Significance
Prions are self-propagating infectious proteins that have been implicated in a number of invariably fatal diseases. Increasing evidence suggests that this self-propagating activity is present in most or all amyloid diseases. To understand the pathology of these diseases, it is imperative to understand the mechanisms involved in self-propagation. To this end, we have used the fungal prion-forming domain HET-s(218–289) as a model system for studying sequence determinants required for structural propagation. We have determined the requirements for folding into the biologically active β-solenoid conformation and characterized the roles of different interactions in the assembly and overall stability of HET-s(218–289) prions. Our results provide insights into the fundamental mechanisms of prion assembly and propagation.
Abstract
The highly ordered and reproducible structure of the fungal prion HET-s makes it an excellent model system for studying the inherent properties of prions, self-propagating infectious proteins that have been implicated in a number of fatal diseases. In particular, the HET-s prion-forming domain readily folds into a relatively complex two-rung β-solenoid amyloid. The faithful self-propagation of this fold involves a diverse array of inter- and intramolecular structural features. These features include a long flexible loop connecting the two rungs, buried polar residues, salt bridges, and asparagine ladders. We have used site-directed mutagenesis and X-ray fiber diffraction to probe the relative importance of these features for the formation of β-solenoid structure, as well as the cumulative effects of multiple mutations. Using fibrillization kinetics and chemical stability assays, we have determined the biophysical effects of our mutations on the assembly and stability of the prion-forming domain. We have found that a diversity of structural features provides a level of redundancy that allows robust folding and stability even in the face of significant sequence alterations and suboptimal environmental conditions. Our findings provide fundamental insights into the structural interactions necessary for self-propagation. Propagation of prion structure seems to require an obligatory level of complexity that may not be reproducible in short peptide models.
Prions are infectious agents consisting of self-propagating, aberrantly folded protein aggregates that contain no nucleic acid component (1). The absence of nucleic acid-based information means that the particular biological activity of a prion is uniquely determined by its structure. Prions form amyloids, long, unbranched fibrils assembled into cross-β architecture (2) consisting of tiers of β-strands running perpendicular to the fiber axis, forming β-sheets that run the length of the fibril. Individual subunits may contain one or more tiers (3–5). This simple architecture allows a diverse array of proteins to form amyloids, each with distinct biological properties.
The most notable examples of prions are associated with the family of diseases known as the transmissible spongiform encephalopathies (TSEs) (1), all of which involve amyloids of the prion protein PrP. Non-PrP amyloids have been implicated in diseases such as Parkinson and Alzheimer’s diseases as well as type II diabetes (6). The common presence of pathological amyloid in these diseases led to the supposition that the prion mechanism of propagation and infectivity may be a common feature of all pathological amyloids (2, 7). Recent experiments have lent credence to this idea; results showing disease pathogenesis from exogenous sources of amyloid (8–11) have been analogous to those that demonstrated the infectivity of TSEs (12, 13). In addition to pathological examples, functional prions and amyloids, such as the fungal prion HET-s (14) and the human amyloid Pmel17 (15), have been found. Functional prions and amyloids seem to exploit the self-assembling properties of cross-β structure but lack pathogenicity, because of robust aggregation control mechanisms such as regulation of environmental conditions (16) or proteolytic cleavage of nonaggregating proprotein (17).
Prions generally propagate with high structural and pathological fidelity. They can, however, be polymorphic; different prions formed from a single protein (strains) can produce distinct pathologies (18). Prion strains generally arise from changing the environment of prions, for example infecting animals with de novo prions formed under different conditions, or inoculating animals with brain homogenates from different species. Serial passaging in a constant environment leads to the formation of stable and reproducible strains (19). Whereas the differentiation of strains indicates structural mutability, the formation of stable strains indicates that reproducible pathologies are a consequence of high-fidelity structural reproduction.
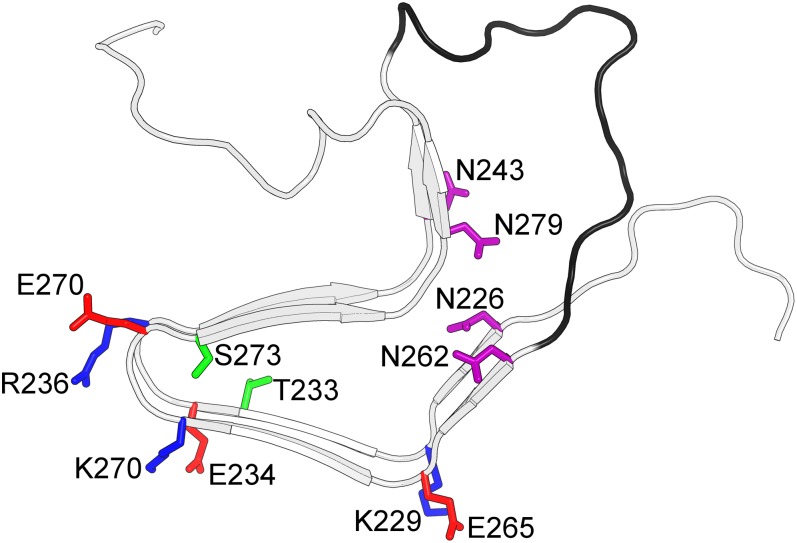
To study the structures and mechanisms underlying reproducible and self-propagating prion folding, we have conducted site-directed mutagenesis studies of the functional fungal prion-forming domain HET-s(218–289). HET-s(218–289) is both necessary and sufficient for biological activity (20, 21) and readily fibrillizes into an infectious, two-rung β-solenoid structure at physiological pH (22, 23) (Fig. 1). Each rung consists of four β-strands, forming four β-sheets along the fibril, three of which form a triangular hydrophobic core. The remaining strand, together with several residues from the C-terminal tail, forms a secondary hydrophobic core. Within the triangular hydrophobic core are two polar residues thought to stabilize the turn between adjacent β-sheets (24). The solvent-exposed surface contains three salt bridges and two asparagine ladders. The two rungs of the β-solenoid structure are connected by a long flexible loop, residues 247–261.
Fig. 1.
Structure of HET-s(218–289) and positions of mutated residues. Model is from the solid-state NMR structure (PDB ID code 2kj3). Green, buried polar residues; red and blue, positive and negative salt-bridge residues, respectively; purple, asparagine ladders; and black, flexible loop.
We have mutated the flexible loop sequences, buried polar interactions, and solvent-exposed salt bridges and asparagine ladders found in HET-s(218–289) to investigate the necessity, cumulative effects, and to some extent redundancy of the different interactions. For mutants that maintained β-solenoid folds, we characterized their fibrillization kinetics and stability in the presence of chemical denaturants to determine the relative importance of each type of interaction as well as the cumulative effects of mutations within an interaction type. We have shown that efficient, faithful self-propagation of the infectious fold of the prion-forming domain requires a complex and diverse array of inter- and intramolecular structural features.
Results
Site-Directed Mutagenesis.
Site-directed mutagenesis was used to replace individual residues involved in salt bridges and asparagine ladders and buried hydroxyl residues, with alanine, effectively abrogating interactions within individual pairs. Combination mutants were also made, replacing both of the buried hydroxyl residues, all positive or negative salt-bridge partners, all permutations of asparagines that abrogate both ladders, and in two mutants, residues that abrogate all solvent-exposed surface interactions. Mutations were also made to reverse or scramble the flexible loop sequence and to remove the loop residues 251–253; the Δ251–253 mutant had previously been shown to form infectious fibrils (25). Mutants are listed with abbreviations in Table 1, and locations of the mutated residues in the solid-state NMR (ssNMR) β-solenoid structure are shown in Fig. 1.
Table 1.
Molecular architecture, fibrillization kinetics, and chemical stability of HET-s(218–289) mutants
| Mutation | Mutation type | Abbreviation | 9.4-Å meridional | Equatorial CC | Architecture | Lag time, min | kapp, min−1 | m1/2, M |
| Wild-type | — | WT | Yes | 1.00 | β-Solenoid | 21.8 ± 0.6 | 0.103 ± 0.002 | 2.77 ± 0.03 |
| Reversed loop | Loop | RL | No | 0.63 | Stacked β-sheet | — | — | — |
| Scrambled loop | Loop | SL | No | 0.66 | Stacked β-sheet | — | — | — |
| Δ251–253 | Loop | Δ251–253 | Yes | 0.96 | β-Solenoid | 147 ± 2 | 0.071 ± 0.004 | 2.30 ± 0.1 |
| T233A | Buried polar | PA | Yes | 0.91 | β-Solenoid | 23.0 ± 1 | 0.133 ± 0.007 | 2.94 ± 0.02 |
| S273A | Buried polar | PB | Yes | 0.83 | β-Solenoid | 77.0 ± 2 | 0.103 ± 0.007 | 1.98 ± 0.02 |
| T233A-S273A | Buried polar | PAB | Yes | 0.99 | β-Solenoid | 38.0 ± 2 | 0.098 ± 0.005 | 2.73 ± 0.06 |
| E234A | Salt bridge | SA− | Yes | 0.99 | β-Solenoid | 28.3 ± 0.9 | 0.151 ± 0.006 | 2.76 ± 0.03 |
| E265A | Salt bridge | SB− | Yes | 0.94 | β-Solenoid | 35.0 ± 2 | 0.120 ± 0.009 | 2.53 ± 0.02 |
| E272A | Salt bridge | SC− | Yes | 0.99 | β-Solenoid | 94.0 ± 2 | 0.106 ± 0.005 | 3.40 ± 0.2 |
| E234A, E265A, E272A | Salt bridge | SABC− | Yes | 0.98 | β-Solenoid | 219 ± 3 | 0.051 ± 0.003 | 2.27 ± 0.06 |
| K229A | Salt bridge | SB+ | Yes | 0.90 | β-Solenoid | 66.0 ± 1 | 0.138 ± 0.006 | 3.20 ± 0.1 |
| K270A | Salt bridge | SA+ | Yes | 0.98 | β-Solenoid | 30.0 ± 1 | 0.170 ± 0.01 | 2.63 ± 0.06 |
| R236A | Salt bridge | SC+ | Yes | 0.99 | β-Solenoid | 59.0 ± 1 | 0.134 ± 0.007 | 2.78 ± 0.07 |
| K229, K270A, R236A | Salt bridge | SABC+ | Yes | — | β-Solenoid | 111 ± 2 | 0.078 ± 0.003 | 1.94 ± 0.08 |
| N226A | N ladder | NA | Yes | 0.94 | β-Solenoid | 95.0 ± 3 | 0.087 ± 0.006 | 1.30 ± 0.2 |
| N243A | N ladder | NB | Yes | 0.96 | β-Solenoid | 91.0 ± 2 | 0.093 ± 0.005 | 1.84 ± 0.05 |
| N262A | N ladder | NC | Yes | 0.89 | β-Solenoid | 124 ± 4 | 0.068 ± 0.005 | 1.40 ± 0.2 |
| N279A | N ladder | ND | Yes | 0.90 | β-Solenoid | 121 ± 2 | 0.066 ± 0.003 | 2.39 ± 0.08 |
| N226A, N243A | N ladder | NAB | Yes | 0.84 | β-Solenoid | 139 ± 6 | 0.054 ± 0.006 | 0.470 ± 0.05 |
| N226A, N279A | N ladder | NAD | Yes | — | β-Solenoid | 97.0 ± 6 | 0.070 ± 0.01 | 0.250 ± 0.02 |
| N243A, N262A | N ladder | NBC | Yes | — | β-Solenoid | 311 ± 4 | 0.039 ± 0.002 | 0.700 ± 0.04 |
| N262A, N279A | N ladder | NCD | Yes | 0.84 | β-Solenoid | 298 ± 1 | 0.048 ± 0.003 | 1.28 ± 0.03 |
| E234A, E265A, E272A, N226A, N243A | Salt bridge, N ladder | SN− | No | — | Stacked β-sheet | — | — | — |
| K229A, K270A, R236A, N262A, N279A | Salt bridge, N ladder | SN+ | No | — | Stacked β-sheet | — | — | — |
Mutation type refers to the type of structure affected by the mutation. Presence of the 9.4-Å meridional reflection indicates a two-rung structure. Equatorial CC is the correlation coefficient between WT and mutant equators; mutants with no listed CC were too disordered to perform quantitative analysis, so qualitative comparisons are shown in Table S1. Lag time is an empirical parameter representing time before exponential growth and kapp is an empirically calculated apparent first-order rate constant. Lag time and kapp for NAB and NAD are for the second aggregation step. m1/2 is the guanidine concentration at the half-denaturation point.
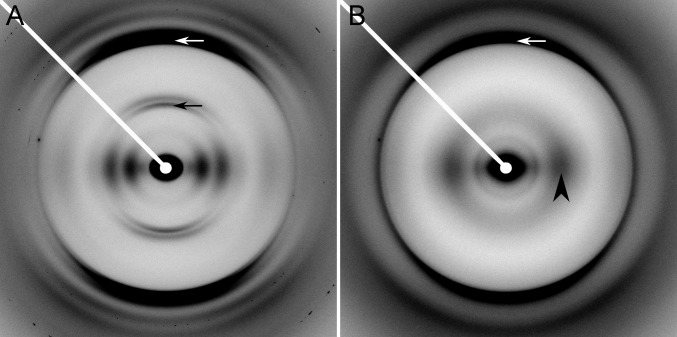
To determine overall molecular architecture, that is, the general configuration of the protein chain, X-ray fiber diffraction data were obtained for each mutant (Fig. S1 and Table 1). Diffraction patterns fell into two distinct classes (examples in Fig. 2). The first class resembled the pattern of WT HET-s(218–289), with strong meridional diffraction close to 4.7 Å, indicating cross-β structure, weaker but distinct meridional diffraction close to 9.4 Å, demonstrating that the structure has a two-strand repeat, and a series of equatorial intensities characteristic of an approximately cylindrical structure at low resolution (26–28) (Fig. 2A). This characteristic pattern has been shown by fiber diffraction and ssNMR (28)(Fig. S2E) to be produced by the two-rung β-solenoidal structure of HET-s(218–289). The second class of patterns was of a different type, with a 4.7-Å meridional cross-β reflection but no trace of 9.4-Å and equatorial intensity concentrated at 8–10 Å (Fig. 2B and Fig. S2 A and B). This class of patterns is characteristic of β-sheets stacked together, with the 8- to 10-Å intensity reflecting the spacing between the sheets (3, 26–29) (Fig. S2 C and D). The equators of mutant diffraction patterns were quantitatively compared with the observed WT equator (Fig. S2B) by calculating correlation coefficients (CC) (Materials and Methods and Table 1). Comparisons with patterns calculated from the ssNMR solenoid structure (Fig. S2E) were not made, because the ssNMR model only correlates well with the observed fiber diffraction pattern to ∼7 Å resolution, perhaps because of errors related to the short-distance nature of NMR restraints. For patterns with insufficient orientation for quantitative analysis, positions of diffraction maxima were compared (Table S1). Mutants with diffraction patterns that matched well with WT were identified as having β-solenoid architecture (28). Those that fit WT diffraction poorly were compared with diffraction calculated from a stacked β-sheet model, quantitatively if well oriented or by positions of diffraction maxima if not (Table S1). Comparison with the stacked β-sheet model provides only a general indicator of architecture, because the simple model used (Fig. S2 C and D) (28) was somewhat arbitrary, reproducing the generic features of a stacked β-sheet but not the particular spacings unique to individual amyloids. Mutants that formed β-solenoids were subjected to fibrillization kinetics (Fig. S3 and Table 1) and guanidine denaturation (Fig. S4 and Table 1) assays to compare the effects of the mutations on the biophysical properties of the β-solenoid structure.
Fig. 2.
Representative X-ray fiber diffraction patterns. (A) HET-s(218–289) WT showing two-rung β-solenoid diffraction. (B) HET-s(218–289) mutant RL showing characteristic stacked β-sheet diffraction. White arrows, 4.7-Å meridional reflection; black arrow, 9.4-Å meridional reflection; and black arrowhead, equatorial ∼10-Å intensity maximum corresponding to the intersheet spacing.
Effects of Loop Mutations on Structure and Assembly.
X-ray fiber diffraction data from the HET-s(218–289) loop mutants RL and SL (Fig. S1 B and C) agreed poorly with WT diffraction data (Fig. S1A and Table 1), but RL data agreed well (CC = 0.84) with the stacked β-sheet model, indicating a stacked β-sheet architecture. SL diffraction showed poor agreement with the stacked β-sheet model (CC = 0.62) and the WT pattern (Fig. S1A and Table 1) but good correlation with RL (CC = 0.88). Comparison of equatorial plots (Fig. S5) shows that the equators of RL and SL are almost identical except for an additional intensity minimum, indicating that SL is also a stacked β-sheet structure, but with sampling in the pattern, perhaps caused by higher-order fibril structure.
The partial loop deletion mutant Δ251–253 exhibited a two-rung β-solenoid diffraction pattern (Fig. S1D and Table 1), as was expected given its confirmed in vivo infectivity. To determine whether the inability of RL and SL to form β-solenoids was due to barriers to fibril nucleation, we obtained fiber diffraction data from fibrils seeded with HET-s(218–289) WT fibrils (Fig. S6 A and B). Seeded diffraction data for RL showed stacked β-sheet architecture, but SL exhibited weak diffraction, consistent with a two-rung β-solenoid structure.
Kinetics assays showed that fibrillization of Δ251–253 had a much longer lag time and lower kapp (apparent first-order rate constant) than WT fibrils (Table 1). Guanidine denaturation assays showed that Δ251–253 had a lower m1/2 (guanidine concentration required for 50% denaturation). These differences suggest that shortening the loop has a particularly large impact on fibril nucleation, as well as a significant effect on growth and stability of fibrils.
Buried Polar Interaction Is Kinetically Favorable.
Fiber diffraction from HET-s(218–289) buried polar mutants PA, PB, and PAB indicated a two-rung β-solenoid structure (Fig. S1 E–G and Table 1). The lag time of PA was roughly the same as that of WT, although kapp and m1/2 were greater. PB showed a significantly longer lag time and lower m1/2 than WT but the same kapp. PAB had the same kapp and m1/2 as WT as well as a longer lag time, although not as long as that of PB.
Salt Bridges Have Cumulative Stabilizing Effects.
Fiber diffraction from all of the single and triple salt-bridge mutants revealed β-solenoid architecture (Fig. S1 H–O and Table 1). Single mutants all had longer lag times, but kapp and m1/2 tended to be the same or greater than those of WT, although SB− and SA+ had lower m1/2 values. There was no obvious correlation in kinetics or stability between mutations affecting the same salt-bridge pairs. The triple salt-bridge mutants all showed longer lag times and lower kapp and m1/2 values than WT or single salt-bridge mutants. SABC− had a much longer lag time and lower kapp than SABC+ but a higher m1/2. The biophysical differences with respect to WT between the triple mutants and their constituent single mutations were decidedly larger, indicating that the stabilizing and kinetics-enhancing effects of salt bridges have a cumulative relationship.
Asparagine Ladders Provide Significant Stability.
Fiber diffraction from single and double asparagine-ladder mutants showed the presence of β-solenoid architecture (Fig. S1 P–W and Table 1). Single asparagine-ladder mutants had longer lag times and lower kapp values than WT, with NC and ND showing significantly longer lag times and lower kapp values than NA and NB (Table 1). The m1/2 for each single mutant was lower than that for WT, but NA, NB, and NC showed m1/2 values lower than those of either triple salt-bridge mutant.
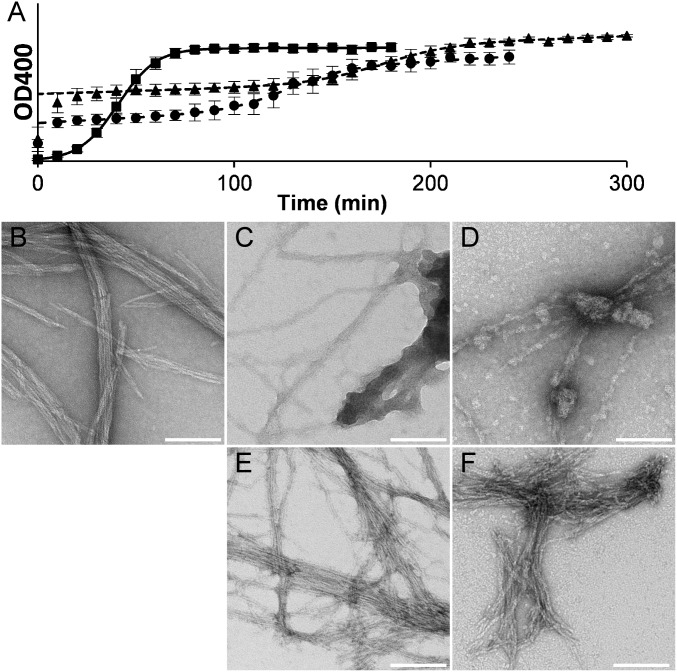
Double asparagine-ladder mutants exhibited two types of kinetics curves: a standard two-state curve for NBC and NCD (Fig. S3 T and U) and an apparent three-state curve for NAB and NAD (Fig. 3A). The three-state curves have a rapid initial aggregation step, followed by a lag phase and a subsequent aggregation phase. To characterize the different stages, negative-stain EM was performed on mature WT fibrils and on NAB and NAD fibrils at the mid and final steady states. Mature WT fibrils are characterized by long cable-like aggregates (Fig. 3B and Fig. S7A), whereas NAB and NAD at 30 min had disperse fibrils associated with large and small amorphous aggregates (Fig. 3 C and D and Fig. S7 B and C). NAB and NAD at 4 h had almost no amorphous aggregates and fibrils were laterally organized similarly to WT fibrils (Fig. 3 E and F and Fig. S7 D and E). The times between initiation and the second aggregation step of NAB and NAD were much longer than the WT lag time, and kapp of the second step for each mutant was also lower than that of WT (Table 1). The m1/2 values for NAB and NAD were the lowest of all of the mutants assayed (Table 1). NBC and NCD had the longest lag times and smallest kapp values of any mutants studied and m1/2 values lower than those of all mutants except NAB and NAD (Table 1).
Fig. 3.
Fibrillization kinetics assays and negative-stain EM of HET-s(218–289) WT and double asparagine-ladder mutants. (A) Fibrillization kinetics assay plots: WT (■ and solid line), NAB (▲ and dotted line), and NAD (● and dashed line). Negative-stain EM for (B) WT, (C) NAB at 30 min, (D) NAD at 30 min, (E) NAB at 4 h, and (F) NAD at 4 h. See also Fig. S5. (Scale bars, 100 nm.)
All four double asparagine-ladder mutants, when moved to a pH 4 buffer for fiber diffraction (Materials and Methods), exhibited stacked β-sheet diffraction patterns (Fig. S8). Diffraction specimens in water from the same fibril preparations, however, exhibited β-solenoid patterns (Fig. S1 V and W). NBC and NCD stacked β-sheets showed very faint 9.4-Å meridional reflections (Fig. S8 C and D), possibly from residual β-solenoid contamination.
Surface Interactions Are Required for β-Solenoid Formation.
X-ray fiber diffraction from the double asparagine ladder/triple salt-bridge mutants SN− and SN+ showed that the fibrils had stacked β-sheet architecture (Fig. S1 X and Y and Table 1). To determine whether the inability of SN− and SN+ to form β-solenoids was due to barriers to fibril nucleation, SN− and SN+ monomer solutions were seeded with preformed WT β-solenoid fibrils. Fiber diffraction data from seeded fibrils (Fig. S6 C and D) showed similar architectures to unseeded patterns, indicating that the abrogation of β-solenoid formation was not related to nucleation.
Discussion
The β-solenoid architecture of HET-s(218–289) proved to be surprisingly robust. Although many of the mutations studied had a significant effect on fibril nucleation, fibrillization rates, and fibril stability, X-ray fiber diffraction showed that in most cases the β-solenoid structure of the WT prion was still formed, rather than a generic stacked β-sheet structure. Only complete scrambling or reversal of the flexible loop connecting the two rungs of the β-solenoid, or removal of both asparagine ladders as well as all three salt bridges, perturbed the structure sufficiently to abolish the β-solenoid.
Some caution is appropriate when considering fibrillization kinetics and denaturation results, because the results may depend on the particular approach. Fibrillization kinetics data were fitted to an empirical function, so the fitted parameters are not necessarily directly related to specific events in the fibrillization process. However, they do allow for convenient comparisons and provide qualitative and relative assessments of the mutational impacts, although the precise parameter values may not be directly relatable to other systems. Similar caution must be used in the interpretation of guanidine denaturation results, because HET-s(218–289) denaturation is dependent on the particular chemical used. For example, urea is insufficient for denaturing HET-s(218–289) β-solenoid fibrils and does not prevent the formation of infectious β-solenoids from monomers (22). This variability with respect to denaturing agent suggests that the relationship between chemical denaturation of HET-s(218–289) and thermodynamic stability may not be a simple one, although denaturation results are nevertheless informative when obtained under comparable conditions.
The only loop mutant that we observed forming β-solenoids was the deletion mutant Δ251–253. The flexible loop joining the two rungs of the β-solenoid structure has previously been shown to tolerate deletions of up to three residues without losing infectivity (25). Our kinetics assays indicate that the loop reduction in Δ251–253 causes reduced fibrillization kinetics, possibly related to the degrees of freedom required to adequately search conformational space and align the two rungs. This deletion also resulted in decreased guanidine stability, which may be due to reduced loop entropy. The stacked β-sheet structure of the RL and SL mutants shows that the amino acid sequence in the loop is important for β-solenoid folding; it may be important for loop flexibility. Residues A247 and A248 are not completely disordered (23, 24), and deletion of residues 244–248 results in loss of infectivity (25), suggesting that these residues are functionally important despite their lack of β-strand structure. The ability of SL to form β-solenoids when seeded indicates that the β-solenoid–abrogating effect of loop scrambling may be kinetic rather than thermodynamic, or at least an effect on fibril nucleation rather than elongation.
Mutations of the buried polar residues T233 and S273 indicate that S273 provides a small benefit to fibrillization rate, but T233 does not. Results from the serine mutant PB demonstrate that T233 interacts unfavorably with the hydrophobic core but is stabilized by the adjacent S273 in WT. S273 does not seem to need stabilization by T233, probably because it interacts with the adjacent turn (24) (Fig. 1); the threonine mutant PA is very like WT. The double-mutant PAB removes the need for polar compensation of T233; kapp and m1/2 are identical to those of WT, but a longer lag time indicates that the buried polar structure is useful during early folding stages.
Salt-bridge mutations affect both folding kinetics and guanidine stability. The nonequivalence of biophysical differences between single mutants, even between partners in the same salt bridge, indicates complex electrostatic interactions during and after fibrillization. The effective removal of all salt bridges in the triple mutants shows that these interactions are cumulative, but as with the single mutants the nonequivalence of the two triple mutants further emphasizes the complexity of the electrostatic interactions.
Single-residue asparagine ladder mutations slowed fibrillization and lowered stabilities in guanidine much more than single salt-bridge mutants. The changes observed for ladder partners were not equivalent, indicating the presence of complex polar interactions, as seen in the salt-bridge mutants. We did observe similar kinetics in single mutants affecting the same rung. Double asparagine-ladder mutants fell into two types: NBC and NCD exhibited extremely slow but otherwise normal fibrillization kinetics, whereas NAB and NAD seemed to have a three-state fibrillization process. The mutants with apparent three-state fibrillization had N226A as a common mutation, but the single N226A mutant had typical kinetics. The three-state mutants seem to form stable fibrillization intermediates or off-path products consisting of amorphous aggregates, oligomers, and protofibrils (Fig. 3 and Fig. S7). Mature β-solenoid fibrils of these mutants have particularly low guanidine stabilities. These observations, taken with fibrillization kinetics data, seem to indicate a flatter energy landscape than that of the two-state double mutants, which have higher guanidine stabilities but much slower fibrillization rates.
Double asparagine mutants refold under acidic conditions to form generic stacked β-sheet amyloids. The equatorial diffraction shows stacked β-sheet architecture and the absence of 9.4-Å meridional reflections in the diffraction patterns of these amyloids suggests denaturation followed by refolding rather than collapse of the triangular hydrophobic core. These observations indicate that without stabilizing surface interactions the cross-β hydrogen bonding network of the β-solenoid can be disassembled by adding a net positive charge to the fibril surface. This emphasizes the particular importance of asparagine ladders, because WT β-solenoid is not only stable at pH 4 (22) but can fibrillize under those conditions (28). It is likely that the resultant stacked β-sheet structure allows for repulsive charge compensation, similarly to noninfectious HET-s(218–289) polymorphs fibrillized under acidic conditions (22, 28).
HET-s has eight homologs in the Fusaria genus, the most studied of which is from Fusaria graminearum (14). The F. graminearum HET-s(218-289) homolog, FgHET-s(218-289), cross-seeds with Podospora anserina HET-s(218–289), which has been attributed to the fact that they form similar β-solenoid structures (30). Within the long flexible loop, sequence alignment (30) shows scattered residues that are identical or similar, which may explain why our RL and SL mutants did not fold into β-solenoids. S273 was highly conserved, whereas T233 was not, consistent with the accelerated fibrillization kinetics associated with S273 but not T233. Although salt bridges in HET-s(218–289) afforded benefits to fibrillization kinetics and guanidine stability, they were not highly conserved, although some salt-bridge residues in the alignment maintained polar or alternating charge interactions, and there were generally no like-charged pairs. Asparagine ladders are very well conserved, with identical residues in nearly all positions for each homolog. Homology comparisons suggest that the conserved features necessary for the formation of a two-rung β-solenoid are a long flexible loop, alternating polar and nonpolar residues to define the buried and exposed surfaces of the hydrophobic core, S273 to stabilize a β-turn in the triangular core structure, and asparagine ladders for the intra- and intermolecular alignment of rungs.
Our results suggest some general principles that may be applicable to other prions and amyloids that, like HET-s, do not have any particular amino acid composition bias. In HET-s(218–289), asparagine ladders play a very important role in reproducible folding and fibril stability, even without the extremely high percentages of asparagine and glutamine found in polyQ/N amyloids such as yeast prions (31) and Huntington-related amyloids (32). In HET-s(218–289), asparagine ladders and salt bridges provide a level of redundancy; β-solenoid formation is not perturbed by mutation of either class of interaction, but only by mutation of both. However, salt bridges are likely to be found only in amyloids with multitier repeats such as β-solenoids and antiparallel β-sheets. Amide ladders formed by asparagines and glutamines do not require properly matched charge pairs and as such are more promiscuous in their interactions, as demonstrated by prion formation from scrambled polyQ/N domains (33, 34). The low number of asparagines in HET-s(218–289) may underlie the absence of in vivo polymorphs, whereas functional polyQ/N amyloids such as yeast prions can take on a number of stable strains (35).
It is clear from these results that solvent-exposed surface interactions play complex but vital roles in high-fidelity amyloid fibrillization and self-propagation. Our inability to induce β-solenoid formation in the double asparagine ladder/triple salt-bridge mutants SN− and SN+ further emphasizes the necessity for surface interactions, demonstrating that the shape of the triangular hydrophobic core alone is insufficient as a structural template. Computational studies have indicated that these solvent-exposed interactions contribute to the high stability of HET-s(218–289) (36), and our observation of acidic denaturation of double asparagine mutants suggests that stability is not due to cross-β hydrogen bonding networks. HET-s(218–289) as a model prion has thus allowed us to elucidate the role of structural features that cannot be probed by short-peptide models (37). The hydrophobic core of HET-s(218–289) is similar to those found in other β-solenoid proteins (38) and does not exhibit the extremely tight, interdigitated packing found in steric zippers (37, 39), although the hydrophobic core and solvent-exposed surfaces of HET-s(218–289) can be thought of as analogous to the dry and wet steric zipper interfaces.
An inverse relationship between incubation time and conformational stability has been observed for the infectious form of PrP, PrPSc (19). It seems likely, therefore, that pathological prions may have a combination of the structural features studied here to allow for high-fidelity propagation, although not necessarily to the extent that provides the robust redundancy found in HET-s(218–289). Overall, our studies with HET-s(218–289) mutants provide a useful basis for understanding the relative contributions of different structural features for high-fidelity fibrillization and structural propagation, two core properties that define a stable prion strain. Prion structure and self-propagation seem to require an obligatory level of complexity not seen in short amyloid models.
Materials and Methods
Site-Directed Mutagenesis.
The initial plasmid was constructed by cloning a synthetic Met-HET-s(218–289)-His6 gene codon optimized for Escherichia coli into a pET-17b vector. Site-directed mutagenesis was carried out using a two-stage QuikChange (Agilent Technologies) protocol (40). Oligonucleotide primers were synthesized by Integrated DNA Technologies. Loop residues (residues 247–261) were mutated to TTQDSIRIGGHLAAA for RL and ATRIGLIQGTHASDA for SL mutants.
Preparation of HET-s(218–289) Fibrils.
All variants of HET-s(218–289) were recombinantly expressed in BL-21 E. coli cells and purified from inclusion bodies under denaturing conditions as previously described (27). To begin fibrillization, purified protein was desalted into 150 mM acetic acid, pH 2.5, and titrated to pH 7.5 with 3 M Tris base. For seeded preparations, 1/100 HET-s(218–289) WT fibrils were added to monomer solutions with the 3 M Tris.
X-Ray Fiber Diffraction.
Fiber diffraction specimens were prepared by hanging 10- to 15-μL droplets of ∼20 mg/mL fibril solution between the sanded tips of silanized glass capillaries and dried under high humidity (∼100% or ∼98% relative humidity). High humidity was maintained throughout the course of the experiments (41). Fibrils were moved into either 5 mM Na acetate, pH 4.0, or water using three cycles of ultracentrifugation and resuspension. Fiber diffraction data were collected at beamline 4-2 at the Stanford Synchrotron Radiation Lightsource, or the BioCAT beamline at the Advanced Photon Source at Argonne National Laboratory. Data were analyzed using WCEN (42). Equators were compared by calculating CCs. The resolution range used for comparisons to WT was ∼33–34 Å (0.03–0.25 Å−1), providing comparisons of architecture up to residue-level resolution, whereas comparisons with the stacked β-sheet model were performed at ∼33–35 Å (0.03–0.20Å−1), providing comparisons of the main chain. For the wide resolution range compared, the CC is a superior measure of agreement to the R factor because it is unaffected by intensity scaling difficulties caused by solvent-contrast effects (43). Correlation coefficients were calculated from background-subtracted equatorial plots as previously described (28). For patterns where disorientation prevented accurate calculations of CCs, equators were compared semiquantitatively by measuring maxima positions. Meridians were indexed using background-subtracted plots; scale factors for each pattern were determined by least-squares fitting of the continuous equatorial diffraction (28).
Fibrillization Kinetics Assays.
Fibrillization assays used a modified version of an assay described elsewhere (28). Briefly, purified HET-s(218–289) monomer solution was desalted into 500 mM acetic acid, divided into working aliquots, lyophilized, incubated with hexafluoroisopropanol, lyophilized, and stored at −20 °C until use. Assays were performed by dissolving monomer powder in 175 mM acetic acid, and fibrillization was started by mixing with 1:1 (vol/vol) 1 M Tris⋅HCl, pH 8.0. Final protein concentration was 60 μM. Assays were monitored by optical density at 400 nm, reading at 10-min intervals. All experiments were carried out in triplicate using disposable plastic cuvettes. Triplicate data were averaged and fitted using an empirical equation (44). For those datasets exhibiting an apparent three-state curve, data from the first 30 min were not included in the fitting. Fitted parameters from the empirical equation were used to determine the lag time and apparent first-order rate constant kapp (44). SDs were determined by a Monte Carlo approach (45), using 100 iterations.
Guanidine Denaturation Assays.
Guanidine denaturation assays were performed in black, nonbinding 96-well plates (Greiner). Guanidine concentrations used were from 0 to 5.75 M in 0.25-M increments over two rows of wells, with each assay performed in triplicate over six rows of wells. The remaining two wells were blanks containing no protein and were used for background subtraction. Final protein concentrations were 10 μM, as determined by bicinchoninic acid assay (Thermo Scientific). To ensure equilibrium, plates were sealed and incubated at room temperature for 7 d; 96-well plates were assayed by reading fluorescence using a Perkin–Elmer EnSpire with λEx = 280 nm and λEm = 340 nm. Fluorescence readings were background-subtracted, averaged, and fit to an empirical equation modified from the equation used for fibrillization kinetics assays:
where yi, mi, yf, and mf are the initial and final intercept and slope and m1/2 is the half-maximal fluorescence point, the guanidine concentration required for 50% denaturation.
Negative-Stain EM.
Fibril suspensions from fibrillization kinetics assays were adsorbed onto freshly glow-discharged carbon-coated copper grids, rinsed with water, and stained with 1% uranyl acetate. Micrographs were recorded using a FEI T-12 transmission electron microscope operating at 80 KeV with a side-mounted 2 k × 2 k charge-coupled device camera.
Supplementary Material
Acknowledgments
We thank Amy Kendall for helpful discussions and comments on the manuscript and the staff of beamline 4-2 at the Stanford Synchrotron Radiation Lightsource (SSRL) and BioCAT at the Advanced Photon Source (APS). This work was supported by National Institutes of Health (NIH) Grants AG002132 (to Principal Investigator G.S.; Program Director Stanley Prusiner) and F31-AG040947 (to W.W.). Negative-stain electron micrographs were collected in the Vanderbilt University Medical Center Cell Imaging Shared Resource, supported by NIH grants. The SSRL is a national user facility operated by Stanford University on behalf of the Department of Energy (DOE). The SSRL Structural Molecular Biology Program is supported by the DOE and NIH. The APS is supported by the DOE. BioCAT is supported by NIH Grant 9 P41 GM103622.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1322933111/-/DCSupplemental.
References
- 1.Prusiner SB. In: Fields Virology. 5th Ed. Knipe DM, Howley PM, editors. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 3059–3091. [Google Scholar]
- 2.Prusiner SB, et al. Scrapie prions aggregate to form amyloid-like birefringent rods. Cell. 1983;35(2 Pt 1):349–358. doi: 10.1016/0092-8674(83)90168-x. [DOI] [PubMed] [Google Scholar]
- 3.Astbury WT, Dickinson S, Bailey K. The X-ray interpretation of denaturation and the structure of the seed globulins. Biochem J. 1935;29(10):2351–2360.1. doi: 10.1042/bj0292351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rudall KM. 1946. The structure of epidermal protein. The Society of Dyers and Colourists: Symposium on Fibrous Proteins (Chorley and Pickersgill, Leeds, U.K.), pp 15–23.
- 5.Eanes ED, Glenner GG. X-ray diffraction studies on amyloid filaments. J Histochem Cytochem. 1968;16(11):673–677. doi: 10.1177/16.11.673. [DOI] [PubMed] [Google Scholar]
- 6.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75(1):333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 7.Frost B, Diamond MI. Prion-like mechanisms in neurodegenerative diseases. Nat Rev Neurosci. 2010;11(3):155–159. doi: 10.1038/nrn2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lundmark K, et al. Transmissibility of systemic amyloidosis by a prion-like mechanism. Proc Natl Acad Sci USA. 2002;99(10):6979–6984. doi: 10.1073/pnas.092205999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo JL, Lee VM-Y. Seeding of normal Tau by pathological Tau conformers drives pathogenesis of Alzheimer-like tangles. J Biol Chem. 2011;286(17):15317–15331. doi: 10.1074/jbc.M110.209296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Volpicelli-Daley LA, et al. Exogenous α-synuclein fibrils induce Lewy body pathology leading to synaptic dysfunction and neuron death. Neuron. 2011;72(1):57–71. doi: 10.1016/j.neuron.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stöhr J, et al. Purified and synthetic Alzheimer’s amyloid beta (Aβ) prions. Proc Natl Acad Sci USA. 2012;109(27):11025–11030. doi: 10.1073/pnas.1206555109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuillé J, Chelle P-L. La maladie dite tremblante du mouton est-elle inoculable. Comptes Rendus Acad Sci. 1936;203:1552–1554. [Google Scholar]
- 13.Gajdusek DC, Gibbs CJ, Alpers M. Experimental transmission of a Kuru-like syndrome to chimpanzees. Nature. 1966;209(5025):794–796. doi: 10.1038/209794a0. [DOI] [PubMed] [Google Scholar]
- 14.Saupe SJ. The [Het-s] prion of Podospora anserina and its role in heterokaryon incompatibility. Semin Cell Dev Biol. 2011;22(5):460–468. doi: 10.1016/j.semcdb.2011.02.019. [DOI] [PubMed] [Google Scholar]
- 15.McGlinchey RP, et al. The repeat domain of the melanosome fibril protein Pmel17 forms the amyloid core promoting melanin synthesis. Proc Natl Acad Sci USA. 2009;106(33):13731–13736. doi: 10.1073/pnas.0906509106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maji SK, et al. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science. 2009;325(5938):328–332. doi: 10.1126/science.1173155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barnhart MM, Chapman MR. Curli biogenesis and function. Annu Rev Microbiol. 2006;60(1):131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morales R, Abid K, Soto C. 2007. The prion strain phenomenon: Molecular basis and unprecedented features. Biochim Biophys Acta 1772(6):681–691.
- 19.Colby DW, et al. Design and construction of diverse mammalian prion strains. Proc Natl Acad Sci USA. 2009;106(48):20417–20422. doi: 10.1073/pnas.0910350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maddelein ML, Dos Reis S, Duvezin-Caubet S, Coulary-Salin B, Saupe SJ. Amyloid aggregates of the HET-s prion protein are infectious. Proc Natl Acad Sci USA. 2002;99(11):7402–7407. doi: 10.1073/pnas.072199199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balguerie A, et al. Domain organization and structure-function relationship of the HET-s prion protein of Podospora anserina. EMBO J. 2003;22(9):2071–2081. doi: 10.1093/emboj/cdg213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sabaté R, et al. Prion and non-prion amyloids of the HET-s prion forming domain. J Mol Biol. 2007;370(4):768–783. doi: 10.1016/j.jmb.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 23.Van Melckebeke H, et al. Atomic-resolution three-dimensional structure of HET-s(218-289) amyloid fibrils by solid-state NMR spectroscopy. J Am Chem Soc. 2010;132(39):13765–13775. doi: 10.1021/ja104213j. [DOI] [PubMed] [Google Scholar]
- 24.Wasmer C, et al. Amyloid fibrils of the HET-s(218-289) prion form a β solenoid with a triangular hydrophobic core. Science. 2008;319(5869):1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 25.Ritter C, et al. Correlation of structural elements and infectivity of the HET-s prion. Nature. 2005;435(7043):844–848. doi: 10.1038/nature03793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wille H, et al. Natural and synthetic prion structure from X-ray fiber diffraction. Proc Natl Acad Sci USA. 2009;106(40):16990–16995. doi: 10.1073/pnas.0909006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wan W, et al. Degradation of fungal prion HET-s(218-289) induces formation of a generic amyloid fold. Biophys J. 2012;102(10):2339–2344. doi: 10.1016/j.bpj.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wan W, et al. Heterogeneous seeding of a prion structure by a generic amyloid form of the fungal prion-forming domain HET-s(218-289) J Biol Chem. 2013;288(41):29604–29612. doi: 10.1074/jbc.M113.505511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jahn TR, et al. The common architecture of cross-β amyloid. J Mol Biol. 2010;395(4):717–727. doi: 10.1016/j.jmb.2009.09.039. [DOI] [PubMed] [Google Scholar]
- 30.Wasmer C, et al. Structural similarity between the prion domain of HET-s and a homologue can explain amyloid cross-seeding in spite of limited sequence identity. J Mol Biol. 2010;402(2):311–325. doi: 10.1016/j.jmb.2010.06.053. [DOI] [PubMed] [Google Scholar]
- 31.Wickner RB, et al. Prions in Saccharomyces and Podospora spp.: Protein-based inheritance. Microbiol Mol Biol Rev. 1999;63(4):844–861. doi: 10.1128/mmbr.63.4.844-861.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perutz MF, Finch JT, Berriman J, Lesk A. Amyloid fibers are water-filled nanotubes. Proc Natl Acad Sci USA. 2002;99(8):5591–5595. doi: 10.1073/pnas.042681399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ross ED, Baxa U, Wickner RB. Scrambled prion domains form prions and amyloid. Mol Cell Biol. 2004;24(16):7206–7213. doi: 10.1128/MCB.24.16.7206-7213.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ross ED, Edskes HK, Terry MJ, Wickner RB. Primary sequence independence for prion formation. Proc Natl Acad Sci USA. 2005;102(36):12825–12830. doi: 10.1073/pnas.0506136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toyama BH, Kelly MJS, Gross JD, Weissman JS. The structural basis of yeast prion strain variants. Nature. 2007;449(7159):233–237. doi: 10.1038/nature06108. [DOI] [PubMed] [Google Scholar]
- 36.Lange A, et al. A combined solid-state NMR and MD characterization of the stability and dynamics of the HET-s(218-289) prion in its amyloid conformation. ChemBioChem. 2009;10(10):1657–1665. doi: 10.1002/cbic.200900019. [DOI] [PubMed] [Google Scholar]
- 37.Sawaya MR, et al. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature. 2007;447(7143):453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 38.Kajava AV, Steven AC. In: Advances in Protein Chemistry. Kajava A, Squire JM, Parry DAD, editors. Vol 73. Amsterdam: Elsevier; 2006. pp. 55–96. [Google Scholar]
- 39.Nelson R, et al. Structure of the cross-β spine of amyloid-like fibrils. Nature. 2005;435(7043):773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang W, Malcolm BA. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange Site-Directed Mutagenesis. Biotechniques. 1999;26(4):680–682. doi: 10.2144/99264st03. [DOI] [PubMed] [Google Scholar]
- 41.McDonald M, Kendall A, Tanaka M, Weissman JS, Stubbs G. Enclosed chambers for humidity control and sample containment in fiber diffraction. J Appl Cryst. 2008;41(1):206–209. [Google Scholar]
- 42.Bian W, Wang H, McCullough I, Stubbs G. WCEN: A computer program for initial processing of fiber diffraction patterns. J Appl Cryst. 2006;39(5):752–756. [Google Scholar]
- 43.Gonzalez A, Nave C, Marvin DA. Pf1 filamentous bacteriophage: Refinement of a molecular model by simulated annealing using 3.3 A resolution X-ray fibre diffraction data. Acta Crystallogr D Biol Crystallogr. 1995;51(Pt 5):792–804. doi: 10.1107/S0907444995003027. [DOI] [PubMed] [Google Scholar]
- 44.Nielsen L, et al. Effect of environmental factors on the kinetics of insulin fibril formation: Elucidation of the molecular mechanism. Biochemistry. 2001;40(20):6036–6046. doi: 10.1021/bi002555c. [DOI] [PubMed] [Google Scholar]
- 45.Lambert RJW, Mytilinaios I, Maitland L, Brown AM. Monte Carlo simulation of parameter confidence intervals for non-linear regression analysis of biological data using Microsoft Excel. Comput Methods Programs Biomed. 2012;107(2):155–163. doi: 10.1016/j.cmpb.2011.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.