Significance
Phosphorylation of eukaryotic protein synthesis initiation factor 2α (eIF2α) is the principal mechanism cells use to regulate translation initiation. Specific kinases phosphorylate eIF2α to inhibit protein synthesis under stress conditions; however, eIF2α dephosphorylation is catalyzed by the general protein phosphatase PP1. In mammalian cells, specific trans-acting targeting subunits direct PP1 to dephosphorylate eIF2α and restore protein synthesis. However, these targeting subunits are missing in the yeast Saccharomyces cerevisiae. We show that a PP1-binding element in a region of the eIF2γ subunit that is unique to yeast functions to target PP1 to dephosphorylate eIF2α. Thus, yeast rely on the recruitment of PP1 in cis to the eIF2 complex to maintain eIF2α phosphorylation at levels appropriate for cellular homeostasis.
Abstract
Whereas the protein kinases GCN2, HRI, PKR, and PERK specifically phosphorylate eukaryotic translation initiation factor 2 (eIF2α) on Ser51 to regulate global and gene-specific mRNA translation, eIF2α is dephosphorylated by the broadly acting serine/threonine protein phosphatase 1 (PP1). In mammalian cells, the regulatory subunits GADD34 and CReP target PP1 to dephosphorylate eIF2α; however, as there are no homologs of these targeting subunits in yeast, it is unclear how GLC7, the functional homolog of PP1 in yeast, is recruited to dephosphorylate eIF2α. Here, we show that a novel N-terminal extension on yeast eIF2γ contains a PP1-binding motif (KKVAF) that enables eIF2γ to pull down GLC7 and target it to dephosphorylate eIF2α. Truncation or point mutations designed to eliminate the KKVAF motif in eIF2γ impair eIF2α dephosphorylation in vivo and in vitro and enhance expression of GCN4. Replacement of the N terminus of eIF2γ with the GLC7-binding domain from GAC1 or fusion of heterologous dimerization domains to eIF2γ and GLC7, respectively, maintained eIF2α phosphorylation at basal levels. Taken together, these results indicate that, in contrast to the paradigm of distinct PP1-targeting or regulatory subunits, the unique N terminus of yeast eIF2γ functions in cis to target GLC7 to dephosphorylate eIF2α.
The reversible phosphorylation of proteins plays a key role in regulating many cellular processes, including cell division, glycogen metabolism, and protein synthesis (1–3). In Saccharomyces cerevisiae, the protein kinase GCN2 regulates protein synthesis under a variety of stress conditions including amino acid starvation (3, 4) by phosphorylating the α subunit of the eukaryotic translation initiation factor 2 (eIF2) on Ser51. The factor eIF2, composed of α, β, and γ subunits, binds GTP and Met-tRNAiMet to form a ternary complex (TC) and then delivers the Met-tRNAiMet to the small ribosomal subunit. After the scanning ribosome selects a start codon for protein synthesis, hydrolysis of the GTP bound to eIF2 is completed and eIF2–GDP is released from the ribosome. To participate in another round of translation initiation, the GDP on eIF2 must be exchanged for GTP by the guanine–nucleotide exchange factor eIF2B (5). This eIF2 recycling step is an important control point in the translation pathway.
Phosphorylation of eIF2α converts eIF2 from a substrate to a competitive inhibitor of eIF2B and thereby blocks TC formation and the subsequent steps in the translation initiation pathway (6). Although eIF2α phosphorylation inhibits general protein synthesis, it also induces the translation of specific mRNAs including a class of mRNAs containing upstream ORFs (uORFs) (3, 7, 8). Included among this class of activated mRNAs is the mRNA encoding yeast GCN4, a transcriptional activator of amino acid biosynthetic genes. The activation of GCN2, phosphorylation of eIF2α, and subsequent induction of GCN4 expression are essential for yeast growth under amino acid starvation conditions (3, 4, 9).
In both yeast and mammalian cells the number of protein kinases is in vast excess to the number of protein phosphatases. Yeast express over 100 different protein kinases, yet only around 30 protein phosphatases (10, 11). Accordingly, the protein phosphatases are likely to display a wider range of substrate preference than the protein kinases. Whereas eIF2α is the sole substrate for the protein kinase GCN2, dephosphorylation of eIF2α in yeast is primarily controlled by GLC7 (12), the essential and sole protein phosphatase 1 (PP1) catalytic subunit in yeast (13, 14). In addition to dephosphorylating eIF2α, GLC7 plays critical roles dephosphorylating substrates in mitosis, meiosis, cell division, glycogen metabolism, and glucose regulation pathways (10).
PP1 is commonly found in association with one or more regulatory subunits that target the phosphatase to different cellular compartments and specify substrate selectivity (15, 16). The best-characterized GLC7 regulatory proteins in yeast are REG1 and GAC1. The REG1-GLC7 complex is responsible for dephosphorylation of the SNF1 kinase, a key regulator of glucose repression pathways (17–21), and the GLC7-GAC1 complex promotes glycogen synthesis by dephosphorylating glycogen synthase (GSY2) (22–24). Many PP1 regulatory subunits contain a degenerate amino acid sequence motif, commonly simplified as RVxF (15, 25, 26). This motif is typically flanked N-terminally by basic residues and C-terminally by acidic residues, and sequence conservation and mutational analyses have established the importance of this consensus sequence (25, 27, 28). Based on the crystal structure of PP1 in complex with a targeting peptide, the RVxF motif binds to a hydrophobic channel that is remote from the catalytic site of PP1 (29, 30). This docking has been proposed either to allosterically affect the activity and/or substrate specificity of PP1 or to more simply target PP1 to its substrates (26, 29, 31–33). In addition to the RVxF motif, several other PP1-binding motifs have been described (25, 27, 30, 31, 34, 35), and in some cases these alternate motifs are present along with the RVxF motif in the PP1 regulatory subunit (27).
In mammalian cells, the regulatory subunits GADD34 (PPP1R15A) and CReP (PPP1R15B), which both contain an RVxF motif, recruit PP1 to dephosphorylate Ser51 on eIF2α (36, 37). Likewise, the herpes simplex virus (HSV) γ34.5 protein, which shares sequence similarity with GADD34 and CReP, binds both PP1 and eIF2α (38) to promote eIF2α dephosphorylation in HSV-infected cells (39). Although GADD34 or CReP homologs have been identified in mammals, chickens, frogs, and zebrafish, a degenerate ortholog is found in flies, and no homologs of these proteins have been identified in other species including worms and fungi. This raises the question of how GLC7 targets eIF2α in the yeast S. cerevisiae. Recently, a mass spectrometric analysis of GLC7-interacting proteins identified eIF2β and eIF2γ in addition to eIF2α (11). Although the eIF2β and eIF2γ interaction with GLC7 might simply reflect the tethering of the former proteins to GLC7 via eIF2α, the relative enrichment in the recovery of eIF2γ versus eIF2α and eIF2β with GLC7 (11) raises the possibility that eIF2γ brings the eIF2 complex to GLC7. Although eIF2γ does not share sequence similarity with GADD34 or CReP, we identified an RVxF motif in an N-terminal extension that is uniquely found in yeast eIF2γ. In this report, using a combination of yeast genetic and biochemical studies, we demonstrate that the N terminus of yeast eIF2γ functionally replaces GADD34 and CReP and recruits GLC7 to dephosphorylate eIF2α in yeast.
Results
Mutations at the N Terminus of Yeast eIF2γ Alter eIF2α Phosphorylation in Vivo.
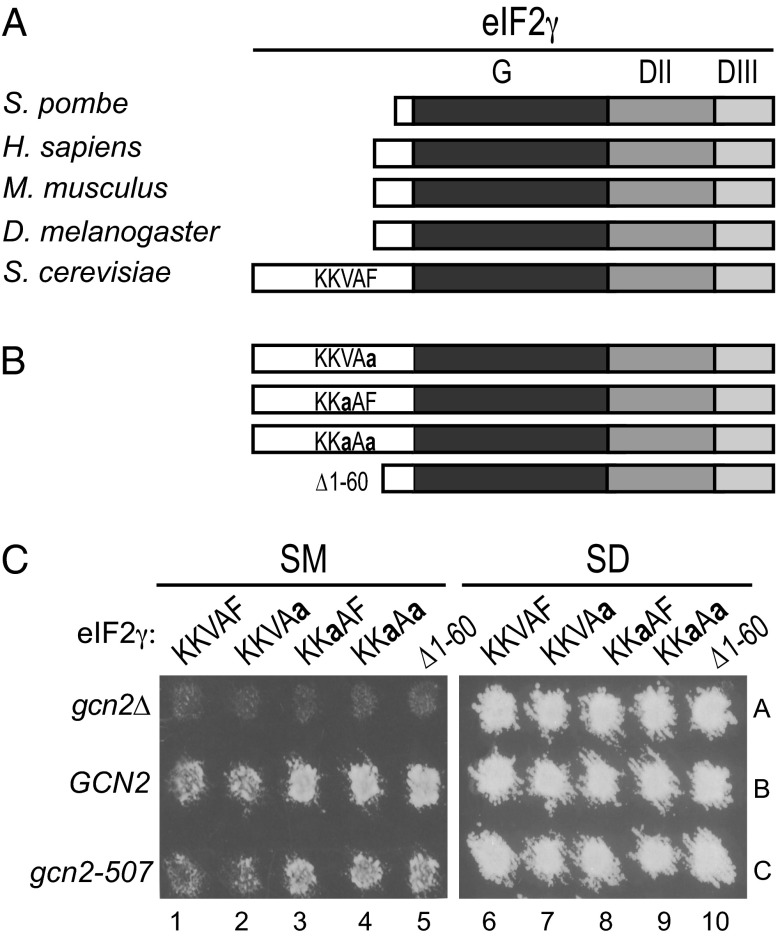
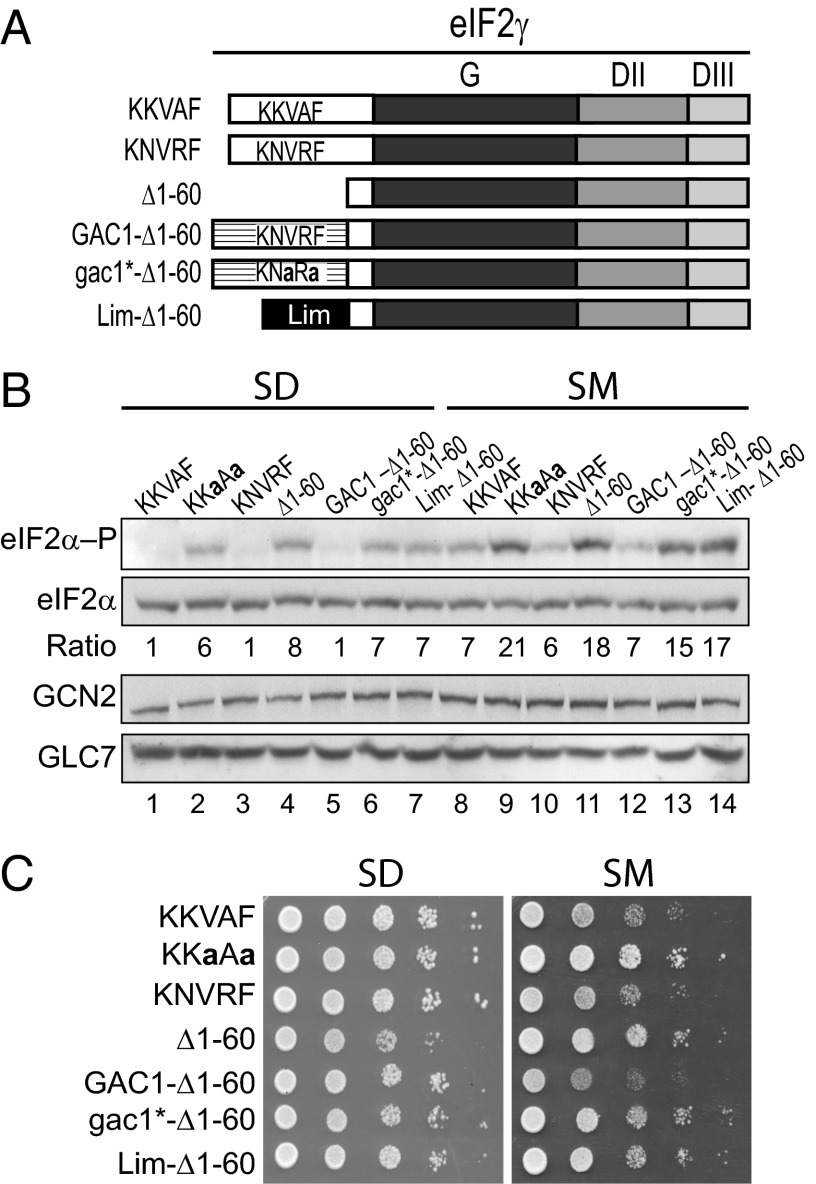
eIF2γ is composed of three domains: a GTP-binding domain (G) and β-barrel domains II and III (Fig. 1A). Comparative sequence analysis of eIF2γ homologs from a variety of single and multicellular eukaryotes revealed the presence of an extension at the N terminus of the G domain that varies in length from 12 residues in Schizosaccharomyces pombe to 89 residues in S. cerevisiae (Fig. 1A, white box). Previously, it was reported that deletion of residues 3–77 in the N-terminal extension of S. cerevisiae eIF2γ does not affect yeast cell growth (40), indicating that this sequence does not play an essential role in eIF2 function. Of interest, however, we identified a PP1-binding motif with the sequence KKVAF (residues 47–51) in this N-terminal extension of eIF2γ (Fig. 1A). Identical or similar PP1-binding motifs were found in eIF2γ from related Saccharomyces species and from several more distantly related fungi including species in the genera Candida, Pichia, and Kluveromyces. However, no PP1-binding motif was easily identified in the N-terminal extension of eIF2γ from S. pombe and several other yeasts in the phylum Ascomycetes. As all three subunits of eIF2 were identified among the pool of proteins interacting with GLC7 (11), and as no orthologs of the mammalian PP1-targeting subunits CReP or GADD34 are present in yeast, we hypothesized that the eIF2γ N-terminal extension functions like a PP1 regulatory subunit and helps recruit GLC7 to dephosphorylate eIF2α. To test this hypothesis, we introduced point mutations or a deletion to eliminate the KKVAF motif in eIF2γ (Fig. 1B) and then expressed the mutants as the sole form of eIF2γ in yeast. Consistent with the results of the previous study (40), deletion of eIF2γ residues 1–60 (hereafter referred to as Δ1–60), eliminating most of the N-terminal extension, did not significantly affect yeast cell growth on minimal synthetic dextrose (SD) medium (Fig. 1C, SD panel, lane 10 versus 6). Likewise, the eIF2γ V49A (KKaAF) and F51A (KKVAa) mutants, as well as the V49A,F51A (KKaAa) double mutant, in the KKVAF motif did not affect yeast growth on minimal medium (Fig. 1C, SD panel, lanes 6–9). Thus, the N-terminal extension on eIF2γ is not critical for eIF2 function in yeast cells.
Fig. 1.
Effects of mutations altering the RVxF motif in eIF2γ on yeast growth under starvation conditions. (A) Schematic diagram showing the relationship among eIF2γ homologs. The RVxF motif (KKVAF sequence) in the N-terminal extension of S. cerevisiae eIF2γ, and the locations of the GTP-binding (G) domain and domains II (DII) and III (DIII), are indicated. (B) eIF2γ mutations designed to alter or eliminate the KKVAF motif. (C) Derivatives of yeast strain YM53 expressing the indicated form of eIF2γ and carrying either an empty vector (gcn2Δ) or a plasmid expressing WT GCN2 or partially active gcn2-507 were patched and grown to confluence on SD plates and then replica-plated to SD plates or SD plates supplemented with 1 μg/mL of SM. Plates were incubated at 30 °C for 2 d.
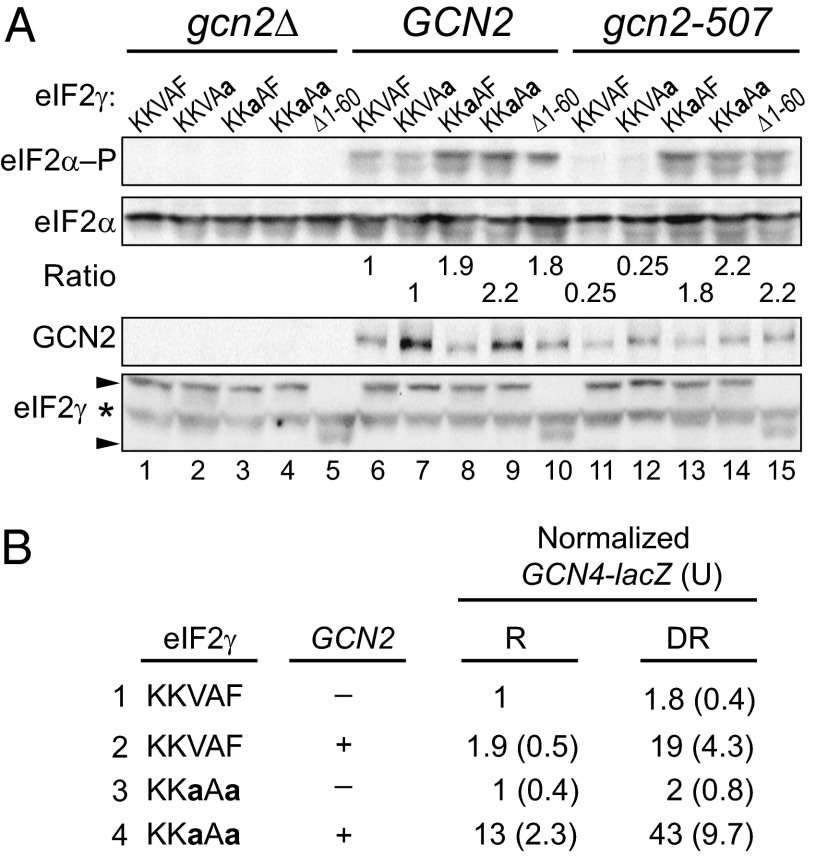
Phosphorylation of eIF2α is critical for the growth of yeast under amino acid starvation conditions. The phosphorylation of eIF2α and the consequent inhibition of eIF2B enable translational derepression of the GCN4 mRNA, which encodes a transcriptional activator of amino acid biosynthesis. Whereas GCN2+ cells are able to derepress GCN4 expression and grow under conditions of Ile and Val starvation imposed by the drug sulfometuron methyl (SM), an inhibitor of the ILV2 enzyme in the branched chain amino acid biosynthetic pathway, gcn2Δ cells lacking the kinase are unable to grow under starvation conditions (Fig. 1C, SM panel, lane 1, rows A and B). Interestingly, yeast strains expressing GCN2 and the eIF2γ mutants KKaAF, KKaAa, or Δ1–60, but not the KKVAa mutant, grew better than strains expressing WT eIF2γ (KKVAF) on medium containing SM (Fig. 1C, SM panel, row B, lanes 1–5). To determine whether this increased growth under starvation conditions was correlated with increased eIF2α phosphorylation, whole cell extracts (WCEs) from strains grown under amino acid starvation conditions were subjected to immunoblot analysis with antibodies specific for the phospho-Ser51 form of eIF2α. As shown in Fig. 2A, yeast expressing the Δ1–60, KKaAF, or KKaAa mutants of eIF2γ showed higher levels of eIF2α phosphorylation than cells expressing WT eIF2γ or the KKaAF mutant (Fig. 2A, Top, lanes 6–10). When normalized for the total amount of eIF2α in the extracts, the Δ1–60, KKaAF, and KKaAa mutants of eIF2γ increased steady-state eIF2α phosphorylation levels under starvation conditions around twofold in the cells expressing GCN2 (Fig. 2A, lanes 6–10). As expected given that GCN2 is the only eIF2α kinase in yeast, no Ser51 phosphorylation was observed in cells lacking GCN2 (Fig. 2A, Top, lanes 1–5), and the eIF2γ mutations did not enhance the growth of the gcn2Δ strain on amino acid starvation medium (Fig. 1C, SM panel, row A). This latter result provides further evidence that the mutations do not significantly impair eIF2 function, as a hallmark of reduced eIF2 activity is the ability of gcn2Δ cells to grow under amino acid starvation conditions (4, 6).
Fig. 2.
Effects of eIF2γ mutations on eIF2α phosphorylation and GCN4 expression. (A) Derivatives of yeast strain YM53 expressing the indicated form of eIF2γ and carrying either an empty vector (gcn2Δ) or a plasmid expressing WT GCN2 or partially active gcn2-507 were grown in SD medium to log phase, treated with 1 μg/mL SM for 1 h, and then equivalent amounts of WCEs were subjected to SDS/PAGE followed by immunoblot analysis using phosphospecific antibodies against phosphorylated Ser51 of eIF2α (eIF2α–P). The membrane was then sequentially stripped and probed with polyclonal antibodies against total yeast eIF2α, eIF2γ, and GCN2. Arrows indicate the positions of full-length eIF2γ and the Δ1–60 mutant; the asterisk indicates a nonspecific protein. The relative level of phosphorylated eIF2α to total eIF2α was determined by quantitative densitometry and normalized to the ratio obtained in the WT control (lane 6). (B) Strains described in A were transformed with GCN4-LacZ reporter plasmids, and β-galactosidase activities (and standard errors) were determined for three independent transformants grown under nonstarvation conditions where GCN4 expression is repressed (R) and under amino acid starvation conditions imposed by SM where GCN4 expression is derepressed (DR).
If, as we propose, the eIF2γ N terminus helps recruit GLC7 to dephosphorylate eIF2α, then a defect in GLC7 recruitment should compensate for a defective GCN2 kinase and enable yeast cell growth under starvation conditions. To test this hypothesis, we took advantage of the leaky gcn2-507 mutant in which a Glu-Leu codon pair was inserted after amino acid 1177 in the histidyl-tRNA synthetase (HisRS) regulatory domain of GCN2 (41). Yeast expressing gcn2-507 and WT eIF2γ show reduced levels of eIF2α phosphorylation compared with strains expressing WT GCN2 (Fig. 2A, lane 11 versus 6), and they are unable to grow on medium containing SM (Fig. 1C, row C, lane 1). Consistent with the idea that the eIF2γ N terminus is required to direct GLC7 to dephosphorylate eIF2α, the Δ1–60, KKaAF, and KKaAa mutants of eIF2γ enhanced eIF2α phosphorylation on Ser51 (Fig. 2A, lanes 13–15) and enabled growth of the gcn2-507 mutant strain under starvation conditions (Fig. 1C, row C). Although GCN2 protein levels varied slightly in the different strains (Fig. 2A), the GCN2 levels did not correlate with the eIF2α phosphorylation levels. Moreover, the eIF2γ mutants were all expressed at similar levels (Fig. 2A). Taken together, these data indicate that loss of the eIF2γ N terminus or mutation of the KKVAF motif interferes with eIF2α dephosphorylation.
As noted above, growth of yeast on starvation medium is dependent on translational induction of GCN4 expression. To determine whether the enhanced growth under starvation conditions of yeast expressing the eIF2γ mutants was associated with increased GCN4 expression, a GCN4-lacZ reporter was introduced into gcn2Δ or GCN2+ strains expressing WT eIF2γ or the eIF2γ KKaAa mutant. As expected, in yeast expressing WT eIF2γ, GCN4-lacZ expression was low in the absence of GCN2 and increased only around twofold when SM was added to induce amino acid starvation (Fig. 2B, row 1). Similar results were obtained in the eIF2γ KKaAa mutant lacking GCN2, consistent with the lack of eIF2α phosphorylation in these strains. In cells expressing GCN2 and WT eIF2γ, GCN4-lacZ expression was repressed under nonstarvation conditions and increased around 10-fold under amino acid starvation conditions (Fig. 2B, row 2). Consistent with the increased eIF2α phosphorylation observed in the KKaAa mutant expressing GCN2 (Fig. 2A, lane 9), GCN4-lacZ expression was increased sixfold under nonstarvation conditions in this mutant compared with the strain expressing WT eIF2γ (Fig. 2B, row 4 versus 2). Finally, GCN4-lacZ expression increased around threefold in the KKaAa mutant under starvation conditions (Fig. 2B, row 4). Based on these various assays, we conclude that the KKVAF motif in yeast eIF2γ is critical for dephosphorylation of eIF2α and proper regulation of GCN4 expression and yeast cell growth under amino acid starvation conditions.
eIF2γ N Terminus is Critical for GLC7 Dephosphorylation of eIF2α in Vitro.
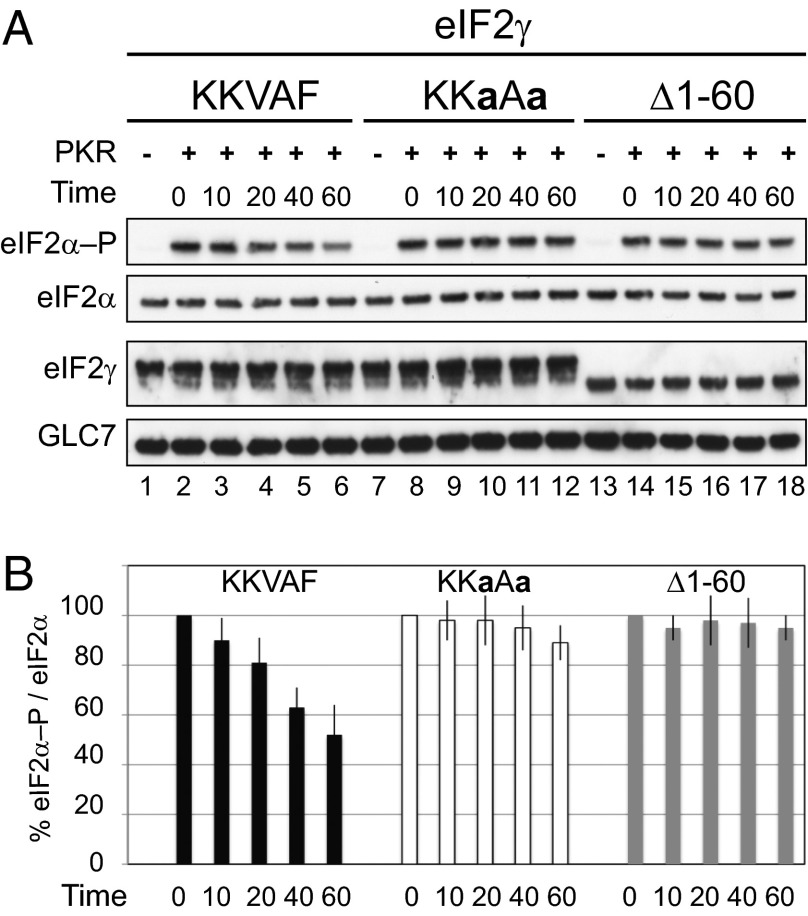
To determine whether the eIF2γ N terminus directly contributes to GLC7 dephosphorylation of eIF2α, purified WT eIF2 complexes consisting of WT eIF2α and eIF2β and either WT eIF2γ or the eIF2γ mutants KKaAa or Δ1–60 were purified from yeast and incubated with purified FLAG epitope-tagged PKR (FLAG-PKR) in the presence of ATP to phosphorylate eIF2α on Ser51. Importantly, PKR readily phosphorylated eIF2α in all three eIF2 complexes indicating that the impact of the eIF2γ mutations on eIF2α phosphorylation was not due to altered kinase recognition of the eIF2 proteins. The phosphorylated eIF2 complexes were incubated with purified, recombinant GST-GLC7-FLAG that was expressed in Escherichia coli in the presence of manganese to enhance phosphatase activity. Consistent with the results of the in vivo assays, GLC7 readily dephosphorylated eIF2α in complexes containing WT eIF2γ with ∼50% of eIF2α dephosphorylated after 60 min (Fig. 3A, lanes 2–6, and B). In contrast, only ∼10% of eIF2α in the eIF2 complex containing the KKaAa mutant of eIF2γ was dephosphorylated after incubation with GLC7 for 60 min (Fig. 3A, lanes 8–12, and B), and no significant dephosphorylation of eIF2α was observed in the complexes containing the Δ1–60 mutant (Fig. 3A, lanes 14–18, and B). These results confirm that eIF2γ plays a direct role in targeting dephosphorylation of eIF2α by the phosphatase GLC7.
Fig. 3.
Dephosphorylation of eIF2α in vitro by purified GLC7. (A) Purified yeast eIF2 complexes containing WT or the indicated mutant forms of eIF2γ were incubated with FLAG-PKR immobilized onto M2-conjugated agarose and ATP to phosphorylate eIF2α on Ser51. Following incubation of the phosphorylated eIF2 complexes with purified GST-GLC7-FLAG for the indicated times (min), the reaction products were subjected to immunoblot analysis using specific polyclonal antibodies to detect eIF2α–P and total eIF2α. Monoclonal antibodies against the His-tag were used to detect eIF2γ, and polyclonal antibodies against GST were used to detect GLC7. (B) The ratio of eIF2α–P to total eIF2α at each time point was quantified from three independent experiments and normalized for each eIF2 complex to the value at time 0.
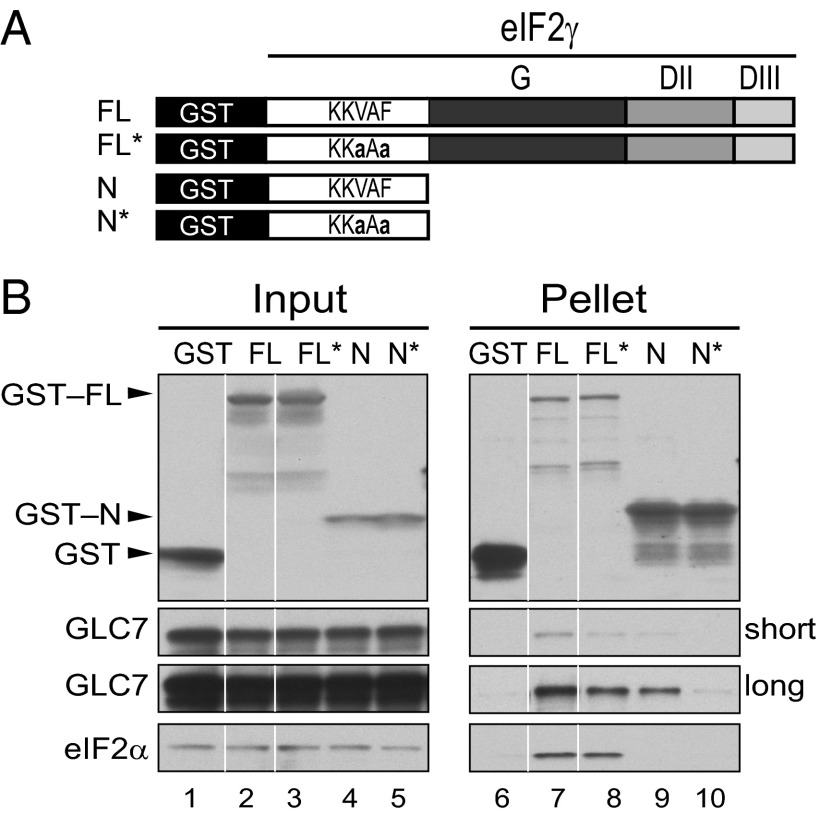
N-Terminal Extension on Yeast eIF2γ Binds to GLC7.
To test whether the eIF2γ N-terminal extension and its KKVAF motif contribute to GLC7 binding to eIF2, a set of GST-eIF2γ fusion proteins (Fig. 4A) was expressed in a yeast strain in which the chromosomal GLC7 gene was tagged at its C terminus with 13 myc epitopes. Yeast WCEs were incubated with glutathione beads, and the products of the pull-down reactions were subjected to immunoblot analysis. As shown in Fig. 4B, GST fusion proteins containing either full-length eIF2γ (FL) or just the N-terminal extension of eIF2γ (N, residues 2–62) readily interacted with GLC7 (Fig. 4B, lanes 7 and 9), pulling down 5 or 0.2%, respectively, of the input GLC7-myc. Consistent with the idea that the KKVAF motif in eIF2γ mediates the interaction with GLC7, the KKaAa mutation in the GST-N fusion protein (N*) substantially impaired the binding of GLC7 (Fig. 4B, compare lanes 9 and 10, ∼97% decrease in binding). Interestingly, introducing the KKaAa mutation in GST-FL (creating FL*) only partially impaired the interaction with GLC7 (Fig. 4B, compare lanes 7 and 8, ∼70% decrease in binding), suggesting that other domain(s) of eIF2γ in addition to the N-terminal extension either directly or indirectly interact with GLC7. As expected, based on the binding of aIF2α to domain II of aIF2γ (42, 43), only the full length GST-eIF2γ fusion bound eIF2α, and the KKaAa mutation did not alter this interaction (Fig. 4B, lanes 7 and 8).
Fig. 4.
The eIF2γ N terminus interacts with GLC7. (A) Schematic depicting GST fusion proteins containing either full-length (FL) eIF2γ or just the N-terminal extension (N) with an intact or mutated (*) KKVAF motif. (B) GST or the indicated GST-eIF2γ fusions proteins were overexpressed in the yeast strain H4476 expressing myc-tagged GLC7. WCEs were mixed with glutathione–Sepharose beads, and after washing, bound proteins were eluted with SDS-loading buffer, separated by SDS/PAGE, and detected by immunoblotting with antibodies against GST, eIF2α, or the myc–epitope on GLC7; 5% of input and 20% of pellet fractions were analyzed. Short and long exposures of the blot for GLC7 are shown; white lines indicate splicing of lanes from the same original blots to generate the final figure.
Functional Replacement of the N-Terminal Extension of Yeast eIF2γ with the GLC7-Binding Domain of Yeast GAC1.
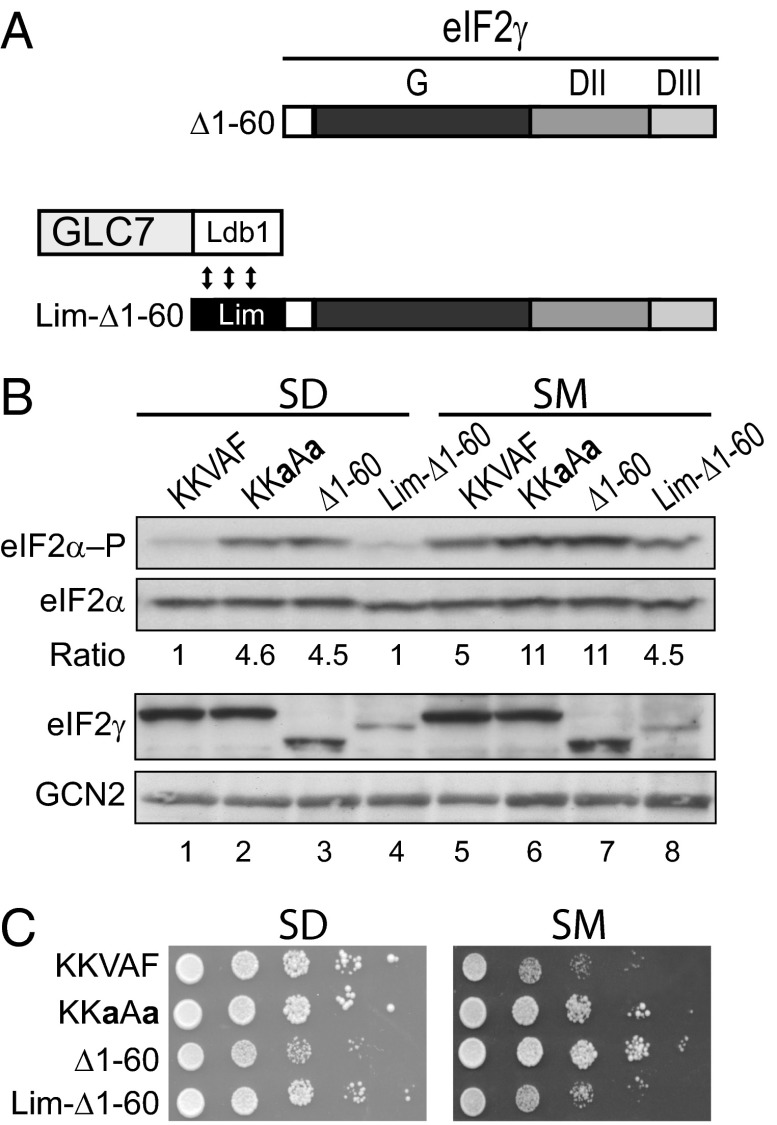
If the primary function of the eIF2γ N-terminal extension is to bind GLC7, then we reasoned it should be possible to replace the eIF2γ N terminus with the GLC7-binding domain from GAC1, a GLC7-targeting protein that directs GLC7 to dephosphorylate glycogen synthase GSY2 (22, 23). Alternatively, if the KKVAF motif and flanking elements of eIF2γ have an additional function to activate the phosphatase activity of GLC7 toward eIF2α, then the GAC1 sequences should not substitute for the N terminus of eIF2γ. The PP1-binding motif in GAC1 (KNVRF) differs at two positions from the KKVAF motif in eIF2γ (Fig. 5A). As shown in Fig. 5B, the KNVRF motif from GAC1 functionally replaced the KKVAF motif of eIF2γ and maintained eIF2α phosphorylation at WT levels under both nonstarvation (SD) and starvation (SM) conditions (Fig. 5B, compare lanes 1, 3, 8, and 10). Having found that the alternate KNVRF motif of GAC1 can substitute in eIF2γ, we next asked if the N-terminal extension of eIF2γ (residues 1–60) could be replaced by the N-terminal 93 amino acids of GAC1, which were previously shown to be sufficient to bind GLC7 (23). As a control, the N-terminal 56 amino acids of the Xenopus protein Xlim-1, which does not contain an RVxF motif, were fused to Δ1–60 (Fig. 5A). In contrast to Δ1–60, which resulted in elevated levels of eIF2α phosphorylation under both nonstarvation and starvation conditions (Fig. 5B, lanes 4 and 11), the GAC1-Δ1–60 fusion protein functioned like full-length eIF2γ, maintained eIF2α phosphorylation at low levels on SD medium (Fig. 5B, lane 5), and prevented the dramatic increase in eIF2α phosphorylation observed in cells expressing the eIF2γ mutants KKaAa, Δ1–60, and Lim-Δ1–60 that lack a functional RVxF motif (Fig. 5B, compare lane 12 with lanes 9, 11, and 14). Consistent with these observations, the ability of the GAC1 segment to functionally substitute for the N-terminal extension of eIF2γ was dependent on the KNVRF motif, as mutation of this motif to KNaRa in gac1*-Δ1–60 resulted in high levels of eIF2α phosphorylation (Fig. 5B, lanes 6 and 13). Notably, GLC7 levels were comparable in the cells expressing the various eIF2γ proteins (Fig. 5B), indicating that the eIF2γ derivatives were likely affecting GLC7 targeting rather than its abundance. Consistent with the requirement for eIF2α phosphorylation to induce GCN4 expression, the yeast strains expressing eIF2γ mutants lacking a functional RVxF motif, and thus having higher levels of phosphorylated eIF2α, showed increased growth under starvation conditions (Fig. 5C, SM panel). These results provide further support for the hypothesis that the eIF2γ N terminus targets GLC7 to dephosphorylate eIF2α. In addition, the ability of the GAC1 N terminus to functionally substitute for the eIF2γ N terminus suggests that the RVxF motif and flanking sequences function by simply tethering GLC7 to its substrate rather than by activating GLC7 to dephosphorylate specific substrates.
Fig. 5.
The N-terminal segment of GAC1 functionally replaces the N-terminal extension of eIF2γ. (A) Schematic diagram of eIF2γ mutants and fusion proteins. The GAC1 element contains residues 1–93; the Lim domain contains residues 1–56 of Xlim-1. (B) Derivatives of yeast strain YM53 expressing the indicated eIF2γ protein and WT GCN2 were grown in SD medium to log phase, incubated for 1 h in the presence or absence of 1 μg/mL SM, and then equivalent amounts of WCEs were subjected to SDS/PAGE followed by immunoblot analysis to detect eIF2α–P, total eIF2α, GCN2, and GLC7-myc. The relative level of phosphorylated to total eIF2α was determined by quantitative densitometry and normalized to the ratio obtained in the nonstarved WT control (lane 1). (C) Yeast transformants described in B were grown to saturation in SD medium, and 4-µL volumes of serial dilutions (optical density; OD600 = 1.0, 0.1, 0.01, 0.001, and 0.0001) were spotted on SD medium or SD medium supplemented with 1 μg/mL SM and incubated 3 d at 30 °C.
Heterodimerization Domains Fused to eIF2γ and GLC7 Functionally Substitute for the eIF2γ N Terminus.
Two models have been proposed for the function of the RVxF motif: (i) to bind PP1 and help direct the phosphatase to its substrate; and (ii) to bind and allosterically activate the phosphatase activity of PP1 (29, 30). The ability of the GAC1 N terminus with an intact RVxF (KNVRF) motif to substitute for the eIF2γ N terminus supports both models, although as described above it does rule out a third possibility that the eIF2γ N terminus contains elements in addition to the KKVAF motif that specifically activate GLC7 to dephosphorylate eIF2α (if this were the case, the GAC1 N terminus would fail to support eIF2α dephosphorylation and would instead specifically promote only GSY2 dephosphorylation).
To test whether the N-terminal domain of eIF2γ promotes eIF2α dephosphorylation by tethering GLC7 to the eIF2 complex, we took advantage of the heterodimerization domains in the Xenopus Xlim-1 and Ldb1 proteins. We previously demonstrated the utility of these domains by showing that they could substitute for the double-stranded RNA-binding domains in PKR and promote the dimerization-dependent activation of the kinase (44). Residues 300–338 of the Xenopus Ldb1 protein, which heterodimerizes with the N terminus of Xlim-1 (45, 46), were fused to the C terminus of GLC7 to generate a GLC7-Ldb1 fusion protein (Fig. 6A). As shown in Fig. 5B, increased levels of eIF2α phosphorylation were observed in yeast expressing native GLC7 and either Δ1–60 or the Lim-Δ1–60 fusion in which the Xlim-1 sequences were inserted at the N terminus Δ1–60 (Fig. 5B, lanes 4, 7, 11, and 14). In contrast, whereas high levels of eIF2α phosphorylation were detected in yeast expressing GLC7-Ldb1 and either Δ1–60 or the KKaAa mutant (Fig. 6B, lanes 2–3 and 6–7), eIF2α phosphorylation levels were maintained at basal levels under both nonstarvation and starvation conditions in strains expressing Lim-Δ1–60 (Fig. 6B, lanes 4 and 8). Consistent with the lower levels of eIF2α phosphorylation under starvation conditions in the GLC7-Ldb1 strain expressing Lim-Δ1–60 versus the Δ1–60 mutant of eIF2γ (Fig. 5B, lanes 7–8), yeast expressing the Lim-Δ1–60 fusion grew like the WT control, and more slowly than strains expressing the Δ1–60 mutant on SM medium (Fig. 6C). The reconstitution of eIF2α dephosphorylation in vivo by fusing heterologous heterodimerization domains to eIF2γ and GLC7 indicates that the primary function of the eIF2γ N terminus is to tether GLC7 to the eIF2 complex where it can then dephosphorylate eIF2α. Moreover, as the Xlim-1 sequences lack an RVxF motif, yet are able to promote eIF2α dephosphorylation by GLC7-Ldb1, these results indicate that any putative allosteric activation of GLC7 by the RVxF motif is not critical for eIF2α dephosphorylation.
Fig. 6.
Reconstitution of eIF2α dephosphorylation in yeast by fusion of the Lim and Ldb1 heterodimerization domains to eIF2γ and GLC7. (A) Schematics of the Δ1–60 eIF2γ mutant and the Lim-Δ1–60 and GLC7-Ldb1 fusion proteins. As indicated by the double-headed arrows, the Ldb1 domain, which contains residues 300–338 of Ldb1, heterodimerizes with the Lim domain. (B) Derivatives of yeast strain YM54 expressing the indicated eIF2γ protein, WT GCN2, and GLC7-Ldb1 were grown in SD medium to log phase, incubated for 1 h in the presence or absence of 1 μg/mL SM, and then equivalent amounts of WCEs were subjected to SDS/PAGE followed by immunoblot analysis to detect eIF2α–P, total eIF2α, eIF2γ, and GCN2. The relative level of phosphorylated to total eIF2α was determined by quantitative densitometry and normalized to the ratio obtained in the nonstarved WT control (lane 1). (C) Yeast transformants described in B were grown to saturation in SD medium, and 4-µL volumes of serial dilutions (OD600 = 1.0, 0.1, 0.01, 0.001, and 0.0001) were spotted on SD medium or SD medium supplemented with 1 μg/mL SM and incubated 3 d at 30 °C.
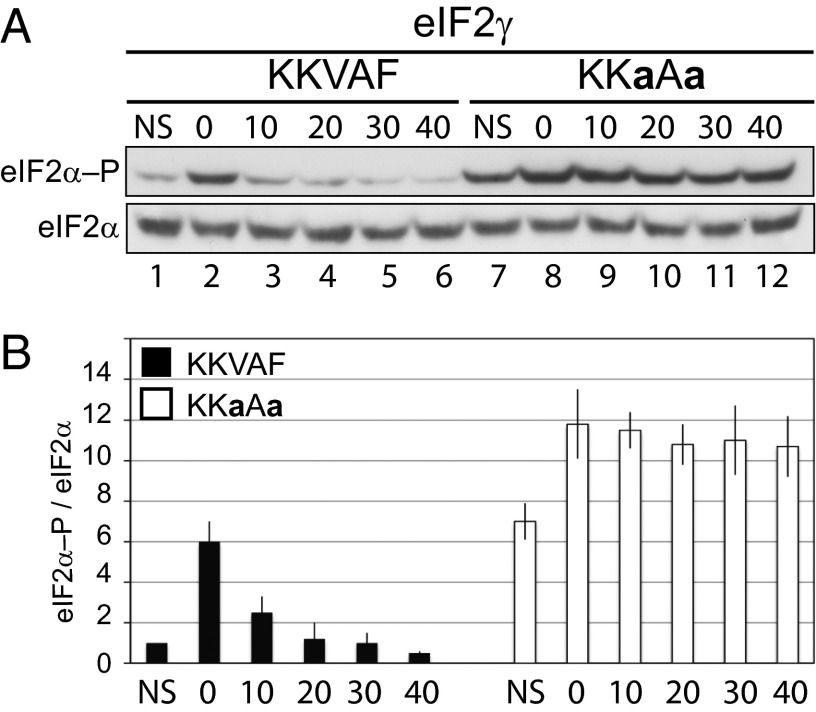
Sustained eIF2α Phosphorylation in Yeast Expressing the eIF2γ KKaAa Mutant.
Although numerous studies have documented the rapid activation of GCN2 and phosphorylation of eIF2α under amino acid starvation and other cellular stress conditions, the dephosphorylation of eIF2α in yeast following recovery from stress has been examined in lesser detail (3, 47–50). We hypothesized that the binding of GLC7 to the eIF2γ N terminus should enable rapid dephosphorylation of eIF2α following relief of the stress conditions, and accordingly, the kinetics of dephosphorylation should be delayed in cells expressing an eIF2γ mutant that fails to recruit GLC7. To test these hypotheses, yeast expressing native GLC7 and either WT eIF2γ or the KKaAa mutant were grown under nonstarvation conditions in minimal SD medium to midlog phase, treated for 1 h with SM to induce amino acid starvation, and then transferred to fresh SD medium. Cells were harvested at various times during the experiment, and eIF2α phosphorylation was monitored by immunoblot analysis. As shown in Fig. 7, low levels of eIF2α phosphorylation were observed in WT cells grown in SD medium (Fig. 7, lane 1), and eIF2α phosphorylation was strongly induced in cells treated with SM (Fig. 7, lane 2). The eIF2α was rapidly dephosphorylated following removal of the cells from the starvation medium with nearly basal levels of phosphorylation detected within 20 min (Fig. 7A, lanes 3–6, and B). As shown in Fig. 7A, yeast expressing the eIF2γ KKaAa mutant exhibited elevated levels of eIF2α phosphorylation compared with the WT strain under both nonstarvation conditions (Fig. 7A, compare lanes 1 and 7) and following amino acid starvation (Fig. 7A, compare lanes 8 and 2). Moreover, eIF2α phosphorylation persisted at high levels for the duration of the experiment (40 min) following transfer of the cells to nonstarvation medium (Fig. 7A, lanes 9–12, and B). Thus, the N terminus of eIF2γ targets GLC7 to the eIF2 complex to maintain eIF2α phosphorylation at low levels under nonstarvation conditions and to allow rapid dephosphorylation of eIF2α following relief of the stress.
Fig. 7.
Sustained eIF2α phosphorylation in yeast cells expressing the KKaAa mutant of eIF2γ. (A) Derivatives of yeast strain YM53 expressing WT GCN2 and the indicated eIF2γ protein were grown in SD medium to log phase, incubated for 1 h in the presence or absence of 1 μg/mL SM, and then washed and incubated in fresh SD medium at 30 °C. After the indicated times (min), cells were lysed, and equivalent amounts of WCEs were subjected to SDS/PAGE followed by immunoblot analysis to detect eIF2α–P and total eIF2α. (B) The relative level of phosphorylated to total eIF2α in three independent experiments was determined by quantitative densitometry and normalized to the ratio obtained in the nonstarved WT control (lane 1).
Discussion
The γ subunit is the keystone of the eIF2 complex. Functioning as a scaffold, eIF2γ has binding sites for eIF2α and eIF2β enabling formation of the eIF2 complex. Moreover, eIF2γ binds GTP and makes the principal contacts with Met-tRNAiMet in the eIF2 ternary complex. In addition, during ribosomal scanning, eIF2γ plays a critical role in start codon recognition; and, as expected of a G protein, eIF2γ directly interacts with the factors eIF5 and eIF2Bε (51), which trigger GTP hydrolysis and promote guanine nucleotide exchange, respectively, on the eIF2 complex. Here, we show that an N-terminal extension on yeast eIF2γ interacts with GLC7 and that this interaction is dependent on the KKVAF motif (Fig. 4B). Interestingly, GLC7 was previously identified as controlling eIF2α phosphorylation in yeast. Expression of a dominant-negative mutant of GLC7 resulted in increased eIF2α phosphorylation in yeast expressing a partially defective form of GCN2, whereas overexpression of functional GLC7 decreased eIF2α phosphorylation (12). Based on the impacts of altering GLC7 activity on eIF2α phosphorylation and the direct binding of GLC7 to eIF2γ, we hypothesized that eIF2γ targeted GLC7 to dephosphorylate eIF2α. Consistent with this hypothesis, mutation or deletion of the KKVAF motif in eIF2γ resulted in higher levels of eIF2α phosphorylation in vivo (Figs. 2, 5, and 7) and disrupted eIF2α dephosphorylation by purified GLC7 in vitro (Fig. 3). Taken together, our results indicate that the interaction between GLC7 and the N-terminal extension of eIF2γ is critical for eIF2α dephosphorylation in yeast. In agreement with this idea, we found that the eIF2γ N terminus could be functionally replaced by the GLC7-binding segment from the protein GAC1 (Fig. 5) or by appending complementary heterodimerization domains to eIF2γ and GLC7.
PP1-targeting subunits have been proposed to activate the phosphatase activity of PP1 or to target the phosphatase to its substrate (29). Consistent with the former model, a truncated derivative of GAC1 consisting of residues 1–93, which retains the ability to bind GLC7 but lacks the binding site for glycogen synthase, is able to partially complement the glycogen storage defect of a gac1Δ mutant (23). It has been proposed that binding of the GAC1 N terminus may activate the GLC7 phosphatase activity or enable GLC7 to adopt a conformation that is required for dephosphorylation of glycogen synthase (24). At odds with the notion that the GAC1 N terminus specifically primes GLC7 to dephosphorylate glycogen synthase, we found that fusion of this same GAC1 fragment to eIF2γ is sufficient to target GLC7 and promote strong dephosphorylation of eIF2α (Fig. 5B). Moreover, binding of an RVxF peptide does not appear to alter the conformation of PP1 (16). These results indicate that binding of an RVxF-containing peptide to GLC7 is not sufficient to activate GLC7 to dephosphorylate a specific substrate. In further support of the scaffolding role of the PP1-targeting sequences, we showed that fusion of the heterodimerization domains from Xlim-1 and Ldb1 to eIF2γ and GLC7, respectively, was sufficient to reconstitute eIF2α dephosphorylation activity in vivo (Fig. 6). Given that the N-terminal 56 residues of Xlim-1 do not contain a potential RVxF motif, our data indicate that simple targeting of GLC7 is sufficient to promote eIF2α dephosphorylation.
Transient phosphorylation of eIF2α functions mostly as a cytoprotective measure through the activation of pathways that promote cell survival. Nevertheless, prolonged eIF2α phosphorylation is proapoptotic (reviewed in ref. 52). In mammalian cells, dephosphorylation of eIF2α by PP1 is regulated by the targeting subunits GADD34 and CReP (reviewed in ref. 7). Both GADD34 and CReP recruit PP1 through their related C-terminal domains (36, 37, 53–56). Aside from the conserved RVxF motif, yeast eIF2γ does not share sequence similarity with GADD34 or CReP. Moreover, the N-terminal extension on yeast eIF2γ is missing in mammalian eIF2γ. Thus, we propose that the eIF2γ N terminus functions in cis within the eIF2 complex to recruit GLC7/PP1, whereas mammalian cells rely on trans-acting PP1-targeting subunits to promote eIF2α dephosphorylation. The expression of different targeting subunits leads to exquisite control of eIF2α phosphorylation in mammals. In contrast to CReP, which is constitutively expressed, GADD34 levels are tightly regulated by stress (36, 37, 53–56). Phosphorylation of eIF2α induces expression of GADD34, which then functions in a feedback-inhibitory manner to promote eIF2α dephosphorylation. Finally, following translational recovery and restoration of cellular homeostasis, GADD34 is rapidly degraded by the proteasome (53).
In conclusion, our results show that eIF2γ is a bona fide GLC7-targeting subunit that promotes dephosphorylation of eIF2α in yeast. Because eIF2γ expression in yeast is not regulated by stress or eIF2α phosphorylation, we propose that eIF2γ functions like mammalian CReP to control the basal levels of eIF2α phosphorylation. It remains to be determined whether yeast express an inducible GLC7-targeting subunit like GADD34 whose expression is regulated by eIF2α phosphorylation. Alternatively, perhaps the presence of a single eIF2α kinase in yeast versus four eIF2α kinases in mammalian cells places a greater demand on mammalian cells to tightly regulate eIF2α phosphorylation. Accordingly, the constitutive eIF2γ-directed dephosphorylation may provide sufficient control of eIF2α phosphorylation in yeast with the regulation centered on GCN2 kinase activity. Finally, it is noteworthy that the RVxF motif in the N-terminal extension of eIF2γ is restricted to Saccharomyces and a few related organisms but is missing from other members of the phylum Ascomycota including S. pombe and Aspergillus. Interestingly, the genome sequences of S. pombe and various Aspergillus strains likewise do not contain recognizable homologs of CReP or GADD34, suggesting that additional proteins or mechanisms have evolved to control eIF2α dephosphorylation in these organisms.
Methods
Plasmid Construction.
Plasmids are listed in Table 1. Standard techniques were used for DNA manipulation. Plasmids p585, p561, pC2872, and pC1722 encoding GCN2, gcn2-507, His8-GCD11 (eIF2γ), and FLAG-PKR were previously described (12, 57–59). Mutations designed to generate the V49A, F51A, V49A/F51A, or K48N/A50R versions of eIF2γ were introduced into pC2872 using a QuikChange site-directed mutagenesis kit (Stratagene) resulting in the plasmids pC4030 (KKaAF), pC4029 (KKVAa), pC4031 (KKaAa), and pC4228 (KNVRF). A BamH1-Sal1 fragment encoding residues 61–527 of eIF2γ was obtained by PCR and was used to replace the cognate fragment in pC2872 generating the His8-eIF2γ-Δ1–60 (Δ1–60) expression plasmid pC4032. A BamH1 fragment encoding GAC1 residues 1–93 or Xlim-1 residues 1–58 was obtained by PCR using either yeast genomic DNA or the vector pC901 (44), respectively, as a template. These PCR products were inserted into pC4032 to create plasmids pC4233 (GAC1-Δ1–60) and pC4234 (Lim-Δ1–60). The V71A/F73A mutations were introduced into pC4233 generating the plasmid pC4538 (gac1*-Δ1–60). Plasmid pC2697 expressing GST fused to full-length eIF2γ (GST-FL) was described previously (51). A PCR fragment encoding the N terminus of eIF2γ (residues 2–62) was cloned in the vector pEGKT (60) between the BamHI and SalI sites to generate the plasmid pC4216 expressing GST-N. The V49A/F51A mutations were introduced into pC2697 and pC4216 generating the plasmids pC4204 and pC4217, respectively. A Sal1-Not1 fragment isolated from p908 (GCN4-lacZ) or p910 (uORF-less GCN4-lacZ) was subcloned to pRS313 creating the plasmids pC4206 and pC4238, respectively. A Not1-BamH1 PCR fragment encoding GLC7-FLAG was amplified using pET-GLC7, kindly provided by Kelly Tatchell (Louisiana State University Health Sciences Center, Shreveport, LA), as a template and then inserted into pGEX-6P-1 (GE Healthcare) to create the plasmid pC4515. Plasmids designed to express and purify the eIF2 complex from yeast were constructed as follows: (i) a BamH1-Nhe1 fragment encoding eIF2α and eIF2β was isolated from p1778 (61) and subcloned between the BamH1 and Xba1 sites of YEplac181 generating pC4546; (ii) a BamH1-BglII fragment encoding eIF2γ was isolated from pC2872, pC4031, and pC4032 and inserted into the BamH1 site of pC4546 to generate the expression vectors pC4558, pC4562, and pC4563, respectively. The sequence of all genes and the presence of the desired mutations were verified by DNA sequencing.
Table 1.
Plasmids used in this study
| Name | Description | Source |
| p561 | lc, URA3, gcn2-507 | (12) |
| p585 | lc, URA3, GCN2 | (58) |
| pC1722 | hc, URA3, FLAG-PKR | (59) |
| pC2697 | hc, URA3, GST-GCD11-(eIF2γ) (FL) | (51) |
| pC2872 | lc, LEU2, His8-GCD11 (eIF2γ) | (57) |
| pC4029 | lc, LEU2, His8-GCD11-KKVAa | This study |
| pC4030 | lc, LEU2, His8-GCD11-KKaAF | This study |
| pC4031 | lc, LEU2, His8-GCD11-KKaAa | This study |
| pC4032 | lc, LEU2, His8-GCD11-Δ1–60 | This study |
| pC4204 | hc, URA3, GST-GCD11-KKaAa (FL*) | This study |
| pC4206 | lc, HIS3, GCN4-LacZ | This study |
| pC4216 | hc, URA3, GST-GCD11- [(2–62)] (N) | This study |
| pC4217 | hc, URA3, GST-GCD11- [(2–62)] (N*) | This study |
| pC4228 | lc, LEU2, His8-GCD11-KNVRF | This study |
| pC4233 | lc, LEU2, His8-GAC1-GCD11-Δ1–60 (GAC1-Δ1–60) | This study |
| pC4234 | lc, LEU2, His8-Lim-GCD11-Δ1–60 (Lim-Δ1–60) | This study |
| pC4238 | lc, HIS3, uORFless GCN4-LacZ | This study |
| pC4515 | GST-GLC7-FLAG | This study |
| pC4538 | lc, LEU2, His8-gac1*(KNaRa)-GCD11-Δ1–60 (gac1*-Δ1–60) | This study |
| pC4546 | hc, LEU2, SUI2 (eIF2α), FLAG-SUI3 (eIF2β) | This study |
| pC4558 | hc, LEU2, SUI2, (eIF2α), FLAG-SUI3 (eIF2β), His8-GCD11 (eIF2γ) | This study |
| pC4562 | hc, LEU2, SUI2, (eIF2α), FLAG-SUI3 (eIF2β), His8-GCD11-KKaAa | This study |
| pC4563 | hc, LEU2, SUI2, (eIF2α), FLAG-SUI3 (eIF2β), His8-GCD11-Δ1–60 | This study |
hc, high copy-number plasmid; lc, low copy-number plasmid; *, mutation altering the KKVAF motif of eIF2γ to KKaAa or altering the KNVRF motif of GAC1 to KNaRa.
Yeast Strains.
Yeast strains are listed in Table 2. H4476, J551, and J292 strains were previously described (57, 61, 62). YM53 and YM54 were constructed by transforming J292 with a GLC7::2myc::NatMX or GLC7::ldb1::NatMX cassette, respectively. The addition of myc or ldb1 coding sequences at the 3′ end of the GLC7 ORF was confirmed by sequencing PCR fragments amplified from genomic DNA. Strains YM61-YM69 and YM70-YM73 were generated by shuffling the indicated plasmids into YM53 and YM54, respectively. The high copy-number LEU2 plasmids pC4558, pC4562, and pC4563 were introduced into J551 by plasmid shuffling to generate the strains YM87, YM91, and YM92, respectively.
Table 2.
Yeast strains used in this study
| Name | Description | Source |
| H2766 | MATa ura3-52 leu2-3 leu2-112 trp1-del'63 gcn2-del' gcd2-del'::hisG p[GCD2-K627T, TRP1] | (64) |
| H4476 | MATa ura3-52 leu2-3 leu2-112 trp1-del63 GLC7-13Myc::KanMX | (62) |
| J551 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 sui2Δ::hisG sui3Δ::KanMX4 gcd11Δ::NAT gcn2Δ::hisG pep4::HIS3 p1780 [URA3, SUI2 (eIF2α), SUI3 (eIF2β), GCD11 (eIF2γ)] | (61) |
| J292 | MATα leu2-3,-112 ura3-52 his3, gcn2Δ::loxP gcd11Δ::KanMX p[URA3, GCD11 (eIF2γ)] | (57) |
| YM53 | MATα leu2-3,-112 ura3-52 his3, gcn2Δ::loxP gcd11Δ::KanMX GLC7::2myc::NatMx p[URA3, GCD11 (eIF2γ)] | This study |
| YM54 | MATα leu2-3,-112 ura3-52 his3, gcn2Δ::loxP gcd11Δ::KanMX GLC7::ldb1::NatMx p[URA3, GCD11 (eIF2γ)] | This study |
| YM61 | MATα leu2-3,-112 ura3-52 his3, gcn2Δ::loxP gcd11Δ::KanMX GLC7::2myc::NatMx p4538 [LEU2, His8-gac1*(KNaRa)-GCD11-Δ1–60 (gac1*-Δ1–60)] | This study |
| YM62 | MATα leu2-3,-112 ura3-52 his3, gcn2Δ::loxP gcd11Δ::KanMX GLC7::2myc::NatMx pC2872 [LEU2, His8-GCD11 (eIF2γ)] | This study |
| YM63A | MATα leu2-3,-112 ura3-52 his3, gcn2Δ::loxP gcd11Δ::KanMX GLC7::2myc::NatMx pC4029[LEU2, His8-GCD11-KKVAa)] | This study |
| YM63B | MATα leu2-3,-112 ura3-52 his3, gcn2Δ::loxP gcd11Δ::KanMX GLC7::2myc::NatMx pC4030[LEU2, His8-GCD11-KKaAF] | This study |
| YM63C | MATα leu2-3,-112 ura3-52 his3, gcn2Δ::loxP gcd11Δ::KanMX GLC7::2myc::NatMx pC4031 [LEU2, His8-GCD11-KKaAa] | This study |
| YM64 | MATα leu2-3,-112 ura3-52 his3, gcn2Δ::loxP gcd11Δ::KanMX GLC7::2myc::NatMx pC4032 [LEU2, His8-GCD11-Δ1–60] | This study |
| YM65 | MATα leu2-3,-112 ura3-52 his3, gcn2Δ::loxP gcd11Δ::KanMX GLC7::2myc::NatMx pC4228 [LEU2, His8-GCD11-KNVRF] | This study |
| YM66 | MATα leu2-3,-112 ura3-52 his3, gcn2Δ::loxP gcd11Δ::KanMX GLC7::2myc::NatMx pC4233 [LEU2, His8-GAC1-GCD11-Δ1–60 (GAC1-Δ1–60)] | This study |
| YM69 | MATα leu2-3,-112 ura3-52 his3, gcn2Δ::loxP gcd11Δ::KanMX GLC7::2myc::NatMx pC4234 [LEU2, His8-Lim-GCD11-Δ1–60 (Lim-Δ1–60)] | This study |
| YM70 | MATα leu2-3,-112 ura3-52 his3, gcn2Δ::loxP gcd11Δ::KanMX GLC7::ldb1::NatMx pC2872 [LEU2, His8-GCD11 (eIF2γ)] | This study |
| YM71 | MATα leu2-3,-112 ura3-52 his3, gcn2Δ::loxP gcd11Δ::KanMX GLC7::ldb1::NatMx pC4031 [LEU2, His8-GCD11-KKaAa] | This study |
| YM72 | MATα leu2-3,-112 ura3-52 his3, gcn2Δ::loxP gcd11Δ::KanMX GLC7::ldb1::NatMx pC4032 [LEU2, His8-GCD11-Δ1–60] | This study |
| YM73 | MATα leu2-3,-112 ura3-52 his3, gcn2Δ::loxP gcd11Δ::KanMX GLC7::ldb1::NatMx pC4234 [LEU2, His8-Lim-GCD11-Δ1–60 (Lim-Δ1–60)] | This study |
| YM87 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 sui2Δ::hisG sui3Δ::KanMX4 gcd11Δ::NAT gcn2Δ::hisG pep4::HIS3, pc4558 [LEU2, SUI2 (eIF2α), FLAG-SUI3 (eIF2β), His8-GCD11 (eIF2γ)] | This study |
| YM91 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 sui2Δ::hisG sui3Δ::KanMX4 gcd11Δ::NAT gcn2Δ::hisG pep4::HIS3, pc4562 [LEU2, SUI2 (eIF2α), FLAG-SUI3 (eIF2β), His8-GCD11-KKaAa] | This study |
| YM92 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 sui2Δ::hisG sui3Δ::KanMX4 gcd11Δ::NAT gcn2Δ::hisG pep4::HIS3, pc4563 [LEU2, SUI2 (eIF2α), FLAG-SUI3 (eIF2β), His8-GCD11-Δ1–60] | This study |
Immunoblot Analysis.
Yeast cells were grown to midlogarithmic phase in synthetic dextrose medium (SD) with minimal supplements, and then 5-mL aliquots were incubated for 1 h in the presence or absence of 1μg/mL of sulfometuron methyl (SM). Cells were harvested by centrifugation, mixed with 2 vol 20% (wt/vol) trichloroacetic acid and then broken by agitation with glass beads. Proteins were extracted with SDS Loading Buffer [2% (wt/vol) SDS, 2 mM EDTA, 50 mM Tris-HC1 (pH 6.8), 10% (vol/vol) glycerol, 0.01% bromophenol blue], and following neutralization with 1 M Tris base, the samples were boiled for 5 min and then subjected to SDS-polyacrylamide gel electrophoresis (SDS/PAGE) and immunoblot analysis using rabbit polyclonal antibodies specific for phospho-Ser51 on eIF2α (BioSource International). Blots were stripped and reprobed with polyclonal antiyeast eIF2α antiserum (3). Specific polyclonal antisera were used to detect yeast eIF2γ (51) or GCN2 (63). Monoclonal anti-His6 (H3), anti-Myc (9E10), and anti-FLAG (F-tag-01) antibodies and polyclonal anti-GST (Z5) antibodies were purchased from Santa Cruz Biotechnology, Inc. Immune complexes were detected using horseradish peroxidase-conjugated anti-rabbit and anti-mouse secondary antibodies (GE Healthcare) and enhanced chemiluminescence.
Yeast GST Pull-Down Assays.
Yeast strain H4476 expressing various GST-eIF2γ fusion proteins was grown in 50 mL SD medium at 30 °C to midlog phase, harvested, and washed with SGal medium (synthetic medium containing 2% galactose). Cells were then seeded in 50 mL of SGal medium and incubated for 6 h to induce expression of the GST fusion protein, harvested, and frozen at −80 °C until further use. Cells were suspended in Lysis Buffer [20 mM Tris⋅HCl (pH 7.4), 100 mM NaCl, 0.2 mM EDTA, 1 mM DTT, 12.5% (vol/vol) glycerol, 1% Triton X-100, containing one tablet of protease inhibitor mixture (Roche) and 2 μM each aprotinin, leupeptin, and pepstatin], and WCEs were prepared by homogenizing the cells by vigorous mixing with glass beads on a vortex. Glutathione–Sepharose 4B beads (Amersham Biosciences) were washed several times with Binding Buffer [20 mM Tris⋅HCl (pH 7.4), 100 mM NaCl, 0.2 mM EDTA, 1 mM DTT, 0.1% Triton X-100, containing protease inhibitor mixture as described above], suspended in 1 mL Binding Buffer plus 5% BSA, incubated with rotation at 4 °C for 1 h, and then washed several times with Binding Buffer. WCEs were mixed with 50 μL of the treated glutathione–Sepharose beads and incubated with rotation at 4 °C for 2 h. Proteins attached to the beads were washed three times with Binding Buffer, resuspended in SDS Loading Buffer, boiled for 5 min, separated by SDS/PAGE, and then analyzed by immunoblotting.
Protein Purification.
To purify the eIF2 complex, yeast strains YM87, YM91, and YM92 were grown in YPD medium and eIF2 was purified as previously described (61). To purify GST-GLC7-FLAG, E. coli cells harboring the vector pC4515 were grown overnight at 37 °C in 50 mL of Luria broth (LB) medium containing 100 mg/L ampicillin and supplemented with 2 mM MnCl2. Then, 25 mL of the overnight culture was used to inoculate 250 mL of LB plus ampicillin medium supplemented with 2 mM MnCl2, 0.1 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG), and 3% ethanol. Following incubation overnight at 18 °C, cells were harvested, washed, and resuspended in 10 mL Buffer A [50 mM Tris⋅HCl (pH 8), 0.2 mM EGTA, 150 mM NaCl, 2 mM MnCl2, 10% (vol/vol) glycerol, 0.1% Triton X-100, 2 mM DTT, 2 mM phenylmethylsulfonyl fluoride, and complete protease inhibitor mixture (Roche)]. Cells were disrupted by sonication, and WCEs were clarified by centrifugation. Glutathione–Sepharose 4B beads (Amersham Biosciences) were washed several times with Buffer A and finally suspended in 1 mL Buffer A containing 5% (wt/vol) BSA, incubated with rotation at 4 °C for 1 h, and then washed several times with Buffer A. WCEs were mixed with the treated Glutathione–Sepharose 4B beads and incubated with rotation at 4 °C overnight. The beads were washed three times with Buffer A, and GST-GLC7-FLAG was eluted with Buffer A containing 10 mM glutathione. The phosphatase activity of the purified GST-GLC7-FLAG fusion protein was confirmed using p-nitrophenylphosphate (New England Biolabs) as substrate. Aliquots of the purified GST-GLC7-FLAG were stored at −80 °C. Plasmid pC1722 was introduced into yeast strain H2766, and FLAG-PKR was overexpressed and purified as described previously (64), except that the FLAG-PKR was not eluted from the resin.
In Vitro Phosphatase Assay.
To generate the substrate for the phosphatase assay, purified eIF2 complex was phosphorylated by PKR. FLAG-PKR immobilized onto M2-agarose was washed three times with Kinase Buffer [20 mM Tris⋅HCl (pH 8.0), 50 mM KCl, 25 mM MgCl2, and 1 μM PMSF], and then a 50-µL aliquot of the immobilized FLAG-PKR was mixed with purified eIF2 complex and 300 μL Kinase Buffer. The phosphorylation reaction was started by adding 0.2 mM ATP, and after 30-min incubation at room temperature with rotation, the FLAG-PKR was removed by centrifugation. Next, 50 μL of the phosphorylated eIF2 complex (final concentration = 20 nM) was mixed with purified GST-GLC7-FLAG (75 nM) and 50 μL of 2× Phosphatase Buffer (100 mM Hepes, 200 mM NaCl, 2 mM DTT, 2 mM MnCl2, 0.01% Brij-35). Reactions were incubated at 30 °C and then stopped after various times by addition of 2× SDS Loading Buffer and boiling of the samples. Reaction products were separated by SDS/PAGE, and eIF2α phosphorylation was monitored by immunoblot analysis as described earlier.
Acknowledgments
We thank Kelly Tatchell for providing reagents and advice for purifying GLC7. We also thank Chune Cao, Meghna Thakur, Byung-Sik Shin, and Jason Murray for technical support and A. Hinnebusch and members of the T.E.D. and Hinnebusch laboratories for helpful discussions. This work was supported, in part, by the Intramural Research Program of the US National Institutes of Health, Eunice Kennedy Shriver National Institute of Child Health and Human Development (to T.E.D.)
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Cohen P. The structure and regulation of protein phosphatases. Annu Rev Biochem. 1989;58:453–508. doi: 10.1146/annurev.bi.58.070189.002321. [DOI] [PubMed] [Google Scholar]
- 2.Fernandez A, Brautigan DL, Lamb NJ. Protein phosphatase type 1 in mammalian cell mitosis: Chromosomal localization and involvement in mitotic exit. J Cell Biol. 1992;116(6):1421–1430. doi: 10.1083/jcb.116.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dever TE, et al. Phosphorylation of initiation factor 2 α by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell. 1992;68(3):585–596. doi: 10.1016/0092-8674(92)90193-g. [DOI] [PubMed] [Google Scholar]
- 4.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 5.Rowlands AG, Panniers R, Henshaw EC. The catalytic mechanism of guanine nucleotide exchange factor action and competitive inhibition by phosphorylated eukaryotic initiation factor 2. J Biol Chem. 1988;263(12):5526–5533. [PubMed] [Google Scholar]
- 6.Hinnebusch AG. Mechanism and regulation of initiator methionyl-tRNA binding to ribosomes. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational Control of Gene Expression. Cold Spring Harbor, NY: Cold Spring Harb Lab Press; 2000. pp. 185–243. [Google Scholar]
- 7.Pavitt GD, Ron D. New insights into translational regulation in the endoplasmic reticulum unfolded protein response. Cold Spring Harb Perspect Biol. 2012;4:a012278. doi: 10.1101/cshperspect.a012278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dever TE. Gene-specific regulation by general translation factors. Cell. 2002;108(4):545–556. doi: 10.1016/s0092-8674(02)00642-6. [DOI] [PubMed] [Google Scholar]
- 9.Dever TE, Yang W, Aström S, Byström AS, Hinnebusch AG. Modulation of tRNA(iMet), eIF-2, and eIF-2B expression shows that GCN4 translation is inversely coupled to the level of eIF-2.GTP.Met-tRNA(iMet) ternary complexes. Mol Cell Biol. 1995;15(11):6351–6363. doi: 10.1128/mcb.15.11.6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stark MJ. Yeast protein serine/threonine phosphatases: multiple roles and diverse regulation. Yeast. 1996;12(16):1647–1675. doi: 10.1002/(SICI)1097-0061(199612)12:16%3C1647::AID-YEA71%3E3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 11.Breitkreutz A, et al. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328(5981):1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wek RC, Cannon JF, Dever TE, Hinnebusch AG. Truncated protein phosphatase GLC7 restores translational activation of GCN4 expression in yeast mutants defective for the eIF-2 α kinase GCN2. Mol Cell Biol. 1992;12(12):5700–5710. doi: 10.1128/mcb.12.12.5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng ZH, et al. The yeast GLC7 gene required for glycogen accumulation encodes a type 1 protein phosphatase. J Biol Chem. 1991;266(35):23796–23801. [PubMed] [Google Scholar]
- 14.Ohkura H, Kinoshita N, Miyatani S, Toda T, Yanagida M. The fission yeast dis2+ gene required for chromosome disjoining encodes one of two putative type 1 protein phosphatases. Cell. 1989;57(6):997–1007. doi: 10.1016/0092-8674(89)90338-3. [DOI] [PubMed] [Google Scholar]
- 15.Cohen PT. Protein phosphatase 1—targeted in many directions. J Cell Sci. 2002;115(Pt 2):241–256. doi: 10.1242/jcs.115.2.241. [DOI] [PubMed] [Google Scholar]
- 16.Bollen M, Peti W, Ragusa MJ, Beullens M. The extended PP1 toolkit: Designed to create specificity. Trends Biochem Sci. 2010;35(8):450–458. doi: 10.1016/j.tibs.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tu J, Carlson M. REG1 binds to protein phosphatase type 1 and regulates glucose repression in Saccharomyces cerevisiae. EMBO J. 1995;14(23):5939–5946. doi: 10.1002/j.1460-2075.1995.tb00282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanz P, Alms GR, Haystead TA, Carlson M. Regulatory interactions between the Reg1-Glc7 protein phosphatase and the Snf1 protein kinase. Mol Cell Biol. 2000;20(4):1321–1328. doi: 10.1128/mcb.20.4.1321-1328.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tu J, Carlson M. The GLC7 type 1 protein phosphatase is required for glucose repression in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14(10):6789–6796. doi: 10.1128/mcb.14.10.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ludin K, Jiang R, Carlson M. Glucose-regulated interaction of a regulatory subunit of protein phosphatase 1 with the Snf1 protein kinase in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1998;95(11):6245–6250. doi: 10.1073/pnas.95.11.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tabba S, Mangat S, McCartney R, Schmidt MC. PP1 phosphatase-binding motif in Reg1 protein of Saccharomyces cerevisiae is required for interaction with both the PP1 phosphatase Glc7 and the Snf1 protein kinase. Cell Signal. 2010;22(7):1013–1021. doi: 10.1016/j.cellsig.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.François JM, et al. GAC1 may encode a regulatory subunit for protein phosphatase type 1 in Saccharomyces cerevisiae. EMBO J. 1992;11(1):87–96. doi: 10.1002/j.1460-2075.1992.tb05031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu X, Hart H, Cheng C, Roach PJ, Tatchell K. Characterization of Gac1p, a regulatory subunit of protein phosphatase type I involved in glycogen accumulation in Saccharomyces cerevisiae. Mol Genet Genomics. 2001;265(4):622–635. doi: 10.1007/s004380100455. [DOI] [PubMed] [Google Scholar]
- 24.Wu X, Tatchell K. Mutations in yeast protein phosphatase type 1 that affect targeting subunit binding. Biochemistry. 2001;40(25):7410–7420. doi: 10.1021/bi002796k. [DOI] [PubMed] [Google Scholar]
- 25.Wakula P, Beullens M, Ceulemans H, Stalmans W, Bollen M. Degeneracy and function of the ubiquitous RVXF motif that mediates binding to protein phosphatase-1. J Biol Chem. 2003;278(21):18817–18823. doi: 10.1074/jbc.M300175200. [DOI] [PubMed] [Google Scholar]
- 26.Bollen M. Combinatorial control of protein phosphatase-1. Trends Biochem Sci. 2001;26(7):426–431. doi: 10.1016/s0968-0004(01)01836-9. [DOI] [PubMed] [Google Scholar]
- 27.Hendrickx A, et al. Docking motif-guided mapping of the interactome of protein phosphatase-1. Chem Biol. 2009;16(4):365–371. doi: 10.1016/j.chembiol.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 28.Meiselbach H, Sticht H, Enz R. Structural analysis of the protein phosphatase 1 docking motif: molecular description of binding specificities identifies interacting proteins. Chem Biol. 2006;13(1):49–59. doi: 10.1016/j.chembiol.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 29.Egloff MP, et al. Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J. 1997;16(8):1876–1887. doi: 10.1093/emboj/16.8.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terrak M, Kerff F, Langsetmo K, Tao T, Dominguez R. Structural basis of protein phosphatase 1 regulation. Nature. 2004;429(6993):780–784. doi: 10.1038/nature02582. [DOI] [PubMed] [Google Scholar]
- 31.Hurley TD, et al. Structural basis for regulation of protein phosphatase 1 by inhibitor-2. J Biol Chem. 2007;282(39):28874–28883. doi: 10.1074/jbc.M703472200. [DOI] [PubMed] [Google Scholar]
- 32.Carmody LC, Baucum AJ, 2nd, Bass MA, Colbran RJ. Selective targeting of the γ1 isoform of protein phosphatase 1 to F-actin in intact cells requires multiple domains in spinophilin and neurabin. FASEB J. 2008;22(6):1660–1671. doi: 10.1096/fj.07-092841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ragusa MJ, et al. Spinophilin directs protein phosphatase 1 specificity by blocking substrate binding sites. Nat Struct Mol Biol. 2010;17(4):459–464. doi: 10.1038/nsmb.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ayllón V, Cayla X, García A, Fleischer A, Rebollo A. The anti-apoptotic molecules Bcl-xL and Bcl-w target protein phosphatase 1α to Bad. Eur J Immunol. 2002;32(7):1847–1855. doi: 10.1002/1521-4141(200207)32:7<1847::AID-IMMU1847>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 35.Huang HB, et al. Characterization of the inhibition of protein phosphatase-1 by DARPP-32 and inhibitor-2. J Biol Chem. 1999;274(12):7870–7878. doi: 10.1074/jbc.274.12.7870. [DOI] [PubMed] [Google Scholar]
- 36.Harding HP, et al. Ppp1r15 gene knockout reveals an essential role for translation initiation factor 2 α (eIF2α) dephosphorylation in mammalian development. Proc Natl Acad Sci USA. 2009;106(6):1832–1837. doi: 10.1073/pnas.0809632106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brush MH, Weiser DC, Shenolikar S. Growth arrest and DNA damage-inducible protein GADD34 targets protein phosphatase 1 α to the endoplasmic reticulum and promotes dephosphorylation of the α subunit of eukaryotic translation initiation factor 2. Mol Cell Biol. 2003;23(4):1292–1303. doi: 10.1128/MCB.23.4.1292-1303.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, et al. ICP34.5 protein of herpes simplex virus facilitates the initiation of protein translation by bridging eukaryotic initiation factor 2α (eIF2α) and protein phosphatase 1. J Biol Chem. 2011;286(28):24785–24792. doi: 10.1074/jbc.M111.232439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He B, Gross M, Roizman B. The gamma(1)34.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1alpha to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc Natl Acad Sci USA. 1997;94(3):843–848. doi: 10.1073/pnas.94.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Erickson FL, Harding LD, Dorris DR, Hannig EM. Functional analysis of homologs of translation initiation factor 2γ in yeast. Mol Gen Genet. 1997;253(6):711–719. doi: 10.1007/s004380050375. [DOI] [PubMed] [Google Scholar]
- 41.Wek RC, Jackson BM, Hinnebusch AG. Juxtaposition of domains homologous to protein kinases and histidyl-tRNA synthetases in GCN2 protein suggests a mechanism for coupling GCN4 expression to amino acid availability. Proc Natl Acad Sci USA. 1989;86(12):4579–4583. doi: 10.1073/pnas.86.12.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yatime L, Mechulam Y, Blanquet S, Schmitt E. Structure of an archaeal heterotrimeric initiation factor 2 reveals a nucleotide state between the GTP and the GDP states. Proc Natl Acad Sci USA. 2007;104(47):18445–18450. doi: 10.1073/pnas.0706784104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stolboushkina E, et al. Crystal structure of the intact archaeal translation initiation factor 2 demonstrates very high conformational flexibility in the α- and β-subunits. J Mol Biol. 2008;382(3):680–691. doi: 10.1016/j.jmb.2008.07.039. [DOI] [PubMed] [Google Scholar]
- 44.Ung TL, Cao C, Lu J, Ozato K, Dever TE. Heterologous dimerization domains functionally substitute for the double-stranded RNA binding domains of the kinase PKR. EMBO J. 2001;20(14):3728–3737. doi: 10.1093/emboj/20.14.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jurata LW, Gill GN. Functional analysis of the nuclear LIM domain interactor NLI. Mol Cell Biol. 1997;17(10):5688–5698. doi: 10.1128/mcb.17.10.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Breen JJ, Agulnick AD, Westphal H, Dawid IB. Interactions between LIM domains and the LIM domain-binding protein Ldb1. J Biol Chem. 1998;273(8):4712–4717. doi: 10.1074/jbc.273.8.4712. [DOI] [PubMed] [Google Scholar]
- 47.Wek SA, Zhu S, Wek RC. The histidyl-tRNA synthetase-related sequence in the eIF-2 α protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol Cell Biol. 1995;15(8):4497–4506. doi: 10.1128/mcb.15.8.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang R, Wek SA, Wek RC. Glucose limitation induces GCN4 translation by activation of Gcn2 protein kinase. Mol Cell Biol. 2000;20(8):2706–2717. doi: 10.1128/mcb.20.8.2706-2717.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zaborske JM, et al. Genome-wide analysis of tRNA charging and activation of the eIF2 kinase Gcn2p. J Biol Chem. 2009;284(37):25254–25267. doi: 10.1074/jbc.M109.000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cherkasova VA, Hinnebusch AG. Translational control by TOR and TAP42 through dephosphorylation of eIF2α kinase GCN2. Genes Dev. 2003;17(7):859–872. doi: 10.1101/gad.1069003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alone PV, Dever TE. Direct binding of translation initiation factor eIF2γ-G domain to its GTPase-activating and GDP-GTP exchange factors eIF5 and eIF2B ε. J Biol Chem. 2006;281(18):12636–12644. doi: 10.1074/jbc.M511700200. [DOI] [PubMed] [Google Scholar]
- 52.Ron D, Harding HP. eIF2α phosphorylation in cellular stress responses and disease. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harb Lab Press; 2007. pp. 345–368. [Google Scholar]
- 53.Brush MH, Shenolikar S. Control of cellular GADD34 levels by the 26S proteasome. Mol Cell Biol. 2008;28(23):6989–7000. doi: 10.1128/MCB.00724-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jousse C, et al. Inhibition of a constitutive translation initiation factor 2α phosphatase, CReP, promotes survival of stressed cells. J Cell Biol. 2003;163(4):767–775. doi: 10.1083/jcb.200308075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Novoa I, Zeng H, Harding HP, Ron D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2α. J Cell Biol. 2001;153(5):1011–1022. doi: 10.1083/jcb.153.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Connor JH, Weiser DC, Li S, Hallenbeck JM, Shenolikar S. Growth arrest and DNA damage-inducible protein GADD34 assembles a novel signaling complex containing protein phosphatase 1 and inhibitor 1. Mol Cell Biol. 2001;21(20):6841–6850. doi: 10.1128/MCB.21.20.6841-6850.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alone PV, Cao C, Dever TE. Translation initiation factor 2γ mutant alters start codon selection independent of Met-tRNA binding. Mol Cell Biol. 2008;28(22):6877–6888. doi: 10.1128/MCB.01147-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wek RC, Ramirez M, Jackson BM, Hinnebusch AG. Identification of positive-acting domains in GCN2 protein kinase required for translational activation of GCN4 expression. Mol Cell Biol. 1990;10(6):2820–2831. doi: 10.1128/mcb.10.6.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dey M, et al. Mechanistic link between PKR dimerization, autophosphorylation, and eIF2α substrate recognition. Cell. 2005;122(6):901–913. doi: 10.1016/j.cell.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 60.Mitchell DA, Marshall TK, Deschenes RJ. Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast. 1993;9(7):715–722. doi: 10.1002/yea.320090705. [DOI] [PubMed] [Google Scholar]
- 61.Shin BS, et al. Initiation factor eIF2γ promotes eIF2-GTP-Met-tRNAi(Met) ternary complex binding to the 40S ribosome. Nat Struct Mol Biol. 2011;18(11):1227–1234. doi: 10.1038/nsmb.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cherkasova V, Qiu H, Hinnebusch AG. Snf1 promotes phosphorylation of the alpha subunit of eukaryotic translation initiation factor 2 by activating Gcn2 and inhibiting phosphatases Glc7 and Sit4. Mol Cell Biol. 2010;30(12):2862–2873. doi: 10.1128/MCB.00183-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Romano PR, et al. Autophosphorylation in the activation loop is required for full kinase activity in vivo of human and yeast eukaryotic initiation factor 2α kinases PKR and GCN2. Mol Cell Biol. 1998;18(4):2282–2297. doi: 10.1128/mcb.18.4.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krishnamoorthy T, Pavitt GD, Zhang F, Dever TE, Hinnebusch AG. Tight binding of the phosphorylated α subunit of initiation factor 2 (eIF2α) to the regulatory subunits of guanine nucleotide exchange factor eIF2B is required for inhibition of translation initiation. Mol Cell Biol. 2001;21(15):5018–5030. doi: 10.1128/MCB.21.15.5018-5030.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]