314 or the re-integrated strain spt20Δ/SPT20 were dead, while only 30% of the worms were dead in the spt20Δ/Δ infection group ( Figure 1A ; p < 0.0001 compared to the wild-type strain or the reintegrated strain). Furthermore, no hyphae were observed in the spt20Δ/Δ infection group ( Figure 1B ).
Figure 1. SPT20 is essential for C. albicans virulence in the C. elegans infection model.
(A) Survival over 120 h (n = 60 worms infected per strain of C. albicans). Compared to infection with C. albicans SC5314, the spt20Δ/Δ mutant was avirulent to C. elegans (p < 0.0001). The reintegration of SPT20 restored the virulence of C. albicans to the wild-type level. (B) The images show C. elegans glp-4;sek-1 nematodes infected by C. albicans strains SC5314, spt20Δ/Δ and spt20Δ/SPT20 for 4 h and then moved into pathogen-free liquid media. On day 4 of the assay, the wild-type and spt20Δ/SPT20 infected worms exhibited a lethal infection with hyphae penetrating the worm cuticle. The hyphae were absent in the spt20Δ/Δ infected worms. The scale bar in Fig.1B represents 20 µm.
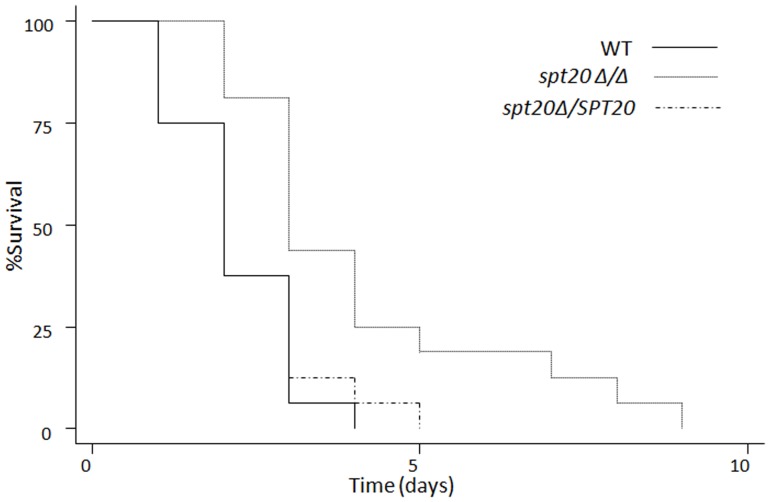
We confirmed the decreased pathogenicity of the spt20Δ/Δ strain in an insect model. In the G. mellonella infection model, the spt20Δ/Δ was significantly less virulent than the wild-type strain SC5314 at 37°C ( Figure 2 ; p < 0.01). All the larvae in the wild-type strain died within 4 days and the re-integrated strain group died within 5 days while the spt20Δ/Δ infection group died within 9 days ( Figure 2 ; p < 0.01).
Figure 2. SPT20 influences virulence of C. albicans in the G. mellonella infection model.
Survival over 216 h (9 days) at 37°C (n = 16 larvae per strain of C. albicans). p < 0.01 compared to the groups infected with the wild-type or the spt20Δ/SPT20 reintegrated strain.
The spt20Δ/Δ mutant has attenuated virulence in the murine candidiasis model
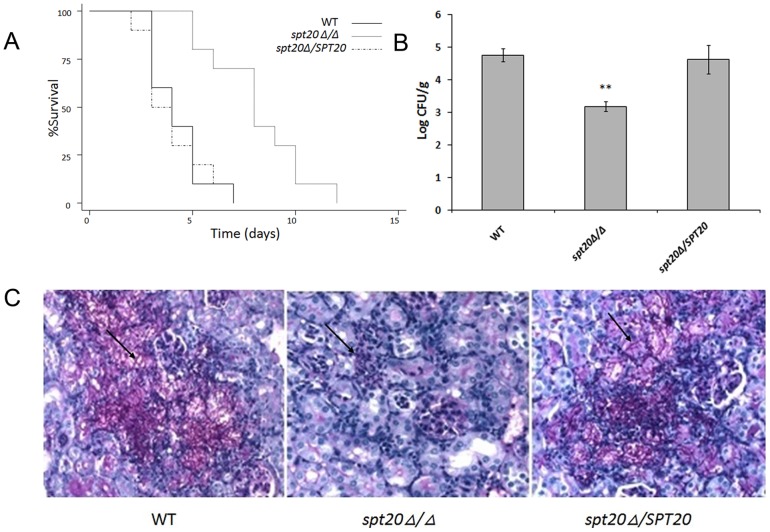
The spt20Δ/Δ strain was significantly less virulent than the wild-type strain SC5314 (p < 0.001) ( Figure 3A ). More specifically, all 12 mice inoculated with the wild-type strains within 7 days post infection by tail vein injection, while only 4 out of 12 mice in the spt20Δ/Δ group were dead at the same period.
Figure 3. SPT20 is required for virulence of C. albicans in the murine candidiasis model.
Mice were inoculated via the tail vein with 1.5 × 106 cfu of the indicated strains of C. albicans. (A) Survival over 21 days (n = 12 mice per strain of C. albicans). p < 0.001 for spt20Δ/Δ strain compared to either the wild-type or spt20Δ/SPT20 reintegrated strain. (B) Fungal burden of the kidneys of mice infected with the wild-type, spt20Δ/Δ and spt20Δ/SPT20 reintegrated strain two days after inoculation (n = 4 mice per strain of C. albicans). *p < 0.01 for spt20Δ/Δ strain compared to either the wild-type or spt20Δ/SPT20 re-integrated strain. (C) Thin sections of kidneys were stained with PAS after 2 days of infection. Arrows indicate C. albicans filaments in the tissues.
We also evaluated the fungal burden in the mouse kidneys as part of the pathogenicity assessment. The fungal burden in the spt20Δ/Δ group was significantly lower than that in the wild-type SC5314 infection group (p < 0.01; Figure 3B ). Evaluation of the kidney tissue by histopathology illustrated the differences in the burden and morphology of the infecting fungi. PAS staining revealed a large number of fungi in the kidneys of mice infected with the wild-type or the re-integrated strains while rare fungal cells were observed in the kidneys from the mice infected with the spt20Δ/Δ strain ( Figure 3C ). We also observed differences in the morphological state of the infecting fungi. Hyphae were visible in the tissues of the wild-type and re-integrated strains infected mice but not in the kidney of mice infected with the spt20Δ/Δ strain ( Figure 3C ). Of note is that the fungal burden of the kidneys of mice infected with spt20Δ/SPT20 re-integrated strain was restored, and the P value was not significantly different compared to the wild type.
The role of SPT20 in C. albicans cell wall integrity
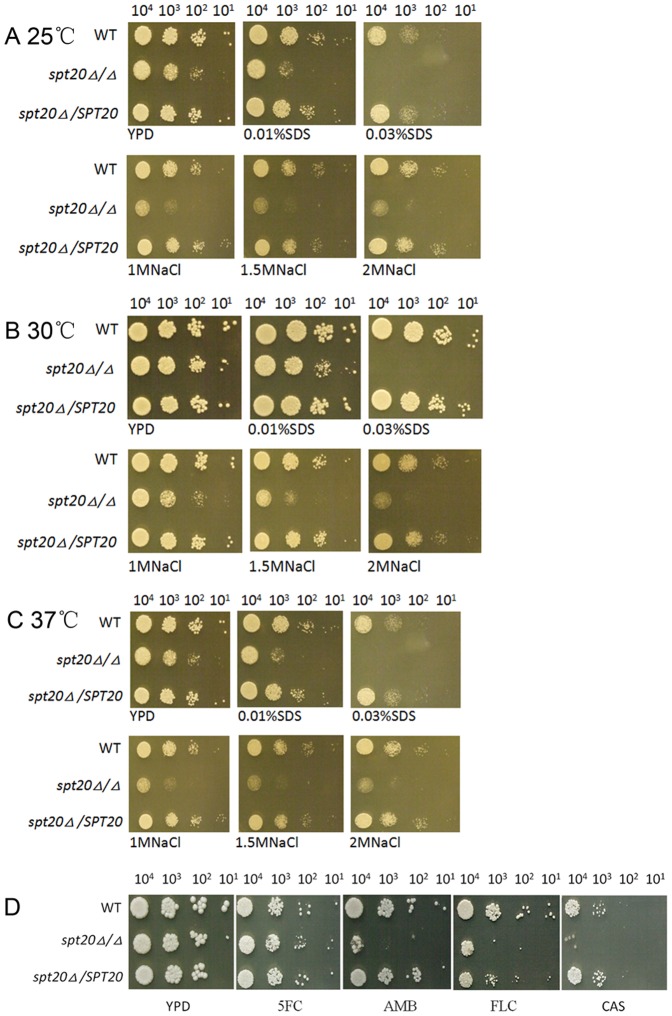
To examine the cell wall integrity, we tested the susceptibility of wild-type strain SC5314, the spt20Δ/Δ mutant and the re-integrated strain spt20Δ/SPT20 to the cell wall stress agent SDS and osmotic stress agent NaCl. Interestingly, the spt20Δ/Δ mutant was susceptible to 0.01%, 0.03% SDS and 1 M,1.5 M, 2 M NaCl at all the temperatures tested, 25°C, 30°C and 37°C ( Figure 4A, 4B and 4C ). Thus, the disruption of SPT20 affected C. albicans stress responses and suggests that SPT20 plays a role in maintaining cell wall integrity.
Figure 4. The disruption of SPT20 affected C. albicans stress responses and antifungal susceptibility.
Ten-fold serial dilutions of the wild-type, spt20Δ/Δ, and spt20/SPT20 were evaluated for susceptibility to cell wall stressors and antifungal agents. The spt20Δ/Δ mutant was specifically susceptible to SDS and NaCl at all the temperatures tested, 25°C (A), 30°C (B) and 37°C(C). The spt20Δ/Δ mutant was hypersensitive to amphotericin B, fluconazole and caspofungin, but was not sensitive to 5-fluorocytosine (D).
Sensitivity to antifungal agents
In our drug susceptibility testing, the wild-type, spt20Δ/Δ and spt20Δ/SPT20 re-integrated cells grew similarly on YPD agar plates and on YPD agar plates supplemented with 5FC, whereas the spt20Δ/Δ mutant was hypersensitive to amphotericin B, fluconazole and caspofungin compared to the wild-type and spt20Δ/SPT20 re-integrated strains ( Figure 4D ).
SPT20 is involved in C. albicans morphology
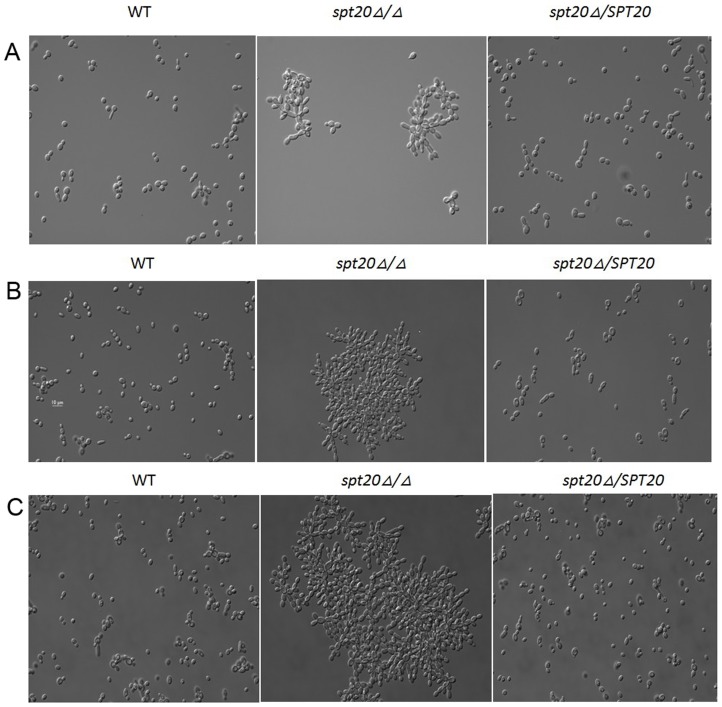
An examination of the cell morphology at 30°C growth temperature was established. C. albicans spt20Δ/Δ has a morphological defect ( Figure 5 ). Micrographs of C. albicans morphology after 2, 8, and 24 h of growth at 30°C are presented. We found that the spt20Δ/Δ cells cluster together and demonstrate a “snow-flake”shape rather than the normal separated and round shape of the wild-type and spt20Δ/SPT20 re-integrated strain. The longer the cells are cultured, the larger the cell cluster.
Figure 5. Micrographs of C. albicans morphology.
Cell morphology was examined. Micrographs after 2(A), 8 h (B), and 24 h (C) of growth at 30°C are presented for the wild-type, spt20Δ/Δ and spt20Δ/SPT20 reintegrated strain. The wild-type and spt20Δ/SPT20 re-integrated cells separate and appear to have a round morphology at all 3 time points while spt20Δ/Δ cells cluster together and demonstrate a “snow-flake” shape.
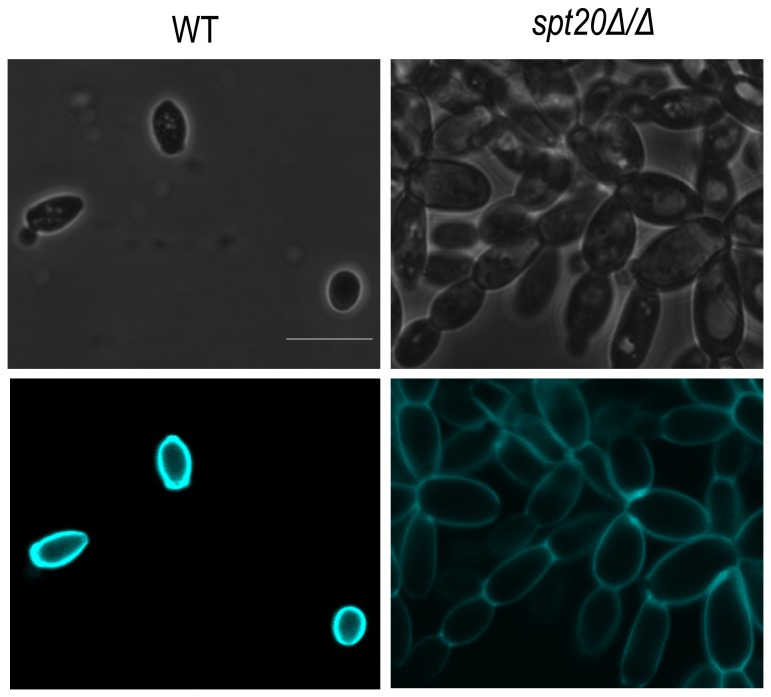
To further validate the clustering morphology, we evaluated the cells for defect in chitin. We analyzed chitin distribution in wild-type strain and spt20Δ/Δ strain using Calcofluor white and tested the expression of CHT3 by RT-PCR. The results showed that spt20Δ/Δ did not exhibit defects in chitin distribution ( Figure 6 ) and the expression of CHT3 between wild-type strain and spt20Δ/Δ strain have no statistical differences (Figure S1).
Figure 6. Calcofluor White staining of the wild-type and spt20Δ/Δ.
Calcofluor White specifically stains chitin. Spt20Δ/Δ did not exhibit defects in chitin distribution. Bar = 5 µm.
SPT20 is associated with hyphal and biofilm formation
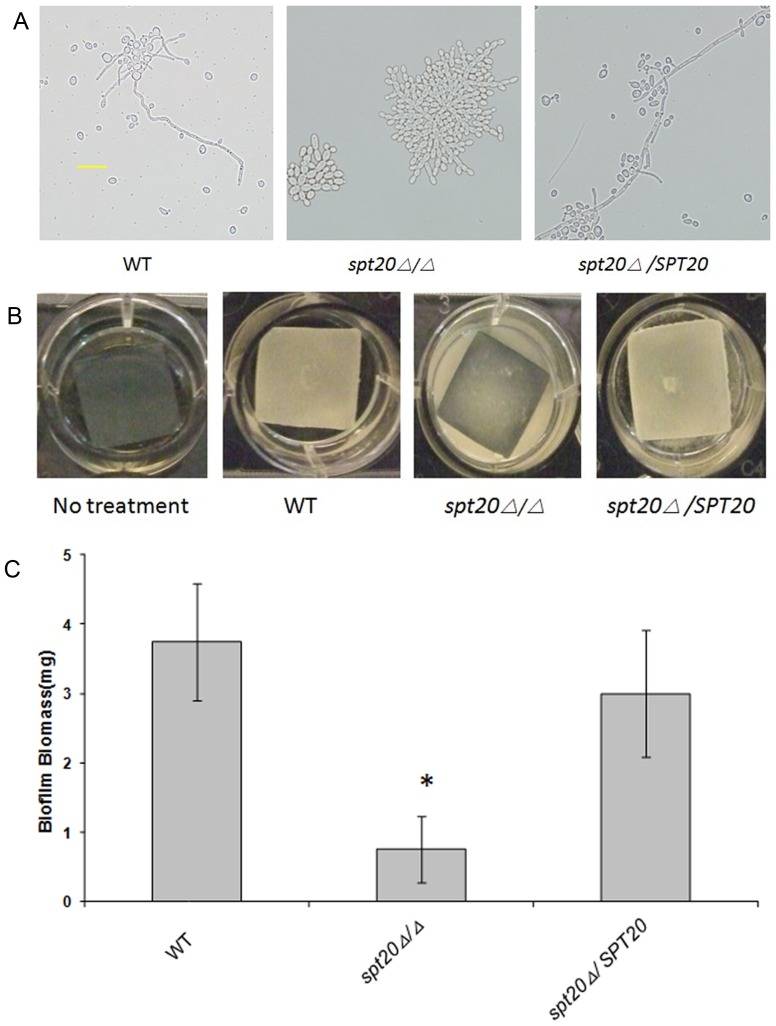
The visible difference in hyphae formation observed in the C. elegans infection assay and the lack of filaments in the infected kidney tissue, suggests that SPT20 is needed for hyphae formation, and plays an important role in biofilm formation. We confirmed this role by studying hyphal development and biofilm formation in vitro. The results showed that the spt20Δ/Δ mutant was defective in hyphal ( Figure 7A ) and biofilm formation (p < 0.05; Figure 7B and 7C ) compared to either the wild-type strain or the re-integrated strain.
Figure 7. Hyphae and biofilm defects of the spt20Δ/Δ.
(A) C. albicans strains SC5314, spt20Δ/Δ and spt20Δ/SPT20 were grown in Spider medium for 20 h at 37°C and examined at ×400 magnification. Compared to the C. albicans SC5314, spt20Δ/Δ showed attenuated hyphae formation in vitro (also showed attenuated hyphal formation in vivo, see Figure 1 ). This phenotype was restored in the spt20Δ/SPT20 reintegrated strain. The scale bar given in panel A represents 20 µm. (B) Strains were grown under in vitro biofilm assay conditions for 60 h. Biofilm formed on the silicone surface of the SC5314 and spt20Δ/SPT20 strains but failed to form on silicone sheets seeded with spt20Δ/Δ. (C) Dry weight of the biofilm biomass. Standard deviations are depicted and based on 6–8 silicone pad measurements. *p < 0.05.
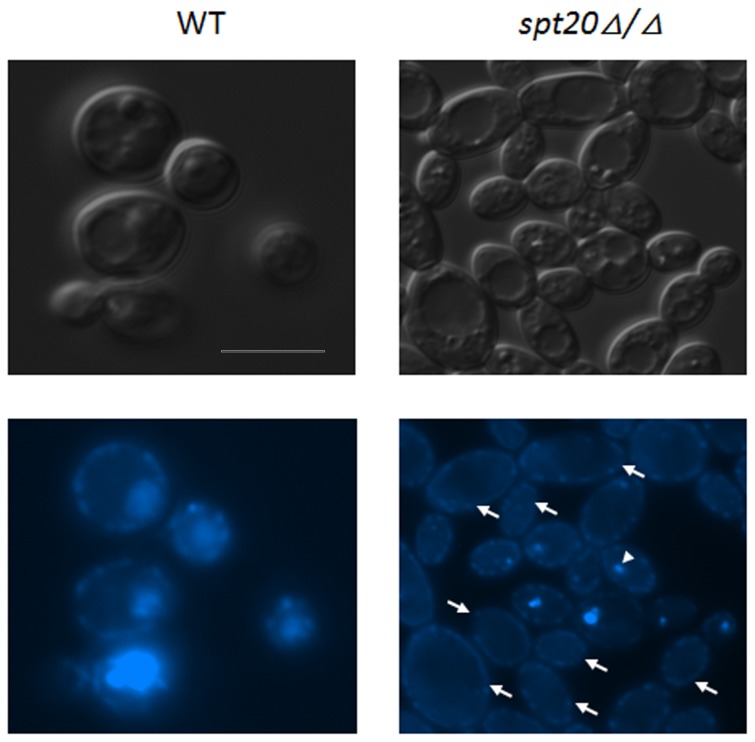
Defects in nuclear localization
In the“snow-flake”morphology, clustered cells appear to have defects in the ability of the daughter cells to separate from the mother cells, thus causing the clustering phenotype. We investigated whether the cell complexes all have nuclei using 6-diamidino-2-phenylindole dihydrochloride (DAPI) to evaluate the presence of nuclei in the complexes of the clustered cells. Interestingly, many gathered cells show defects in the location of nuclei ( Figure 8 ). This indicates that the nucleus is not properly dividing or being distributed to the dividing cells. Overall, the lack of nuclei in some gathered compartments indicates a role for SPT20 in nuclear division. The lack of nuclear material in complexes of the clustered cells may also be associated with the cytokinesis between the cell complexes of the clustered SPT20 mutant cells.
Figure 8. Nuclear defects in the spt20Δ/Δ cells.
The light field image on the upper right shows a cluster of cells. The image on the lower right is a DAPI staining of the same cell cluster complex. The arrowhead indicates the location of the nucleus. Arrows indicate the cell complexes that lack a nucleus. Bar = 5 µm.
Discussion
In this report, we show that in C. albicans, SPT20 plays a role in maintaining cell wall integrity. The defects in the cell wall are coupled with altered morphology and the inability of spt20Δ/Δ cells to form hyphae and generate a biofilm. Strikingly, these defects translate into reduced pathogenicity of the strain in multiple infection models.
The defects associated with cell morphology are characterized by a clustering effect, suggesting that spt20Δ/Δ cells are defective for cytokinesis, the mother and daughter cells do not separate properly. Moreover, some of the spt20Δ/Δ cells lacked nuclei therefore it is more accurate to describe some of the cells as “compartments” or “complexes”. These clusters are able to enlarge in the number of compartments that comprise the overall entity over time through the continuation of cell proliferation without complete separation. Interestingly, the SPT20 mutant daughter cells of S. cerevisiae do not separate well from the mother cell [26], but the cells appears to have a nucleus [27], an aspect that differs in the C. albicans clustered complexes.
In S. cerevisiae, the growth rate of the deletion mutant is slow, and the SPT20 mutant displays an elongated and irregularly bud morphology instead of the normal round morphology of wild-type cells [14], [28]. Although both fungi display morphological defects they differ in the rate of growth. Our examination of growth on solid media indicated that the SPT20 mutant strain grew at rates comparable to the wild-type strain.
Adapting to variations in the environment is a basic feature for microorganism survival and, through adaptive evolutionary mechanisms, this process is under strict regulation and control. In C. albicans, we found that SPT20 is involved with the ability of the fungal cells to adapt to compounds such as SDS that are toxic to the cell wall as well as to high osmolarity caused by sodium chloride. Similarly, in S. cerevisiae SPT20 is essential for cell survival during environmental stress conditions [26], [29]. It's worth noting that SPT20 maintains the structural integrity and the function of SAGA complex[30]–[33]. SPT20 and SAGA complex in S. cerevisiae and C. albicans are also highly conserved [29]. In S. cerevisiae, HOG1 signal transduction pathways can regulate intracellular synthesis and storage of glycerol, which resulted in increased intracellular osmotic pressure to improve the adaption to high osmolarity environments [34], [35]. SAGA is necessary for cell survival at hyperosmotic conditions [36]. In the hyperosmotic environment, it activates the HOG1 MAP kinase and is transferred from the cytoplasm to the nucleus, phosphorylating Sko1, and forms the Sko1-Cyc8-Tup1-Hog complex that recruits SAGA to the modified chromosome so that the RNA polymerase II binding to the promoter to initiate transcription [36].
Cell wall sensitivity to stress appears to be a conserved attribute between SPT20 of S. cerevisiae and C. albicans. In S. cerevisiae, SPT20 is hypersensitive to caspofungin [37], a drug that targets the fungal glucan synthase, important for the cell wall. Amphotericin B and azole target (directly or indirectly) the cell membrane and not the cell wall. In C. albicans, Vandeputte and colleagues found that SPT20 the C. albicans mutant is more susceptible to 3 antifungal agents: amphotericin B, fluconazole and caspofungin than the wild-type strain, no alteration in sensitivity to flucytosine, a drug that targets the nuclear membrane [38]. This finding prompted us to confirm the spt20Δ/Δ mutation sensitivity to the antifungal agents using our generated strain. As shown in Figure 4D, our results were consistent with what was report by Vandeputte et al. The spt20Δ/Δ strain was more susceptible to compounds that targeted that cell membrane or wall than nuclear targeted 5FC. This suggests that the spt20Δ/Δ membrane defects are specifically associated with the cell wall or cell membrane and not to membranes in general and does not affect the nuclear membrane.
In S. cerevisiae, SPT20 is believed to be required for the overall structural integrity and function of SAGA, a complex that facilitates the binding of TATA-binding proteins, because there is no intact SAGA complex in SPT20 deletion mutants[39]-[42].SAGA controls 10% of all adjustable genes, and is known to be stress inducible [43]. Researches have shown that the SAGA complex of C. albicans is similar to the SAGA complex of S. cerevisiae [26]. Environmental stress conditions that influence gene expression that tends to be SAGA dominated include: heat, oxidation, acidity and nitrogen starvation, conditions that also influence the cell wall integrity pathway. In S. cerevisiae, SAGA regulates expression of FKS1 and ERG11, involved in cell wall glucan synthase and ergosterol synthesis, respectively [43].
An important part of C. albicans' pathogenicity is its ability to form hyphae and generate biofilm, which play important roles in the colonization of medical devices. The hyphal form belongs to the overall factor of C. albicans' virulence by invading epithelial and endothelial cells and evading phagocytes that cause the release of hydrolytic enzymes, forming biofilms that colonize medical devices, and lead to tissue damage. This process requires active changes to the cell wall in order to accommodate expanding volume, relying on actin to maintain the cytoskeletal structure during the process of hyphae and pseudohyphae formation. Our investigation of Spt20 suggests that it plays a role in biofilm formation and filamentation. The spt20Δ/Δ showed attenuated hyphal formation within the clustered morphology. The defective filaments were unable to support the formation of a biofilm, as assessed by the reduced biofilm mass. Importantly, the inability to form hyphae was reflected in the pathogenicity studies by the inability to produce hyphae that could penetrate the worm cuticle in the C. elegans infection model or the lack of hyphae penetration the kidney tissue in the murine model. The aspect of pathogenicity is a differing attribute between S. cerevisiae and C. albicans, where in the former does not cause disease but the latter can be an opportunistic pathogen. Thus the role that SPT20 plays in C. albicans can contribute to virulence, a marked difference from the contribution made to S. cerevisiae.
Although S. cerevisiae do not form hyphae for the invasion of host tissues, filaments are formed during other biological functions. In S. cerevisiae, the ability of SPT20 mutant to form filaments is impaired [14], [26], [44]–[46]. Combined with our results, the data strongly indicates that SPT20 acts as an activator of filamentous and biofilm growth. It may affect the expression of hyphal-specific genes. In addition to SPT20, several SAGA related genes such as SPT6, ADA2, SPT3 have shown to be related for the filamentous growth of C. albicans. The genes SPT6 and ADA2 play a positive role in filamentous growth [18], [47], while SPT3 plays a negative role [26]. Both ADA2 and SPT3 play an important role in pathogenicity [18], [26].
In this work, we show that C. albicans SPT20 is associated with virulence. It is likely that the defects in hyphae and biofilm formation of the spt20Δ/Δ mutant and the sensitivity to cell wall and osmotic stresses may explain the loss of virulence in both invertebrates and mammals. Moreover, SPT20 plays a vital role in cell morphology and distribution of nuclear material, which may lead to the defects in filamentation and biofilm formation directly when this gene is deleted.
Supporting Information
The expression of CHT3 was not affected by spt20Δ/Δ . CHT3 mRNA levels were tested by RT-PCR in wild-type strain and spt20Δ/Δ strain. No significant difference between the two strains was found(P>0.05).
(TIF)
Primers for the disruption and reconstitution of SPT20 .
(DOCX)
Primers used to identify replacements of the SPT20 allels.
(DOCX)
Acknowledgments
We would like to thank Dr. Joachim Morschhauser (Institut fur Molekulare Infektionsbiologie, University of Wurzburg, Germany) for kindly providing the plasmid pSFS2. The Candida albicans mutant library was obtained from the Fungal Genetics Stock Center (Kansas City, MO).
Funding Statement
This work was supported by the National Institutes of Health through an R01 award (AI075286), and an R21 award (AI070569) to Eleftherios Mylonakis, and Science and Technology Planning Project of Guangdong Province, P. R. China (2011B080701091) to Xiaojiang Tan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Spanakis EK, Kourkoumpetis TK, Livanis G, Peleg AY, Mylonakis E (2010) Statin therapy and decreased incidence of positive Candida cultures among patients with type 2 diabetes mellitus undergoing gastrointestinal surgery. Mayo Clin Proc 85: 1073–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Douglas LJ (2003) Candida biofilms and their role in infection. Trends Microbiol 11: 30–36. [DOI] [PubMed] [Google Scholar]
- 3. Wenzel RP, Gennings C (2005) Bloodstream infections due to Candida species in the intensive care unit: identifying especially high-risk patients to determine prevention strategies. Clin Infect Dis 41 Suppl 6S389–S393. [DOI] [PubMed] [Google Scholar]
- 4. Colombo AL, Nucci M, Park BJ, Nouer SA, Arthington-Skaggs B, et al. (2006) Epidemiology of candidemia in Brazil: a nationwide sentinel surveillance of candidemia in eleven medical centers. J Clin Microbiol 44: 2816–2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sudbery P, Gow N, Berman J (2004) The distinct morphogenic states of Candida albicans. Trends Microbiol 12: 317–324. [DOI] [PubMed] [Google Scholar]
- 6. Berman J, Sudbery PE (2002) Candida Albicans: a molecular revolution built on lessons from budding yeast. Nat Rev Genet 3: 918–930. [DOI] [PubMed] [Google Scholar]
- 7. Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, et al. (1997) Nonfilamentous C. albicans mutants are avirulent. Cell 90: 939–949. [DOI] [PubMed] [Google Scholar]
- 8. Kobayashi SD, Cutler JE (1998) Candida albicans hyphal formation and virulence: is there a clearly defined role? Trends Microbiol 6: 92–94. [DOI] [PubMed] [Google Scholar]
- 9. Liu H, Kohler J, Fink GR (1994) Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266: 1723–1726. [DOI] [PubMed] [Google Scholar]
- 10. Fanning S, Mitchell AP (2012) Fungal biofilms. PLoS Pathog 8: e1002585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kelly MT, MacCallum DM, Clancy SD, Odds FC, Brown AJ, et al. (2004) The Candida albicans CaACE2 gene affects morphogenesis, adherence and virulence. Mol Microbiol 53: 969–983. [DOI] [PubMed] [Google Scholar]
- 12.Brown A HKGN (2012) Stress responses in Candida. Candida and Candidiasis.
- 13. Marcus GA, Horiuchi J, Silverman N, Guarente L (1996) ADA5/SPT20 links the ADA and SPT genes, which are involved in yeast transcription. Mol Cell Biol 16: 3197–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Roberts SM, Winston F (1996) SPT20/ADA5 encodes a novel protein functionally related to the TATA-binding protein and important for transcription in Saccharomyces cerevisiae. Mol Cell Biol 16: 3206–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Horiuchi J, Silverman N, Pina B, Marcus GA, Guarente L (1997) ADA1, a novel component of the ADA/GCN5 complex, has broader effects than GCN5, ADA2, or ADA3. Mol Cell Biol 17: 3220–3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee TI, Causton HC, Holstege FC, Shen WC, Hannett N, et al. (2000) Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 405: 701–704. [DOI] [PubMed] [Google Scholar]
- 17. Reuss O, Vik A, Kolter R, Morschhauser J (2004) The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341: 119–127. [DOI] [PubMed] [Google Scholar]
- 18. Pukkila-Worley R, Peleg AY, Tampakakis E, Mylonakis E (2009) Candida albicans hyphal formation and virulence assessed using a Caenorhabditis elegans infection model. Eukaryot Cell 8: 1750–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Walther A, Wendland J (2003) An improved transformation protocol for the human fungal pathogen Candida albicans. Curr Genet 42: 339–343. [DOI] [PubMed] [Google Scholar]
- 20. Nobile CJ, Andes DR, Nett JE, Smith FJ, Yue F, et al. (2006) Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog 2: e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fuchs BB, O'Brien E, Khoury JB, Mylonakis E (2010) Methods for using Galleria mellonella as a model host to study fungal pathogenesis. Virulence 1: 475–482. [DOI] [PubMed] [Google Scholar]
- 22. Fuchs BB, Eby J, Nobile CJ, El KJ, Mitchell AP, et al. (2010) Role of filamentation in Galleria mellonella killing by Candida albicans. Microbes Infect 12: 488–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang Y, Jia XM, Jia JH, Li MB, Cao YY, et al. (2009) Ascorbic acid decreases the antifungal effect of fluconazole in the treatment of candidiasis. Clin Exp Pharmacol Physiol 36: e40–e46. [DOI] [PubMed] [Google Scholar]
- 24. Kopecka M, Gabriel M, Takeo K, Yamaguchi M, Svoboda A, et al. (2001) Microtubules and actin cytoskeleton in Cryptococcus neoformans compared with ascomycetous budding and fission yeasts. Eur J Cell Biol 80: 303–311. [DOI] [PubMed] [Google Scholar]
- 25. Fuchs BB, Tang RJ, Mylonakis E (2007) The temperature-sensitive role of Cryptococcus neoformans ROM2 in cell morphogenesis. PLoS One 2: e368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Laprade L, Boyartchuk VL, Dietrich WF, Winston F (2002) Spt3 plays opposite roles in filamentous growth in Saccharomyces cerevisiae and Candida albicans and is required for C. albicans virulence. Genetics 161: 509–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chekanova JA, Abruzzi KC, Rosbash M, Belostotsky DA (2008) Sus1, Sac3, and Thp1 mediate post-transcriptional tethering of active genes to the nuclear rim as well as to non-nascent mRNP. RNA 14: 66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Watanabe M, Watanabe D, Nogami S, Morishita S, Ohya Y (2009) Comprehensive and quantitative analysis of yeast deletion mutants defective in apical and isotropic bud growth. Curr Genet 55: 365–380. [DOI] [PubMed] [Google Scholar]
- 29. Sellam A, Askew C, Epp E, Lavoie H, Whiteway M, et al. (2009) Genome-wide mapping of the coactivator Ada2p yields insight into the functional roles of SAGA/ADA complex in Candida albicans. Mol Biol Cell 20: 2389–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cosma MP, Tanaka T, Nasmyth K (1999) Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97: 299–311. [DOI] [PubMed] [Google Scholar]
- 31. Bhaumik SR, Green MR (2002) Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol Cell Biol 22: 7365–7371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Daniel JA, Grant PA (2007) Multi-tasking on chromatin with the SAGA coactivator complexes. Mutat Res 618: 135–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Koutelou E, Hirsch CL, Dent SY (2010) Multiple faces of the SAGA complex. Curr Opin Cell Biol 22: 374–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Saito H, Tatebayashi K (2004) Regulation of the osmoregulatory HOG MAPK cascade in yeast. J Biochem 136: 267–272. [DOI] [PubMed] [Google Scholar]
- 35. Schwartz MA, Madhani HD (2004) Principles of MAP kinase signaling specificity in Saccharomyces cerevisiae. Annu Rev Genet 38: 725–748. [DOI] [PubMed] [Google Scholar]
- 36. Proft M, Struhl K (2002) Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol Cell 9: 1307–1317. [DOI] [PubMed] [Google Scholar]
- 37. Lesage G, Sdicu AM, Menard P, Shapiro J, Hussein S, et al. (2004) Analysis of beta-1,3-glucan assembly in Saccharomyces cerevisiae using a synthetic interaction network and altered sensitivity to caspofungin. Genetics 167: 35–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vandeputte P, Pradervand S, Ischer F, Coste AT, Ferrari S, et al. (2012) Identification and functional characterization of Rca1, a transcription factor involved in both antifungal susceptibility and host response in Candida albicans. Eukaryot Cell 11: 916–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Daniel JA, Torok MS, Sun ZW, Schieltz D, Allis CD, et al. (2004) Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J Biol Chem 279: 1867–1871. [DOI] [PubMed] [Google Scholar]
- 40. Hall DB, Struhl K (2002) The VP16 activation domain interacts with multiple transcriptional components as determined by protein-protein cross-linking in vivo. J Biol Chem 277: 46043–46050. [DOI] [PubMed] [Google Scholar]
- 41. Larschan E, Winston F (2001) The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev 15: 1946–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, et al. (2003) Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev 17: 2648–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Huisinga KL, Pugh BF (2004) A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell 13: 573–585. [DOI] [PubMed] [Google Scholar]
- 44. Lo WS, Dranginis AM (1996) FLO11, a yeast gene related to the STA genes, encodes a novel cell surface flocculin. J Bacteriol 178: 7144–7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Madhani HD, Styles CA, Fink GR (1997) MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell 91: 673–684. [DOI] [PubMed] [Google Scholar]
- 46. Morillon A, Benard L, Springer M, Lesage P (2002) Differential effects of chromatin and Gcn4 on the 50-fold range of expression among individual yeast Ty1 retrotransposons. Mol Cell Biol 22: 2078–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Al-Rawi N, Laforce-Nesbitt SS, Bliss JM (2010) Deletion of Candida albicans SPT6 is not lethal but results in defective hyphal growth. Fungal Genet Biol 47: 288–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The expression of CHT3 was not affected by spt20Δ/Δ . CHT3 mRNA levels were tested by RT-PCR in wild-type strain and spt20Δ/Δ strain. No significant difference between the two strains was found(P>0.05).
(TIF)
Primers for the disruption and reconstitution of SPT20 .
(DOCX)
Primers used to identify replacements of the SPT20 allels.
(DOCX)