Significance
Membranes of eukaryotic cells retain an essential asymmetric distribution of phospholipids in the two leaflets, which depends on the translocation by P4-ATPases (flippases) of specific phospholipids across the lipid bilayer. Severe diseases result from flippase mutations. Flippases are closely related to ion pumps like Na+,K+-ATPase and Ca2+-ATPase, yet the large size of the lipid substrate compared with inorganic cations raises the fundamental issue of how the flippases accomplish lipid transport. We characterized the mammalian flippase ATP8A2, applying mutagenesis and specific functional assays in combination with computational studies. We identify a unique putative lipid transport pathway through the flippase protein and propose a plausible translocation mechanism, which is both distinct from and related to the well-characterized cation transport mechanisms.
Keywords: P-type ATPase, phosphatidylserine transport, flippase mechanism, flippase structure, CAMRQ syndrome
Abstract
P4-ATPases (flippases) translocate specific phospholipids such as phosphatidylserine from the exoplasmic leaflet of the cell membrane to the cytosolic leaflet, upholding an essential membrane asymmetry. The mechanism of flipping this giant substrate has remained an enigma. We have investigated the importance of amino acid residues in transmembrane segment M4 of mammalian P4-ATPase ATP8A2 by mutagenesis. In the related ion pumps Na+,K+-ATPase and Ca2+-ATPase, M4 moves during the enzyme cycle, carrying along the ion bound to a glutamate. In ATP8A2, the corresponding residue is an isoleucine, which recently was found mutated in patients with cerebellar ataxia, mental retardation, and dysequilibrium syndrome. Our analyses of the lipid substrate concentration dependence of the overall and partial reactions of the enzyme cycle in mutants indicate that, during the transport across the membrane, the phosphatidylserine head group passes near isoleucine-364 (I364) and that I364 is critical to the release of the transported lipid into the cytosolic leaflet. Another M4 residue, N359, is involved in recognition of the lipid substrate on the exoplasmic side. Our functional studies are supported by structural homology modeling and molecular dynamics simulations, suggesting that I364 and adjacent hydrophobic residues function as a hydrophobic gate that separates the entry and exit sites of the lipid and directs sequential formation and annihilation of water-filled cavities, thereby enabling transport of the hydrophilic phospholipid head group in a groove outlined by the transmembrane segments M1, M2, M4, and M6, with the hydrocarbon chains following passively, still in the membrane lipid phase.
Members of the P4 subfamily of P-type ATPases are known as flippases, because they transport (flip) specific phospholipids from the exoplasmic to the cytoplasmic leaflet of the plasma membrane bilayer (1–3). Thus, they establish and maintain a physiologically essential asymmetry between the two leaflets, with phosphatidylserine (PS) and phosphatidylethanolamine (PE) being concentrated in the cytoplasmic leaflet and phosphatidylcholine (PC) and sphingomyelin in the exoplasmic leaflet. There are 14 human P4-ATPases, and mutations in many of these are linked to severe disorders (4–7). On the basis of amino acid sequence alignment, the P4-ATPases are predicted to structurally resemble the classic P-type ATPase cation pumps Na+,K+-ATPase and Ca2+-ATPase, possessing a transmembrane domain with 10 helices (M1–M10) and three cytoplasmic domains, P (phosphorylation), N (nucleotide binding), and A (actuator) (Fig. 1A), known from crystal structures to undergo large movements during the catalytic cycle (8, 9). Recently, we showed that the P4-ATPase ATP8A2, like the cation pumps, forms an aspartyl-phosphorylated intermediate and undergoes a catalytic cycle involving the conformations E1, E1P, E2P, and E2 similar to the Post-Albers scheme originally proposed for the Na+,K+-ATPase (Fig. 1 B and C) (10, 11). We found that the dephosphorylation of E2P is activated by the transported substrates PS and PE, similar to K+ activation of dephosphorylation in Na+,K+-ATPase (Fig. 1C), and that K873, located in transmembrane helix M5 of ATP8A2 at a position equivalent to that of a K+-binding serine in Na+,K+-ATPase, is critical for the sensitivity of the phosphoenzyme to PS and PE (12), which seems to argue for a high degree of mechanistic similarity of the P4-ATPases to Na+,K+-ATPase and Ca2+-ATPase, not only in relation to the catalysis of ATP hydrolysis, but also in the transduction of the liberated energy into substrate movement. The M5 lysine might function directly or indirectly in binding of the phospholipid (12). It is, however, an open question how the flippase is able to move a phospholipid, which is ∼10-fold larger than the ions transported by Na+,K+-ATPase and Ca2+-ATPase and which has to reorient during the translocation. This enigma has been referred to as the “giant substrate problem” (13, 14). On the other hand, it is now clear that no protein other than the P4-ATPase catalytic chain in association with its small CDC50 subunit (β-subunit) is required for the flippase function, because the flipping of specific phospholipids is retained after purification and reconstitution of these proteins in lipid vesicles (2, 3, 12). In Na+,K+-ATPase and Ca2+-ATPase, the central helices, M4, M5, and M6, play major roles in cation binding and occlusion. However, in recent studies using mutagenesis to exchange residues between the two yeast flippases Dnf1 and Drs2 of differing phospholipid head group specificity, side-chains critical to the PS specificity of Drs2 have been pinpointed at two interfacial regions flanking transmembrane helices M1–M3, outside the central core region of the protein, thus leading to the suggestion that the transport pathway for the lipid is unique compared with the canonical pathway used by the P-type cation pumps (13, 15). From these results, it also appears that a tyrosine at the cytoplasmic border of M4 (not present in Drs2 or mammalian PS flippases) participates in selection against PS in Dnf1 (13, 16).
Fig. 1.
Topology and reaction schemes of flippase and Na+,K+-ATPase. (A) Diagram of P4-ATPase topology indicating P-type ATPase domains and residues studied here by mutagenesis, as well as K873 of M5 studied previously (12). (B) Proposed flippase reaction cycle. (C) Reaction cycle of Na+,K+-ATPase [Post-Albers scheme (10, 11)]. E1, E1P, E2P, and E2 are the major enzyme forms, the P indicating phosphorylation.
Transmembrane segment M4 is a key element in the mechanism of the cation-transporting P-type ATPases. In the well-documented catalytic cycles of Na+,K+-ATPase and Ca2+-ATPase, the glutamate of M4 binds Na+/K+ and Ca2+/H+ and is alternately exposed to the two sides of the membrane during the cycle (9, 17, 18). Mutations of nearby located residues in M4 have been shown to block the E1P→E2P or E2P→E2 transition (19–21). Crystal structures of the Ca2+-ATPase indicate that the E1P→E2P conformational change involves a vertical movement of M4, like a pump rod, corresponding to a whole turn of the helix, allowing delivery of Ca2+ bound at the M4 glutamate to the lumen. During the E2→E1 transition of the dephosphoenzyme, M4 moves a similar distance in the opposite direction toward the cytosol (8, 9). In ATP8A2, the residue located at the position equivalent to the ion binding glutamate in M4 of Ca2+-ATPase and Na+,K+-ATPase is an isoleucine, which is highly conserved among P4-ATPases (Fig. S1). Intriguingly, a missense mutation of this isoleucine to methionine was recently identified as the cause of cerebellar ataxia, mental retardation, and dysequilibrium (CAMRQ) syndrome in a Turkish family (22).
Here, we investigated the functional consequences of mutations of isoleucine-364 (I364) and adjacent residues of ATP8A2 (see Fig. 1A for overview). The specific interaction with phospholipids and the individual partial reactions of the enzyme cycle were studied, and we provide evidence that I364 is crucial to the lipid transport, with mutations affecting the affinity for the phospholipid or the dissociation of the translocated lipid toward the cytoplasmic leaflet. In addition, N359 of M4 appears to play an important functional role. Molecular modeling suggests a mechanistic explanation of our observations, showing a remarkable movement of large, water-filled cavities during the transition between the E2P and E2 conformations. Moreover, the water flow appears to be gated by a central hydrophobic cluster composed of the side-chains of I364 and nearby conserved hydrophobic residues of M1, M2, and M6. Among these, I115 of M2 is found to be as mutation sensitive as I364 and N359. These data suggest a unique water-mediated transport pathway for the phospholipid head group along a groove outlined by M1, M2, M4, and M6, which could allow the lipid tail to follow passively, still in the membrane lipid phase, jutting out between M2 and M6. In such a mechanism, movement of M4 and in particular I364 would be as crucial to energy transduction and movement of the transported substrate as in the cation-translocating P-type ATPases.
Results
Lipid Flipping and ATPase Activity of I364 Mutants.
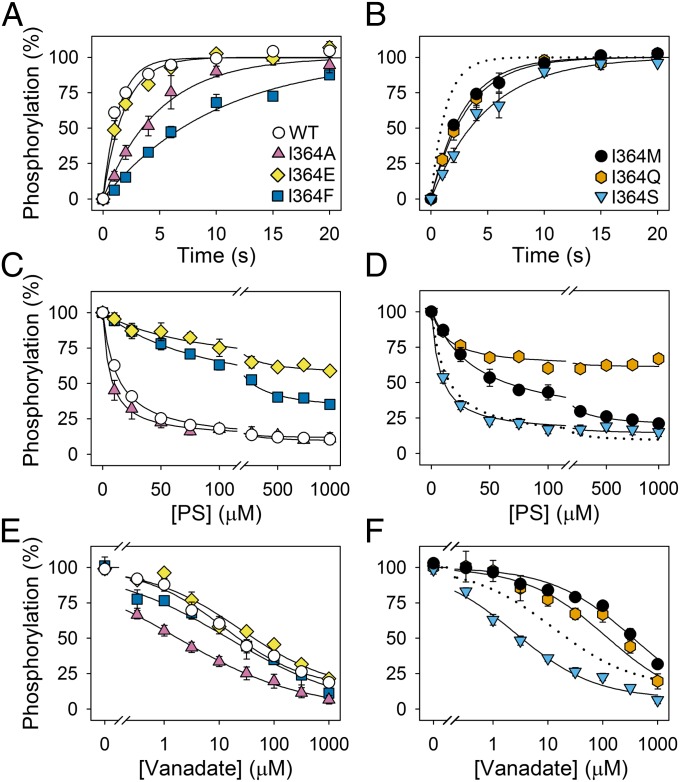
We analyzed a series of bovine ATP8A2 mutants, in which I364 was replaced by alanine, glutamate (present in Na+,K+-ATPase and Ca2+-ATPase), phenylalanine, methionine (CAMRQ syndrome mutant), glutamine, or serine. The mutants were well expressed in HEK293T cells (Table 1) together with CDC50A, and this protein complex was purified and reconstituted with either PC or a mixture of PC with fluorescent (7-nitro-2–1,3-benzoxadiazol-4-yl)amino-labeled PS probe (NBD-PS) to determine the lipid flipping activity as previously described (12, 23). As seen in Fig. 2A, only mutants I364A and I364S displayed significant NBD-PS flipping activity (80% and 30% of WT ATP8A2, respectively; these numbers are corrected for variation in expression level to reflect specific activity).
Table 1.
Analysis of expression and ATPase activity
| Mutant | Expression, % of WT | ATPase activity stimulated by PS | ATPase activity stimulated by PE | ||||||
| Vmax with PS*, μmol/min/mg | K0.5 for PS†, μM | Inhibition at high PS concentration‡ | n | Vmax with PE*, μmol/min/mg | K0.5 for PE†, μM | Inhibition at high PE concentration‡ | n | ||
| WT | 100 | 160 ± 6 (100%) | 38 ± 1 | − | 1,185 | 67 ± 3 (100%) | 371 ± 13 | − | 369 |
| F354A | 44 ± 4 | 150 ± 7 (94%) | 42 ± 3 (1.1×↑) | − | 60 | ND | ND | ND | ND |
| Y358A | 12 ± 3 | 78 ± 4 (49%) | 36 ± 3 (1.1×↓) | − | 60 | ND | ND | ND | ND |
| N359A | 124 ± 25 | 17 ± 6 (11%) | 220 ± 22 (5.8×↑) | − | 96 | 6 ± 1 (9%) | 2215 ± 1317 (6.0×↑) | − | 35 |
| N360A | 23 ± 2 | 75 ± 11 (47%) | 23 ± 2 (1.7×↓) | (+) | 80 | ND | ND | ND | ND |
| L361A | 30 ± 8 | 53 ± 8 (33%) | 58 ± 5 (1.5×↑) | − | 70 | ND | ND | ND | ND |
| I362A | 32 ± 8 | 64 ± 7 (40%) | 24 ± 2 (1.6×↓) | (+) | 97 | 32 ± 1 (48%) | 149 ± 8 (2.5×↓) | + | 30 |
| P363A | –§ | ND | ND | ND | ND | ND | ND | ND | ND |
| I364A | 99 ± 15 | 95 ± 26 (59%) | 16 ± 3 (2.4×↓) | + | 60 | 11 ± 2 (16%) | 131 ± 10 (2.8×↓) | + | 60 |
| I364E | 58 ± 6 | 6 ± 1 (4%) | 111 ± 12 (2.9×↑) | − | 80 | 3 ± 1 (4%) | 292 ± 89 (1.3×↓) | − | 79 |
| I364F | 104 ± 9 | 9 ± 2 (6%) | 178 ± 26 (4.7×↑) | − | 57 | 3 ± 1 (4%) | 1120 ± 457 (3.0×↑) | − | 59 |
| I364M | 118 ± 3 | 18 ± 10 (11%) | 148 ± 15 (3.9×↑) | − | 50 | 2 ± 1 (3%) | 369 ± 80 (1.0×) | − | 56 |
| I364Q | 98 ± 19 | 6 ± 2 (4%) | 19 ± 3 (2.0×↓) | + | 74 | 3 ± 0.4 (4%) | 288 ± 45 (1.3×↓) | (+) | 59 |
| I364S | 97 ± 14 | 68 ± 1 (43%) | 16 ± 1 (2.4×↓) | + | 79 | 15 ± 4 (22%) | 105 ± 11 (3.5×↓) | + | 60 |
| S365A | 48 ± 9 | 118 ± 12 (74%) | 104 ± 9 (2.7×↑) | − | 77 | 10 ± 1 (15%) | 862 ± 330 (2.3×↑) | − | 59 |
| L367A | 69 ± 8 | 54 ± 11 (34%) | 68 ± 3 (1.8×↑) | − | 118 | 20 ± 7 (30%) | 379 ± 33 (1.0×) | − | 70 |
| T369A | 26 ± 9 | 55 ± 7 (34%) | 20 ± 2 (1.9×↓) | (+) | 119 | ND | ND | ND | ND |
| E371Q | 14 ± 5 | 57 ± 18 (36%) | 49 ± 3 (1.3×↑) | − | 90 | ND | ND | ND | ND |
| K374A | 5 ± 1 | 68 ± 10 (43%) | 49 ± 4 (1.3×↑) | − | 50 | ND | ND | ND | ND |
| F88A | 38 ± 29 | 80 ± 9 (50%) | 27 ± 1 (1.4×↓) | (+) | 70 | 14 ± 3 (21%) | 250 ± 17 (1.5×↓) | (+) | 50 |
| L112A | 26 ± 19 | 199 ± 8 (124%) | 45 ± 1 (1.2×↑) | − | 29 | 75 ± 7 (112%) | 394 ± 42 (1.1×↑) | − | 30 |
| I115A | 147 ± 15 | 14 ± 5 (9%) | 73 ± 4 (1.9×↑) | + | 39 | 1 ± 0.4 (1%) | 376 ± 61 (1.0×) | (+) | 56 |
| V906A | 85 ± 8 | 40 ± 4 (25%) | 20 ± 1 (1.9×↓) | (+) | 79 | 18 ± 4 (27%) | 103 ± 14 (3.6×↓) | + | 40 |
SEs and number of data points n, on which the analysis is based, are indicated. ND, not determined.
Numbers in parentheses denote mutant Vmax values as percent of the WT value.
K0.5 values (concentration at half Vmax) for activation of ATP hydrolysis were determined from the rising phase of the lipid concentration dependence (Figs. 2, 4, and 8). Fold change relative to the average of the WT is shown in parentheses.
+ and − indicate that an inhibitory effect is present or absent, respectively, parentheses indicate that only a slight effect is observed.
Expression below detectable level.
Fig. 2.
Flippase activity and ATPase activity of WT ATP8A2 and I364 mutants. (A) Transport of NBD-labeled PS (NBD-PS) was measured by reducing the fluorophore in the outer leaflet of the reconstituted vesicles with dithionite following incubation with ATP. The ordinate indicates the percentage of NBD-PS that was transported during the 2.5-min incubation with ATP, normalized to a flippase protein level equal to the average of the WT levels. (B–F) ATPase activity in the presence of PS, PE, and PC, and apparent affinity for PS or PE of WT and mutants, measured in the presence of PC and CHAPS and various concentrations of PS or PE. Values for the maximal activities per mg ATP8A2 protein, Vmax (μmol ATP/min/mg), obtained for each lipid are shown in B. In C–F, activities at the various lipid concentrations indicated on the abscissa are shown relative to Vmax. Error bars show SEs when larger than symbols. Dotted lines reproduce the data for WT. Refer to Table 1 for statistical analysis. Symbols for mutants are the same in E and F as in C and D.
The ATP hydrolysis activity was examined following solubilization of the PC-reconstituted vesicles with CHAPS detergent, allowing addition of various concentrations of PS or PE to activate the enzyme. Fig. 2B depicts the specific ATPase activity (ATP hydrolysis rate per mg ATP8A2 protein) determined in the presence of PC alone or with the PS or PE concentration giving maximum activity (Vmax). With PC alone, the ATPase activity of both WT and mutants was low, and a considerable stimulation of the activity was obtained by addition of PS or PE. Hence, PS stimulated the ATPase activity of the WT ATP8A2 ∼20-fold, consistent with previous findings (12, 23). For I364A and I364S, PS stimulated the ATPase activity to maximum levels of 59% and 43% of the WT level, respectively. The PE-stimulated maximum activity was only 16% and 22%, respectively, relative to that of the WT. For the other I364 mutants, both PS- and PE-stimulated activity was very low. Nevertheless, because the enzyme was highly purified with no background activity from other ATPases present, it was feasible to obtain accurate and reliable measurements of the activation profiles for increasing PS or PE concentration for all of the mutants, as shown on a relative scale in Fig. 2 C–F. Mutants I364E, I364F, and I364M showed standard monophasic PS activation profiles, the apparent affinities being three- to fivefold reduced (K0.5 values three- to fivefold increased), relative to that of the WT (Table 1). By contrast, a reduced apparent affinity for PE was found only for I364F (K0.5 threefold increased relative to that of the WT), whereas I364E and I364M displayed WT-like apparent affinities for PE. Mutants I364A, I364S, and to a lesser extent, I364Q, displayed a complex substrate dependence not previously described for a flippase, in which the usual activation phase was followed by an inhibition phase at high PS concentration (Fig. 2 C and D). Furthermore, for I364A, I364S, and I364Q, the activation occurred at lower PS concentration than in the WT, the K0.5 for PS activation being approximately twofold reduced, corresponding to an increased apparent affinity for PS (Table 1). For these three mutants, the same pattern of biphasic substrate concentration dependence was seen for PE as for PS, and for I364A and I364S there was also a significant increase of the apparent affinity for PE, relative to that of the WT (Fig. 2 E and F and Table 1).
Partial Reactions of I364 Mutants.
The utilization of ATP leads to formation of the E1P phosphoenzyme intermediate, which subsequently is converted to the PS- and PE-reactive E2P species (Fig. 1B) (12). The time dependence of phosphorylation from [γ-32P]ATP was studied for WT ATP8A2 and I364 mutants, reconstituted in pure PC vesicles to allow phosphoenzyme to accumulate. Several I364 mutants displayed reduced phosphorylation rates relative to the WT (Fig. 3 A and B and Table 2). An approximate fourfold reduction was seen for I364A and I364S, and I364F was even slower. The phosphorylation rates of I364M and I364Q were twofold reduced, whereas I364E was rather close to WT. Because we speculated that the phosphorylation rate of I364E might be dependent on protonation of the glutamate, we investigated the effect of increasing the pH from 7.5 to 9.0. However, as previously reported for WT ATP8A2 (12), the phosphorylation rate of I364E was not dependent on the pH in this pH range (Fig. S2).
Fig. 3.
Phosphorylation, dephosphorylation, and vanadate sensitivity of WT and I364 mutants. (A and B) Enzyme reconstituted in PC was phosphorylated at 0 °C with [γ-32P]ATP in standard phosphorylation medium (SPM) without CHAPS, quenched after the indicated time intervals, and the phosphoenzyme was quantified by phosphoimaging following acidic SDS/PAGE. The 100% value corresponds to maximum phosphorylation, defined by the exponential curve fitting. (C and D) Following phosphorylation of PC-reconstituted enzyme at 0 °C with [γ-32P]ATP in SPM containing CHAPS for 30 s, PS mixed with PC dissolved in CHAPS was added to provide the indicated PS concentrations, and dephosphorylation was terminated 5 s later. The 100% value represents the phosphorylation level 5 s after addition of PC dissolved in CHAPS without PS. No dephosphorylation was observed in the latter condition (12). (E and F) Enzyme reconstituted in PC was incubated at 25 °C with the indicated concentrations of orthovanadate in SPM containing CHAPS for 30 min and subsequently phosphorylated with [γ-32P]ATP at 0 °C. The phosphorylation obtained following incubation in the absence of vanadate was taken as 100%. In C–F, the symbols for mutants are the same as in A and B. Error bars show SEs. Dotted lines reproduce the data for WT. Refer to Table 2 for statistical analysis.
Table 2.
Analysis of phosphorylation and dephosphorylation
| Mutant | Phosphorylation rate* | Activation of dephosphorylation by PS† | Inhibition of phosphorylation by vanadate‡ | |||
| k, s−1 | n | K0.5, μM | n | K0.5, μM | n | |
| WT | 0.72 ± 0.04 | 230 | 13 ± 1 | 387 | 13 ± 3 | 70 |
| N359A | 1.88 ± 0.26 (2.6×↑) | 27 | 78 ± 23 (6.0×↑) | 47 | 103 ± 58 (7.9×↑) | 29 |
| N360A | 0.65 ± 0.07 (1.1×↓) | 19 | 12 ± 2 (1.1×↓) | 20 | ND | ND |
| I362A | ND | ND | 8.4 ± 1.1 (1.5×↓) | 20 | ND | ND |
| I364A | 0.20 ± 0.02 (3.6×↓) | 18 | 6.7 ± 1.2 (1.9×↓) | 38 | 1.8 ± 0.5 (7.2×↓) | 36 |
| I364E | 0.53 ± 0.03 (1.4×↓) | 30 | 90 ± 30 (6.9×↑) | 30 | 24 ± 9 (1.8×↑) | 29 |
| I364F | 0.10 ± 0.01 (7.2×↓) | 20 | 103 ± 18 (7.9×↑) | 40 | 15 ± 13 (1.2×↑) | 39 |
| I364M | 0.34 ± 0.01 (2.1×↓) | 20 | 39 ± 6 (3.0×↑) | 30 | 358 ± 305 (27.5×↑) | 27 |
| I364Q | 0.31 ± 0.02 (2.3×↓) | 30 | ND | 37 | 117 ± 101 (9.0×↑) | 37 |
| I364S | 0.20 ± 0.02 (3.6×↓) | 36 | 7.6 ± 1.1 (1.7×↓) | 58 | 2.5 ± 0.4 (5.2×↓) | 34 |
| L367A | ND | ND | 20 ± 1 (1.5×↑) | 29 | ND | ND |
| L112A | 0.54 ± 0.04 (1.3×↓) | 19 | ND | ND | ND | ND |
| I115A | 0.21 ± 0.01 (3.4×↓) | 20 | ND | ND | ND | ND |
SEs and number of data points n, on which the analysis is based, are indicated. ND, not determined.
Rate constants k were obtained by fitting a monoexponential rise to max function to the data (Figs. 3 and 8).
K0.5 values were determined by fitting a single site hyperbolic function plus a constant to the data (Figs. 3, 4, and 8).
K0.5 values were obtained by fitting a Hill equation for an inhibitory ligand EP = EPmax × (1 − [vanadate]h/(K0.5h + [vanadate]h) to the data (Fig. 3).
Phosphoenzyme decay of enzyme phosphorylated from [γ-32P]ATP in the presence of PC and CHAPS detergent for 30 s was analyzed following addition of PS to activate the dephosphorylation. In this assay, the presence of detergent was needed to allow the added PS to access the protein (12). Fig. 3 C and D shows the phosphoenzyme fraction remaining after 5-s incubation with PS at various concentrations. Fitting of a hyperbolic function to the PS dependence allowed estimation of an apparent affinity for PS (12). I364A and I364S displayed an approximate twofold enhancement of the apparent affinity for PS (reduced K0.5 value) relative to WT ATP8A2, in accordance with the data for PS activation of ATPase activity described above. In further agreement with the ATPase activity results, I364E, I364F, and I364M showed substantial reductions of the apparent affinity for PS, relative to that of the WT, the K0.5 for PS activation being increased three- to eightfold (Table 2). For I364E, I364F, and I364Q, the plateau level reached at high PS concentration was considerably higher than for WT ATP8A2, indicating that the maximum PS-stimulated dephosphorylation rate was reduced. For I364Q, the plateau level was as high as 64% (compare with 9% for the WT), precluding an accurate estimation of the apparent affinity for PS.
To further address the influence of the I364 mutations on the partial reaction steps, we determined the sensitivity to inhibition by vanadate (Fig. 3 E and F). Vanadate inhibits P-type ATPases by reacting exclusively with the E2 form and is believed to mimic the penta-coordinated transition state of the phosphoryl group during E2P dephosphorylation (24). A change in apparent vanadate affinity occurs if the E1-E2 conformational equilibrium is displaced, less E2 resulting in lower apparent affinity and vice versa, or if the intrinsic affinity of E2 is altered, which due to the transition state-like nature of vanadate might be related to a change in E2P dephosphorylation rate (a slowing of dephosphorylation corresponds to lower affinity for the phosphoryl transition state). WT and I364 mutants were equilibrated with various concentrations of vanadate before addition of [γ-32P]ATP to determine the fraction of the enzyme that had not bound vanadate by measuring the phosphorylation, vanadate binding and phosphorylation being mutually exclusive. I364A and I364S showed significantly increased apparent vanadate affinity relative to WT. I364M and I364Q displayed markedly reduced vanadate affinity, whereas I364F displayed WT-like vanadate affinity and I364E showed only a slight reduction (Fig. 3 E and F and Table 2).
Among Several Other M4 Mutants, N359A Displayed the Most Dramatically Reduced ATPase Activity and Apparent Substrate Affinity.
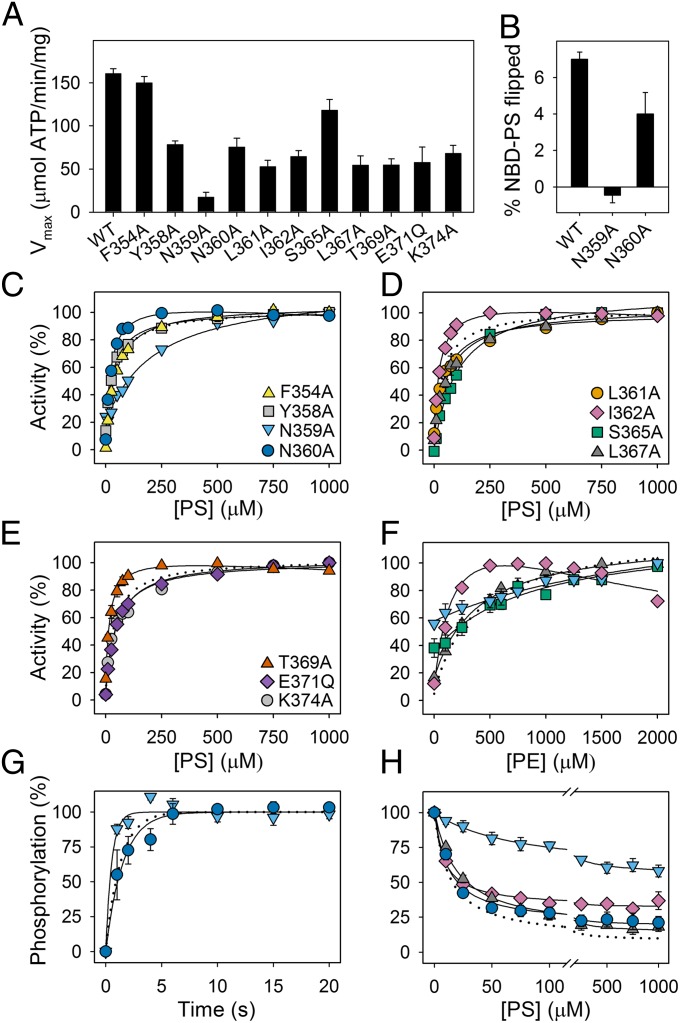
To identify other M4 residues of functional importance, alanine scanning mutagenesis was performed for F354, Y358, N359, N360, L361, I362, P363, S365, L367, T369, and K374, whereas E371 was replaced by glutamine, removing only the negative charge (for location of mutated residues, see Fig. 1A and Fig. S1). These mutants could be expressed at levels sufficient for functional characterization, except for P363A (Table 1). This proline is conserved in all P-type ATPases and is considered crucial to local unwinding of the M4 helix (25). Although the Vmax of most of these mutants was reduced relative to that of the WT, only N359A displayed a dramatic reduction of Vmax (to 11% that of the WT for PS and 9% for PE; Fig. 4A and Table 1). N359A furthermore displayed an approximate sixfold reduction of the apparent affinity for PS and PE. Conversely, the adjacent mutation N360A, and also I362A and T369A, led to an increase of the apparent affinity for PS. It should also be noted that N360A, I362A, and T369A all were inhibited slightly at high PS concentration (Fig. 4). For I362A, a strong inhibition was seen at high PE concentration (Fig. 4F). Mutants F354A, Y358A, L361A, E371Q, and K374A exhibited WT-like apparent affinities for PS, whereas S365A and L367A displayed significant reductions. S365A displayed a reduced apparent affinity for PE as well, whereas the apparent affinity of L367A for PE was WT-like. L367 is located at the position equivalent to the tyrosine that participates in selection against PS in the yeast flippase Dnf1 (13, 16). In line with the strongly reduced ATPase activity of N359A, the PS flipping by this mutant was undetectably low, whereas the PS flipping rate of N360A was only approximately twofold reduced, relative to that of the WT (Fig. 4B). In further analyses, the phosphorylation rate was found approximately threefold enhanced for N359A and was WT-like for N360A (Fig. 4G and Table 2). For N359A, dephosphorylation studies showed a high plateau, indicating that the maximum dephosphorylation rate corresponding to saturating PS concentration was markedly reduced and a sixfold reduction of the apparent affinity for PS activation of dephosphorylation (Fig. 4H), explaining the low Vmax and reduced apparent PS affinity in the ATPase activity analysis. Furthermore, we found the apparent affinity for vanadate highly reduced in N359A relative to that of the WT (Table 2). For I362A and L367A, the apparent PS affinity for activation of dephosphorylation was 1.5-fold increased and reduced, respectively (Fig. 4H), again in good agreement with the results of the analysis of the overall ATPase reaction.
Fig. 4.
Functional analysis of other M4 mutants. Maximal ATPase activities in the presence of PC and CHAPS and the optimal concentration of PS (A), flippase activity (B), PS and PE concentration dependences of the ATPase activity (C–F), phosphorylation (G), and dephosphorylation (H) of WT and mutant enzymes, determined as described for Figs. 2 and 3. Error bars show SEs when larger than symbols. Dotted lines reproduce the data for WT. Refer to Tables 1 and 2 for statistical analysis. Symbols for mutants are the same in F–H as in C–E.
Homology Models of ATP8A2 Display a Groove Outlined by M1, M2, M4, and M6.
To examine the structural basis for the functional importance of I364, in the absence of a crystal structure of a P4-ATPase, we built two homology-based models of ATP8A2. These models were based on extracts of a precise multiple sequence alignment (Fig. S3) and high-resolution templates, the crystal structures of sarco(endo)plasmic reticulum Ca2+-ATPase (SERCA) isoform 1a in the E2P [Protein Data Bank (PDB) ID 3B9B] and E2 (PDB ID 3AR4) states. The models presented the distinct cytoplasmic domains P, N, and A characteristic of P-type ATPases and 10 transmembrane helices as present in Ca2+-ATPase, Na+,K+-ATPase, and H+-ATPase (Fig. S4). Both models were refined at 27 °C using molecular dynamics (MD). The membrane domains were inserted into a lipid bilayer system consisting of 90% PC and 10% PS. Unrestrained MD refinement relaxed both the E2P and E2 models to a root mean square deviation of 3.6 ± 0.1 Å from the starting structures. Strikingly, for both models, this conformational change mainly originated from the opening of a large groove bordered by the M1, M2, M4, and M6 transmembrane segments, although all helices were still stably associated (Fig. 5). Hence, this region, deviating from the ion transporting P-type ATPases with known structures, might be directly involved in lipid transport by the flippases. Intriguingly, the cytoplasmic end of this groove coincides with a bound PE molecule present in several SERCA E2 crystal structures (Fig. S4D).
Fig. 5.
Refined model membrane domains. Refined models of E2P and E2 states are shown in cartoon with helices M1–M2 in purple, M3–M4 in green, M5–M6 in ochre, and M7–M10 as silver molecular isosurface representation. Partial A- and P-domains are shown in yellow and blue, respectively. Unrefined models are shown as transparent gray cartoons. The models were aligned using M7–M10 helix backbones as reference. Black lines denote membrane borders, defined as average phospholipid head group-phosphate positions.
Plausible Water-Filled Phospholipid Translocation Pathway.
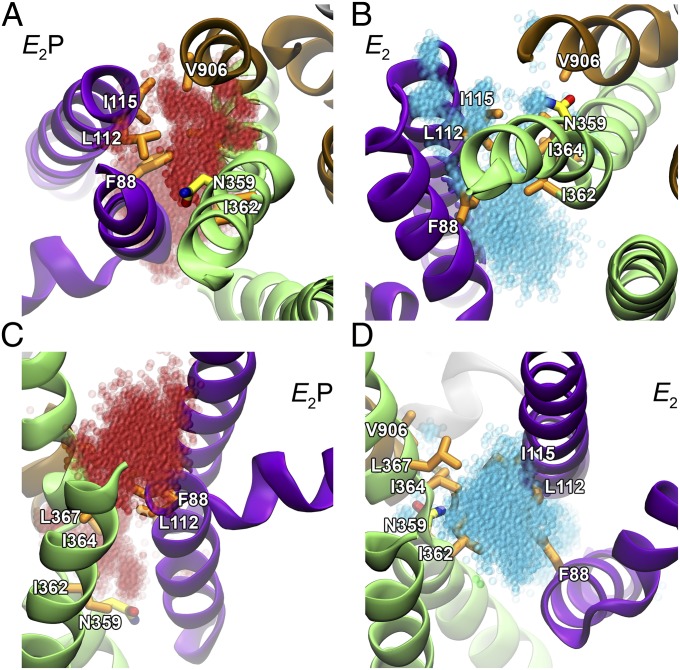
Attention has recently been drawn to the potential of water-filled cavities in proteins as transport pathways for lipid head groups in phospholipid transporting membrane proteins (26). Remarkably, for both of the E2P and E2 models, substantial amounts of water entered the groove between M1, M2, M4, and M6 during the MD refinement, manifesting as two distinct water-filled pockets placed in the exoplasmic and cytoplasmic ends of the groove (Figs. 6 and 7 and Fig. S5). By quantifying water permeation through the groove along the z axis (perpendicularly to the membrane surface) using kernel density estimation, it became evident that the exoplasmic water cluster was buried deeper toward the center of the protein in the E2P model than in the E2 model, whereas the opposite was true for the cytoplasmic water cluster (Fig. 6C). Notably, N359 was fully hydrated by the exoplasmic water pocket of the E2P model, but placed in between the water pockets and poorly hydrated in the E2 model. Separating the two water pockets, a central cluster of hydrophobic residues was observed, including the CAMRQ-associated I364 residue as well as I362 and L367 of M4, also reported above to be critical for the functional characteristics related to interaction of the transported lipid with the protein. In addition, the hydrophobic cluster separating the two water pockets encompassed F88 of M1, L112 and I115 of M2, and V906 of M6 (Figs. 6 and 7). With the exception of L367, the hydrophobicity of these residues is highly conserved among flippases (Table S1). Among these residues, I115 is the one with the shortest distance to I364 (4.60 ± 0.05 Å) in the E2 model.
Fig. 6.
Distribution of water in groove between M1, M2, M4, and M6 in equilibrated models. (A and B) Proteins are visualized as in Fig. 5 with selected hydrophobic side-chains shown in licorice representation with carbons in orange for hydrophobic residues and in yellow for N359, nitrogen in blue, and oxygen in red. Water molecules present throughout a 20-ns MD simulation are red transparent spheres in the E2P model (A) and cyan in the E2 model (B). The two models were aligned with the M7–M10 helix backbones as reference. (C) Kernel density function of water along the z axis. The same position and scale is used across all three panels.
Fig. 7.
Water in groove between M1, M2, M4, and M6 at membrane interfaces of equilibrated models. Models and water are displayed as in Fig. 6. (A and B) View from exoplasmic side, showing an opening to water cavities with N359 at the entrance in E2P but not in E2. (C and D) View from cytoplasmic side, showing a large opening to the water cavity in E2, but not in E2P. In all cases, water clusters are gated by hydrophobic residues.
Hydrophobic Residues Adjacent to I364 Display Mutational Sensitivity Similar to I364.
The modeling results encouraged us to explore the possible mechanistic role of this hydrophobic cluster in additional mutagenesis studies. The residues F88, L112, I115, and V906 were each replaced by alanine. These mutants could be expressed at levels sufficient for functional characterization, with I115A at a higher level than the WT (Table 1), and ATPase activity analyses were performed (Fig. 8 and Table 1). I115A, like some of the I364 mutants, exhibited distinct biphasic PS concentration dependence with decreasing activity at high PS concentration. Furthermore, the Vmax of I115A was less than 10% that of the WT, for PS as well as PE, and the apparent affinity of I115A for PS was approximately twofold reduced, whereas the apparent affinity for PE was WT-like, similar to the observation with I364M. The phosphorylation rate of I115A was also reduced to an extent similar to the most affected I364 mutants (Fig. 8D and Table 2). Hence, the characteristics of I115A bear a clear resemblance to the I364 mutants. Mutation L112A did not appreciably affect Vmax, the apparent affinities for the substrates, or the phosphorylation rate. F88A showed a slight tendency for inhibition at high substrate concentration, and V906A showed pronounced inhibition at high PE concentration with only a slight inhibition at high PS concentration, as also reported above for I362A. Both F88A and V906A showed enhanced substrate affinities like I364A and I364S, and a considerable reduction of Vmax.
Fig. 8.
Functional analyses of other hydrophobic gate mutants. Dependence of ATPase activity on PS (A) and PE (B) concentrations, maximum activity (C), and phosphorylation rate (D), determined as described for Figs. 2 and 3. Error bars show SEs when larger than symbols. Dotted lines reproduce the data for WT. Refer to Tables 1 and 2 for statistical analysis. Symbols for mutants are the same in B and D as in A.
Discussion
ATP8A2 is mainly expressed in the central nervous system, including the cerebrum, cerebellum, spinal cord, and retina, as well as in testis. It is speculated that PS flippase activity is critical for developmental processes of the central nervous system (7). Mutation of ATP8A2 was recently identified as the cause of the CAMRQ syndrome in a Turkish family (22), who are the first reported patients with a disease-causing point mutation in ATP8A2. Previously, a patient with severe mental retardation and major hypotonia had been reported to carry a de novo balanced translocation of chromosomes 10 and 13 disrupting the coding sequence of the ATP8A2 gene (27). Moreover, wabbler-lethal mice with inactivating deletion or insertion mutations of ATP8A2 have been found to develop severe neurological abnormalities such as ataxia and body tremors, due to distal axonal degeneration in spinal neurons (28).
The point mutation found to cause CAMRQ syndrome is a methionine substitution of the isoleucine present in transmembrane segment M4 at the position corresponding to the glutamate central to binding and translocation of the ions in Na+,K+-ATPase and Ca2+-ATPase. At the same position, most heavy-metal transporting P-type ATPases possess either a cysteine or a histidine, also believed to be involved in binding of the metal ion during translocation (29, 30), whereas plasma membrane H+-ATPases of yeast and plants like the flippases possess a hydrophobic residue at this position (Fig. S1). The present results clearly demonstrate that the methionine substitution is compatible with a WT-like expression level of the protein in HEK293T cells but inhibits both lipid flipping and the associated ATPase activity (Fig. 2 A and B). The I364M mutant was found capable of undergoing phosphorylation from ATP, albeit at a reduced rate relative to WT. The dephosphorylation of I364M was significantly less sensitive to PS compared with the WT, the apparent affinity for PS being threefold reduced in the I364M mutant, thus indicating that PS binding to E2P is disturbed by the mutation. Correspondingly, the PS activation of the overall ATPase reaction occurred with an approximate fourfold reduced apparent affinity for PS. Furthermore, the apparent affinity for vanadate inhibition was greatly reduced in I364M, thus attesting to the E2 state being modified and destabilized.
The examined I364 mutants fall into two distinct categories: one group, consisting of I364M, I364E, I364F, and I364Q, that showed little or no flippase and ATPase activity, and the other group, consisting of I364A and I364S, that showed a considerable level of lipid flipping and ATPase activity. I364E, I364F, and I364Q all displayed a marked reduction of the maximum dephosphorylation rate corresponding to saturating PS concentration, as seen from the plateau levels reached after 5-s dephosphorylation. Altered properties of the E2P state with bound PS and/or a reduced rate of the E1P to E2P transition may be responsible for this. Generally, the mutations studied here were found to similarly influence the PS activation of ATP hydrolysis and dephosphorylation, with a good correlation between the changes in K0.5 values observed in the two assays, thus suggesting that rate limitation of the overall reaction under the prevailing conditions is imposed by the PS-activated dephosphorylation of E2P (Fig. 1B). The change of apparent lipid affinity therefore seems to reflect a change to the interaction of the E2P state with the lipid rather than a kinetic change to a partial reaction step remote from the step where the lipid binds. I364F and I364E, like I364M, showed reduced apparent affinity for PS in activation of both dephosphorylation and ATPase activity. By contrast, mutants I364A and I364S with the smallest substituents displayed a normal maximum dephosphorylation rate corresponding to saturating PS concentration and enhanced apparent affinity for PS and PE, indicating that the size of the side-chain is important. Only the I364 mutant with the largest substituent of all, I364F, showed reduced apparent affinity for both PS and PE (Table 1), which may be related to the smaller size of the head group of PE relative to PS, thus generally causing less steric problems with PE than with PS. The I364Q mutant has the enhanced apparent affinity for PS and PE in common with I364A and I364S, which may be due to the smaller size of the glutamine side-chain relative to isoleucine (31). Polarity and charge also contribute to the disruptive effects of the larger substituents. Hence, the methionine side-chain has about the same van der Waals volume as that of isoleucine, but is considerably more polar. The negative charge of the glutamate seems important, because I364E showed a significant reduction of the apparent affinity for PS, whereas the isosteric replacement with the uncharged, but polar and oxygen-containing glutamine did not reduce the apparent PS affinity. Hence, the negatively charged head group of PS might be repelled by the glutamate. The apparent affinity of I364E for PE was WT-like, which may be related to the fact that the phospholipid head group of PE is neutral. Taken together, these data suggest that during the transport process, the head group of the lipid passes near I364, the PS head group perhaps closer than that of PE.
Remarkably, the highest concentrations of PS and PE inhibited the ATPase activity of mutants I364A, I364S, and I364Q, the most pronounced effect being seen for I364A and I364S. This complex substrate dependence, characterized by activation at low substrate concentration and inhibition at high substrate concentration, is reminiscent of the K+ dependence of the Na+,K+-ATPase, where K+ activates the enzyme by binding at the externally facing sites of E2P and inhibits at higher concentration by binding at the internally facing sites of E1 in competition with Na+, driving the enzyme into the K+-occluded E2 state (10, 32, 33). On stabilizing the E2 state by mutation, the K+ inhibition of the Na+,K+-ATPase activity becomes very pronounced, occurring at rather low K+ concentration (34). In direct analogy, the inhibition of the ATP8A2 mutants at high PS or PE concentration could arise from stabilization of the E2 state with bound lipid substrate (compare the reaction schemes for flippase and Na+,K+-ATPase; Fig. 1 B and C). Such stabilization would also explain the higher apparent PS and PE affinities of I364A, I364S, and I364Q in activation of the E2P to E2 dephosphorylation reaction. Because the dissociation of the transported lipid depends on the transformation of E2 into E1, a stabilization of E2 would slow the release of the lipid into the cytoplasmic leaflet, thus inhibiting ATP hydrolysis as seen for I364A, I364S, and I364Q. In accordance with this interpretation of the complex substrate dependence even the WT becomes slightly inhibited at very high PS concentrations (Fig. S6). Furthermore, we did not observe inhibition at high PS concentrations for the I364 mutants I364M, I364E, and I364F with significantly reduced apparent affinity for PS activation of dephosphorylation, which is in line with the proposed destabilization of lipid-protein interaction in these mutants. Our interpretation is furthermore supported by the vanadate inhibition results, showing a higher apparent affinity for vanadate in I364A and I364S relative to the WT and other I364 mutants (Fig. 3 E and F). Because the E2 form is the vanadate-reactive species, accumulation of E2 gives rise to an increased apparent affinity for vanadate. For I364Q, it should, however, also be taken into consideration that the transition state of the E2P dephosphorylation reaction may be destabilized, as indicated by the strongly reduced maximum rate of dephosphorylation. As a phosphoryl transition state analog, vanadate is therefore expected to bind more weakly to I364Q, which is in accordance with the actual finding (Fig. 3F).
Most of the I364 mutants furthermore displayed a reduced rate of phosphorylation from ATP, relative to WT. For I364A and I364S, it is tempting to relate the significant reduction of the phosphorylation rate to the stabilization of E2, displacing the E2-E1 equilibrium away from the phosphorylatable E1 form. For the other I364 mutants, the vanadate data do not provide similar evidence of a stabilization of E2, because the apparent vanadate affinity was reduced or WT-like. As mentioned above in relation to I364Q, a simultaneous destabilization of the transition state of E2P dephosphorylation that reduces the intrinsic vanadate affinity could play a role here. The finding that mutation of I364 impedes phosphorylation from ATP supports a general mechanistic resemblance of the flippase to the cation-transporting P-type ATPases, where the intramolecular communication between the M4 glutamate, interacting with the bound cation, and the cytoplasmic domains is decisive for the activation of the phosphorylation reaction (9, 17, 18). Thus far, no ion or other substrate has been shown to be required for phosphorylation of the flippase, even though several possibilities have been examined (12). That abundance of H+ ions is not required was confirmed here by the close to equal phosphorylation rates observed at pH 7.5 and 9.0 for both the WT and I364E (Fig. S2). For I364E, one might wonder whether protonation of the glutamate side-chain is required for phosphorylation. This scenario would only be consistent with the WT-like phosphorylation rate of I364E at pH 9.0, if the pKa of this glutamate was unusually high in the E1 state (well above 9.0).
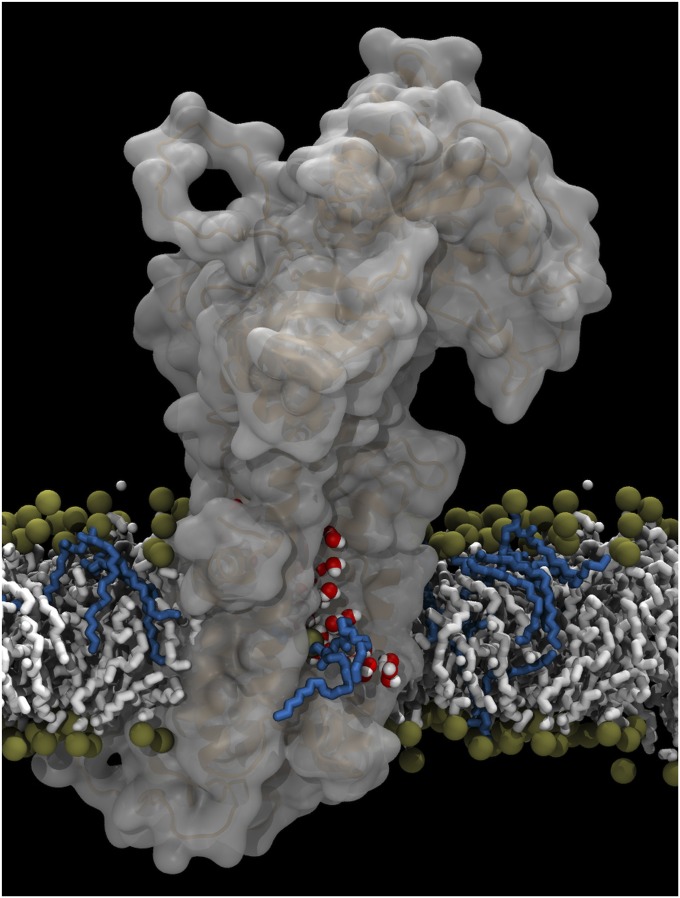
Our modeling provides a structural basis for explaining the functional behavior of the I364 mutants. We obtained a clear indication of a groove bordered by the transmembrane segments M1, M2, M4, and M6, which likely contains water-filled pockets, as shown by MD simulations. The cytoplasmic end of this groove coincides with a bound PE molecule present in several SERCA E2 crystal structures (Fig. S4D). As previously noted, in the flippases, this primordial phospholipid-binding site might have evolved into a site playing a role in the lipid transport (35). Therefore, it is suggested that the groove we see is a pathway for transport of lipid from the exoplasmic side to a cytoplasmically facing exit site situated approximately at the location of the PE molecule in SERCA. MD simulations have indicated that lipid translocation (flip-flop) across a lipid bilayer is enhanced by the presence of packing defects creating water pores in the bilayer (36). In analogy, it has been proposed that the phospholipid scramblase activity of serpentine receptors, such as rhodopsin and the β-adrenergic receptor, owes to the presence of a string of water molecules in the protein interior that interact with the hydrophilic head groups of phospholipids during transfer across the bilayer interior (26). This idea is readily extended to the flippases based on our modeling studies. As visualized in Fig. 9 and Fig. S7, the water-filled pockets in the groove observed in our MD analysis can accommodate the hydrophilic head group of PS (hence also the smaller PE), allowing the hydrocarbon chains to jut out into the membrane lipid phase. This hypothesis is referred to as the “credit card model,” because the protein’s preference to interact with the head group of the lipid (via the water in the groove), and not with its hydrophobic tail, is comparable to a credit card payment terminal, which touches the card only at the magnetic stripe. Water pockets located differentially between the conformational states E2P and E2 (Fig. 6 and Fig. S5) might allow for the phospholipid head group to move during the transfer of the lipid across the bilayer. In the E2 and E2P flippase models, the water pockets nearest to the cytoplasmic surface coincide with a hydrated Ca2+ entry pathway of SERCA in stable MD (root mean square deviation ∼ 1 Å), which stretches from above the M1 kink, passes the crystallized PE-binding site, and extends all of the way down to E309, corresponding to I364 in ATP8A2 (37). However, the hydrated grooves of the refined flippase models are considerably wider than that of SERCA and large enough to accommodate a phospholipid (Fig. S7), in line with the hypothesis that the annular lipid binding site and hydrated Ca2+ entry path of SERCA have evolved into the exit path for hydrated phospholipid head group of the flippases. Together with other hydrophobic residues including I362 and L367 of M4, F88 of M1, I115 of M2, and V906 of M6, I364 seems to form a hydrophobic gate between the water pockets in the groove. In yeast flippases, the M4 residue corresponding to L367 has been shown to be critical to lipid head group specificity; however, mechanistic studies of these flippases have focused primarily on possible exit and entry sites of the lipid (13, 15). Our data suggest that the hydrophobic gate, which is situated in between these sites, directs the sequential formation and annihilation of water-filled cavities, thereby enabling transport of the hydrophilic phospholipid head group. According to the proposed flippase reaction cycle (Fig. 1B), the E2P model represents the enzyme conformation ready to bind the lipid from the exoplasmic side, whereas dephosphorylation to E2 and the ensuing transition to E1 leads to lipid translocation and release. On the basis of the functional analysis described above, in combination with the structural modeling, we propose that the hydrophobic amino acids in the gate, in particular I364, play a role in releasing the phospholipid into the cytoplasmic leaflet during the transformation of E2 into E1. The nonfavorable interaction of the hydrophobic side-chain with the polar lipid head group may be instrumental here. Hence, the overall function of M4 in the flippase would be reminiscent of the functioning of M4 in the ion pumps (8, 9), as a pump rod moving up and down to bring about the translocation of the substrate.
Fig. 9.
Visualization of transport hypothesis. ATP8A2, reconstructed based on the equilibrated E2 model membrane domain, is represented as a cartoon under transparent isosurface, embedded in the phospholipid membrane, where ochre spheres denote phosphate groups and hydrocarbon chains are shown in white (PC) or blue (PS) licorice representation. Water molecules in the proposed transport groove are shown. The hydrophilic head group of the phospholipid is moved upwards along the groove, partly solvated by water molecules. The hydrocarbon chains of the lipid project into the membrane lipid phase.
The functional data clearly indicate that, besides I364, I115 is an essential part of the hydrophobic gate. I115A showed biphasic PS concentration dependence with inhibition at the highest concentrations, indicative of stabilization of E2 with retarded release of the lipid, and a preferential lowering of apparent PS affinity, the affinity for PE being WT-like. I362A and V906A, on the other hand, showed inhibition at high PE concentration, thus differentiating the roles of the various parts of the putative hydrophobic gate in the interaction with PS and PE. The very strong resemblance of the characteristics of I115A to those of I364 mutants is in accordance with I115 being the hydrophobic residue in the cluster with the shortest distance to I364 in our model. Likewise, I362 and V906 are close to I364 (Fig. 6). I362 is positioned corresponding to an isoleucine/valine in Ca2+-ATPase/Na+,K+-ATPase that donates its backbone carbonyl to cation binding, and V906 corresponds to a leucine in Ca2+-ATPase/Na+,K+-ATPase M6 positioned right after the cation binding asparagine/aspartate (Fig. S1).
Also worth noting is the participation of F88 in the hydrophobic gate. This residue is positioned corresponding to L99 of the Na+,K+-ATPase and L65 of Ca2+-ATPase, which are known to function as a gatekeeper that, together with other hydrophobic residues in the vicinity, hold the glutamic acid residue in M4 in correct position for the binding and occlusion of the transported ions (38, 39). Hence, there are distinct similarities between the gate control in relation to ion translocation by Na+,K+-ATPase and Ca2+-ATPase and the hydrophobic gate control mechanism of the flippase. In the flippase, some of the hydrophobic residues clustering around I364 may be instrumental in positioning I364 correctly for functioning in lipid translocation.
The modeling results further indicated that N359 of M4 changes its relation to the water pockets during the dephosphorylation of E2P to E2 (Figs. 6 and 7), which coincides with the functional analysis showing that N359 is a key residue in the phospholipid-activated dephosphorylation. The maximum dephosphorylation rate and the apparent affinity for both PS and PE were strongly affected in the N359A mutant (Fig. 4). N359 is not conserved in all P4-ATPases, but a medium-sized polar residue is generally found at the corresponding position (Fig. S1). The polarity and hydrogen bonding potential of the side-chain might allow N359 to interact favorably with the polar head group of a phospholipid. Because the N359 side-chain projects into the exoplasmic water pocket of the E2P model, but is placed in between the water pockets and is poorly hydrated in the E2 model (Figs. 6 and 7), N359 might bind the phospholipid head group preferentially in the E2P form, before the lipid is moved further toward the cytoplasmic side in E2, which would be consistent with its critical role in determining the apparent affinity in the interaction of the lipid with E2P. Furthermore, the increased rate of phosphorylation from ATP and the reduced apparent vanadate affinity of N359A may be taken as evidence of a shift of the E2-E1 equilibrium in favor of E1 in N359A. It is further noteworthy that the residue in Na+,K+-ATPase at the position corresponding to N359 in ATP8A2 is a valine that contributes its backbone carbonyl oxygen to binding of the transported K+ (Fig. S1).
In conclusion the present study has demonstrated that I364 and adjacent hydrophobic residues, seconded by N359, play critical roles in the mechanism of ATP8A2. On the basis of the functional analysis of mutants in combination with modeling studies, we are able to propose a hydrophobic gating mechanism that makes use of a hydrated groove between M1, M2, M4, and M6 for translocation of the phospholipid substrate. It is intriguing how the pump rod function of M4 known from the cation transporting P-type ATPases may be brought into play also in this P4 ATPase to accomplish lipid translocation. The central role of the M4 movements in the transport process is likely a general feature of all P-type ATPases.
Materials and Methods
Mutagenesis, expression, purification, and reconstitution in lipid vesicles of WT and mutant ATP8A2 with its accessory CDC50 subunit and the procedures used for functional analysis and data processing have previously been described in detail (12). The details of the flippase assay and the computational procedures are described in SI Materials and Methods. A brief overview of the methods is given here. ATP hydrolysis was measured at 37 °C over a period of 15 min in the presence of 2.5 mg/mL lipid (PC, PC with PS, or PC with PE) contained in 50 mM Hepes-NaOH, pH 7.5, 150 mM NaCl, 12.5 mM MgCl2, 10 mM CHAPS, and 1 mM DTT with 7.5 mM ATP. The amount of enzyme in the reaction was adjusted to keep the colorimetric measures within the linear phase and yet sufficiently high for an accurate determination. The ATPase activity was normalized to ATP8A2 protein levels to obtain micromoles of ATP hydrolyzed per minute per milligram ATP8A2 protein. Phosphorylation was generally carried out at 0 °C with 2 μM [γ-32P]ATP in standard phosphorylation medium (SPM) consisting of 50 mM Hepes-NaOH (pH 7.5), 150 mM NaCl, 1 mM MgCl2, and 1 mM DTT, modifications being described in figure legends. When indicated, CHAPS was added at a concentration of 10 mM to solubilize protein and lipid. Quenching of the phosphorylation reaction was performed with 1–2.5 volumes of 25% (wt/vol) trichloroacetic acid (TCA) containing 100 mM H3PO4. The TCA-precipitated enzyme was washed by centrifugation and was analyzed by SDS/PAGE in 5.8% (wt/vol) gels under acidic conditions and quantified by phosphorimaging (12). Flippase activity was determined by the fluorescence-dithionite assay at 23 °C, essentially as previously described (12) except that excess EDTA was added for termination of flipping by removal of Mg2+, and NBD fluorescence was measured in a plate reader (SI Materials and Methods). A 2.5-min incubation time with ATP was chosen based on experiments showing that flipping was linear with time during this interval. The values for %NBD-PS flipped were normalized to represent a protein amount equal to the average protein expression level of the WT.
For computational analysis and modeling, a multiple sequence alignment of bovine ATP8A2 and 2782 homologous P-type ATPases was performed using HHblits (40) and HHpred (41). A modeling alignment of ATP8A2 and SERCA protein sequences was extracted, and gaps of SERCA were patched with structures of secondary structure identical to the prediction for ATP8A2. Homology models of E2P and E2 were produced with MODELLER 9v11 (42). The models of the membrane domains were then inserted into a membrane patch and refined using MD simulations. All simulations were performed using the NAMD (43) engine, with the CHARMM27 force field (44), including cross-term energy dihedral angle corrective map corrections for the protein and the united-atom CHARMM27 force field for the membrane (45). The two systems containing the E2P and E2 models, respectively, were first energy minimized for 5,000 steps, followed by an equilibration phase of 10 ns with the whole protein backbone restrained to let side-chains and lipids pack, and then 10 ns with only the well-defined protein secondary structure elements being restrained to allow loops to relax, and finally 80 ns of unrestrained production run, both systems reaching a stable plateau phase of at least 20 ns.
Supplementary Material
Acknowledgments
We thank Marco Biasini and Torsten Schwede (Biozentrum, University of Basel and Swiss Institute of Bioinformatics) for expert guidance on multiple alignment and homology modeling. We thank Lene Jacobsen and Karin Kracht (Department of Biomedicine, Aarhus University) for expert technical assistance. This study was funded in part by grants from the Danish Medical Research Council, the Novo Nordisk Foundation (Fabrikant Vilhelm Pedersen og Hustrus Legat), the Lundbeck Foundation, Canadian Institutes for Health Research (Grant MOP-106667), and National Institutes of Health (Grant EY02422). T.L. and M.D.P. were supported by Swiss National Science Foundation Grant 200020_138013. J.A.C. was supported by a National Sciences and Engineering Council predoctoral studentship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321165111/-/DCSupplemental.
References
- 1.Auland ME, Roufogalis BD, Devaux PF, Zachowski A. Reconstitution of ATP-dependent aminophospholipid translocation in proteoliposomes. Proc Natl Acad Sci USA. 1994;91(23):10938–10942. doi: 10.1073/pnas.91.23.10938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou X, Graham TR. Reconstitution of phospholipid translocase activity with purified Drs2p, a type-IV P-type ATPase from budding yeast. Proc Natl Acad Sci USA. 2009;106(39):16586–16591. doi: 10.1073/pnas.0904293106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coleman JA, Kwok MCM, Molday RS. Localization, purification, and functional reconstitution of the P4-ATPase Atp8a2, a phosphatidylserine flippase in photoreceptor disc membranes. J Biol Chem. 2009;284(47):32670–32679. doi: 10.1074/jbc.M109.047415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Folmer DE, Elferink RP, Paulusma CC. P4 ATPases - lipid flippases and their role in disease. Biochim Biophys Acta. 2009;1791(7):628–635. doi: 10.1016/j.bbalip.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Stapelbroek JM, et al. ATP8B1 is essential for maintaining normal hearing. Proc Natl Acad Sci USA. 2009;106(24):9709–9714. doi: 10.1073/pnas.0807919106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levano K, et al. Atp8a1 deficiency is associated with phosphatidylserine externalization in hippocampus and delayed hippocampus-dependent learning. J Neurochem. 2012;120(2):302–313. doi: 10.1111/j.1471-4159.2011.07543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coleman JA, Quazi F, Molday RS. Mammalian P4-ATPases and ABC transporters and their role in phospholipid transport. Biochim Biophys Acta. 2013;1831(3):555–574. doi: 10.1016/j.bbalip.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toyoshima C. How Ca2+-ATPase pumps ions across the sarcoplasmic reticulum membrane. Biochim Biophys Acta. 2009;1793(6):941–946. doi: 10.1016/j.bbamcr.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 9.Møller JV, Olesen C, Winther A-ML, Nissen P. The sarcoplasmic Ca2+-ATPase: Design of a perfect chemi-osmotic pump. Q Rev Biophys. 2010;43(4):501–566. doi: 10.1017/S003358351000017X. [DOI] [PubMed] [Google Scholar]
- 10.Post RL, Hegyvary C, Kume S. Activation by adenosine triphosphate in the phosphorylation kinetics of sodium and potassium ion transport adenosine triphosphatase. J Biol Chem. 1972;247(20):6530–6540. [PubMed] [Google Scholar]
- 11.Albers RW. Biochemical aspects of active transport. Annu Rev Biochem. 1967;36:727–756. doi: 10.1146/annurev.bi.36.070167.003455. [DOI] [PubMed] [Google Scholar]
- 12.Coleman JA, Vestergaard AL, Molday RS, Vilsen B, Andersen JP. Critical role of a transmembrane lysine in aminophospholipid transport by mammalian photoreceptor P4-ATPase ATP8A2. Proc Natl Acad Sci USA. 2012;109(5):1449–1454. doi: 10.1073/pnas.1108862109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baldridge RD, Graham TR. Identification of residues defining phospholipid flippase substrate specificity of type IV P-type ATPases. Proc Natl Acad Sci USA. 2012;109(6):E290–E298. doi: 10.1073/pnas.1115725109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stone A, Williamson P. Outside of the box: Recent news about phospholipid translocation by P4 ATPases. J Chem Biol. 2012;5(4):131–136. doi: 10.1007/s12154-012-0078-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baldridge RD, Graham TR. Two-gate mechanism for phospholipid selection and transport by type IV P-type ATPases. Proc Natl Acad Sci USA. 2013;110(5):E358–E367. doi: 10.1073/pnas.1216948110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baldridge RD, Xu P, Graham TR. Type IV P-type ATPases distinguish mono- versus diacyl phosphatidylserine using a cytofacial exit gate in the membrane domain. J Biol Chem. 2013;288(27):19516–19527. doi: 10.1074/jbc.M113.476911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vilsen B, Andersen JP. Mutation to the glutamate in the fourth membrane segment of Na+,K+-ATPase and Ca2+-ATPase affects cation binding from both sides of the membrane and destabilizes the occluded enzyme forms. Biochemistry. 1998;37(31):10961–10971. doi: 10.1021/bi9802925. [DOI] [PubMed] [Google Scholar]
- 18.Olesen C, et al. The structural basis of calcium transport by the calcium pump. Nature. 2007;450(7172):1036–1042. doi: 10.1038/nature06418. [DOI] [PubMed] [Google Scholar]
- 19.Vilsen B, Andersen JP, Clarke DM, MacLennan DH. Functional consequences of proline mutations in the cytoplasmic and transmembrane sectors of the Ca2+-ATPase of sarcoplasmic reticulum. J Biol Chem. 1989;264(35):21024–21030. [PubMed] [Google Scholar]
- 20.Andersen JP, Vilsen B, MacLennan DH. Functional consequences of alterations to Gly310, Gly770, and Gly801 located in the transmembrane domain of the Ca2+-ATPase of sarcoplasmic reticulum. J Biol Chem. 1992;267(4):2767–2774. [PubMed] [Google Scholar]
- 21.Vilsen B. Leucine 332 at the boundary between the fourth transmembrane segment and the cytoplasmic domain of Na+,K+-ATPase plays a pivotal role in the ion translocating conformational changes. Biochemistry. 1997;36(43):13312–13324. doi: 10.1021/bi971030q. [DOI] [PubMed] [Google Scholar]
- 22.Emre Onat O, et al. Missense mutation in the ATPase, aminophospholipid transporter protein ATP8A2 is associated with cerebellar atrophy and quadrupedal locomotion. Eur J Hum Genet. 2013;21(3):281–285. doi: 10.1038/ejhg.2012.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coleman JA, Molday RS. Critical role of the beta-subunit CDC50A in the stable expression, assembly, subcellular localization, and lipid transport activity of the P4-ATPase ATP8A2. J Biol Chem. 2011;286(19):17205–17216. doi: 10.1074/jbc.M111.229419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cantley LC, Jr, Cantley LG, Josephson L. A characterization of vanadate interactions with the (Na,K)-ATPase. Mechanistic and regulatory implications. J Biol Chem. 1978;253(20):7361–7368. [PubMed] [Google Scholar]
- 25.Morth JP, et al. A structural overview of the plasma membrane Na+,K+-ATPase and H+-ATPase ion pumps. Nat Rev Mol Cell Biol. 2011;12(1):60–70. doi: 10.1038/nrm3031. [DOI] [PubMed] [Google Scholar]
- 26.Williamson P. Phospholipid transport: Sighting a new face of an old friend. Curr Biol. 2011;21(4):R168–R169. doi: 10.1016/j.cub.2011.01.030. [DOI] [PubMed] [Google Scholar]
- 27.Cacciagli P, et al. Disruption of the ATP8A2 gene in a patient with a t(10;13) de novo balanced translocation and a severe neurological phenotype. Eur J Hum Genet. 2010;18(12):1360–1363. doi: 10.1038/ejhg.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu X, et al. Mutations in a P-type ATPase gene cause axonal degeneration. PLoS Genet. 2012;8(8):e1002853. doi: 10.1371/journal.pgen.1002853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gourdon P, et al. Crystal structure of a copper-transporting PIB-type ATPase. Nature. 2011;475(7354):59–64. doi: 10.1038/nature10191. [DOI] [PubMed] [Google Scholar]
- 30.Argüello JM, Eren E, González-Guerrero M. The structure and function of heavy metal transport P1B-ATPases. Biometals. 2007;20(3-4):233–248. doi: 10.1007/s10534-006-9055-6. [DOI] [PubMed] [Google Scholar]
- 31.Creighton TE. 1993. Proteins: Structures and Molecular Properties (W H Freeman & Co, New York), 2nd Ed, pp 140–156.
- 32.Skou JC. The influence of some cations on an adenosine triphosphatase from peripheral nerves. Biochim Biophys Acta. 1957;23(2):394–401. doi: 10.1016/0006-3002(57)90343-8. [DOI] [PubMed] [Google Scholar]
- 33.Toustrup-Jensen MS, et al. The C terminus of Na+,K+-ATPase controls Na+ affinity on both sides of the membrane through Arg935. J Biol Chem. 2009;284(28):18715–18725. doi: 10.1074/jbc.M109.015099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vilsen B. Mutant Phe788 —> Leu of the Na+,K+-ATPase is inhibited by micromolar concentrations of potassium and exhibits high Na+-ATPase activity at low sodium concentrations. Biochemistry. 1999;38(35):11389–11400. doi: 10.1021/bi990951t. [DOI] [PubMed] [Google Scholar]
- 35.Lenoir G, Williamson P, Holthuis JCM. On the origin of lipid asymmetry: The flip side of ion transport. Curr Opin Chem Biol. 2007;11(6):654–661. doi: 10.1016/j.cbpa.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 36.Gurtovenko AA, Anwar J, Vattulainen I. Defect-mediated trafficking across cell membranes: Insights from in silico modeling. Chem Rev. 2010;110(10):6077–6103. doi: 10.1021/cr1000783. [DOI] [PubMed] [Google Scholar]
- 37.Musgaard M, Thøgersen L, Schiøtt B, Tajkhorshid E. Tracing cytoplasmic Ca2+ ion and water access points in the Ca2+-ATPase. Biophys J. 2012;102(2):268–277. doi: 10.1016/j.bpj.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Einholm AP, Andersen JP, Vilsen B. Roles of transmembrane segment M1 of Na+,K+-ATPase and Ca2+-ATPase, the gatekeeper and the pivot. J Bioenerg Biomembr. 2007;39(5-6):357–366. doi: 10.1007/s10863-007-9106-x. [DOI] [PubMed] [Google Scholar]
- 39.Einholm AP, Andersen JP, Vilsen B. Importance of Leu99 in transmembrane segment M1 of the Na+, K+ -ATPase in the binding and occlusion of K+ J Biol Chem. 2007;282(33):23854–23866. doi: 10.1074/jbc.M702259200. [DOI] [PubMed] [Google Scholar]
- 40.Remmert M, Biegert A, Hauser A, Söding J. HHblits: Lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods. 2012;9(2):173–175. doi: 10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- 41.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33(Web Server issue):W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eswar N, et al. 2007. Comparative Protein Structure Modeling Using MODELLER. Curr Protoc Protein Sci (John Wiley & Sons, New York), Chap 2, Unit 2.9.
- 43.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brooks BR, et al. CHARMM: The biomolecular simulation program. J Comput Chem. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hénin J, Shinoda W, Klein ML. United-atom acyl chains for CHARMM phospholipids. J Phys Chem B. 2008;112(23):7008–7015. doi: 10.1021/jp800687p. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.