Significance
Polycystic liver disease (PCLD) is an autosomal dominantly inherited disorder characterized by multiple fluid-filled hepatic cysts that may cause an extremely enlarged liver. PCLD is genetically heterogeneous, and mutations in PRKCSH and SEC63 are present in ∼25% of PCLD patients. This research identifies four unique LRP5 mutations in four independent families that were all located at highly conserved protein domains. Functional activity analyses suggest that mutant LRP5 reduces wingless (Wnt) signal activation. This study suggests that imbalanced Wnt signaling is related to hepatic cyst formation.
Keywords: gene identification, β-catenin pathway
Abstract
Polycystic livers are seen in the rare inherited disorder isolated polycystic liver disease (PCLD) and are recognized as the most common extrarenal manifestation in autosomal dominant polycystic kidney disease. Hepatic cystogenesis is characterized by progressive proliferation of cholangiocytes, ultimately causing hepatomegaly. Genetically, polycystic liver disease is a heterogeneous disorder with incomplete penetrance and caused by mutations in PRKCSH, SEC63, PKD1, or PKD2. Genome-wide SNP typing and Sanger sequencing revealed no pathogenic variants in hitherto genes in an extended PCLD family. We performed whole-exome sequencing of DNA samples from two members. A heterozygous variant c.3562C > T located at a highly conserved amino acid position (p.R1188W) in the low density lipoprotein receptor-related protein 5 (LRP5) gene segregated with the disease (logarithm of odds score, 4.62) but was not observed in more than 1,000 unaffected individuals. Screening of LRP5 in a PCLD cohort identified three additional mutations in three unrelated families with polycystic livers (p.V454M, p.R1529S, and p.D1551N), again all undetected in controls. All variants were predicted to be damaging with profound structural effects on LRP5 protein domains. Liver cyst tissue and normal hepatic tissue samples from patients and controls showed abundant LRP5 expression by immunohistochemistry. Functional activity analyses indicated that mutant LRP5 led to reduced wingless signal activation. In conclusion, we demonstrate that germ-line LRP5 missense mutations are associated with hepatic cystogenesis. The findings presented in this study link the pathophysiology of PCLD to deregulation of the canonical wingless signaling pathway.
Polycystic liver diseases (PLD) consist of a group of inherited disorders characterized by abnormal proliferation and differentiation of the bile duct epithelium (1). Ductal plate malformation results in development of multiple fluid-filled cysts spread throughout the liver parenchyma. Progressive hepatic cystogenesis and cyst growth cause hepatomegaly and symptoms such as abdominal distension and pain, pyrosis, anorexia, and dyspnea.
Congenital PLD are clinically heterogeneous, and two major types can be distinguished. Autosomal dominant polycystic kidney disease (ADPKD) is a potentially lethal condition afflicting about 1:400–1:1,000 persons in the United States (2). Patients may develop end-stage renal disease resulting from polycystic kidneys, and 83% have the simultaneous presence of multiple hepatic cysts (3). Formal diagnostic criteria for ADPKD include age, number of renal cysts for each kidney, and family history of renal disease (4, 5). Isolated polycystic liver disease (PCLD) shares the phenotype of a polycystic liver but is distinct from ADPKD because renal disease is absent (6). Some patients may, however, have sporadic renal cysts. Diagnosis of PCLD is made by a family history consistent with autosomal dominant inheritance and the presence of at least one (<40 y) or more than three (>40 y) hepatic cysts (7).
The natural course of PCLD and ADPKD depends on a spectrum of factors that include age, sex, anticonceptives, pregnancy, and mutation(s) (8–10). Classical linkage analysis has identified four genes that underlie both inherited disorders. Mutations in polycystic kidney disease 1 or 2 (PKD1 or PKD2) genes (NM_601313 and NM_173910, respectively), which encode polycystin-1 and polycystin-2, are responsible for almost all ADPKD cases (11–13). Both proteins are located in the cilium and are important in renal tubular cell morphogenesis and signaling functions, including the wingless (Wnt) signaling pathway (β-catenin pathway). Mutations in protein kinase C substrate 80K-H (PRKCSH) and Saccharomyces cerevisiae homolog 63 (SEC63) are linked to PCLD (NM_002743.2 and NM_007214.4, respectively), and both gene products are located to the endoplasmic reticulum and are involved in processing and folding of glycosylated proteins (14–16). In addition, the canonical Wnt signaling may be deregulated by the interaction partner nucleoredoxin of the Sec63 protein during oxidative stress (17). In contrast to ADPKD, only 25% of PCLD cases can be explained by mutations in already known genes (18), indicating that mutations in several other as yet unidentified genes are involved in this disease. We hypothesized that another PCLD gene is involved in an extended PCLD family negative for the known disease genes and performed whole-exome sequencing. This unique approach has proven to be successful for the identification of the genetic cause of other Mendelian disorders (19–21).
Results
Extended Polycystic Liver Disease Family.
We assessed a 49-y-old female (III/18) by clinical history taking and physical and radiological examination. She presented to us with a severe symptomatic polycystic liver without renal cysts and complained of abdominal distension, pain, dyspnea, and pyrosis. There was no history of renal disease, hypertension, or intracranial aneurysms. She used contraceptives for a total of 30 y and had two pregnancies. CT scanning revealed numerous cysts with diameters of 5–18 cm in liver segments I–III and VII. Despite aspiration sclerotherapy and laparoscopic deroofing of the liver cysts, her symptoms persisted, and she enrolled in a clinical trial for long-acting octreotide treatment (22). We expanded our clinical analysis and identified a 40-member Dutch family with three generations of early- and late-onset cystogenesis inherited in an autosomal dominant fashion (PCLD-1). Next, we studied 19 individuals with cystogenesis, of which 16 were affected with a polycystic liver according to the Reynolds criteria (5, 7), and three members were affected with renal cysts that came from the 40-member Dutch family (Fig. 1A). The proband (III/18) possessed no pathogenic variants affecting any of the known PCLD or ADPKD genes.
Fig. 1.
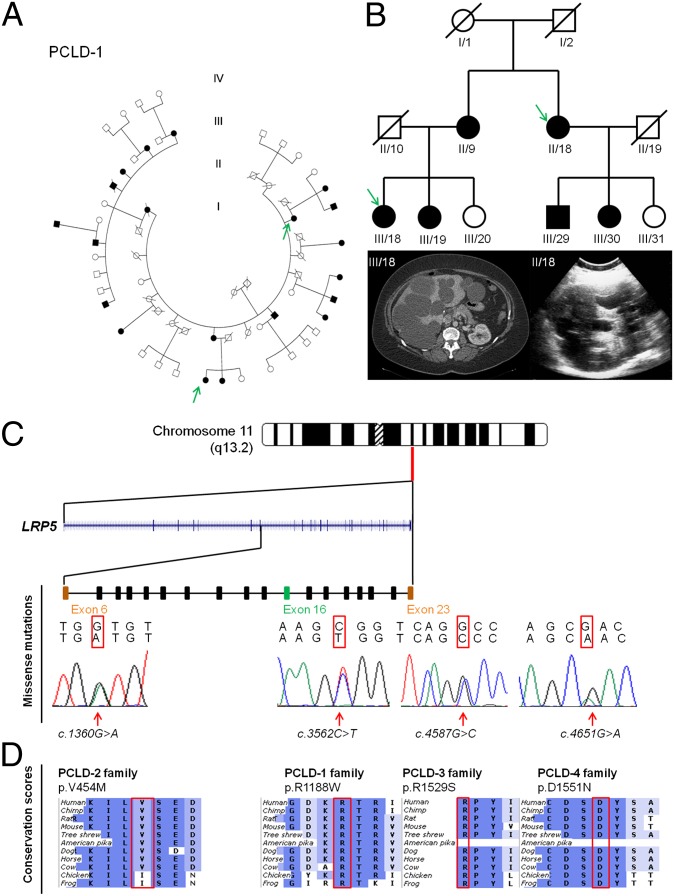
Identification of LRP5 variants p.R1188W in an extended Dutch PCLD-1 family (A) and three additional LRP5 variants in three unrelated PCLD families. Generations are denoted with Roman numerals, and individuals are numbered in a counterclockwise way. Squares indicate male sex, and circles indicate female sex. Solid symbols denote affected individuals, and open symbols are individuals without or unknown for PCLD. A slash indicates that the individual is deceased. (B) Simplified pedigree from PCLD-1 family with the clinical features by abdominal CT scanning and ultrasound of the liver of both probands (III/18 and II/18) in which whole-exome sequencing was performed (green arrow). (C) LRP5 is located at chromosome 11q13.2, and the sequence electropherogram shows heterozygous germ-line mutations. (D) All LRP5 missense mutations were located at highly evolutionary conserved amino acid regions with ortholog proteins from human to frog.
Whole-Exome Sequencing Identifies Pathogenic LRP5 Variant.
We performed exome sequencing on genomic DNA in two members (III/18 and II/18) from the PCLD-1 family with an advanced polycystic liver phenotype (Fig. 1B). Exome capture and sequencing were performed using the Agilent SureSelect target enrichment system with SOLiD4 sequencing. We identified 24,178 and 25,332 genetic variants per proband, respectively (Tables S1 and S2). Variants were annotated by a bioinformatics pipeline as described previously (19, 20). Variant follow-up required the presence of at least five unique variant reads (different start sites), and the variant had to be present in at least 20–80% of all reads suggestive of heterozygous changes in a dominant model of disease. Next, we prioritized variants based on predicted amino acid consequences and overlap with common variation (presence in dbSNP v134 and/or an in-house database containing 1,300 analyzed exomes of predominantly European ancestry). We identified 11 unique nonsynonymous variants present in both affected relatives (Table S3). All 11 variants were tested for segregation in both probands and all other family members by Sanger sequencing. The only private nonsynonymous variant that cosegregated completely with the disease was a missense mutation (c.3562C > T with evolutionary conserved amino acid change p.R1188W) located on chromosome 11q13.2 in the low-density lipoprotein receptor-related protein 5 (LRP5) gene (NM_002335.2; Fig. 1 C and D). We checked sequence coverage and excluded the presence of potential pathogenic variants in the 2 Mb surrounding LRP5. All 19 individuals with cystogenesis possessed LRP5 c.3562C > T (Table S4 and Fig. S1). Analysis of the sequence data revealed linkage at the LRP5 locus with the disease with a significant logarithm of odds (LOD) score of 4.62 (Fig. S2). This LRP5 variant (c.3562C > T) was not detected in genome-wide sequence data from the 1000 Genomes Project (23), 6,500 individuals from the National Heart, Lung, and Blood Institute Exome Sequencing Project (24), or exome data from 1,300 individuals of predominantly European ancestry sequenced in-house (19, 20) and excluded by Sanger sequencing in a control set of 1,000 Dutch DNAs of healthy, unrelated individuals. This LRP5 mutation affected a highly conserved amino acid and was predicted to be damaging by PolyPhen-2, MutPred, and sorting intolerant from tolerant (SIFT) models (Table 1).
Table 1.
Summary and in silico analysis of four LRP5 variants in polycystic liver disease
| Family ethnicity | Mutation (c.DNA) NM_002335.2 | Predicted effect on protein | PolyPhen2 | MutPred | SIFT |
| PCLD-1 Caucasian (Dutch) | c.3562C > T | p.R1188W | Probably damaging 1.00 | 0.552 | Deleterious 0.00 |
| PCLD-2 Caucasian (Dutch) | c.1360G > A | p.V454M | Possibly damaging 0.872 | 0.520 | Deleterious 0.01 |
| PCLD-3 Moroccan | c.4587G > C | p.R1529S | Possibly damaging 0.610 | 0.288 | Deleterious 0.00 |
| PCLD-4 Caucasian (Dutch) | c.4651G > A | p.D1551N | Probably damaging 0.999 | 0.235 | Deleterious 0.00 |
LRP5 is a known disease gene causing severe skeletal bone or retinal disorders and is associated with metabolic disease (Fig. S3). Therefore, we actively investigated the possible presence of subtle clinical signs in our family by slit-lamp eye examination. These investigations excluded familial exudative vitreoretinopathy (FEVR) in any of the family members (SI Text). We assessed bone density of the lumbar spine and left hip in 13 patients and 9 unaffected relatives. The lumbar T-score was lower in LRP5 mutation carriers but within the normal range, and no member had a bone density disorder. Routine laboratory testing, including renal parameters, did not reveal differences between individuals with and without the LRP5 mutation (Table 2). Therefore, other LRP5-related disorders were ruled out in this index family.
Table 2.
Baseline characteristics of PCLD-1 family
| Characteristics | No hepatic cystogenesis and no LRP5 mutation (n = 9) | Hepatic cystogenesis and LRP5 mutation (n = 13) | P value |
| Age (y) | 55 ± 12 | 56 ± 13 | NS |
| Female sex, no. (%) | 6 (67) | 9 (69) | NS |
| Bone density: Hip | |||
| T-score | −0.19 ± 0.78 | −0.08 ± 0.93 | NS |
| Z-score | 0.69 ± 0.68* | 0.95 ± 0.94 | NS |
| Bone density: L1-L2-L3-L4 | |||
| T-score | 1.18 ± 0.95* | 0.19 ± 0.88 | 0.042 |
| Z-score | 1.81 ± 0.89* | 1.18 ± 1.03 | NS |
| Creatinin (µmol/L) | 69 ± 14 | 73 ± 20 | NS |
| Cholesterol (mmol/L) | 5.39 ± 0.71 | 5.44 ± 1.01 | NS |
| GFR (MDRD) (ml/min/1.73 m2) | 87 ± 13 | 85 ± 25 | NS |
| BMI (kg/m2) | 25.7 ± 2.5 | 26.4 ± 5.2 | NS |
| Triglycerides (mmol/L) | 1.35 ± 0.71 | 1.80 ± 0.93 | NS |
| HDL (mmol/L) | 1.34 ± 0.40 | 1.38 ± 0.32 | NS |
| LDL (mmol/L) | 3.44 ± 0.82 | 3.25 ± 0.78 | NS |
| Non HDL (mmol/L) | 4.06 ± 0.93 | 4.07 ± 0.93 | NS |
| HbA1C (%) | 5.5 ± 0.6 | 5.7 ± 0.8 | NS |
| HbA1C (mmol/mol) | 37 ± 6 | 39 ± 9 | NS |
NS, not significant.
There was one missing value because of degenerative abnormalities of the lumbar spine in an 85-y-old woman.
Additional LRP5 Variants in Polycystic Liver Disease.
To confirm our results, we sequenced LRP5 in a cohort of 150 unrelated PCLD probands without PRKCSH, SEC63, or PKD2 gene mutations. We identified two additional PCLD families (one Dutch and one Moroccan) and one Dutch PCLD singleton case with private LRP5 missense variants. Mutations c.1360G > A (p.V454M) and c.4587G > C (p.R1529S) segregated with the disease in both families, and unaffected relatives of the singleton case (c.4651G > A; p.D1551N) did not carry the mutation (Fig. S4).
Mutation c.1360G > A (p.V454M) was present in an 86-y-old polycystic liver patient. Her mutation-positive daughter had several bilateral renal cysts and small hepatic cysts on CT scanning. The daughter died as a result of a metastasized liposarcoma at the age of 49. A 43-y-old Moroccan female had multiple dominant hepatic cysts without renal disease. Her 71-y-old mother had several hepatic cysts without renal cysts. Both carried the c.4587G > C (p.R1529S) mutation in the LRP5 gene. A fourth mutation [c.4651G > A (p.D1551N)] was present in a 65-y-old male with a polycystic liver and three renal cysts. There was no evidence for renal failure. The phenotype of his deceased parents was unknown, and both his healthy children were without hepatic or renal cysts and did not harbor the LRP5 mutation. None of the LRP5 variants were detected in chromosomes from healthy unrelated and ethnically matched controls (Dutch, n = 1,000; Moroccan, n = 525) nor were they present in the in-house or online exome sequencing datasets. In addition, all four LRP5 missense variants affected highly conserved amino acids and again were predicted to be damaging or deleterious by PolyPhen-2, MutPred, and SIFT (Table 1).
LRP5 Expression in Liver Cyst Tissues.
LRP5 has a wide tissue distribution, including liver and kidney, and expression has been reported in Kupffer cells, macrophages, stellate cells, cholangiocytes, and hepatocytes (25). In line with Northern blotting experiments (26), we found abundant LRP5 protein presence in normal liver tissues and relevant for PCLD. In the affected proband (III/18), we observed intense LRP5 staining of cyst-lining epithelium and bile ducts. The intensity of LRP5 expression was comparable in cyst tissue sections derived from a PRKCSH mutant and to bile duct epithelium (Fig. 2 A and B). This abundant LRP5 expression in cyst tissue indicates that there is no significant loss of structural LRP5 protein from relevant tissue in LRP5 carriers.
Fig. 2.
Functional and structural analyses of LRP5 variants in polycystic liver disease. (A) Immunohistochemistry of liver cyst tissue from proband III/18 of PCLD-1 family with LRP5 mutation c.3562C > T (p.R1188W). The cyst lining cholangiocytes present positive staining for LRP5 compared with the negative control next. (B) PCLD patient with a PRKCSH c.1341–2A > G mutation shows similar staining of liver cyst tissue and expression of LRP5 compared with A. (C) Localization of LRP5 protein was analyzed by immunofluorescence microscopy. CHO cells were transfected with constructs expressing LRP5WT, (D) LRP5R1188W, or LRP5D1551N and compared with negative controls. No differences in LRP5 localization between all constructs was detected. (E) Presentation of the human LRP5 protein and homology modeling of the β-propeller domains highlighting the amino acid changes to emphasize the impact on the configuration and surrounding protein domains. (F) Homology modeling of the LRP5 domains and detailed view of the amino acid changes shows extracellular LRP5 mutation (*indicated) p.R1188W located at the sixth blade of the fourth β-propeller domain, and LRP5 mutation p.V454M at the third blade of the second β-propeller domain. Both intracellular mutations p.R1529S and p.D1551N are located between PPPSP motifs A and B and PPPSP motifs B and C, respectively. (G) CHO cells were transiently transfected with empty, WT, or mutant LRP5 vector. Whole cell lysate was analyzed by Western blotting using the V5 antibody and anti–β-actin. LRP5 protein expression levels (normalized to β-actin) are similar between LRP5WT and both mutant constructs LRP5R1188W and LRP5D1551N. (H) Canonical Wnt signaling activity was analyzed by firefly luciferase activity and normalized to renilla luciferase activity with (in gray) or without addition of 250 ng/mL Wnt3a. All LRP5 constructs showed a significant increase in Wnt signaling activity compared with the empty vector (P < 0.0001). Both LRP5 mutants showed a decreased Wnt3a-induced signal activity (*P < 0.001; **P < 0.0001). Activity of LRP5D1551N without Wnt3a was significantly decreased compared with LRP5WT.
For analysis of structural effects of the LRP5 variants, we generated separate models for these domains (Fig. 2 E and F and Table S5) (27). The human LRP5 protein contains 1,615 amino acids and includes a long extracellular region, a single-span transmembrane region, and a relatively short (208 amino acids) intracellular region. The extracellular region consists of four β-propeller domains with subsequent epidermal growth factor (EGF)-like domains. Near the transmembrane region are three LDL receptor class A repeats, whereas on the cytoplasmatic side, five PPPSP motifs are present. The β-propeller domains all consist of six segments of which most carry a characteristic YWTD motif. Arginine 1188 creates hydrogen bonds and ionic interactions in the core of the β-propeller structure, which is predicted to be lost by the tryptophan mutation (p.R1188W). Additionally, the larger tryptophan side-chain will cause steric clashes, which will most likely disturb the whole β-propeller domain. Valine 454 induces several hydrophobic interactions but is also partly exposed to the solvent. Mutant methionine is predicted not to have major structural effects as both amino acids are hydrophobic, and the methionine side-chain appears to fit at this position (p.V454M). Both p.R1529S and p.D1551N mutations are intracellularly located between PPPSP motifs, and an intracellular homology model is absent.
Reduced Activation of Canonical Wnt Signaling.
To identify the underlying mechanism of mutated LRP5 on the Wnt pathway, we conducted luciferase activity assays with LRP5WT, one extracellularly (LRP5R1188W) and one intracellularly located (LRP5D1551N) mutant, and an empty expression vector as a control. Immunofluorescence imaging in WT and both mutant constructs presented similar localization of LRP5 (Fig. 2 C and D). Western blots of the cell lysates showed comparable protein expression of all constructs (Fig. 2G). Overexpression of the LRP5 constructs in CHO cells increased basal and Wnt3a-induced luciferase activity compared with the empty vector (P < 0.0001). In the presence of Wnt3a, signal activation was significantly down-regulated by 30% and 45%, taking into account the basal activity in both LRP5 mutants compared with the LRP5WT. We also detected a significant decreased (P < 0.001) activity of LRP5 mutant p.D1551N without the presence of Wnt3a (Fig. 2H). Luciferase activity assays were repeated in HEK293 and human liver-derived H69 cells, where comparable significant results were obtained (Fig. S5).
Altered Expression Levels of Wnt Target Genes.
Subsequently, we conducted quantitative PCR (qPCR) experiments of transducers and transcription factors associated with the canonical Wnt signaling. Our analyses show that HEK293 cells transfected with mutant LRP5 led to altered expression levels of target genes compared with LRP5WT (Fig. S6). There was a significant increased gene expression of transducers adenomatous polyposis coli, glycogen synthase kinase 3β (GSK3β), and leucine-rich repeat-containing G-protein-coupled receptor 5 (LGR5) and transcription factor v-myc avian myelocytomatosis viral oncogene homolog (c-Myc) in LRP5R1188W or LRP5D1551N, compared with LRP5WT. Similarly, axis inhibitor-1 (AXIN-1), axis inhibitor-2 (AXIN-2), lymphoid enhancer-binding factor 1 (LEF1), SRY-box 9 (SOX9), fibroblast growth factor 18 (FGF18), and cyclin D1 (CCND1) were also up-regulated. In the presence of the extracellular regulator Wnt3a, expression levels of several Wnt signaling components changed (Fig. S7). Decreased expression levels were found for GSK3β, AXIN-1, AXIN-2, LGR5, c-Myc, CCND1, and LEF1 compared with LRP5WT in both mutated LRP5 constructs. These results are in line with the functional consequences of these LRP5 mutations as shown with luciferase activity assays (Figs. S8 and S9).
Discussion
This study identifies LRP5 as a novel gene associated with hepatic cystogenesis in patients clinically diagnosed with PCLD. The initial discovery was made in two affected relatives from an extended Dutch family by exome sequencing (21). A private missense mutation c.3562C > T (p.R1188W) in the LRP5 gene segregated with 18 affected relatives (>40 y), with a significant LOD score of 4.62. These findings are corroborated by the presence of three additional private missense LRP5 variants in two PCLD families and one PCLD singleton. All unique variants identified in LRP5 affected highly conserved amino acids and were predicted to be damaging or deleterious. The identification of LRP5 as a causative gene follows that of PRKCSH (15%) and SEC63 (6%) for isolated polycystic liver disease in a PCLD cohort (18).
LRP5 is a single-span transmembrane protein that acts as a coreceptor with Frizzled protein family members for transducing signals by Wnt proteins. Wnt signaling directs a number of fundamental physiological mechanisms such as cell proliferation, cell polarity, and cell fate determination during embryonic development (28). Until now, LRP5 variants were linked to a spectrum of Mendelian genetic diseases. LRP5-related disorders include autosomal dominant conditions with abnormal bone density, such as endosteal hyperostosis and osteosclerosis (29–32), but also eye disorders such as recessive and dominant forms of FEVR (29, 33, 34). Our findings expand the clinical spectrum of LRP5-associated phenotypes because there were no extrahepatic features in patients with LRP5 mutations in our studies. Specifically metabolic disorders, diabetes mellitus, skeletal bone, and retinal diseases were absent in the index family, and these were not reported for the other three families. We performed specific clinical investigations in extended family 1 and excluded FEVR in individuals with LRP5 germ-line mutations by slit-lamp eye examination. Bone density measurements ruled out bone diseases in individuals with and without PCLD. Hepatic or renal cystogenesis has not been observed in association with FEVR or with bone diseases.
Why different LRP5 mutations can result in such a wide spectrum of complex diseases that targets different tissues remains to be determined. LRP5 is detected by Northern blot analysis, immunohistochemistry, and in situ hybridization studies in several tissues including the liver and kidney (25, 26). Until now, LRP5 mutations were linked to pathological retina or bone development. In the extended PCLD-1 family, we identified the LRP5 c.3562C > T mutation in 22 individuals, of which 19 had hepatic and/or renal cystogenesis. Two members were too young (<40 y) to develop hepatic cysts, and one individual is an example of incomplete penetrance. Indeed, there was considerable clinical heterogeneity in affected members without complaints or patients with severe abdominal discomfort in the PCLD-1 family. This clinical heterogeneity has been described in families with PRKCSH- and SEC63-associated PCLD (14, 18), and penetrance of PRKCSH-associated PCLD is estimated at ∼80%. Intrafamilial phenotypic variability suggests that modifier genes and/or environmental factors play a major role in PCLD disease expression. Similarly, the clinical expression of LRP5-associated bone and eye diseases is highly variable (27).
There have been efforts to recapitulate the involvement of LRP5 mutations in a number of bone and eye disorders in mice. A targeted KO Lrp5tm1Jsak/Lrp5tm1Jsak mouse suffers from unspecified hepatobiliary abnormalities that might implicate phenotypic overlap with our observed human phenotypes (35). Human and mouse LRP5 share a high degree of amino acid identity to coreceptor low density lipoprotein receptor-related protein 6 (LRP6) and have a similar domain structure (36). Furthermore, there has been speculation about functional redundancy between both transmembrane coreceptors of the canonical Wnt signaling pathway (37). The homozygous KO Lrp6 mouse is embryonic lethal and possesses polycystic kidneys (37) compatible with the ADPKD phenotype. There is further experimental data to suggest that renal cystogenesis in ADPKD is linked to defective canonical Wnt signal transduction. Perturbations of polycystins cause inappropriate levels of β-catenin, and activation and inactivation of the Wnt signaling pathway are reported in different polycystic kidney disease mice models during embryonic or postnatal development (38–42). Our findings provide a direct link between canonical Wnt signaling and polycystic liver disease as Wnt signaling is reduced in mutant LRP5 compared with WT LRP5. A moderate reduction of signaling activity is in line with known LRP5 missense mutations in retina and bone disorders (43, 44). These observations support a role of an imbalanced canonical and noncanonical Wnt signal transduction in the pathogenesis of LRP5-associated polycystic diseases. We speculate that LRP5, along with other genes implicated in hepatic cystogenesis, PKD1, PKD2, PRKCSH, and SEC63, is part of the functional genetic network. Experimental studies have observed that reduced dosage of these gene products, and in particular polycystin-1, is required to cause cyst formation (45, 46).
The identification of LRP5 associated with hepatic cystogenesis affords a better understanding of the pathophysiology of PCLD. Positivity for immunoreactive LRP5 in cyst epithelium suggests that the protein is structurally intact. Functional assays in three different cell systems indicate that the mutation renders the protein to be less functional and that leads to inhibition of the canonical Wnt signaling (41, 42).
In conclusion, PCLD is a genetically heterogeneous disorder that may be caused by LRP5 gene mutations. Our results link hepatic cystogenesis to dysregulated canonical Wnt signaling and are in line with data that were generated from ADPKD models (41, 42). Polycystins modulate Wnt signaling during organ development and contribute to hepatic and renal cystogenesis (42, 47, 48). Our study provides evidence that mutations in LRP5 are related to hepatic cyst development and fuels the hypothesis that canonical Wnt signal transduction is important in polycystic diseases. It is possible that disruption of other (downstream) members of the Wnt signaling pathway may be associated with initiation of hepatic and renal cyst formation.
Materials and Methods
Human Subjects.
Ultrasound images of liver and kidneys were acquired using a 3.6-MHz general purpose clinical echo system (Acuson ×150; SiemensAG) equipped with a curved linear array transducer. Blood samples were collected from all 40 subjects, and DNA was extracted from blood leukocytes using the HP-PCR Template Preparation kit (Roche Applied Science). Formalin-fixed paraffin-embedded liver cyst tissue specimens were available from proband III/18 after laparoscopic cyst fenestration. DNA samples from 1,000 Dutch and 525 Moroccan healthy unrelated individuals were used as controls. All subjects provided written informed consent for all investigations. This study was approved by the Medical Ethics Committee of the Radboud university medical center, The Netherlands.
Sanger Sequencing.
PCR products of proband III/18 for all exons of the PRKCSH, SEC63, PKD1, and PKD2 genes were analyzed by traditional Sanger sequencing on ABI310 or ABI3100 Genetic Analyzers (Applied Biosystems). Complete sequence analysis for PKD1 variants was conducted by an experienced Center (Bioscientia). Exon and exon-intron boundaries of LRP5 were determined using the Genome Bioinformatics Group of University of California, Santa Cruz (UCSC) Genome Browser, and unique primers were designed (Primer3). Heterozygous changes in LRP5 were detected by high-resolution melting curves (RotorGene-Q; Qiagen) and validated by Sanger sequencing.
Genome-Wide Copy Number Variation Analysis.
DNA from whole blood was analyzed on the CytoScanHD, which contains 2.6 million probes (Affymetrix). Hybridizations were performed according to the manufacturer’s protocol. Genotype calls and copy number variation analysis were made using Affymetrix Chromosome Analysis Suite v1.2.0.225.
Whole-Exome Sequencing.
Exome enrichment was performed using the SureSelect Human All Exon 50-Mb Kit (Agilent), covering ∼23,000 genes. Emulsion PCR and bead preparation were made by using the EZbead system according to the manufacturer's instructions followed by SOLiD4 sequencing (Life Technologies). Reads were mapped to the hg19 reference genome using SOLiD Bioscope software v1.3 and annotated as described previously (19, 20). All nonsynonymous variants shared in both affected individuals, absent or with very low frequency in dbSNP, were tested and validated in healthy and affected members of PCLD-1 family for cosegregation by high-resolution melting curve analysis and Sanger sequencing.
Linkage Analysis.
We calculated the two-point LOD score for linkage in the extended family using the SuperLink-v1.6 program in EasyLinkage-v5.08 software package. To determine the actual two-point LOD score for the LRP5 mutation that was detected in this family, the mutation was considered to be a microsatellite marker in close proximity of LRP5 (D11S4117). An autosomal dominant mode of inheritance was assumed with a penetrance of ∼80%, and the disease allele frequency was estimated at 0.0001 (Fig. S2 and SI Text).
Clinical Investigations.
Because LRP5 mutations are associated with bone density disorder, metabolic, and ocular developmental diseases, we performed additional clinical investigations of members from the PCLD-1 family with a clear genotype-phenotype relation. An ophthalmologist (C.E.N.) performed an eye examination by a slit lamp to exclude retinal disorders such as FEVR. At the same time, we analyzed metabolic and renal parameters in blood from these 22 individuals of the PCLD-1 family. We assessed bone density of the lumbar spine and left hip by dual energy X-ray absorptiometry (DXA scan; Hologic Discovery A). Results are reported as T- and Z-scores, which reflect the number of SDs by which a patient's value differs from the mean of a group of young or age-matched normal controls, respectively.
In Silico Analysis and Homology Modeling.
We used three computational tools for the prediction of the functional effect of mutational variants (PolyPhen-2, Mutpred, and SIFT). LRP5 protein structure was created by using an LRP6 template as a start homology model and reconstruction by the YASARA&WHAT-IF Twinset. PDB files were available as templates for homology modeling of WD40 domains (β-propeller subdomains) to incorporate the identified extracellularly located LRP5 mutations. Separate models were visualized for analysis of structural effects by YASARA.
Immunohistochemistry Studies.
Formalin-fixed paraffin-embedded liver cyst tissue from proband III/18, four unrelated PCLD patients with PRKCSH c.1341–2A > G mutations, and normal human liver tissue were available for immunohistochemistry studies. Staining intensity for the presence of LRP5 was compared between LPR5 and PRKCSH mutants.
Expression Constructs.
Total RNA was isolated from liver tissue using TRIzol Reagent (Invitrogen), and oligodT cDNA was obtained by RT Transcriptor First Strand cDNA synthesis kit (Roche Applied Sciences). Full-length WT LRP5 was obtained using the Faststart High Fidelity PCR System (Roche). LRP5 was cloned into the mammalian expression vector pcDNA3.1_V5_His TOPO-TA (Invitrogen) and checked by Sanger sequencing. An extracellularly c.3562C > T (p.R1188W) and an intracellularly located c.4651G > A (p.D1551N) LRP5 construct were generated by mutating the pcDNA.LRP5.WT vector using the Quick Change-II-XL Site-Directed Mutagenesis Kit (Agilent Technologies).
Luciferase Activity Assays.
We used three cell lines to assess the functional effects of WT and mutant LRP5. For the activity assay, 5.0 × 103 CHO cells, 5.0 × 103 HEK293 cells, and 5.0 × 103 human cholangiocyte 69 (H69) cells per well were seeded in a 96-well plate in triplicate. After 24 h, cells were transiently transfected using X-tremeGeneHD (Roche) with 100 ng LRP5 construct (LRP5WT; LRP5R1188W; LRP5D1551N) or empty vector and 100 ng of TCF/LEF1 Reporter or 100 ng negative control according to the manufacturers’ instructions (Cignal TCF/LEF1 Reporter Assay Kit; Qiagen). Sixteen hours after transfection, medium was replaced by medium with or without 250 ng/mL human Wnt3a (5036-WN; R&D Systems) to initiate Wnt signaling. Cells were cultured for another 24 h, and luciferase activity was detected using the Dual-Glo Luciferase Assay System (Cat. No. E2920; Promega) in an InfiniteM200-Pro plate reader (Tecan). Firefly luciferase activity was normalized to Renilla luciferase activity for variations in transfection efficiencies. Values are reported as means ± SD. These experiments were conducted in triplicate and performed three times. Western blotting was also performed to analyze possible differences in expression levels between the WT and mutant LRP5.
qPCR.
We conducted transient transfections of HEK293 cells with LRP5 constructs as previously described. The signaling was activated by addition of Wnt3a for 24 h. Total RNA was extracted with TRIzol (Invitrogen). Template cDNA was obtained using the iScript cDNA synthesis kit (Biorad). Expression levels of Wnt target genes were assessed twice by qPCR experiments (in triplicate) using the CFX96 real-time detection system (Biorad).
Immunofluorescence Imaging.
A total of 4.0 × 104 CHO cells per well were seeded on poly-l-lysine–coated Ø12-mm cover glasses on a 24-well plate and transiently transfected with 100 ng of LRP5WT or mutant LRP5 construct. After 24 h, medium was refreshed, and cells were cultured for another 24 h followed by paraformaldehyde fixation and immunofluorescence staining.
Statistics.
Groups were compared using descriptive statistics and compared by the Student t test. Both groups were compared by using the nonparametric Mann–Whitney U test. P < 0.05 was considered statistically significant. All analyses were calculated by SPSS software v18.0.
Supplementary Material
Acknowledgments
We thank the patients and their families for participation, providing samples, and assessment of clinical investigations for this study. We thank the Genomic Disorders Group Nijmegen for technical support in performing the whole-exome sequencing; Dr. C. Erik van Nouhuys (C.E.N.), ophthalmologist, for his professional contribution; Ms. Ing. Marjo T. P. van de Ven, Department of Nuclear Medicine; Mrs. Irene Otte-Höller, Department of Pathology; Dr. Rob W. J. Collin, Dr. Rolph Pfundt, and Mrs. Ing. Saskia van der Velde-Visser, Department of Human Genetics, Radboud university medical center Nijmegen; and Mr. Adriaan S. Grainger for technical support. This study was supported by a grant from the Institute of Genetic and Metabolic Diseases of the Radboud university medical center and by a grant from the Dutch Foundation of Digestive Diseases.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. X.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1309438111/-/DCSupplemental.
References
- 1.Strazzabosco M, Somlo S. Polycystic liver diseases: Congenital disorders of cholangiocyte signaling. Gastroenterology. 2011;140(7):1855–1859. doi: 10.1053/j.gastro.2011.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torres VE, Harris PC. Autosomal dominant polycystic kidney disease: The last 3 years. Kidney Int. 2009;76(2):149–168. doi: 10.1038/ki.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bae KT, et al. Consortium for Radiologic Imaging Studies of Polycystic Kidney Disease (CRISP) Magnetic resonance imaging evaluation of hepatic cysts in early autosomal-dominant polycystic kidney disease: The Consortium for Radiologic Imaging Studies of Polycystic Kidney Disease cohort. Clin J Am Soc Nephrol. 2006;1(1):64–69. doi: 10.2215/CJN.00080605. [DOI] [PubMed] [Google Scholar]
- 4.Ravine D, et al. Evaluation of ultrasonographic diagnostic criteria for autosomal dominant polycystic kidney disease 1. Lancet. 1994;343(8901):824–827. doi: 10.1016/s0140-6736(94)92026-5. [DOI] [PubMed] [Google Scholar]
- 5.Pei Y, et al. Unified criteria for ultrasonographic diagnosis of ADPKD. J Am Soc Nephrol. 2009;20(1):205–212. doi: 10.1681/ASN.2008050507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qian Q. Isolated polycystic liver disease. Adv Chronic Kidney Dis. 2010;17(2):181–189. doi: 10.1053/j.ackd.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reynolds DM, et al. Identification of a locus for autosomal dominant polycystic liver disease, on chromosome 19p13.2-13.1. Am J Hum Genet. 2000;67(6):1598–1604. doi: 10.1086/316904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapman AB. Cystic disease in women: Clinical characteristics and medical management. Adv Ren Replace Ther. 2003;10(1):24–30. doi: 10.1053/jarr.2003.50005. [DOI] [PubMed] [Google Scholar]
- 9.Van Keimpema L, et al. Patients with isolated polycystic liver disease referred to liver centres: Clinical characterization of 137 cases. Liver Int. 2011;31(1):92–98. doi: 10.1111/j.1478-3231.2010.02247.x. [DOI] [PubMed] [Google Scholar]
- 10.Gabow PA, et al. Risk factors for the development of hepatic cysts in autosomal dominant polycystic kidney disease. Hepatology. 1990;11(6):1033–1037. doi: 10.1002/hep.1840110619. [DOI] [PubMed] [Google Scholar]
- 11.The European Polycystic Kidney Disease Consortium The polycystic kidney disease 1 gene encodes a 14 kb transcript and lies within a duplicated region on chromosome 16. Cell. 1994;77(6):881–894. doi: 10.1016/0092-8674(94)90137-6. [DOI] [PubMed] [Google Scholar]
- 12.Mochizuki T, et al. PKD2, a gene for polycystic kidney disease that encodes an integral membrane protein. Science. 1996;272(5266):1339–1342. doi: 10.1126/science.272.5266.1339. [DOI] [PubMed] [Google Scholar]
- 13.Rossetti S, et al. CRISP Consortium Comprehensive molecular diagnostics in autosomal dominant polycystic kidney disease. J Am Soc Nephrol. 2007;18(7):2143–2160. doi: 10.1681/ASN.2006121387. [DOI] [PubMed] [Google Scholar]
- 14.Drenth JP, te Morsche RH, Smink R, Bonifacino JS, Jansen JB. Germline mutations in PRKCSH are associated with autosomal dominant polycystic liver disease. Nat Genet. 2003;33(3):345–347. doi: 10.1038/ng1104. [DOI] [PubMed] [Google Scholar]
- 15.Li A, et al. Mutations in PRKCSH cause isolated autosomal dominant polycystic liver disease. Am J Hum Genet. 2003;72(3):691–703. doi: 10.1086/368295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davila S, et al. Mutations in SEC63 cause autosomal dominant polycystic liver disease. Nat Genet. 2004;36(6):575–577. doi: 10.1038/ng1357. [DOI] [PubMed] [Google Scholar]
- 17.Müller L, Funato Y, Miki H, Zimmermann R. An interaction between human Sec63 and nucleoredoxin may provide the missing link between the SEC63 gene and polycystic liver disease. FEBS Lett. 2011;585(4):596–600. doi: 10.1016/j.febslet.2011.01.024. [DOI] [PubMed] [Google Scholar]
- 18.Waanders E, te Morsche RH, de Man RA, Jansen JB, Drenth JP. Extensive mutational analysis of PRKCSH and SEC63 broadens the spectrum of polycystic liver disease. Hum Mutat. 2006;27(8):830. doi: 10.1002/humu.9441. [DOI] [PubMed] [Google Scholar]
- 19.Hoischen A, et al. De novo mutations of SETBP1 cause Schinzel-Giedion syndrome. Nat Genet. 2010;42(6):483–485. doi: 10.1038/ng.581. [DOI] [PubMed] [Google Scholar]
- 20.Gilissen C, et al. Exome sequencing identifies WDR35 variants involved in Sensenbrenner syndrome. Am J Hum Genet. 2010;87(3):418–423. doi: 10.1016/j.ajhg.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilissen C, Hoischen A, Brunner HG, Veltman JA. Unlocking Mendelian disease using exome sequencing. Genome Biol. 2011;12(9):228. doi: 10.1186/gb-2011-12-9-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chrispijn M, et al. Everolimus does not further reduce polycystic liver volume when added to long acting octreotide: Results from a randomized controlled trial. J Hepatol. 2013;59(1):153–159. doi: 10.1016/j.jhep.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 23.Abecasis GR, et al. 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.National Institute of Environmental Health Sciences 2012. Environmental Genome Project Exome Variant Server. Available at http://evs.gs.washington.edu/niehsExome/. Accessed September 28, 2012.
- 25.Figueroa DJ, et al. Expression of the type I diabetes-associated gene LRP5 in macrophages, vitamin A system cells, and the Islets of Langerhans suggests multiple potential roles in diabetes. J Histochem Cytochem. 2000;48(10):1357–1368. doi: 10.1177/002215540004801006. [DOI] [PubMed] [Google Scholar]
- 26.Hey PJ, et al. Cloning of a novel member of the low-density lipoprotein receptor family. Gene. 1998;216(1):103–111. doi: 10.1016/s0378-1119(98)00311-4. [DOI] [PubMed] [Google Scholar]
- 27.Nikopoulos K, et al. Overview of the mutation spectrum in familial exudative vitreoretinopathy and Norrie disease with identification of 21 novel variants in FZD4, LRP5, and NDP. Hum Mutat. 2010;31(6):656–666. doi: 10.1002/humu.21250. [DOI] [PubMed] [Google Scholar]
- 28.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149(6):1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 29.Gong Y, et al. Osteoporosis-Pseudoglioma Syndrome Collaborative Group LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107(4):513–523. doi: 10.1016/s0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- 30.Boyden LM, et al. High bone density due to a mutation in LDL-receptor-related protein 5. N Engl J Med. 2002;346(20):1513–1521. doi: 10.1056/NEJMoa013444. [DOI] [PubMed] [Google Scholar]
- 31.Little RD, et al. A mutation in the LDL receptor-related protein 5 gene results in the autosomal dominant high-bone-mass trait. Am J Hum Genet. 2002;70(1):11–19. doi: 10.1086/338450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Wesenbeeck L, et al. Six novel missense mutations in the LDL receptor-related protein 5 (LRP5) gene in different conditions with an increased bone density. Am J Hum Genet. 2003;72(3):763–771. doi: 10.1086/368277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toomes C, et al. Mutations in LRP5 or FZD4 underlie the common familial exudative vitreoretinopathy locus on chromosome 11q. Am J Hum Genet. 2004;74(4):721–730. doi: 10.1086/383202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin M, et al. Complexity of the genotype-phenotype correlation in familial exudative vitreoretinopathy with mutations in the LRP5 and/or FZD4 genes. Hum Mutat. 2005;26(2):104–112. doi: 10.1002/humu.20191. [DOI] [PubMed] [Google Scholar]
- 35.Fujino T, et al. Low-density lipoprotein receptor-related protein 5 (LRP5) is essential for normal cholesterol metabolism and glucose-induced insulin secretion. Proc Natl Acad Sci USA. 2003;100(1):229–234. doi: 10.1073/pnas.0133792100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacDonald BT, Semenov MV, Huang H, He X. Dissecting molecular differences between Wnt coreceptors LRP5 and LRP6. PLoS ONE. 2011;6(8):e23537. doi: 10.1371/journal.pone.0023537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pinson KI, Brennan J, Monkley S, Avery BJ, Skarnes WC. An LDL-receptor-related protein mediates Wnt signalling in mice. Nature. 2000;407(6803):535–538. doi: 10.1038/35035124. [DOI] [PubMed] [Google Scholar]
- 38.Saadi-Kheddouci S, et al. Early development of polycystic kidney disease in transgenic mice expressing an activated mutant of the beta-catenin gene. Oncogene. 2001;20(42):5972–5981. doi: 10.1038/sj.onc.1204825. [DOI] [PubMed] [Google Scholar]
- 39.Happé H, et al. Toxic tubular injury in kidneys from Pkd1-deletion mice accelerates cystogenesis accompanied by dysregulated planar cell polarity and canonical Wnt signaling pathways. Hum Mol Genet. 2009;18(14):2532–2542. doi: 10.1093/hmg/ddp190. [DOI] [PubMed] [Google Scholar]
- 40.Qian CN, et al. Cystic renal neoplasia following conditional inactivation of apc in mouse renal tubular epithelium. J Biol Chem. 2005;280(5):3938–3945. doi: 10.1074/jbc.M410697200. [DOI] [PubMed] [Google Scholar]
- 41.Miller MM, et al. T-cell factor/β-catenin activity is suppressed in two different models of autosomal dominant polycystic kidney disease. Kidney Int. 2011;80(2):146–153. doi: 10.1038/ki.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lal M, et al. Polycystin-1 C-terminal tail associates with beta-catenin and inhibits canonical Wnt signaling. Hum Mol Genet. 2008;17(20):3105–3117. doi: 10.1093/hmg/ddn208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mao W, Wordinger RJ, Clark AF. Functional analysis of disease-associated polymorphism LRP5.Q89R. Mol Vis. 2011;17:894–902. [PMC free article] [PubMed] [Google Scholar]
- 44.Kiel DP, et al. Genetic variation at the low-density lipoprotein receptor-related protein 5 (LRP5) locus modulates Wnt signaling and the relationship of physical activity with bone mineral density in men. Bone. 2007;40(3):587–596. doi: 10.1016/j.bone.2006.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fedeles SV, et al. A genetic interaction network of five genes for human polycystic kidney and liver diseases defines polycystin-1 as the central determinant of cyst formation. Nat Genet. 2011;43(7):639–647. doi: 10.1038/ng.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fedeles SV, Gallagher AR, Somlo S. 2014. Polycystin-1: A master regulator of intersecting cystic pathways. Trends Mol Med, 10.1016/j.molmed.2014.01.004. [DOI] [PMC free article] [PubMed]
- 47.Kim E, et al. The polycystic kidney disease 1 gene product modulates Wnt signaling. J Biol Chem. 1999;274(8):4947–4953. doi: 10.1074/jbc.274.8.4947. [DOI] [PubMed] [Google Scholar]
- 48.Gao H, et al. PRKCSH/80K-H, the protein mutated in polycystic liver disease, protects polycystin-2/TRPP2 against HERP-mediated degradation. Hum Mol Genet. 2010;19(1):16–24. doi: 10.1093/hmg/ddp463. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.