Significance
Complex I is the largest membrane protein complex of the respiratory chain. In addition to 14 central subunits conserved from bacteria to man, the mitochondrial enzyme comprises some 30 so-called accessory subunits of largely unknown structure and function. We show that accessory subunit NB4M [human ortholog NDUFA6, LYR motif containing protein 6 (LYRM6)], a member of the LYR protein superfamily, anchors an acyl carrier protein to complex I, thus forming a structurally well-defined subdomain essential for complex I activity. Our study offers structural insights into the functional interplay of several central and accessory subunits that seem to be specifically involved in controlling the catalytic activity of mitochondrial complex I. This also provides the structural basis for human complex I inactivation in HIV-linked LYRM6 deficiency.
Abstract
Mitochondrial complex I is the largest and most complicated enzyme of the oxidative phosphorylation system. It comprises a number of so-called accessory subunits of largely unknown structure and function. Here we studied subunit NB4M [NDUFA6, LYR motif containing protein 6 (LYRM6)], a member of the LYRM family of proteins. Chromosomal deletion of the corresponding gene in the yeast Yarrowia lipolytica caused concomitant loss of the mitochondrial acyl carrier protein subunit ACPM1 from the enzyme complex and paralyzed ubiquinone reductase activity. Exchanging the LYR motif and an associated conserved phenylalanine by alanines in subunit NB4M also abolished the activity and binding of subunit ACPM1. We show, by single-particle electron microscopy and structural modeling, that subunits NB4M and ACPM1 form a subdomain that protrudes from the peripheral arm in the vicinity of central subunit domains known to be involved in controlling the catalytic activity of complex I.
Mitochondrial complex I (proton pumping NADH:ubiquinone oxidoreductase, EC 1.6.5.3) is a 1-MDa membrane protein complex with a central function in cellular energy conversion (1). Redox-linked proton translocation by complex I contributes to the electrochemical proton gradient across the inner mitochondrial membrane that drives ATP synthesis by ATP synthase. Complex I dysfunction caused by mutations or toxins is associated with a number of neuromuscular and neurodegenerative human disorders, such as Parkinson’s disease. Fourteen central subunits are conserved from bacteria to humans that harbor the core function of energy conversion. In eukaryotes, a substantial fraction of the mass of the holoenzyme is contributed by so-called accessory subunits (2). A broad range of functions from structural reinforcement to regulation of the enzyme complex has been suggested for the accessory subunits, but in most cases, their specific role remains unresolved. Electron microscopy (EM) (3) and X-ray crystallographic analysis of mitochondrial complex I at 6.3 Å resolution (4) revealed the arrangement of functional modules within the l-shaped complex of a highly hydrophobic membrane arm consisting of the proximal and distal pump-modules (PP and PD modules) and a hydrophilic peripheral arm extruding into the mitochondrial matrix that comprises the NADH oxidation and ubiquinone reduction modules (N and Q modules). However, information on the structure and position of individual accessory subunits is still very limited.
In this study, we focused on accessory complex I subunit NB4M (alternative designations NDUFA6 and B14). NB4M/NDUFA6 belongs to the Complex1_LYR family of LYRM proteins (5–7) that is characterized by a motif comprising an N-terminal leucine-tyrosine-arginine sequence located upstream of several conserved arginines and an invariant phenylalanine. The human genome contains at least 11 proteins of the LYRM superfamily, and the mammalian complex I subunit NDUFA6 is identical to LYR motif containing protein 6 (LYRM6). Several other LYRM proteins were shown to be associated with the maintenance of mitochondrial homeostasis. LYRM4 (alternative designation ISD11, iron-sulfur protein biogenesis desulfurase-interacting protein 11) plays an essential role in mitochondrial iron–sulfur cluster biogenesis, and it has been implied that it functions as an adaptor protein for anchoring frataxin to the complex of the cysteine desulfurase Nfs1 and the iron-sulfur protein assembly protein ISCU (8). LYRM7 and LYRM8 are assembly factors of respiratory complexes III and II, respectively (5, 6). The function of LYRM6 (i.e., the NB4M/NDUFA6 subunit of complex I) is unknown, but it is interesting to note that it was identified as a hot spot for peroxynitrite-induced tyrosine nitration (9). Down-regulation of the NDUFA6 subunit has been linked with inactivation of complex I, leading to induction of apoptosis and, ultimately, T-cell depletion in HIV infections (10).
The aerobic yeast Yarrowia lipolytica is a useful yeast genetic model to study eukaryotic complex I in health and disease (11). We have deleted the gene encoding the NDUFA6 ortholog NB4M in this organism and observed formation of a stable and nearly complete complex I subassembly. Proteomic analysis revealed that the absence of NB4M caused detachment of the accessory mitochondrial acyl carrier protein subunit ACPM1 from the enzyme complex, and EM of single particles permitted assignment of subunits NB4M and ACPM1 to a specific domain of the peripheral arm of complex I. Remarkably, despite complete assembly of all central subunits, ubiquinone reductase activity of complex I was abolished in the NB4M deletion strain.
Results
Complex I from Strain nb4mΔ Lacks Two Accessory Subunits.
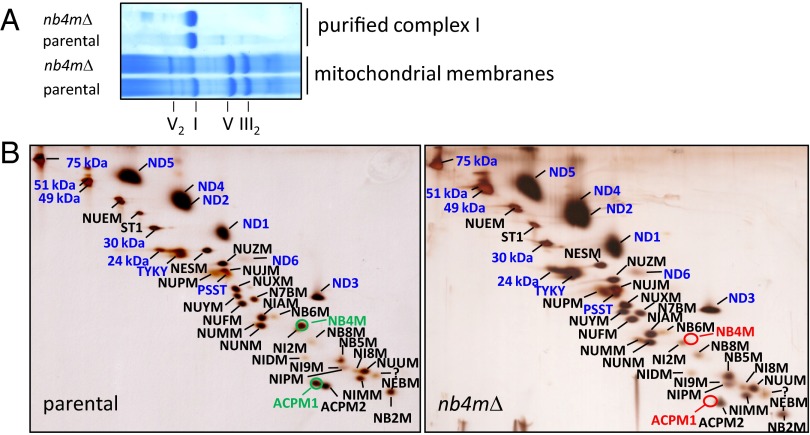
Blue native (BN)-PAGE analysis of mitochondrial membranes prepared from Y. lipolytica strain nb4mΔ, in which we had deleted the entire NB4M ORF by homologous recombination, indicated a somewhat reduced amount of complex I compared with the parental strain (Fig. 1A). To obtain a comprehensive picture of the migration profiles of the complex I subunits in a native gel and the composition of possible subassemblies, we applied our recently developed Complexome Profiling approach (Fig. S1) (12). With the notable exception of subunit ST1, which is known to dissociate from complex I during BN-PAGE (13) and was indeed found exclusively as free subunit, in both strains, the major fraction of all detected subunits was present in monomeric complex I and in the supercomplexes formed by its association with complexes III and IV (14). Of several complex I subunits, a small fraction also migrated as free proteins, and some subcomplexes corresponding to the membrane arm and its PD module were observed even in the parental strain. However, the presence of complex I fragments was much more evident in mitochondria from strain nb4mΔ, in which subcomplexes of the peripheral arm also were observed (Fig. S1). Similar to the lower complex I abundance in mitochondrial membranes (Fig. 1A), this indicated a somewhat reduced stability of complex I in strain nb4mΔ. Most notably, subunit ACPM1 was found almost exclusively as free protein in strain nb4mΔ, and none of it was associated with complex I or its supercomplexes. Because the hydrophobic subunits ND2, ND4L, and ND6 are notoriously difficult to detect via mass spectrometry (15), we could not find these proteins in the Complexome Profiling data sets.
Fig. 1.
Proteomic analysis of complex I from the parental strain and strain nb4mΔ. (A) Coomassie-stained 4–16% blue native (BN)-PAGE of dodecylmaltoside-solubilized mitochondrial membranes and affinity-purified complex I from parental strain and strain nb4mΔ. Roman numerals indicate complexes of oxidative phosphorylation in monomeric or dimeric form. (B) Silver-stained dSDS-PAGE of affinity-purified complex I from parental strain and strain nb4mΔ; blue captions, central subunits; black captions, accessory subunits; NB4M and ACPM1 are highlighted in green. NB4M and the acyl carrier protein ACPM1 are absent in complex I from strain nb4mΔ (red circles). Please note that the assignment of subunits NUNM and NUMM was corrected and differs from that in ref. 15.
For further analysis, we therefore isolated complex I from strain nb4mΔ, taking advantage of the histidine tag attached to the 30-kDa subunit of complex I in our laboratory strains of Y. lipolytica. Purification was straightforward, and the elution profile of the final size-exclusion column was indistinguishable from a parental strain preparation (Fig. S2), as was the mobility of the isolated complexes in BN-PAGE (Fig. 1A). Doubled (d) SDS-PAGE analysis confirmed the absence of subunits NB4M and ACPM1 of complex I from strain nb4mΔ and also showed that the membrane integral subunits ND2 and ND6 were present (Fig. 1B). No spot could be assigned unambiguously to subunit ND4L in the dSDS gels, but mass spectrometric analysis of purified complex I (Table S1) did allow the detection of a single peptide of this small, 9.8-kDa subunit in both the parental and the deletion strains, suggesting that subunit ND4L was present in complex I from strain nb4mΔ.
Taken together, our data clearly show that subunit ACPM1 was the only additional subunit not present in complex I from strain nb4mΔ. This indicated that the binding of this protein depended on the presence of subunit NB4M. The combined mass of the two missing subunits is 24 kDa. This is less than 3% of the total mass of complex I and explains why neither via BN-PAGE nor in the elution profile from the size exclusion column was a significant difference between complex I from the parental strain and strain nb4mΔ observed.
Because the position of subunit NB4M had remained ambiguous in a topological model for the arrangement of the subunits in different functional modules of complex I (15), we used controlled dissociation of purified complex I with the detergent lauryldimethylamine-oxide to biochemically prepare subcomplex Iλ that corresponds essentially to the peripheral arm. dSDS-PAGE (Fig. S3) and mass spectrometric analysis (Table S1) of the subcomplex unambigously revealed that subunits NB4M and ACPM1 were associated with the peripheral arm of complex I.
All Iron–Sulfur Clusters Are Present in Complex I from Strain nb4mΔ.
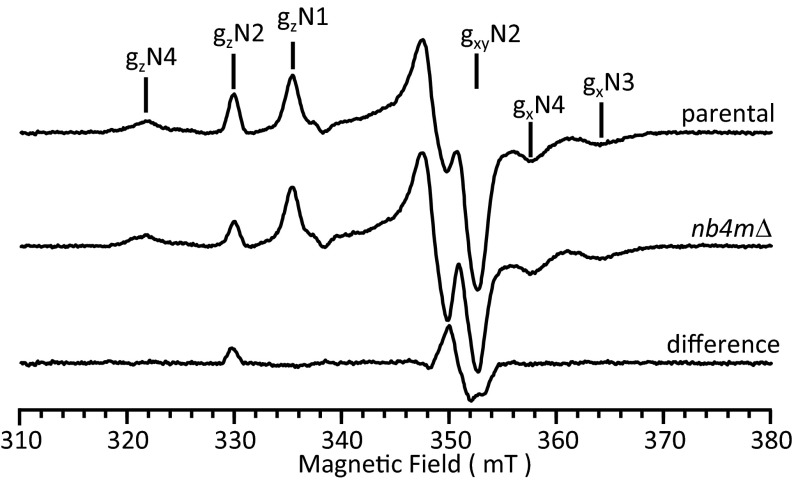
EPR spectroscopy of purified complex I from strain nb4mΔ at 12 K showed that the signals originating from NADH-reducible iron–sulfur clusters N1, N3, and N4 were essentially unchanged (Fig. 2). The spectrum of cluster N5 was recorded at 8 K and was indistinguishable from the corresponding spectrum of complex I from the parental strain (Fig. S4). However, the signal intensity for the 4Fe–4S cluster N2 was decreased to about 60% compared with parental complex I, whereas line shape and field position remained identical (Fig. 2). EPR spectra of the oxidized complex from strain nb4mΔ, but not of the parental strain, revealed a signal characteristic for a 3Fe–4S cluster (Fig. S4). This signal most likely represented defective iron–sulfur cluster N2, but the intensity was far too low to account for the missing 40% in signal intensity. Nevertheless, this observation suggested that iron–sulfur cluster N2 was incompletely assembled or somewhat less stable in the deletion strain, whereas the unchanged spectral signature rendered major structural alterations around the fraction of intact clusters unlikely.
Fig. 2.
EPR spectrum of purified complex I from strain nb4mΔ. EPR spectra of complex I purified from parental and strain nb4mΔ were recorded using the following parameters: microwave frequency, 9.47 GHz; microwave power, 1 mW; modulation amplitude, 0.64 mT; and modulation frequency, 100 kHz. Samples were reduced with NADH (2 mM), and the spectra were recorded at 12 K. Characteristic signal contributions of the four EPR visible iron–sulfur clusters of complex I (N1, N2, N3, N4) detectable under these conditions are indicated above the spectra. For comparison, spectra were normalized on signal intensities of clusters N3 and N4. The difference spectrum largely shows the pure signature of the terminal iron–sulfur cluster N2.
To assess whether the remaining EPR-silent iron–sulfur clusters were present in complex I of the deletion strain, we determined the specific iron content of the purified complex by total reflection X-ray fluorescence spectroscopy. The number of iron atoms found per complex was 29 ± 1 (n = 3) for the parental control and 29 (n = 2) for the enzyme lacking subunits NB4M and ACPM1. Both values are in agreement with the theoretical value of 28 irons per complex, indicating that all iron–sulfur clusters were present in complex I from strain nb4mΔ.
The LYR Motif of Subunit NB4M Is Required for Ubiquinone Reductase Activity of Complex I.
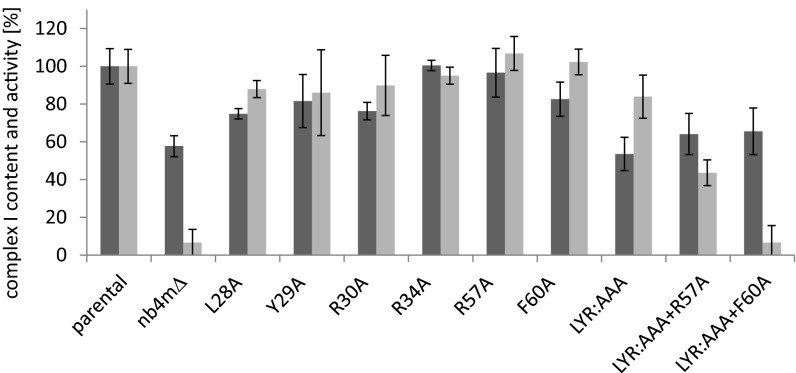
Although the effect of deleting the gene encoding subunit NB4M was rather limited on the assembly and structural integrity of complex I, we found that specific inhibitor-sensitive ubiquinone reductase activity was almost completely abolished in mitochondrial membranes from strain nb4mΔ (Fig. 3). To investigate whether this loss of activity was related to the Complex1_LYR family motif present in subunit NB4M, we analyzed the N-terminal sequence LYR (position 28–30), the conserved arginines at position 34 and 57, and the invariant phenylalanine residue at position 60 by site-directed mutagenesis, which are the hallmarks of this motif (7) (Fig. 3). BN-PAGE of mitochondrial membranes showed that all mutant strains expressed high amounts of assembled complex I (Fig. S5). Quantitative assessment of the complex I content using our established approach to measure activity with the nonphysiological electron acceptor hexaamineruthenium(III) indicated that except for mutations R34A and R57A, the amount of assembled complex I was somewhat reduced (Fig. 3). Although replacement of each individual amino acid of the LYR sequence by alanine reduced complex I content by 10–20%, the effect of an exchange of all three amino acid residues (LYR:AAA) was essentially the same as for the complete deletion of subunit NB4M, reducing the content of complex I in the mitochondrial membranes to about 60% compared with the parental strain. Additional exchange of R57 and F60 did not lower the abundance of complex I any further. In contrast, only a combination of these mutations with the complete exchange of the LYR sequence markedly affected the normalized ubiquinone reductase activity (Fig. 3). Although mutation LYR:AAA alone reduced quinone reductase activity by less than 20%, it was down to about 40% of the parental strain when combined with R57A. Mutations LYR:AAA, together with F60A, abolished complex I activity almost completely to the level of the deletion of the entire subunit NB4M. Notably, exchange of R57 and F60 alone had no effect at all on the specific complex I activity, and all other single amino acid exchanges reduced it only by about 5–15%.
Fig. 3.
Ubiquinone reductase activities and complex I content of mitochondrial membranes from parental strain, strain nb4mΔ, and site-directed mutants. Complex I content (dark gray bars) assessed as nonphysiological NADH:hexaammineruthenium(III) oxidoreductase activity of mitochondrial membranes (100% = 1.38 µmol⋅min−1⋅mg−1). Relative inhibitor sensitive deamino-NADH:decylubiquinone oxidoreductase activities (light gray bars) normalized to complex I content (100% = 0.61 µmol⋅min−1⋅mg−1).
After purification and lipid reactivation, the ubiquinone reductase activity of complex I from the LYR:AAA mutant was found to be decreased to about 25%, whereas the activity for the Y29A mutant remained close to the wild-type level (Table 1). Quantification of the protein spots after dSDS-PAGE revealed that the amounts of subunits NB4M and ACPM1 were reduced about half in complex I from the LYR:AAA mutant (Table 1). This was also true for subunit NB4M in the LYR:AAA+F60A mutant, whereas subunit ACPM1 was completely lacking from complex I purified from this strain. In complex I from mutant Y29A, the amounts of both subunits were unchanged. Although the EPR spectrum of mutant Y29A showed no deviation from the wild-type control, we observed a reduction to ∼80% signal intensity for cluster N2 in mutants LYR:AAA and LYR:AAA+F60A (Fig. S6).
Table 1.
Inhibitor-sensitive NADH:decyl-ubiquinone oxidoreductase activities of purified complex I and relative abundance of subunits NB4M and ACPM1
| Complex I | Catalytic activity, µmol⋅min−1⋅mg−1 | Relative abundance | |
| NB4M | ACPM1 | ||
| Parental | 4.2 | 1 | 1 |
| nb4mΔ | <0.2 | 0 | 0 |
| Y29A | 4.2 | 1 | 1 |
| LYR:AAA | 1.1 | 0.6 | 0.4 |
| LYR:AAA+F60A | <0.2 | 0.6 | 0 |
Subunits NB4M and ACPM1 Form Domain 6 of the Peripheral Arm.
To investigate structural differences between complex I from the parental strain and strain nb4mΔ, we analyzed both complexes by single-particle 2D EM, averaging, and difference imaging (Fig. 4). The previously described subdomain architecture of the peripheral arm (3) was clearly visible. A comparison with average images of the holoenzyme clearly showed that subdomain 6 was lacking in complex I from strain nb4mΔ (Fig. 4). To confirm that domain 6 is missing, independent of possible variability in the data, the aligned images of the holocomplex and subcomplex were combined, analyzed as a single set by correspondence analysis, and classified. The images contributing to each class average were split into two sets, and two averages were calculated: one of the parental strain only and one of the subcomplex only. All of the class averages show that domain 6 is present in the holoenzyme and missing in complex I from strain nb4mΔ (Fig. S7). In addition to domain 6, some weak interior differences can be seen. These are most probably caused by minor differences in the imaging conditions for both data sets, as the images of the holoenzyme were recorded at a slightly higher defocus (1.0–1.5 μm) and corrected for the microscope transfer-function, whereas the subcomplex images were obtained close to focus (<1 μm), so that contrast transfer function correction was not needed.
Fig. 4.
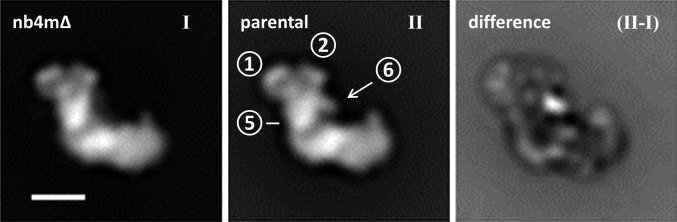
Single-particle 2D electron microscopic analysis of complex I purified from parental strain and strain nb4mΔ. Average images of the subcomplex (I) and the holocomplex (II), followed by the difference image (II-I). The circled numbers indicate the previously described subdomains 1, 2, 5, and 6 of the peripheral arm (3). (Scale bar, 10 nm.)
Extensive structural consistency between the central subunits of eukaryotic and prokaryotic complex I can be assumed on the basis of high sequence conservation and was demonstrated by fitting three subunits of the bacterial Q module into the low-resolution X-ray electron density map of Y. lipolytica complex I (4). An overlay of the EM envelope of Y. lipolytica complex I (3) with the X-ray structure of complete complex I from Thermus thermophilus (16) clearly showed that subdomain 6 was missing in the simpler bacterial enzyme (Fig. 5A). For the mitochondrial enzyme, a close spatial relationship of subdomain 6 with the Q module was evident (Fig. 5A, blue). Sequence conservation among the small acyl carrier proteins is generally high (e.g., ACP Escherichia coli/ACPM1 Y. lipolytica 33% identity and 47% similarity), and a large number of related structures were solved at high resolution, exhibiting a characteristic four-helix arrangement. Calculation of a structural model for ACPM1 using MODELLER (17) was thus straightforward. Inspection of the 6.3-Å X-ray electron density map of Y. lipolytica complex I immediately identified the characteristic acyl carrier protein fold at the distal end of subdomain 6, and we manually fitted the calculated structural model for ACPM1, as well as the NMR structure of the human ortholog NDUFAB1 (PDB code 2DNW) and the X-ray structure of ACP from E. coli (PDB code 2FAE) (18) (Fig. 5 B and C and Fig. S8).
Fig. 5.
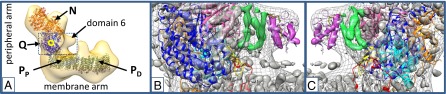
Structural modeling of subunits NB4M and ACPM1. (A) Overlay of the X-ray structure of T. thermophilus complex I (ribbon representation, PDB file 4HEA) (16) and the envelope of Y. lipolytica complex I determined by single-particle EM (3). NADH oxidation module (N, orange), ubiquinone reduction module (Q, blue), proton pump modules (green, PP and PD), and iron-sulfur cluster N2 (yellow circle). The blue frame indicates the zoom window for B and C. (B) Overlay of X-ray electron density map (4) (gray surface, contour level 1.9) with an envelope of Y. lipolytica complex I (3) (gray mesh), and X-ray structure of T. thermophilus complex I; 49-kDa subunit (NQO4, blue, N-terminal β-sheet highlighted in yellow), PSST subunit (NQO6, cyan), TYKY subunit (NQO9, orange), 30-kDa subunit (NQO5, hot pink), loop connecting helices 1 and 2 of subunit ND3 (NQO7, red); structure of ACP from E. coli [PDB file 2FAE) (18), pink; decanoyl phosphopantetheine, yellow stick representation] fitted to electron density in the distal part of subdomain 6 (magenta), electron density features tentatively assigned to subunit NB4M are highlighted in green (cf. Fig. S8). (C) Alternative view of B rotated by about 180°.
In contrast, generation of a 3D structural model for NB4M was compromised by the lack of clearly related protein structures. Secondary structure prediction indicated that NB4M is predominantly α-helical. Consistently, the proximal part of subdomain 6 comprises three long tubular electron density features very likely representing α-helices (Fig. 5 B and C; green). The fit of the different acyl carrier protein structures indicated that helix α-2 of mitochondrial ACPM1 is mainly responsible for its binding to NB4M. This arrangement is highly similar to the interaction reported for E. coli ACP and the enoyl reductase Fabl of the type 2 fatty acid synthesis pathway of this organism (19). Likewise, the ACP domain of the cytosolic eukaryotic type 1 fatty acid synthesis complex interacts with its binding partner via the corresponding recognition helix 8 (20). In the ACP structure from E. coli that we fitted into the electron density map of the mitochondrial enzyme, the phosphopantetheine prosthetic group carrying a decanoyl modification is resolved. Assuming a similar conformation for the prosthetic group of ACPM1, the phosphate moiety would bridge the tip of ACPM1 helix α-2 and the adjacent helical feature assigned to NB4M. A strong binding interaction, probably via a salt bridge, is supported by a clearly visible connectivity in the X-ray electron density map (Fig. S8 and Fig. 5 B and C).
Discussion
We investigated the structure and function of the mitochondrial complex I accessory subunit NB4M, a member of the LYRM-family of proteins (LYRM6), by generating a strain of Y. lipolytica carrying a chromosomal deletion of the NB4M gene. Mitochondrial membranes of the resulting strain nb4mΔ contained moderately reduced amounts of assembled complex I that, in addition to subunit NB4M, lacked only one additional small subunit; namely, the mitochondrial acyl carrier protein ACPM1 (Fig. 1 and Fig. S1). Remarkably, the absence of only a minute fraction of the holoenzyme representing less than 3% of its total mass completely abolished ubiquinone reductase activity.
Other LYRM proteins, namely, Mzm1 (LYRM7) and SDHAF1 (LYRM8), were identified as assembly factors of respiratory chain complexes III and II, respectively (5, 6). Moreover, the yeast homolog ISD11 of human LYRM4 is associated with the machinery for iron–sulfur cluster synthesis, and a general function for insertion or retention of iron–sulfur clusters was proposed for LYRM proteins, including the mammalian ortholog of complex I subunit NB4M/NDUFA6 (6). Indeed, we observed that the EPR signal intensity of cluster N2 in the central PSST subunit was diminished (Fig. 2). However, the majority of the iron–sulfur cluster was properly inserted, and considering that Mzm1 (LYRM7) was shown to be an important chaperone for assembly of the central Rip1 subunit of complex III, but not for insertion of the Rieske iron–sulfur cluster, the strict link of LYRM proteins with iron–sulfur cluster assembly seems less compelling.
At this stage, the reason for the lower N2 EPR signal remains unclear because the residual signal was completely normal, the additional signal in the EPR spectrum of the mutant indicating an incomplete 3Fe–4S cluster was minute, and there was no indication of a lower iron stoichiometry or an impaired assembly of the PSST subunit in complex I of the deletion strain. However, considering that cluster N2 contributes only 14% of the total iron content of complex I, a fractional loss of the cluster cannot be excluded on the basis of the measurements of total iron content. Therefore, it remains undecided whether a minor part of cluster N2 was physically absent or was converted to a state that could not be monitored by EPR spectroscopy. In any case, as 60% of the N2 clusters showed normal spectral properties on reduction by NADH, partial modification of this redox center cannot explain the total loss of activity observed in the deletion mutant.
To evaluate whether the LYR motif in subunit NB4M was critical for keeping complex I catalytically active, we characterized a series of site-directed mutants (Fig. 3). None of the single-amino acid exchanges had major consequences, suggesting that functionality of the LYR motif was rather robust and relied on an ensemble of residues. In contrast, a single mutation of the LYR tyrosine in the Mzm1 assembly factor of complex III caused a major loss of function comparable with deletion of the mzm1 gene (5). Complete exchange of the N-terminal LYR triad with three alanines decreased the amount of complex I in mitochondrial membranes somewhat, but the specific activity of the enzyme was hardly affected. Only when, in addition, mutation R57A was introduced did ubiquinone reductase activity decline significantly; it was abolished completely in mutant LYR:AAA+F60A. Notably, complex I purified from this mutant was essentially inactive but still contained about 60% of subunit NB4M, whereas subunit ACPM1, the second subunit missing in strain nb4mΔ, was completely absent (Table 1). In comparison, the purified LYR:AAA mutant retained about 40% of ACPM1 and about 25% of activity, whereas the purified Y29A mutant displayed a change in neither subunit stoichiometry nor activity. The correlation between loss of ACPM1 and catalytic activity in the LYR:AAA and LYR:AAA+F60A mutants seems remarkable. However, even though the EPR spectra of the two mutants were virtually identical, it remains difficult to unambiguously dissect the effect of NB4M mutation and partial or complete detachment of ACPM1. In Y. lipolytica, ACPM1 is an essential gene, precluding generation of a deletion strain and selective evaluation of the function of ACPM1 for complex I activity. In any case, our data suggest that a functional NB4M subunit was required for binding subunit ACPM1 to mitochondrial complex I and that binding of subunit ACPM1 to complex I was required to maintain catalytically active complex I. These observations constitute structural and functional links between the two accessory subunits. In this context, it is interesting to note that LYRM4 has also been proposed to act as an adaptor protein that links the small protein frataxin to a larger protein complex (8).
ACPMs have been proposed to be components of the bacterial type fatty acid synthesis system of mitochondria involved in formation of lipoic acid, a prosthetic group of mitochondrial dehydrogenases (21). The presence of a single acyl carrier protein subunit was first discovered in bovine complex I (22). In contrast, Y. lipolytica was shown to harbor two highly similar but mutually nonexchangeable ACPM subunits, ACPM1 and ACPM2 (23). In agreement with our previous study (23), our data show unequivocally that ACPM1 is almost completely associated with complex I in the parental strain (Fig. S1). However, in strain nb4mΔ, ACPM1 was completely detached from complex I. Because the NB4M deletion strain was nevertheless viable, we conclude that ACPM1 can carry out its essential function independent of the structural context of complex I. This finding is in agreement with the situation reported for bovine mitochondria, in which the acyl carrier protein subunit SDAP of complex I was detected in significant excess in free form in the mitochondrial matrix (24), and with Arabidopsis thaliana, in which none of the three known ACPMs were found to be associated with complex I (25).
In conclusion, the yet-unspecified essential function of ACPM1 is independent of its association with complex I and is most likely linked to mitochondrial fatty acid synthesis required, for example, in biosynthesis of lipoic acid. We show that integrity of the domain consisting of NB4M and ACPM1 together is essential for complex I activity. However, the ultimate cause for the association of the acyl carrier protein with complex I remains unresolved.
Single-particle EM of complex I purified from strain nb4mΔ clearly showed that the two missing subunits corresponded to the small but characteristic subdomain 6 protruding from the peripheral arm (Fig. 4) that had been previously defined by electron microscopic analysis of complex I from Y. lipolytica (3). Electron microscopic models of bovine complex I also show subdomain 6 in the peripheral arm (26). Remarkably, however, the mammalian enzyme features a thin connection between the tip of the domain and the membrane arm. The identity of this stalk-like structure is unknown, but it might explain the unusual association of the single bovine acyl carrier protein subunit with two different subcomplexes, one of which represents the distal domain of the membrane arm (27).
The assignment of the two proteins to subdomain 6 was further corroborated by proteomic analysis of a biochemically prepared peripheral arm of complex I (Table S1 and Fig. S3) and tentative modeling of subunits NB4M and ACPM1 into the X-ray crystallographic 6.3-Å map of Y. lipolytica complex I (4) (Fig. 5 and Fig. S8). According to this structural model, ACPM1 docks to complex I via subunit NB4M to form a nose-like domain extending from the inner side of the peripheral arm approximately at the level of iron–sulfur cluster N2 and the binding site of the ubiquinone headgroup. In line with the loss of ACPM1 on exchange of the conserved arginine in the LYR:AAA mutant in conjunction with F60A (Table 1), the model also suggests that the phosphate moiety of the phosphopantetheine group of ACPM1 is critically involved in binding subunit NB4M (Fig. S8). Subunit NB4M itself seems to be bound primarily to the 30-kDa subunit and to have also some contacts to the 49-kDa subunit (Fig. 5). This position provides some clues as to why the catalytic activity of mitochondrial complex I required an intact subdomain 6. The domain within the 49-kDa predicted to be in contact with subunit NB4M is the N-terminal β-sheet (Fig. 5, yellow) comprising two histidines known to be critical for complex I activity (28, 29). They are part of the large ubiquinone and inhibitor binding pocket at the interface between the 49-kDa and PSST subunits that has been characterized in detail by site-directed mutagenesis and X-ray structure determination (4, 28). Moreover, the contact site between NB4M and the 49-kDa subunit is in the vicinity of another structural element known to control complex I activity.
Mitochondrial complex I from many species, including Y. lipolytica, is known to undergo a reversible transition between an active (A) form and a deactive (D) form (30). Structural changes linked to this A/D interconversion of mammalian complex I involve a specific cysteine residue in the long loop connecting transmembrane helices 1 and 2 of subunit ND3 (31). The recently solved X-ray structure of complex I from the bacterium T. thermophilus shows that the ND3 loop resides adjacent to the N-terminal β sheet of the 49-kDa subunit near the interface between the Q and P modules (Fig. 5C, red) (16). The A/D transition has been studied in the context of reperfusion-induced tissue damage, and conversion into the D form (e.g., during hypoxia) is discussed to prevent excessive formation of reactive oxygen species on reoxygenation. It was shown recently in vivo that S-nitrosylation of the cysteine in subunit ND3 prevented or retarded return of complex I to the A form (31, 32). Our results show that subunits NB4M and ACPM1 are an important part of this regulatory domain, and we speculate that an interaction of NB4M with the ND3 loop is critically involved in the A/D conversion of mitochondrial complex I. Although the two small proteins constituting subdomain 6 may contribute merely by passively maintaining the proper structure, considering their exposed position, it is also tempting to speculate that they may bind so-far-unidentified factors that take part in the regulation of complex I activity. Finally, our data provide insight into the structural basis of the inactivation of human complex I because of specific down-regulation of subunit NDUFA6/NB4M during HIV infection that results in T-cell depletion by induction of apoptosis (10).
Methods
Deletion Strain nb4mΔ and Site-Directed Mutagenesis of Subunit NB4M from Y. lipolytica.
The complete NB4M gene (YALI0A01419g) was deleted in Y. lipolytica strain GB20 (mus51Δ, nugm-Htg2, ndh2i, lys11−, leu2−, ura3−, MatB). This strain carries a deletion of the MUS51 gene that has a function in nonhomologous end-joining of DNA strands. This modification greatly enhanced the efficiency of gene deletion by homologous recombination. Site-directed mutagenesis was done essentially as described previously (11).
Preparation of Intact Mitochondria, Mitochondrial Membranes, and Purification of Complex I.
After breaking Y. lipolytica cells in a blender, intact mitochondria were prepared by several differential centrifugation steps, including separation on a sucrose density gradient. The details of this method, the preparation of mitochondrial membranes, and the purification and fragmentation of complex I are described in SI Methods. Specific NADH:hexaammineruthenium and deamino-NADH:decylubiquinone oxidoreductase activity was measured for mitochondrial membranes and for purified enzymes after lipid activation, as described (33).
Electrophoresis.
Mitochondrial membranes or intact mitochondria from Y. lipolytica were solubilized in 1.5 g/g n-dodecyl-β-d-maltoside or 3 g/g digitonin, respectively, and separated by BN-PAGE with a 4–16% acrylamide gradient. The subunit composition of purified complex I was analyzed by dSDS-PAGE (34). To determine the relative abundance of individual complex I subunits, fluorescence signals of cyanine 3-labeled protein were detected after dSDS-PAGE separation by laser scanner Typhoon 9400 (GE Healthcare) and quantified by the DeCyder 2D software 7.0 (GE Healthcare). Signals from subunits NB4M and ACPM1 were normalized to subunit ND3 in complex I from parental and mutant strains.
Mass Spectrometry.
Proteins from gel spots were identified essentially as described in ref. 35. Complexome Profiling analysis was done according to ref. 12. Complexes I, III, and IV and monomeric and dimeric ATP synthase in the Complexome Profile were used for mass calibration of each slice according to ref. 14.
EPR Spectroscopy and Iron Determination.
X-band EPR spectra were recorded as described previously (29) and as detailed in the Fig. 2 legend. Iron-to-protein ratios were determined by total-reflection X-ray fluorescence, as described in SI Methods.
EM.
EM of single particles in deep stain was done essentially as described previously (3). Details are summarized in SI Methods.
Structure Prediction and Modeling.
Secondary structure prediction was carried out using the Quick2D package at http://toolkit.tuebingen.mpg.de/quick2_d. For modeling, we used the bioinformatic tools HHpred (36), and MODELLER (17). For manual fitting and display of structures, we used Chimera (37).
Supplementary Material
Acknowledgments
We thank Christophe Wirth for helpful discussions, Martina Ding for supportive information on acyl carrier proteins, and Stefan Kerscher for strain GB20. Excellent technical assistance by Karin Siegmund, Andrea Duchene, Gudrun Beyer, and Claudia Rittmeyer is gratefully acknowledged. This work was supported by the Deutsche Forschungsgemeinschaft Grant ZI 552/3-1, Sonderforschungsbereich 815, Project Z1 (Redox-Proteomics), and the Cluster of Excellence Frankfurt “Macromolecular Complexes,” as well as by National Institutes of Health Grant R01 GM068650.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1322438111/-/DCSupplemental.
References
- 1.Hirst J. Mitochondrial complex I. Annu Rev Biochem. 2013;82:551–575. doi: 10.1146/annurev-biochem-070511-103700. [DOI] [PubMed] [Google Scholar]
- 2.Kmita K, Zickermann V. Accessory subunits of mitochondrial complex I. Biochem Soc Trans. 2013;41(5):1272–1279. doi: 10.1042/BST20130091. [DOI] [PubMed] [Google Scholar]
- 3.Radermacher M, et al. The three-dimensional structure of complex I from Yarrowia lipolytica: A highly dynamic enzyme. J Struct Biol. 2006;154(3):269–279. doi: 10.1016/j.jsb.2006.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hunte C, Zickermann V, Brandt U. Functional modules and structural basis of conformational coupling in mitochondrial complex I. Science. 2010;329(5990):448–451. doi: 10.1126/science.1191046. [DOI] [PubMed] [Google Scholar]
- 5.Atkinson A, et al. The LYR protein Mzm1 functions in the insertion of the Rieske Fe/S protein in yeast mitochondria. Mol Cell Biol. 2011;31(19):3988–3996. doi: 10.1128/MCB.05673-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghezzi D, et al. SDHAF1, encoding a LYR complex-II specific assembly factor, is mutated in SDH-defective infantile leukoencephalopathy. Nat Genet. 2009;41(6):654–656. doi: 10.1038/ng.378. [DOI] [PubMed] [Google Scholar]
- 7.Angerer H. The superfamily of mitochondrial Complex1_LYR motif-containing (LYRM) proteins. Biochem Soc Trans. 2013;41(5):1335–1341. doi: 10.1042/BST20130116. [DOI] [PubMed] [Google Scholar]
- 8.Shan Y, Napoli E, Cortopassi G. Mitochondrial frataxin interacts with ISD11 of the NFS1/ISCU complex and multiple mitochondrial chaperones. Hum Mol Genet. 2007;16(8):929–941. doi: 10.1093/hmg/ddm038. [DOI] [PubMed] [Google Scholar]
- 9.Murray J, Taylor SW, Zhang B, Ghosh SS, Capaldi RA. Oxidative damage to mitochondrial complex I due to peroxynitrite: Identification of reactive tyrosines by mass spectrometry. J Biol Chem. 2003;278(39):37223–37230. doi: 10.1074/jbc.M305694200. [DOI] [PubMed] [Google Scholar]
- 10.Ladha JS, Tripathy MK, Mitra D. Mitochondrial complex I activity is impaired during HIV-1-induced T-cell apoptosis. Cell Death Differ. 2005;12(11):1417–1428. doi: 10.1038/sj.cdd.4401668. [DOI] [PubMed] [Google Scholar]
- 11.Kerscher S, Dröse S, Zwicker K, Zickermann V, Brandt U. Yarrowia lipolytica, a yeast genetic system to study mitochondrial complex I. Biochim Biophys Acta. 2002;1555(1-3):83–91. doi: 10.1016/s0005-2728(02)00259-1. [DOI] [PubMed] [Google Scholar]
- 12.Heide H, et al. Complexome profiling identifies TMEM126B as a component of the mitochondrial complex I assembly complex. Cell Metab. 2012;16(4):538–549. doi: 10.1016/j.cmet.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 13.Morgner N, et al. Subunit mass fingerprinting of mitochondrial complex I. Biochim Biophys Acta. 2008;1777(10):1384–1391. doi: 10.1016/j.bbabio.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 14.Nübel E, Wittig I, Kerscher S, Brandt U, Schägger H. Two-dimensional native electrophoretic analysis of respiratory supercomplexes from Yarrowia lipolytica. Proteomics. 2009;9(9):2408–2418. doi: 10.1002/pmic.200800632. [DOI] [PubMed] [Google Scholar]
- 15.Angerer H, et al. A scaffold of accessory subunits links the peripheral arm and the distal proton-pumping module of mitochondrial complex I. Biochem J. 2011;437(2):279–288. doi: 10.1042/BJ20110359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baradaran R, Berrisford JM, Minhas GS, Sazanov LA. Crystal structure of the entire respiratory complex I. Nature. 2013;494(7438):443–448. doi: 10.1038/nature11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 18.Roujeinikova A, et al. Structural studies of fatty acyl-(acyl carrier protein) thioesters reveal a hydrophobic binding cavity that can expand to fit longer substrates. J Mol Biol. 2007;365(1):135–145. doi: 10.1016/j.jmb.2006.09.049. [DOI] [PubMed] [Google Scholar]
- 19.Rafi S, et al. Structure of acyl carrier protein bound to FabI, the FASII enoyl reductase from Escherichia coli. J Biol Chem. 2006;281(51):39285–39293. doi: 10.1074/jbc.M608758200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leibundgut M, Jenni S, Frick C, Ban N. Structural basis for substrate delivery by acyl carrier protein in the yeast fatty acid synthase. Science. 2007;316(5822):288–290. doi: 10.1126/science.1138249. [DOI] [PubMed] [Google Scholar]
- 21.Hiltunen JK, Chen Z, Haapalainen AM, Wierenga RK, Kastaniotis AJ. Mitochondrial fatty acid synthesis—an adopted set of enzymes making a pathway of major importance for the cellular metabolism. Prog Lipid Res. 2010;49(1):27–45. doi: 10.1016/j.plipres.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 22.Runswick MJ, Fearnley IM, Skehel JM, Walker JE. Presence of an acyl carrier protein in NADH:ubiquinone oxidoreductase from bovine heart mitochondria. FEBS Lett. 1991;286(1-2):121–124. doi: 10.1016/0014-5793(91)80955-3. [DOI] [PubMed] [Google Scholar]
- 23.Dobrynin K, et al. Characterization of two different acyl carrier proteins in complex I from Yarrowia lipolytica. Biochim Biophys Acta. 2010;1797(2):152–159. doi: 10.1016/j.bbabio.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 24.Cronan JE, Fearnley IM, Walker JE. Mammalian mitochondria contain a soluble acyl carrier protein. FEBS Lett. 2005;579(21):4892–4896. doi: 10.1016/j.febslet.2005.07.077. [DOI] [PubMed] [Google Scholar]
- 25.Meyer EH, Heazlewood JL, Millar AH. Mitochondrial acyl carrier proteins in Arabidopsis thaliana are predominantly soluble matrix proteins and none can be confirmed as subunits of respiratory Complex I. Plant Mol Biol. 2007;64(3):319–327. doi: 10.1007/s11103-007-9156-9. [DOI] [PubMed] [Google Scholar]
- 26.Clason T, et al. The structure of eukaryotic and prokaryotic complex I. J Struct Biol. 2010;169(1):81–88. doi: 10.1016/j.jsb.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirst J, Carroll J, Fearnley IM, Shannon RJ, Walker JE. The nuclear encoded subunits of complex I from bovine heart mitochondria. Biochim Biophys Acta. 2003;1604(3):135–150. doi: 10.1016/s0005-2728(03)00059-8. [DOI] [PubMed] [Google Scholar]
- 28.Tocilescu MA, Zickermann V, Zwicker K, Brandt U. Quinone binding and reduction by respiratory complex I. Biochim Biophys Acta. 2010;1797(12):1883–1890. doi: 10.1016/j.bbabio.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 29.Grgic L, Zwicker K, Kashani-Poor N, Kerscher S, Brandt U. Functional significance of conserved histidines and arginines in the 49-kDa subunit of mitochondrial complex I. J Biol Chem. 2004;279(20):21193–21199. doi: 10.1074/jbc.M313180200. [DOI] [PubMed] [Google Scholar]
- 30.Vinogradov AD. Catalytic properties of the mitochondrial NADH-ubiquinone oxidoreductase (complex I) and the pseudo-reversible active/inactive enzyme transition. Biochim Biophys Acta. 1998;1364(2):169–185. doi: 10.1016/s0005-2728(98)00026-7. [DOI] [PubMed] [Google Scholar]
- 31.Galkin A, et al. Identification of the mitochondrial ND3 subunit as a structural component involved in the active/deactive enzyme transition of respiratory complex I. J Biol Chem. 2008;283(30):20907–20913. doi: 10.1074/jbc.M803190200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chouchani ET, et al. Cardioprotection by S-nitrosation of a cysteine switch on mitochondrial complex I. Nat Med. 2013;19(6):753–759. doi: 10.1038/nm.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dröse S, Zwicker K, Brandt U. Full recovery of the NADH:ubiquinone activity of complex I (NADH:ubiquinone oxidoreductase) from Yarrowia lipolytica by the addition of phospholipids. Biochim Biophys Acta. 2002;1556(1):65–72. doi: 10.1016/s0005-2728(02)00307-9. [DOI] [PubMed] [Google Scholar]
- 34.Rais I, Karas M, Schägger H. Two-dimensional electrophoresis for the isolation of integral membrane proteins and mass spectrometric identification. Proteomics. 2004;4(9):2567–2571. doi: 10.1002/pmic.200400829. [DOI] [PubMed] [Google Scholar]
- 35.Wittig I, et al. Assembly and oligomerization of human ATP synthase lacking mitochondrial subunits a and A6L. Biochim Biophys Acta. 2010;1797(6-7):1004–1011. doi: 10.1016/j.bbabio.2010.02.021. [DOI] [PubMed] [Google Scholar]
- 36.Söding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21(7):951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 37.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.