Significance
Rapid evolution of drug resistance associated with secondary kinase domain (KD) mutations is the best characterized mechanism of acquired resistance to effective tyrosine kinase inhibitor (TKI) therapy. Medicinal chemistry efforts have largely been devoted toward synthesizing type II TKIs that, by targeting an inactive kinase conformation, are believed to afford greater selectivity than type I TKIs that bind an active kinase conformation. The only previously described TKI with the ability to successfully suppress all resistance-conferring KD mutants (i.e. “pan-kinase” inhibitor) is the type II multikinase TKI ponatinib. Here, we demonstrate that a type I TKI can be potent, selective, and invulnerable to resistance-conferring KD mutation as a mechanism of resistance. Efforts to develop potent, selective type I pan-kinase inhibitors are warranted.
Keywords: sorafenib, activation-loop mutations, D835 mutations
Abstract
Tyrosine kinase inhibitors (TKIs) represent transformative therapies for several malignancies. Two critical features necessary for maximizing TKI tolerability and response duration are kinase selectivity and invulnerability to resistance-conferring kinase domain (KD) mutations in the intended target. No prior TKI has demonstrated both of these properties. Aiming to maximize selectivity, medicinal chemists have largely sought to create TKIs that bind to an inactive (type II) kinase conformation. Here we demonstrate that the investigational type I TKI crenolanib is a potent inhibitor of Fms tyrosine kinase-3 (FLT3) internal tandem duplication, a validated therapeutic target in human acute myeloid leukemia (AML), as well as all secondary KD mutants previously shown to confer resistance to the first highly active FLT3 TKI quizartinib. Moreover, crenolanib is highly selective for FLT3 relative to the closely related protein tyrosine kinase KIT, demonstrating that simultaneous FLT3/KIT inhibition, a prominent feature of other clinically active FLT3 TKIs, is not required for AML cell cytotoxicity in vitro and may contribute to undesirable toxicity in patients. A saturation mutagenesis screen of FLT3–internal tandem duplication failed to recover any resistant colonies in the presence of a crenolanib concentration well below what has been safely achieved in humans, suggesting that crenolanib has the potential to suppress KD mutation-mediated clinical resistance. Crenolanib represents the first TKI to exhibit both kinase selectivity and invulnerability to resistance-conferring KD mutations, which is unexpected of a type I inhibitor. Crenolanib has significant promise for achieving deep and durable responses in FLT3–mutant AML, and may have a profound impact upon future medicinal chemistry efforts in oncology.
Pioneering studies demonstrated that imatinib, the first small molecule tyrosine kinase inhibitor (TKI), binds to an inactive kinase conformation of the Abelson protein tyrosine kinase (ABL) (1). Compounds that bind to kinases in this manner have been termed “type II”, whereas “type I” inhibitors bind to an active conformation. Type II inhibitors typically target the ATP binding region and an adjacent allosteric site available only in an inactive conformation. Because this region is less well conserved among kinases than the ATP binding region, interactions with this area allow for greater selectivity.
Despite the clinical success of imatinib for the treatment of chronic myeloid leukemia (CML), durability of response is compromised by secondary kinase domain (KD) mutations in BCR–ABL (1), many of which destabilize the inactive conformation required for imatinib binding (1). Second-generation ABL inhibitors, dasatinib (type I) and nilotinib (type II), were subsequently developed to retain efficacy against most imatinib-resistant BCR–ABL KD mutants, and each are vulnerable to only approximately five resistance-conferring mutations (2, 3). In contrast, the third-generation type II inhibitor ponatinib has proven largely invulnerable to single nucleotide resistance-conferring KD mutations in vitro (4), and no single nucleotide substitution within BCR–ABL has been associated with acquired resistance in patients (5). Therefore, ponatinib has justifiably been termed a “pan-BCR–ABL” inhibitor and holds significant promise for achieving prolonged remissions. However, ponatinib potently inhibits a number of kinases, which may be responsible for unanticipated serious cardiovascular toxicities that have emerged with this agent. The ability to selectively suppress resistance-conferring mutations remains a highly desirable quality that no TKI has yet demonstrated.
Internal tandem duplication (ITD) mutations in the class III receptor tyrosine kinase (RTK) Fms tyrosine kinase-3 (FLT3) juxtamembrane domain (FLT3–ITD) occur in ∼30% of acute myeloid leukemia (AML) patients (6), and are associated with poor outcomes. An additional subset of AML patients has activating point mutations within the activation loop (AL) of FLT3 (7) that presumably stabilize an active (type I) kinase confirmation. Although initial attempts to target FLT3–ITD with first-generation TKI monotherapy were largely unsuccessful (8, 9), the highly potent second-generation TKI quizartinib (AC220), which primarily targets FLT3, KIT, and RET in the inactive (type II) conformation, has recently demonstrated clearance of bone marrow blasts in a large proportion of chemotherapy-resistant FLT3–ITD+ AML patients (10, 11). Moreover, the demonstration that drug-resistant point mutations in the KD of FLT3–ITD evolve at the time of relapse on quizartinib has formally validated FLT3–ITD as a therapeutic target (12) and rekindled interest in developing potent FLT3 inhibitors. To date, clinically relevant quizartinib-resistant KD mutations in FLT3–ITD are restricted to amino acids F691 and D835 (12). F691 represents the “gatekeeper” position in FLT3, analogous to residues in other kinases that are hotspots for drug-resistance to TKIs (both type I and type II), such as BCR–ABL/T315 (1). Mutations at D835 within the FLT3 AL have been found in the majority of FLT3–ITD+ AML patients with acquired resistance to quizartinib. These substitutions are postulated to destabilize the inactive conformation required for efficient binding by type II TKIs such as quizartinib. Analogous substitutions in KIT (KIT/D816V) and platelet-derived growth factor receptor (PDGFR) (PDGFR/D842V) confer a high degree of resistance to type II TKIs (13, 14). Although a number of potent second-generation type II FLT3 TKIs are currently undergoing clinical development for the treatment of AML and/or have reported single agent activity (e.g., sorafenib) (15), these agents, including the pan-BCR–ABL inhibitor ponatinib, which has potent activity against FLT3, are uniformly ineffective at inhibiting quizartinib-resistant FLT3–ITD/D835 mutants (12, 16). These data suggest that a type I FLT3 TKI will be required to effectively inhibit FLT3–ITD/D835 mutants. However, previous monotherapy clinical experience with type I FLT3 TKIs midostaurin and lestaurtinib demonstrated minimal clinical activity (8, 9), presumably due to a lack of selectivity resulting in dose-limiting toxicities that preclude potent FLT3 inhibition.
Crenolanib is an investigational TKI that was initially developed as a highly selective PDGFR inhibitor (17). A phase I study documented the safe achievement of low micromolar trough plasma concentrations without significant myelosuppression (17), suggesting limited off-target effects in human hematopoietic cells. In kinome assays, crenolanib has demonstrated high selectivity for class III RTKs (including FLT3 and PDGFR) at clinically achievable drug concentrations (18), with activity against only a limited number of other kinases (19). Notably, crenolanib retains activity against the PDGFR/D842V mutant, analogous to the quizartinib-resistant FLT3–ITD/D835V substitution (18). Two recent reports suggest that crenolanib is a type I TKI that harbors activity against a subset of quizartinib-resistant FLT3–ITD/D835 mutants in vitro (19, 20) and in vivo (20), although these reports differed as to the degree of sensitivity of FLT3–ITD/D835 mutations compared with FLT3–ITD. Based upon our previous molecular docking studies with quizartinib and our proposed model of resistance conferred by D835 substitutions, we hypothesized that if crenolanib is a type I inhibitor, D835 substitutions would be expected to be equivalently sensitive to crenolanib compared with FLT3–ITD. Along with D835 mutations, activating substitutions at FLT3 AL residue Y842 have also been described in AML patients (21) and have been found to confer resistance to both quizartinib and sorafenib preclinically when found in the context of the FLT3–ITD (12). We therefore sought to assess the activity of crenolanib against a full panel of known TKI-resistant FLT3–ITD KD substitutions, including Y842 mutations. Additionally, because clinical trials of crenolanib in FLT3-mutant AML are ongoing, we aimed to prospectively identify secondary mutations in FLT3–ITD that can cause crenolanib resistance.
Results
Type I TKI Crenolanib Retains Activity Against FLT3 D835 Mutations.
Confirming the activity of crenolanib against FLT3–ITD, we observed that crenolanib treatment prolonged survival in a FLT3–ITD+ murine model of AML previously found to be sensitive to FLT3 TKI treatment (22) (Fig. S1). As crenolanib has been reported to bind kinases in a type I manner as evidenced by its relative affinity for phosphorylated versus nonphosphorylated ABL (20), we assessed the ability of crenolanib to inhibit FLT3 phosphorylation and downstream signaling in a D835–mutant acute-lymphoblastic-leukemia-patient–derived cell line, HB119, which harbors a FLT3 D835H activating mutation in the absence of an ITD. Crenolanib treatment abolished phosphorylation of FLT3 and ERK in these cells, as well as in the AML-patient–derived FLT3–ITD+ cell line Molm14 (Fig. 1A). Both cell lines underwent apoptosis in the presence of crenolanib, whereas control cell lines were minimally affected (Fig. 1B). Additionally, a quizartinib-resistant subclone of the Molm14 cell line that acquired a D835Y mutation through exposure to gradually escalating concentrations of quizartinib retained sensitivity to crenolanib equivalent to parental Molm14 cells (Fig. S2). Fifty nanomolar crenolanib suppressed phosphorylation of FLT3 in primary isolates, including in leukemic blasts from a quizartinib-resistant patient whose disease had evolved a FLT3–ITD/D835Y mutation (Fig. 1C). The clonogenic potential of primary AML cells from a patient with FLT3–ITD/D835Y was significantly reduced (Fig. S3). These data corroborate the findings of recent reports that described the activity of crenolanib in vivo against FLT3–ITD/D835 mutant leukemias in murine models (20) and ex vivo activity against patient blasts containing FLT3 D835 mutations with and without an ITD (19, 20).
Fig. 1.
Activity of crenolanib in cells expressing FLT3 D835 mutations. (A) Western blot analysis using 4G10 and anti-FLT3 antibody after immunoprecipitation with anti-FLT3 antibody and Western blot analysis of phospho-ERK (pERK) and ERK performed on whole cell lysates from HB119 and Molm14 cells. Cells were exposed to 100 nM crenolanib for 60 min. (B) Average normalized percentage of live cells as assessed by caspase-3 activation at 72 h and 96 h following treatment with 100 nM crenolanib for HB119, HL60, K562, and Molm14 cells (error bars represent SD of triplicate experiments). (C) Western blot analysis using 4G10 and anti-FLT3 antibody after immunoprecipitation with anti-FLT3 antibody on lysates prepared from MV4;11 cells or primary patient blasts exposed for 60 min to DMSO or 50 nM crenolanib as indicated: 2830, patient FLT3–ITD+/D835Y+, refractory quizartinib; 1727, newly diagnosed patient FLT3–ITD+. (D) Western blot analysis as described in A, including phospho-STAT5 (pSTAT5), phospho-S6 (pS6), STAT5, and S6 on whole cell lysates from parental Molm14 cells and Molm14 cells expressing the D835Y mutation. Cells were exposed to crenolanib in human plasma for 120 min.
Because extensive plasma protein binding has been previously shown to compromise the activity of FLT3 TKIs (23), we assessed the biochemical activity of crenolanib in Molm14 and Molm14–D835Y cells in the presence of human plasma. High nanomolar crenolanib concentrations, well below those achieved clinically (17), successfully inhibited phosphorylation of FLT3 and downstream targets STAT5 and S6 (Fig. 1D).
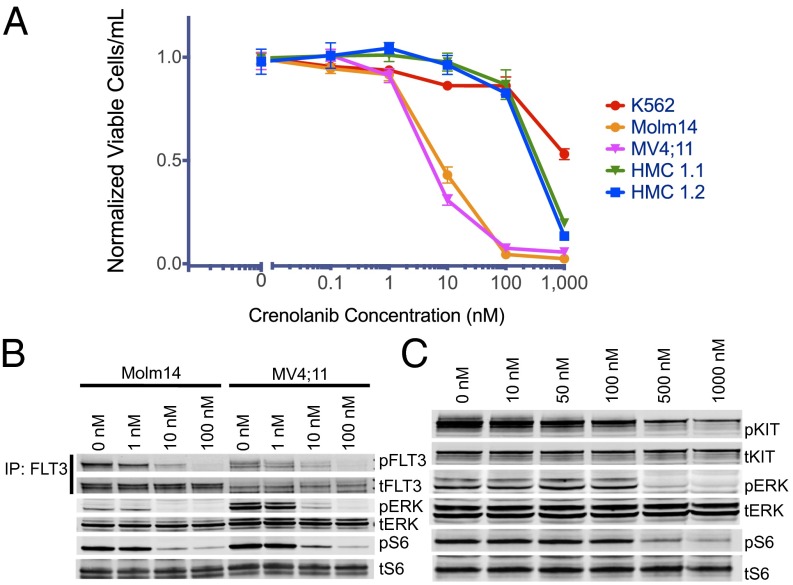
Crenolanib Is ∼100-Fold More Selective for FLT3 than KIT.
All clinically active FLT3 TKIs [quizartinib (10, 11), ponatinib (24), and sorafenib (15)] are multikinase inhibitors that also inhibit KIT. Because crenolanib has been reported to bind selectively to class III RTKs (18), we assessed its activity in a cell line that harbors an activating KIT D816 mutation (analogous to D835 in FLT3). Although crenolanib inhibited proliferation and induced cytotoxicity at low nanomolar concentration (IC50 ∼10 nM) in two FLT3–ITD-dependent patient-derived AML cell lines (Molm14 and MV4;11), no impact on proliferation was observed even at substantially higher concentrations (100 nM) in KIT-driven cell lines, HMC 1.1 (V560G+, D816V-) and HMC 1.2 (V560G, D816V+) (Figs. 1B and 2A). While low nanomolar concentrations of crenolanib led to a substantial reduction of phospho-FLT3 as well as diminished phosphorylation of downstream signaling effectors ERK and S6 (Fig. 2B), minimal effect was seen in KIT-dependent lines (Fig. 2C) in the absence of much higher drug concentrations. Crenolanib is therefore highly selective toward FLT3 relative to KIT. These data strongly suggest that potent FLT3 inhibition (without KIT inhibition) is sufficient to effect cytotoxicity in FLT3–ITD+ AML cells in vitro.
Fig. 2.
Crenolanib Is 100-fold more selective for FLT3 compared with KIT. (A) Normalized cell viability of Molm14, MV4;11, HMC 1.1, HMC 1.2, and K562 cells after 48 h in various concentrations of crenolanib (error bars represent SD of triplicates from the same experiment). (B) Western blot analysis using 4G10 and anti-FLT3 antibody after immunoprecipitation with anti-FLT3 antibody and Western blot analysis of pSTAT5, pERK, anti-STAT5, and anti-ERK antibody performed on whole cell lysates from MV4;11 and Molm14 cells. Cells were exposed to crenolanib for 60 min. (C) Western blot analysis of pKIT (Y703), pERK, pS6, KIT, ERK, and S6 antibody performed on whole cell lysates from HMC 1.2 cells. Cells were exposed to crenolanib for 60 min.
Crenolanib Retains Activity Against TKI-Resistant FLT3 Mutants.
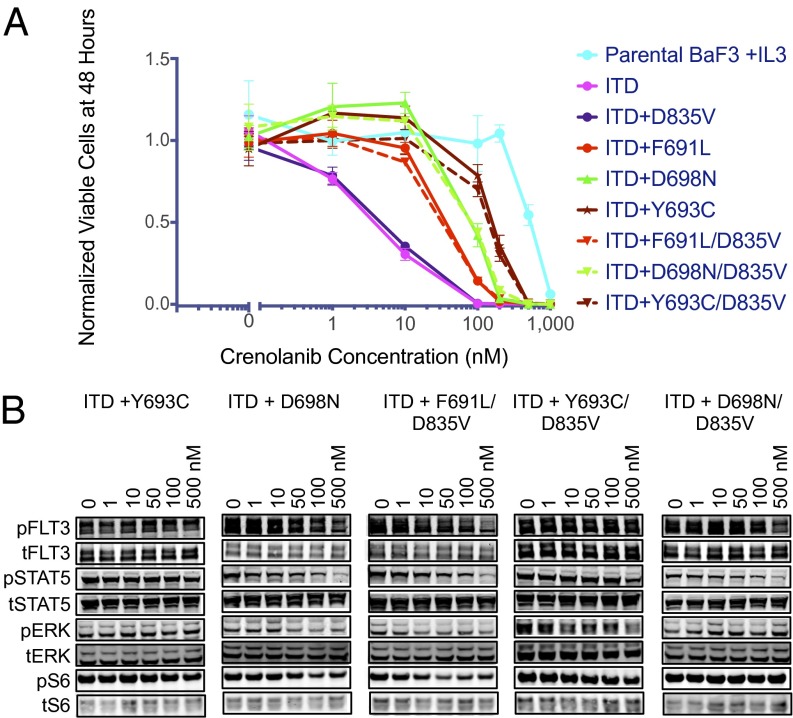
Based on the activity of crenolanib against quizartinib-resistant FLT3–ITD/D835 mutants, we next assessed the activity of crenolanib in cell-based assays against an expanded panel of FLT3–ITD mutants known to confer quizartinib or sorafenib resistance in preclinical and/or in clinical studies (12, 15, 25). We found that cells expressing differing substitutions at the FLT3–ITD D835 residue were uniformly sensitive to crenolanib, whereas the FLT3–ITD/F691L mutant demonstrated only mildly reduced sensitivity (Fig. 3A and Table S1). Similar results were observed in Molm14–D835Y and F691L mutant cell lines (Fig. S2). In the absence of an ITD mutation, FLT3 AL mutants D835V and D835Y were highly sensitive to crenolanib (Fig. 3A and Table S1), indicating that crenolanib may be effective in treating the subset of AML patients with activating point mutations in the FLT3 AL in the absence of an ITD. Crenolanib also inhibited the proliferation of FLT3–ITD Y842 mutants, which have been associated with preclinical resistance to quizartinib and sorafenib (12), at concentrations equivalent to those effective against FLT3–ITD D835 mutants (Fig. 3A and Table S1). In all cases, crenolanib-mediated cell growth inhibition was associated with a reduction of FLT3 phosphorylation and downstream signaling (Fig. 3B).
Fig. 3.
Activity of crenolanib against quizartinib resistance-causing FLT3–ITD KD mutations. (A) Normalized cell viability of Ba/F3 populations stably expressing FLT3–ITD mutant isoforms after 48 h in various concentrations of crenolanib (error bars represent SD of triplicates from the same experiment). (B) Western blot analysis using pFLT3, pSTAT5, pERK, pS6, FLT3, STAT5, ERK, and S6 performed on lysates from IL-3–independent Ba/F3 populations expressing the FLT3–ITD mutant isoforms indicated. Cells were exposed to crenolanib for 90 min.
Saturation Mutagenesis Reveals No Crenolanib-Resistant KD Mutations.
Anticipating that a significant proportion of patients initially treated with crenolanib will have previously acquired resistance to quizartinib and sorafenib due to evolution of D835 mutations, we sought to prospectively identify secondary mutations in the KD of FLT3–ITD that can confer resistance to crenolanib, both in the presence or absence of a preexisting D835 mutation. We used a well-validated saturation mutagenesis assay (12) to randomly mutagenize FLT3–ITD and FLT3–ITD/D835V. We initially selected for crenolanib-resistant clones in the presence of 200 nM crenolanib, which represents ∼20× the IC50 for unmutated FLT3–ITD in cells (similar to the relative concentration of quizartinib used in our previous screen) (12) and is well below the concentration safely achieved in humans (17). Surprisingly, we detected no drug-resistant colonies at this concentration. A second mutagenesis screen was then performed using 100 nM crenolanib (∼10× IC50 against native FLT3–ITD). Despite infecting 300 × 106 cells per construct under these conditions (compared with 120 × 106 in our prior screen for quizartinib resistance, which generated ∼100 independent resistant clones), we identified only two mutations recurrently in multiple clones: F691L and D698N. Of these, the D698N substitution conferred the greater degree of resistance (∼10-fold) in proliferation and biochemical assays when independently created and reintroduced into Ba/F3 cells, both in the presence and absence of the D835V mutation (Fig. 4 A and B and Table S2). We also identified single clones containing Y693C, F729L, and N841H mutations. Of these, only Y693C conferred resistance (∼15-fold) when independently created and introduced into Ba/F3 cells, both in the setting of FLT3–ITD and FLT3–ITD/D835V (Fig. 4 A and B and Table S2). In aggregate, these data suggest that at clinically achievable concentrations, crenolanib is invulnerable to resistance-conferring secondary KD mutations in FLT3–ITD. These results mirror those of ponatinib with BCR–ABL, where no single mutations were found to confer resistance at concentrations achievable in human plasma (4).
Fig. 4.
Activity of crenolanib against FLT3–ITD KD mutations identified in an in vitro mutagenesis screen. (A) Normalized cell viability of Ba/F3 populations stably expressing FLT3–ITD mutant isoforms after 48 h in various concentrations of crenolanib (error bars represent SD of triplicates from the same experiment). (B) Western blot analysis of pFLT3, pSTAT5, pERK, pS6, FLT3, STAT5, ERK, and S6 performed on lysates from IL-3–independent Ba/F3 populations expressing the FLT3–ITD mutant isoforms indicated. Cells were exposed to crenolanib for 90 min.
Although crenolanib is highly selective for FLT3 (18, 19), it has been reported to bind a limited number of other kinases at the 100 nM concentration used in our screen, including Unc-51–like kinase 2 (ULK2), SNARK, JAK3, Trk system potassium uptake protein (TRKA), ROCK2, CDK7, mixed-lineage kinase 1 (MLK1), and TYK2 (19). To test whether our inability to recover highly resistant clones in crenolanib could be due to off-target toxicity at this drug concentration, we assessed the ability of crenolanib to inhibit the biochemical activity of these kinases in vitro. As expected, native and D835Y–mutant FLT3 kinase activity was potently inhibited at 100 nM crenolanib, but of the other targets tested, only PDGFRα D842V, ULK2, MLK1, and TRKA were inhibited to <50% of control (Fig. S4). Importantly, crenolanib failed to induce apoptosis in non-FLT3–driven cell lines, including parental and BCR–ABL-transformed Ba/F3 cells at concentrations of crenolanib as high as 500 nM (Fig. S5), arguing that our inability to select highly resistant substitutions is not a consequence of off-target toxicity.
Crenolanib-Resistant Mutations Confer Cross-Resistance to Other Type I FLT3 Inhibitors.
Although the type II inhibitors quizartinib, sorafenib, and ponatinib have all demonstrated a high degree of vulnerability to FLT3 AL mutations (12, 15, 16), of the few crenolanib-resistant mutations identified, only the F691L mutant conferred cross-resistance to quizartinib and sorafenib. Ponatinib retained activity against all three mutants (F691L, Y693C, and D698N) (Table S3). Interestingly, the type I FLT3 inhibitors (PKC412 and sunitinib) exhibited vulnerability to the crenolanib-resistant Y693C and D698N mutants, although they largely retained activity against the F691L mutant (Table S3).
Molecular Docking Studies Reveal Molecular Interaction of Crenolanib with FLT3.
As binding data support that crenolanib is a type I kinase inhibitor that binds preferentially to the active kinase conformation (20), we modeled the binding of crenolanib to the active conformation of FLT3 in an effort to understand the structural basis of FLT3 inhibition by crenolanib as well as how select mutants confer modest resistance. Although the active conformation of FLT3 has not yet been reported, the crystal structure of KIT, which shares 64.8% sequence identity with FLT3 KD, has been determined in an active conformation (26). In this KIT conformation, the AL adopts an extended conformation (loop-out conformation) that is compatible with substrate binding. The DFG motif at the amino-terminal end of the AL adopts the DFG-in conformation, in which the Asp side chain is in position to coordinate a magnesium ion bound to ATP. We constructed a model for FLT3 using the KIT structure as a template (Fig. 5A) and used this to dock crenolanib into the ATP-binding site. The docking studies revealed nine different binding poses of crenolanib at the ATP-binding site. Although the top scoring docked model is not well separated from the other docked models in terms of the docking scores, we favor this model for several reasons. In this pose, crenolanib makes hydrogen bonds with the hinge region of the kinase, a classical attribute of kinase inhibitors. A hydrophobic face of crenolanib packs against the gatekeeper region of FLT3, forming interactions with F691. This pose also involves minimal distortion of side chains in the active site of FLT3. In this pose, F691 and Y693 form aromatic interactions with the benzimidazole–quinolin moiety in crenolanib (Fig. 5B). Substitution of these residues to nonaromatic residues such as leucine and cystine might therefore be energetically unfavorable. D698 interacts with the positively charged amine group in the piperidin moiety of crenolanib (Fig. 5B). Mutation of D698 to a neutral residue such as asparagine is expected to destabilize the interaction between crenolanib and FLT3. The 3′ nitrogen in the crenolanib–benzimidazole moiety interacts with the main-chain amide group of C694. In addition, L616 and L818 each form hydrophobic packing interactions with the benzimidazole–quinolin moiety, further stabilizing the binding between crenolanib and FLT3 (Fig. 5B).
Fig. 5.
Modeling of FLT3–crenolanib interactions. (A) Cartoon presentation of modeled FLT3 KD in an active conformation. Two orthogonal views are shown. The AL, Helix-C, and P-loop are colored in yellow, pink, and green, respectively. Blue-colored crenolanib is presented in stick and surface mode. The figures were made by PyMOL. (B) Two orthogonal views of coordination of crenolanib in the top scored docking pose. Crenolanib is in blue. The FLT3 KD residues that coordinate crenolanib binding are colored in yellow and white.
Discussion
Two of the most highly prized attributes in the design of kinase inhibitors are kinase selectivity and invulnerability to secondary resistance-conferring KD mutations. Unusual for a type I inhibitor, crenolanib has been found to display a high degree of selectivity for FLT3 and other class III RTKs (18, 19). Here, we provide evidence that crenolanib, currently undergoing evaluation in phase II clinical trials of FLT3–mutant AML, also appears invulnerable to resistance-conferring KD mutations in vitro. To increase our chances of identifying crenolanib-resistant mutations in Ba/F3 cells infected with randomly mutagenized FLT3–ITD-expressing retrovirus, we screened ∼2.5× the number of cells that we assessed in a previous successful screen for quizartinib-resistant mutations (which yielded ∼100 independent resistant clones), yet we still failed to identify any highly crenolanib-resistant mutants. We nonetheless cannot formally rule out the possibility that our mutagenesis screen was not fully saturating, and translational studies will be necessary to confirm our findings. Efforts to understand the molecular interactions of crenolanib with the KD of FLT3 are ongoing. These studies will be essential to inform our understanding of how this molecule achieves its selectivity and appears to retain activity against all resistance-conferring KD mutants.
Although the type II TKI ponatinib provides the only prior example of a clinically active “pan-kinase” inhibitor for BCR–ABL, it is noteworthy that ponatinib is not a pan-inhibitor of FLT3, as evidenced by its lack of activity against AL mutants in FLT3 (16). In addition, ponatinib is not selective. Other kinases are associated with highly TKI-resistant AL mutations, including KIT (13) and PDGFR (27), and translational studies (12, 15, 25) demonstrate that these mutations pose a major barrier to achieving durable clinical responses. Our data demonstrate that it is not only possible but preferable to develop potent and selective type I inhibitors for these kinases. As expected of a type I inhibitor, we have found that crenolanib retains full activity against AL mutants in FLT3–ITD, particularly those at the D835 residue, which favor the active conformation of FLT3 and represent the most common cause of acquired resistance to quizartinib (12). These FLT3 AL mutations notably confer a high degree of cross-resistance to all type II FLT3 inhibitors (15, 16). The ability of crenolanib to successfully inhibit the kinase activity of all AL mutants holds therapeutic promise for AML patients who acquire resistance to quizartinib and other investigational type II FLT3 inhibitors through FLT3–ITD/D835 substitutions, and also for AML patients with FLT3/D835 mutations in the absence of an ITD, which are associated with clinical resistance to quizartinib (28).
As a pan-BCR–ABL inhibitor (4), ponatinib represented a major therapeutic advance for the treatment of TKI-resistant CML (29). Of note, the BCR–ABL/E255V KD mutant confers 72-fold resistance to ponatinib relative to native BCR–ABL in Ba/F3 cells, yet this mutant retains clinical sensitivity (29). In contrast, the least sensitive FLT3–ITD KD mutant we identified in our in vitro mutagenesis screen conferred only ∼15-fold relative resistance to crenolanib. Importantly, although we identified substitutions at three residues (F691, Y693, and D698) that confer mild resistance to crenolanib, the safe achievement of low micromolar plasma concentrations in humans (17) suggests that, similar to ponatinib, these mutants will remain sensitive in patients, although translational studies of clinical samples obtained from patients undergoing treatment will be required to definitively confirm this prediction. In the event that these mutants are found to confer clinical resistance to crenolanib, their in vitro sensitivity to clinically active type II FLT3 inhibitors suggests that they will be responsive to these agents (Table S3). Because resistance-conferring KD mutations account for the majority of acquired clinical resistance to quizartinib and sorafenib, response durability observed with crenolanib may be greater than that achieved with current FLT3 TKIs.
Crenolanib has several appealing characteristics for the treatment of FLT3–mutant AML. Whereas the type II FLT3 TKIs quizartinib, sorafenib, and ponatinib efficiently target the related class III RTK KIT, we demonstrate that crenolanib elicits cytotoxicity in FLT3–mutant AML while largely sparing KIT inhibition (100-fold offset in both viability and biochemical assays). Furthermore, crenolanib has limited activity against other kinases. As a result, crenolanib may be associated with less toxicity than the available type II FLT3 TKIs. Combined FLT3 and KIT knockout has been shown to yield profound hematopoietic defects in mice (30), and quizartinib, which potently inhibits both FLT3 and KIT, is associated with a substantial incidence of severe myelosuppression in clinical experience (10, 11). Encouragingly, severe myelosuppression was not observed in a phase I study of crenolanib in patients with nonhematologic malignancies (17). Additionally, cardiac ventricular repolarization abnormalities have hampered the clinical development of quizartinib, but do not appear to be associated with crenolanib treatment (17). Previous work has suggested that binding of FLT3 TKIs by plasma proteins may negatively impact the their clinical efficacy (23). However, crenolanib potently inhibits FLT3–ITD and the common quizartinib-resistant isoform FLT3–ITD/D835Y in the presence of human plasma at concentrations below those safely achieved in the first-in-human trial. Encouragingly, Galanis et al. recently reported that plasma obtained from AML patients treated with crenolanib strongly inhibited FLT3 phosphorylation in Molm14 cells and in D835V+ patient blasts (19).
As the first potent FLT3 TKI to retain activity against D835 mutants and other FLT3 mutant isoforms at concentrations well below those safely achieved in clinical trial experience, crenolanib has substantial promise to effect durable clinical responses and may represent a critical advance in the treatment of FLT3–mutant AML. More broadly, as the first example of a potent and selective pan-kinase inhibitor that binds in a type I manner, crenolanib may have a significant impact upon the future direction of medicinal chemistry in oncology.
Materials and Methods
Inhibitors.
Crenolanib, sunitinib, PKC412, and quizartinib were purchased from Selleckchem. Sorafenib was purchased from LC Labs.
Cell Lines.
Stable Ba/F3 lines were generated by retroviral spinfection with the appropriate mutated plasmid as previously described (12).
Cell-Viability Assay.
Exponentially growing cells (5 × 103 cells per well) were plated in each well of a 96-well plate with 0.1 mL of RPMI 1640 + 10% (vol/vol) FCS containing the appropriate concentration of drug in triplicate, and cell viability was assessed after 48 h as previously described (16).
Assessment of Caspase-3 Activation.
Exponentially growing cells were plated in the presence of crenolanib in RPMI + 10% (vol/vol) FCS for 48 h. Cells were fixed with 4% (vol/vol) paraformaldehyde (Electron Microscopy Sciences) and permeabilized with 100% (vol/vol) methanol (Electron Microscopy Sciences) followed by staining with a FITC-conjugated antiactive caspase-3 antibody (BD Pharmingen). Cells were run on a BD LSRFortessa cell analyzer, and data were analyzed using FlowJo (Tree Star Inc.). Percentage of live cells was determined by negative staining for activated caspase-3.
Immunoblotting.
Exponentially growing Molm14, HB119, or Ba/F3 cells stably expressing mutant isoforms were plated in RPMI medium 1640 + 10% (vol/vol) FCS supplemented with crenolanib at the indicated concentration. HMC1.2 cells were cultured and treated in IMDM + 10% (vol/vol) FCS. After a 90-min incubation, the cells were washed in PBS and lysed and processed as previously described (16). Immunoblotting was performed using anti–phospho-FLT3, anti–phospho-KIT, anti–phospho-STAT5, anti-STAT5, anti–phospho-ERK, anti-ERK, anti–phospho-S6, anti-S6, anti-KIT (Cell Signaling), and anti-FLT3 S18 antibody (Santa Cruz Biotechnology).
Plasma Inhibitory Assay.
Plasma inhibitory assay was performed as previously described (16). All samples were collected under the University of California, San Francisco institutional review board (IRB)-approved cell banking protocol (CC#112514). Informed consent was obtained in accordance with the Declaration of Helsinki.
Assessment of FLT3 Phosphorylation and Colony Assays in Primary Patient Blasts.
Primary AML blood samples and/or marrow aspirates were obtained on an IRB-approved protocol at the University of Pennsylvania. Informed consent was obtained in accordance with the Declaration of Helsinki. Mononuclear cells were purified by density centrifugation (Ficoll-Paque Plus, GE Healthsciences) before cryopreservation in 10% (vol/vol) DMSO and FCS or immediate use in assays. For protein analysis, thawed samples were incubated for 1 h in RPMI medium 1640 + 10% (vol/vol) FCS and crenolanib or DMSO before immunoprecipitation and immunoblotting as above with the following modifications: cell pellets from 10 million cells per condition were lysed in ice-cold lysis buffer (50 mM Tris⋅HCl, pH 7.6, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Nonidet P-40) containing a mixture of phosphatase inhibitors and protease inhibitors (Pierce). After clarification by centrifugation, a minimum of 500 μg of lysate was immunoprecipitated at 4 °C overnight with anti-FLT3 S18 antibody and subjected to Western blot with 4G10 and anti-FLT3 S18 antibody. Clonogenic assays were performed as previously described (31).
DNA Constructs, Mutagenesis, and Resistance Screen.
Random mutagenesis was performed as previously described (12). Cells were selected in 100 nM and 200 nM crenolanib in soft agar. After 10–21 d, visible colonies were plucked and expanded in 100 nM crenolanib.
Sequencing and Alignments.
Sequencing was performed from amplified genomic DNA from colonies expanded from soft agar as previously described (12).
Generation of Mutants.
Mutations isolated in the screen were engineered into pMSCVpuroFLT3–ITD by QuikChange mutagenesis (Stratagene) as previously described (12).
Homology Modeling of the Active Conformation of FLT3.
Homology modeling was performed using Modeler (32) via program Chimera (33). The sequence of the KDs of FLT3 and KIT were aligned by ClustalW2 (www.ebi.ac.uk/Tools/msa/clustalw2/). There are two molecules in the asymmetric unit of the KIT crystal structure (Protein Data Bank ID code 1PKG) (26), and both were used as templates for modeling the structure of FLT3. We chose the model with lowest Discrete Optimized Protein Energy (-0.86) as representative of the active conformation of FLT3 for the docking studies.
Molecular Docking of Crenolanib.
Molecular docking was performed using Autodock 4.2 (34). Hydrogens were added to the modeled FLT3 KD, and partial atomic charges were assigned using AutoDockTools (ADT) (34). The coordinates of crenolanib were generated using the Dundee PROGRD2 server (35), and its initial conformation was energy minimized using the GROMACS force field. The Gasteiger partial charges (36) were then assigned to the ligand using ADT. Six torsion angles were defined as flexible during the docking procedure. The ligand was put into the kinase ATP-binding pocket and aligned manually to avoid atom clashes. A 3D grid box (dimensions, 60 × 60 × 60 unit in number of grid points; grid spacing, 0.375 Å) centered at the ATP-binding pocket was then created by AutoGrid4.2. Fifteen hundred runs of Larmarckian Genetic Algorithm were performed to optimize the ligand–protein interactions. The solutions were ranked by the calculated binding free energy.
We first performed docking calculations with FLT3 treated as rigid body. Residues L616, V624, K644, F691, Y693, C695, Y696, D698, N816, L818, and D829 were found to interact with crenolanib in close proximity to it in the results of the docking calculation. We then performed docking runs with each of these residues defined as flexible, one at a time. The dockings studies revealed that the optimized conformations of C695, N816, E829, and K644 in the docked structures are similar to their initial conformations, and so only L616, V624, F691, Y693, Y696, D698, and L818 were chosen as flexible residues. The top 5% of the docking solutions (80 out of 1,500) as ranked by the calculated binding free energy were clustered into nine distinct binding poses. The average calculated energy of these nine poses is −6.98 kcal/mol, and the lowest calculated energy is −7.75 kcal/mol. All nine docking poses were analyzed.
Supplementary Material
Acknowledgments
The authors acknowledge Daniel Treiber, Kevin Shannon, and Mark Levis for helpful discussions. C.C.S. is an American Society of Hematology Faculty Scholar, Leukemia and Lymphoma Society (LLS) Special Fellow in Clinical Research, and recipient of a Hellman Family Foundation Early Career Faculty Award. N.P.S. is an LLS Scholar in Clinical Research. This work was supported in part by National Cancer Institute Grants 1R01 CA176091-01 (to N.P.S.), 5R01 CA095274 (to S.C.K.), and 5T32CA108462-08 (to E.A.L.), and by LLS Grant TRP 6360-13 (to N.P.S.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1320661111/-/DCSupplemental.
References
- 1.Shah NP, et al. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2(2):117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 2.Burgess MR, Skaggs BJ, Shah NP, Lee FY, Sawyers CL. Comparative analysis of two clinically active BCR-ABL kinase inhibitors reveals the role of conformation-specific binding in resistance. Proc Natl Acad Sci USA. 2005;102(9):3395–3400. doi: 10.1073/pnas.0409770102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradeen HA, et al. Comparison of imatinib mesylate, dasatinib (BMS-354825), and nilotinib (AMN107) in an N-ethyl-N-nitrosourea (ENU)-based mutagenesis screen: High efficacy of drug combinations. Blood. 2006;108(7):2332–2338. doi: 10.1182/blood-2006-02-004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Hare T, et al. AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer Cell. 2009;16(5):401–412. doi: 10.1016/j.ccr.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cortes JE, et al. PACE Investigators A phase 2 trial of ponatinib in Philadelphia chromosome-positive leukemias. N Engl J Med. 2013;369(19):1783–1796. doi: 10.1056/NEJMoa1306494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kottaridis PD, et al. The presence of a FLT3 internal tandem duplication in patients with acute myeloid leukemia (AML) adds important prognostic information to cytogenetic risk group and response to the first cycle of chemotherapy: Analysis of 854 patients from the United Kingdom Medical Research Council AML 10 and 12 trials. Blood. 2001;98(6):1752–1759. doi: 10.1182/blood.v98.6.1752. [DOI] [PubMed] [Google Scholar]
- 7.Yamamoto Y, et al. Activating mutation of D835 within the activation loop of FLT3 in human hematologic malignancies. Blood. 2001;97(8):2434–2439. doi: 10.1182/blood.v97.8.2434. [DOI] [PubMed] [Google Scholar]
- 8.Stone RM, et al. Patients with acute myeloid leukemia and an activating mutation in FLT3 respond to a small-molecule FLT3 tyrosine kinase inhibitor, PKC412. Blood. 2005;105(1):54–60. doi: 10.1182/blood-2004-03-0891. [DOI] [PubMed] [Google Scholar]
- 9.Knapper S, et al. A phase 2 trial of the FLT3 inhibitor lestaurtinib (CEP701) as first-line treatment for older patients with acute myeloid leukemia not considered fit for intensive chemotherapy. Blood. 2006;108(10):3262–3270. doi: 10.1182/blood-2006-04-015560. [DOI] [PubMed] [Google Scholar]
- 10.Levis MJ, et al. Final results of a phase 2 open-label, monotherapy efficacy and safety study of quizartinib (AC220) in patients with FLT3-ITD positive or negative relapsed/refractory acute myeloid leukemia after second-line chemotherapy or hematopoietic stem cell transplantation. ASH Annual Meeting Abstracts. 2012;120(21):673. [Google Scholar]
- 11.Cortes JE, et al. Final results of a phase 2 open-label, monotherapy efficacy and safety study of quizartinib (AC220) in patients >= 60 years of age with FLT3 ITD positive or negative relapsed/refractory acute myeloid leukemia. ASH Annual Meeting Abstracts. 2012;120(21):48. [Google Scholar]
- 12.Smith CC, et al. Validation of ITD mutations in FLT3 as a therapeutic target in human acute myeloid leukaemia. Nature. 2012;485(7397):260–263. doi: 10.1038/nature11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gajiwala KS, et al. KIT kinase mutants show unique mechanisms of drug resistance to imatinib and sunitinib in gastrointestinal stromal tumor patients. Proc Natl Acad Sci USA. 2009;106(5):1542–1547. doi: 10.1073/pnas.0812413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weisberg E, et al. Effects of PKC412, nilotinib, and imatinib against GIST-associated PDGFRA mutants with differential imatinib sensitivity. Gastroenterology. 2006;131(6):1734–1742. doi: 10.1053/j.gastro.2006.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Man CH, et al. Sorafenib treatment of FLT3-ITD(+) acute myeloid leukemia: Favorable initial outcome and mechanisms of subsequent nonresponsiveness associated with the emergence of a D835 mutation. Blood. 2012;119(22):5133–5143. doi: 10.1182/blood-2011-06-363960. [DOI] [PubMed] [Google Scholar]
- 16.Smith CC, et al. Activity of ponatinib against clinically-relevant AC220-resistant kinase domain mutants of FLT3-ITD. Blood. 2013;121(16):3165–3171. doi: 10.1182/blood-2012-07-442871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis NL, et al. Phase I study of the safety, tolerability, and pharmacokinetics of oral CP-868,596, a highly specific platelet-derived growth factor receptor tyrosine kinase inhibitor in patients with advanced cancers. J Clin Oncol. 2009;27(31):5262–5269. doi: 10.1200/JCO.2009.21.8487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heinrich MC, et al. Crenolanib inhibits the drug-resistant PDGFRA D842V mutation associated with imatinib-resistant gastrointestinal stromal tumors. Clin Cancer Res. 2012;18(16):4375–4384. doi: 10.1158/1078-0432.CCR-12-0625. [DOI] [PubMed] [Google Scholar]
- 19.Galanis A, et al. Crenolanib is a potent inhibitor of FLT3 with activity against resistance-conferring point mutants. Blood. 2014;123(1):94–100. doi: 10.1182/blood-2013-10-529313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zimmerman EI, et al. Crenolanib is active against models of drug-resistant FLT3-ITD-positive acute myeloid leukemia. Blood. 2013;122(22):3607–3615. doi: 10.1182/blood-2013-07-513044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kindler T, et al. Identification of a novel activating mutation (Y842C) within the activation loop of FLT3 in patients with acute myeloid leukemia (AML) Blood. 2005;105(1):335–340. doi: 10.1182/blood-2004-02-0660. [DOI] [PubMed] [Google Scholar]
- 22.Sohal J, et al. A model of APL with FLT3 mutation is responsive to retinoic acid and a receptor tyrosine kinase inhibitor, SU11657. Blood. 2003;101(8):3188–3197. doi: 10.1182/blood-2002-06-1800. [DOI] [PubMed] [Google Scholar]
- 23.Levis M, et al. Plasma inhibitory activity (PIA): A pharmacodynamic assay reveals insights into the basis for cytotoxic response to FLT3 inhibitors. Blood. 2006;108(10):3477–3483. doi: 10.1182/blood-2006-04-015743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shah NP, et al. Ponatinib in patients with refractory acute myeloid leukaemia: Findings from a phase 1 study. Br J Haematol. 2013;162(4):548–552. doi: 10.1111/bjh.12382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baker SD, et al. Emergence of polyclonal FLT3 tyrosine kinase domain mutations during sequential therapy with sorafenib and sunitinib in FLT3-ITD-positive acute myeloid leukemia. Clin Cancer Res. 2013;19(20):5758–5768. doi: 10.1158/1078-0432.CCR-13-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mol CD, et al. Structure of a c-kit product complex reveals the basis for kinase transactivation. J Biol Chem. 2003;278(34):31461–31464. doi: 10.1074/jbc.C300186200. [DOI] [PubMed] [Google Scholar]
- 27.Corless CL, et al. PDGFRA mutations in gastrointestinal stromal tumors: Frequency, spectrum and in vitro sensitivity to imatinib. J Clin Oncol. 2005;23(23):5357–5364. doi: 10.1200/JCO.2005.14.068. [DOI] [PubMed] [Google Scholar]
- 28.Smith CC, et al. Constitutively activating mutations at the FLT3 activation loop residue D835 are associated with clinical resistance to AC220. ASH Annual Meeting Abstracts. 2012;120(21):674. [Google Scholar]
- 29.Cortes JE, et al. Ponatinib in refractory Philadelphia chromosome-positive leukemias. N Engl J Med. 2012;367(22):2075–2088. doi: 10.1056/NEJMoa1205127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mackarehtschian K, et al. Targeted disruption of the flk2/flt3 gene leads to deficiencies in primitive hematopoietic progenitors. Immunity. 1995;3(1):147–161. doi: 10.1016/1074-7613(95)90167-1. [DOI] [PubMed] [Google Scholar]
- 31.Thompson JE, Conlon JP, Yang X, Sanchez PV, Carroll M. Enhanced growth of myelodysplastic colonies in hypoxic conditions. Exp Hematol. 2007;35(1):21–31. doi: 10.1016/j.exphem.2006.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 33.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 34.Morris GM, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schüttelkopf AW, van Aalten DM. PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 8):1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 36.Gasteiger J, Marsili M. Iterative partial equalization of orbital electronegativity—A rapid access to atomic charges. Tetrahedron. 1980;36(22):3219–3228. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.