Significance
Apoptosis, or programmed cell death, is critical for maintaining tissue homeostasis and development in multicellular organisms. This article describes a unique molecularly engineered cyclic herpes simplex virus type 1–thymidine kinase (cTK) reporter for real-time apoptosis PET imaging. This reporter enables quantitative and real-time monitoring of caspase-3 activity in response to extracellular stimuli. As a PET reporter, cTK shows low background and high sensitivity in response to caspase-3 activation. The application of this particular caspase reporter system would be of significance in assisting preclinical high-throughput screening of different apoptosis-inducing drugs and monitoring their therapeutic efficacy in a noninvasive and quantitative manner.
Keywords: reporter probe, positron-emission tomography
Abstract
The coordination of cell proliferation and programmed death (apoptosis) is essential for normal physiology, and imbalance in these two opposing processes is implicated in various diseases. Objective and quantitative noninvasive imaging of apoptosis would significantly facilitate rapid screening as well as validation of therapeutic chemicals. Herein, we molecularly engineered an apoptosis switch-on PET-based cyclic herpes simplex virus type 1–thymidine kinase reporter (cTK266) containing a caspase-3 recognition domain as the switch. Translation of the reporter and protein splicing in healthy mammalian cells produce an inactive cyclic chimera. Upon apoptosis, caspase-3–specific cleavage of the circular product occurs, resulting in the restoration of the thymidine kinase activity, which can be detected in living cells and animals by noninvasive PET imaging. Our results showed the high sensitivity of this reporter in dynamic and quantitative imaging of apoptosis in living subjects. This reporter could be applied as a valuable tool for high-throughput functional screening of proapoptotic and antiapoptotic compounds in preclinical models in drug development, and monitoring the destination of therapeutic cells in clinical settings.
The apoptotic pathway plays a crucial role in many biological processes, and is involved in various diseases as well (1, 2). Most of the chemotherapy agents cause tumor cell death primarily by induction of apoptosis. Resistance to anticancer treatment is widely believed to involve mutations that lead to deregulated cellular proliferation and suppression of mechanisms that control apoptosis (3–6). Therefore, methods that can evaluate apoptosis as an early assessment of tumor response are urgently required to manage patients in terms of quality of life versus intensive chemotherapy (7, 8). Development of imaging probes to accurately monitor the process of apoptosis in living organisms has extensive implications for better understating the pathological changes during therapeutic interventions and screening apoptosis-targeted drugs.
To date, a number of strategies have been attempted to monitor the progress of apoptosis using various imaging modalities that target the myriad of changes involving membrane composition, protein synthesis, enzyme activation, and energy levels throughout the apoptotic program (9). Among them, phosphatidylserine (PS) exposure and its high-affinity binder Annexin V has been most intensively investigated for in vivo apoptosis detection, especially with radionuclide labeling (10–12). However, this strategy is unable to discriminate apoptotic from necrotic cells because PS exposure also occurs in necrotic cells (13).
Alternatively, visualization and evaluation of apoptosis can be achieved by targeting more specific apoptotic mediators, such as caspases, which are a family of intracellular cysteine–aspartate–specific proteases that play central roles in the execution of cell death (14, 15). Among the spectrum of various caspases, caspase-3 has been identified as the primary executioner of cell apoptosis (16) and is an attractive target molecule for apoptosis imaging (17). Several synthetic activatable probes have been developed to image caspase-3 activity directly, but most of these probes suffer from low cell membrane penetration and relatively high background due to the nonspecific cleavage (18, 19). Consequently, fluorescence and bioluminescence reporter systems have been developed based on a small peptide aspartylglutamylvalanyl aspartic acid (DEVD), which can be recognized and cleaved by caspase-3 (17, 20–22). Although optical imaging with these reporter probes has been demonstrated to be useful for in vivo monitoring of apoptosis especially in cell culture or s.c. animal models (23–28), further applications of these approaches are thwarted by tissue attenuation of photons and lack of fully quantitative and tomographic capabilities. Therefore, there is an urgent need to develop inherently more sensitive, quantitative, and translatable reporter systems for noninvasive apoptosis imaging.
Being fully quantitative and tomographic, positron-emission tomography (PET) is the most sensitive and specific technique for imaging molecular pathways in vivo (29). The herpes simplex virus type 1–thymidine kinase (HSV1-TK) is the most frequently exploited reporter for PET imaging (30). Upon expression of the HSV1-TK gene, the resulting enzyme can phosphorylate certain radiolabeled purine and pyrimidine nucleoside analogs. This TK-catalyzed phosphorylation results in trapping of the radiolabeled entity within cells, and thereby allowing the visualization by single-photon emission computed tomography (SPECT) or PET (31). In this study, we genetically modified human head and neck squamous carcinoma UM-SCC-22B (University of Michigan squamous cell carcinoma 22B) cells with the cyclic truncated HSV1-TK. After treatment with a chemotherapeutic agent, doxorubicin (Dox), to induce apoptosis, this apoptosis reporter probe could effectively and quantitatively sense caspase-3 activation in both cultured cells and tumor tissues.
Results
Design of the Cyclic HSV1-TK Reporter Probe.
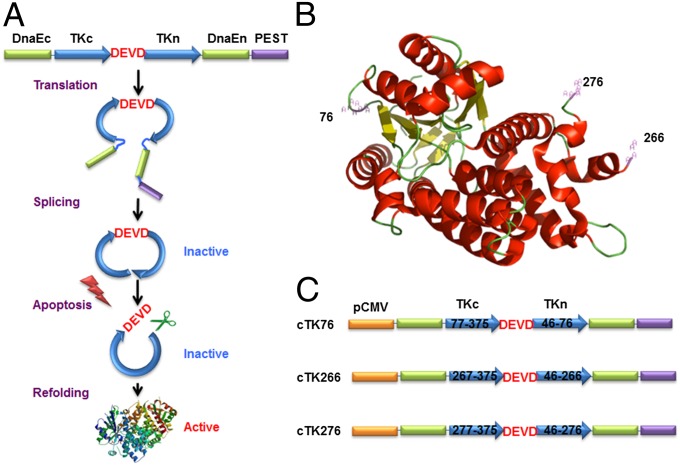
As shown in Fig. 1A, the cyclic PET reporter was developed by fusing two fragments of DnaE intein to neighboring ends of HSV1-TK. The substrate sequence DEVD was used for caspase-3 recognition, flanked by the N and C termini of TK. After translation into a single polypeptide, the two fragments of TK were ligated by protein splicing, producing a closed circular polypeptide. We started with a truncated version of HSV1-TK by deletion of the first 45 amino acids from the N terminus, which showed reduced cytotoxicity in vivo compared with the full-length HSV1-TK (32). Truncation of this flexible region also helps to keep the tension of the protein structure after cyclization. A PEST sequence, a peptide sequence that is rich in proline (P), glutamic acid (E), serine (S), and threonine (T) (33), was included to guarantee the degradation of the uncyclized polypeptide. Upon caspase-3 activation, DEVD sequence will be cleaved, and termini of cyclic HSV1-TK (cTK) are released to restore the TK activity. With the corresponding radiolabeled substrates, PET imaging could be performed to measure the caspase-3 activity.
Fig. 1.
The cTK reporter probe for apoptosis imaging. (A) Schematic overview of the principle for monitoring apoptosis. The N and C termini of HSV1-TK are linked with DEVD, a substrate peptide of caspase-3. Upon caspase-3 activation during apoptosis, the DEVD sequence is cleaved and the cyclized TK restores its activity. (B) The crystal structure of truncated HSV1-TK with 1–45 aa deletion. The subunits of TK display a general αβ folding pattern, each of which consists of 13 α-helices (in red) and seven β-sheets (in yellow). The numbers represent the split sites. (C) Schematic structure of the variants of cTK constructs. Each construct is made by cloning into a pcDNA3.1 (+) plasmid backbone, under the control of a CMV promoter.
To create the cTK reporter probe, the key step is to identify the specific sites where the primary polypeptide of TK can be disrupted into two fragments and then ligated to form the cyclic probe. The crystal structure of TK (34, 35) indicates that the constituent subunits of TK display the general αβ folding pattern, each of which consists of 13 α-helices and seven β-sheets (Fig. 1B). Based on the structural information of TK, we identified three potential split sites that are within the disordered loop regions but not within periodic secondary structure regions (Fig. S1). Among them, one split site (between Asp76 and Asp77) is within the active site of TK, and the other two split sites (between Ala266 and Val267, and Ser276 and Asn277) are outside of its active sites. Accordingly, three engineered variants of cTK reporters, designated as cTK76, cTK266, and cTK276, were constructed with linkage of the N and C termini of TK by the DEVD substrate sequence (Fig. 1C).
Detection of TK Enzyme Activity upon Apoptosis.
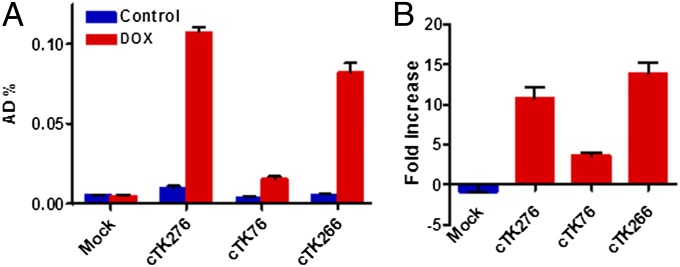
To assess whether the enzyme activity of the three cTK reporters were restored in apoptotic cells, we transiently transfected UM-SCC-22B cells with cTK76, cTK266, and cTK276. A representative wide spectrum anticancer drug (36, 37), Dox, was chosen as the apoptosis-inducing agent. Western blots of apoptosis signaling pathway study in UM-SCC-22B cells (Fig. S2A) showed Dox dose-dependent apoptosis induction to caspase-3 activity, a key factor in mediating cTK fuction in our study. After treatment with Dox (1 µg/mL) for 24 h, all cTK cells showed increased 9-(4-18F-fluoro-3-[hydroxymethyl]butyl)guanine (18F-FHBG) uptake, and the cTK266 was chosen for further study because it had the highest fold increase (Fig. 2).
Fig. 2.
The 18F-FHBG cell uptake assay for the cTK variants. (A) The 18F-FHBG uptake of three cTK variants (cTK76, cTK266, and cTK276) transiently transfected UM-SCC-22B cells, with untransfected cells as the control (mock). The cells were stimulated with Dox (1 μg/mL) for 24 h to induce apoptosis. (B) Fold change of FHBG uptake in treated cells compared with untreated cells. Results were representative data collected from independent duplicate experiments.
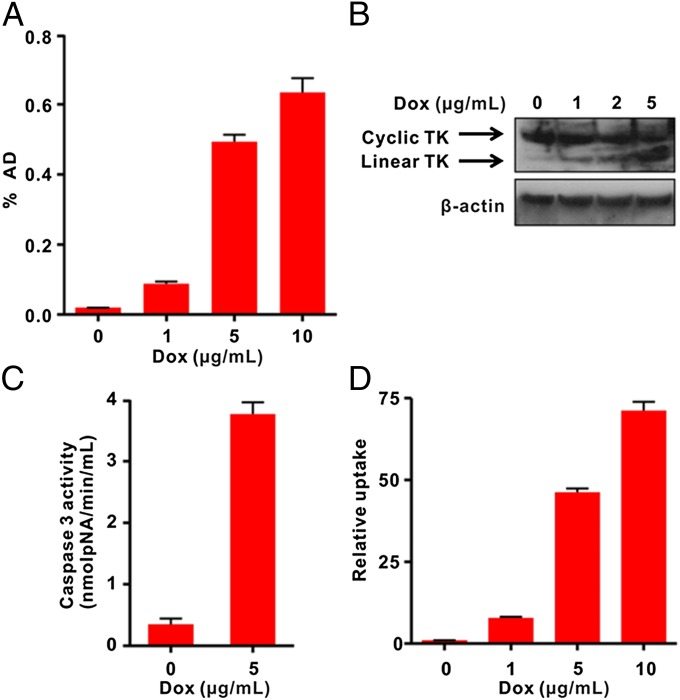
Next, we genetically engineered the cTK266 reporter into firefly luciferase expressing UM-SCC-22B cells, generating a stable cell line with dual reporters. The cells, designated as cTK266-22B-Fluc, retained the similar morphology and growth kinetics to their parental cells. With increased dose of Dox, the 18F-FHBG uptake in treated cells was also increased. With 10 µg/mL Dox, the treated cells showed the highest FHBG uptake (32.8-fold) compared with the control cells (Fig. 3A). It is highly likely that the increased FHBG uptake is related to apoptosis, as increased green fluorescence signal on the cell membrane after staining with FITC–Annexin V and flow cytometry were also observed in cells with the same treatment (Fig. S2).
Fig. 3.
In vitro characterization of cTK266 reporter. (A) The 18F-FHBG uptake assay of cTK266-22B-Fluc cells treated with different doses of Dox for 24 h (n = 4). (B) Western blot analysis using HSV1-TK and β-actin antibodies for cTK266-22B-Fluc cell lysates. The cells were treated with 1, 2, and 5 μg/mL of Dox for 24 h. (C) The activity of caspase-3 determined by a caspase colorimetric assay kit (n = 3). (D) Ratiometric analysis of the relative FHBG uptake. The FHBG uptake was normalized with the bioluminescence signal from control and treated cells. Results were representative data collected from independent triplicate experiments.
Indeed, the cTK266-22B-Fluc cells treated with increasing doses of Dox showed apparent double bands in Western blot using a TK-specific antibody (Fig. 3B). The upper band corresponds to the cyclic form of TK, as it is dominant in untreated cells and tends to decrease in a dose-dependent manner. The lower band corresponds to the linear TK counterpart, which is increased along with increased doses of Dox, resulting from the cleavage of DEVD by the activated caspase-3 protease. The caspase-3 activation was confirmed using the caspase-3 substrate, Ac-DEVD-pNA, with a dramatic increase in absorbance at 450 nm in Dox-treated cells (Fig. 3C). The Fluc signal, on the other hand, decreased with increasing Dox concentrations (Fig. S3), due to the cytotoxicity from Dox. After normalizing the FHBG uptake with Fluc signal, a much higher increase in FHBG uptake (71.2-fold) in the treated cells was observed (Fig. 3D).
In Vitro Real-Time Sensing of Apoptosis.
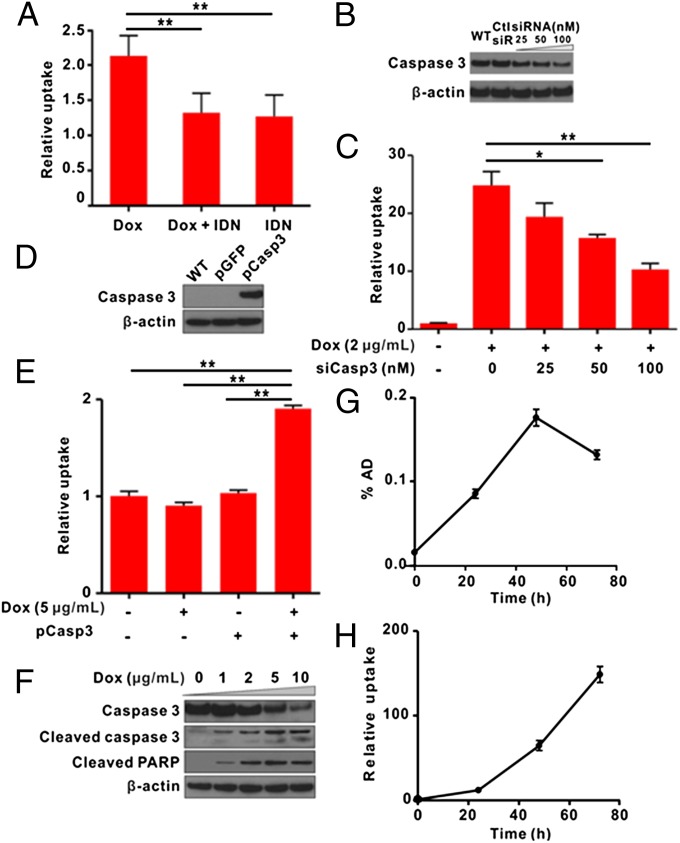
To confirm that the linearization of cyclic TK was due to caspase-3 activation, first we used a chemical caspase inhibitor (IDN6556) in combination with Dox to detect FHBG uptake. The result showed that FHBG uptake in apoptotic cells was significantly (P < 0.01) reduced in the presence of caspase inhibitor (Fig. 4A). By introducing siRNA against caspase-3 (si–caspase-3) in cells (Fig. 4B), the FHBG uptake was inhibited in a dose-dependent manner (Fig. 4C). To further ensure that the observed increase of FHBG uptake was indeed mediated by caspase-3 activation, MCF-7 breast carcinoma cells, deficient of functional caspase-3 (38), were transiently cotransfected with cTK266 and caspase-3 plasmids (Fig. 4D). The FHBG uptake increased significantly (P < 0.01) upon Dox stimulation (Fig. 4E). Western blot studies of cotransfected MCF7 cells indicated that the caspase-3 activity in apoptotic MCF7 cells were in a Dox dose-dependent manner (Fig. 4F). Collectively, our results confirm that the restoration of TK activity is caspase-3 specific.
Fig. 4.
Real-time monitoring of apoptosis induced by Dox in vitro. (A) The 18F-FHBG uptake by cTK266-22B-Fluc cells stimulated by Dox (0.5 μg/mL, 24 h) in the presence of IDN6556 (20 μM). Data were normalized to control cells in culture medium. IDN, IDN6556; **P < 0.01 (n = 4). (B) Western blot analysis of cTK266-22B-Fluc transfected with different doses of anti–caspase-3 siRNA or scramble siRNA. (C) 18F-FHBG uptake in cells treated with caspase-3–specific siRNA. After transfection of cTK266-22B-Fluc cells with different concentrations of siRNA against caspase-3 or scramble siRNA for 24 h, the cells were then treated with 2 μg/mL of Dox for 24 h. *P < 0.05, **P < 0.01 (n = 4). (D) Western blot analysis of MCF-7 cell caspase-3 expression level after being transiently transfected with caspase-3 plasmid or GFP control plasmid. (E) Caspase-3–dependent 18F-FHBG uptake in MCF-7 cells. The cells were transiently transfected with cTK266 only or with both cTK266 and pCaspase-3. Twenty-four hours later, the cells were treated with 5 μg/mL of Dox for another 24 h and then evaluated by 18F-FHBG uptake assay. The data were normalized to the same total protein amount of cell lysate. **P < 0.01 (n = 3). (F) Western bolt analysis of apoptotic MCF-7 cell for caspase-3 and polyADP ribose polymerase (PARP) cleavage. (G) The 18F-FHBG uptake assay for the treated cells at different time points. The cells were seeded in a 24-well plate and treated with Dox (5 μg/mL) for 3 h. Then the plate was washed and refilled with fresh medium (n = 4). (H) Ratiometric analysis of relative FHBG uptake, which was normalized with the BLI signal at different time points. Results were representative data collected from independent duplicate experiments.
To check the efficacy of this biosensor for real-time apoptosis monitoring, we further investigated the time course of Dox-induced caspase-3 activation. After being treated with 5 µg/mL of Dox for 3 h, the Fluc signal decreased continuously over time (Fig. S4). In contrast, the FHBG uptake initially increased with time and peaked at 48 h after Dox treatment, then decreased (Fig. 4G). After normalizing the FHBG uptake with Fluc signal, a continuous increase in FHBG uptake was observed over time, and the maximum fold increase was as high as 148.6 at the 72 h time point (Fig. 4H).
In Vivo Micro-PET Imaging of Apoptosis.
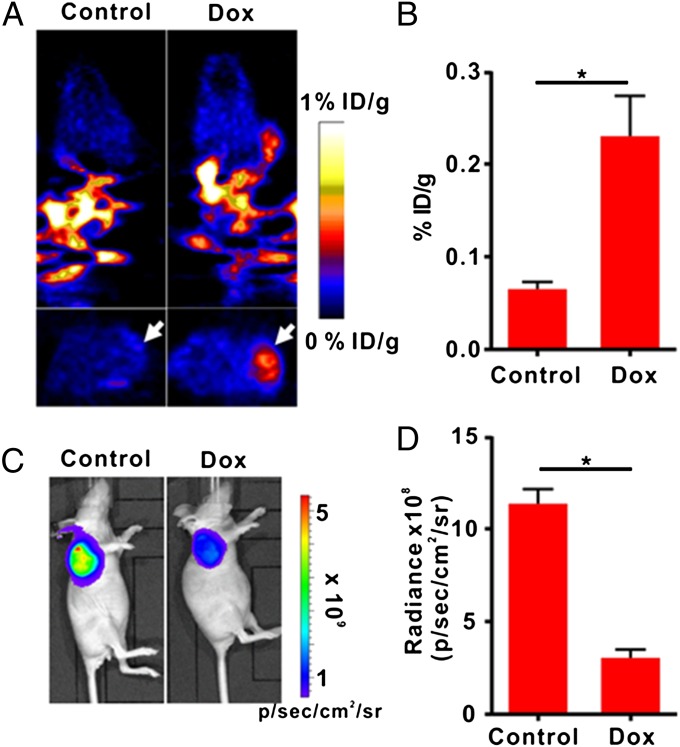
To apply the apoptosis biosensor developed here for apoptosis monitoring in vivo, we developed a tumor model with cTK266-22B-Fluc cells and treated the tumor-bearing mice with either Dox or PBS. PET imaging was performed after i.v. administration of 18F-FHBG (11.1 MBq per mouse, n = 6 per group). Treatment with Dox induced a remarkable 18F-FHBG accumulation inside the tumor (Fig. 5A). Quantification of PET images revealed a significant increase of signal level in Dox-treated tumors compared with PBS-treated control tumors (Fig. 5B).
Fig. 5.
In vivo PET imaging and BLI of apoptosis after Dox treatment. (A) Representative coronal (Upper) and transaxial (Lower) micro-PET images of mice bearing cTK266-22B-Fluc tumors at 2 h postinjection of 18F-FHBG (11.1 MBq per mouse) after treatment with 10 mg/kg of Dox for 72 h (n = 6 per group). The white arrows point to tumors. (B) Quantification of the PET signal over the tumor region, represented as a mean percentage injected dose per gram (%ID/g). (C) Representative BLI images of the same groups of mice after Dox treatment.*P < 0.05. (D) Quantification of tumor BLI signal. *P < 0.05. Results were representative data collected from independent duplicate experiments.
In contrast, the bioluminescence imaging (BLI) signal from the treated tumors decreased as much as 3.72-fold compared with that from the control tumors, indicating that Dox inhibited the tumor growth (Fig. 5 C and D). The Dox-treated group also showed a slight reduction in tumor volume in comparison with the control group (Fig. S5A). With BLI signal for standardization, PET signal in treated tumors increased by 13.3-fold compared with that in the control tumors (Fig. S5B). A set of tumors from each group were then removed for TUNEL staining. Sporadic apoptotic cells were indentified within the treated tumors, whereas no apoptotic cells were found in the control tumors (Fig. S6). To ensure the apoptosis cascade in the tumor did not lead to severe loss of cell membrane integrity, we conducted in vitro FHBG uptake assay, followed by mass spectroscopy analysis of phosphorylated FHBG (FHBG-P) to free FHBG ratio (Fig. S7). The reuslt showed almost no FHBG-P leakage upon apoptosis induction by Dox.
Discussion
Noninvasive imaging and real-time evaluation of apoptosis is crucial to drug development and early stage therapy response monitoring (39). In this study, we presented an apoptosis responsive cTK reporter for the detection of caspase-3 activity by PET imaging. This probe enabled quantitative and real-time monitoring of caspase-3 activity in response to extracellular stimuli. As a PET reporter, cTK showed low background and high sensitivity in response to caspase-3 activation. The in vivo results confirmed that PET is fully quantitative and tomographic, which is particularly advantageous over optical imaging-based approaches. Moreover, the trace amount and relatively short half-life of the imaging substrate also facilitate repetitive scans over a short period of intervals, which would be very helpful to decipher the extremely dynamic nature of apoptosis.
So far, there are several caspase-3 responsive reporters available for in vivo imaging of apoptosis, most of which use a luciferase protein (23–26) or a fusion protein (27, 28) separated by the DEVD sequence. In these linear designs, the deactivation or “silence” of the probes is usually achieved by fusing other proteins or protein domains on each side of the reporter proteins. The background signal is relatively high, as the reporter protein may still keep its original structure. Moreover, it needs double cleavage to fully release the “blockers.” For example, when HSV1-TK was used in such a fusion biosensor for multimodal imaging of apoptosis (27), only a moderate twofold increase in TK activity upon apoptosis was found for both in vitro measurement and in vivo PET imaging after normalization to cell viability. In this study, the deactivation of TK reporter was through structure disruption after terminal ligation and transsplicing. The DEVD sequence was exposed to the surface to facilitate caspase-3 enzyme recognition and cleavage. More importantly, there is a significant difference in the FHBG uptake between the active linear and inactive cyclic forms, although they have the similar amino acid sequences and molecular weights. All these features make the cTK reporter much more sensitive to caspase-3 activation. Indeed, with the cTK266 reporter, we observed a much higher signal increase (148.6-fold in vitro and 13.3-fold in vivo after normalization) during apoptosis.
To design the cTK reporter, the first and most critical step is to identify the optimal site in the primary amino acid sequence of TK for transsplicing. Unlike the firefly luciferase (40) or glycinamide ribonucleotide (GAR) transformylase (41), there is no long cleft-like peptide loop segment in the backbone of TK amenable to single site cleavage. Based on the crystal structure of TK (35), we investigated three potential cleavage sites within the disordered loop regions and showed that one of the variants (where the split site was between Ala266 and Val267) exhibited the highest capability to restore TK enzymatic activity after apoptosis induction (Fig. 2B). In fact, the transsplicing site we identified is consistent with previous findings. Massoud et al. (42) performed a partial circular permutation screen of the TK molecule and found that the circularly permuted variant with the split site between Thr265 and Ala266 was the only mutation to retain TK enzymatic activity.
Because only the cyclic form of the TK reporter probe is responsive to caspase-3 activation, another speed-limiting step is transsplicing. Intein-mediated cyclization of peptide has been investigated for the analysis of the protein folding of GFP and dihydrofolate reductase (DHFR) (43, 44). The DnaE intein, derived from Synechocystis sp. PCC6803, was reported to give a high yield of cyclic peptides or proteins by transsplicing (45). Therefore, in the current cTK reporter, we fused two fragments of DnaE intein to the neighboring ends of TK to generate cyclic TK proteins. This cTK reporter platform is sensitive to detect apoptosis. This merit may arise from the efficient caspase-3 cleavage and the high sensitivity of PET imaging. Moreover, we observed that even though the early stage of apoptosis can activate cTK function on FHBG phosphorylation in cells, the late stage of apoptosis presents favorable FHBG uptake due to extensive caspase-3 activity. Although with great sensitivity, the absolute uptake is much lower than wild-type TK, indicating a more efficient transsplicing strategy may be needed for further investigation.
Taken together, the development and comprehensive validation of our engineered reporter system allows quantitative detection of apoptosis from cultured cells to in vivo tumor tissues. The application of this particular caspase reporter system would significantly assist in preclinical high-throughput screening of different apoptosis-inducing agents and monitoring the therapeutic efficacy noninvasively and quantitatively by PET imaging. Further application in clinical cell therapy may also be possible, for instance, when using this reporter to track the apoptotic cells after cell implantation.
Materials and Methods
Plasmids Construction and Reagents.
To construct three variants of cTK with three different split sites, corresponding N-terminal (TKn) and C-terminal (TKc) fragments of the truncated HSV1-TK gene were amplified by PCR using the pCMV–HSV1-TK as the template. For convenient cloning, the primers for the C-terminal fragment of DnaE (DnaEc) were introduced with a start codon and the restriction enzyme sites, Hind III and BamH I. The TKc was modified to connect with the TKn by introducing a PstI site, and BamH I and NotI sites were introduced at the 5′ and 3′ ends of the fragments, respectively. The N-terminal fragment of DnaE (DnaEn) was modified by PCR to introduce a PEST sequence and the enzyme sites, NotI and XhoI. All these fragments were connected and subcloned into an expression vector pcDNA 3.1/Zeo (+) (Invitrogen) at the unique enzyme sites of Hind III and XhoI. All cassettes were driven by a CMV promoter. All of the final plasmids were sequenced to ensure fidelity in sequence (MacroGen). The cDNA of DnaE and PEST were a gift from Takeaki Ozawa (University of Tokyo, Tokyo, Japan). The caspase-3 expression plasmid pCaspase-3 was purchased from Addgene. The siRNA against caspase-3 and control scramble siRNA were purchased from Cell Signaling Inc. Goat anti–HSV1-TK antibody was obtained from Santa Cruz Biotechnology. All other antibodies used in this study were purchased from Cell Signaling Inc. Corresponding secondary antibodies were obtained from Jackson ImmunoResearch Inc.
Cell Culture and Stable Cell Line Establishment.
The human head and neck squamous carcinoma UM-SCC-22B cells were cultured in DMEM supplemented with 10% (vol/vol) FBS, 100 U/mL penicillin, and 100 mg/mL streptomycin (Invitrogen). To establish the cell lines that stably express cTK266, the plasmid cTK266 was transfected into UM-SCC-22B cells with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Twenty-four hours later, the cells were selected with 0.5 mg/mL zeocin antibiotic (Sigma-Aldrich) every 2–3 d. Two weeks later, the single clones were picked up in 24-well plates and allowed to grow. When reaching 80% confluence, the cells were treated with Dox and subjected to 18F-FHBG cell uptake assay. The highest expressing clone, named cTK266-22B, was identified and used for further study. The cells were also transfected and selected with plasmids expressing firefly luciferase (Fluc) with 1 mg/mL of G418 (Sigma-Aldrich) as screening antibiotics. The final selected cells were named cTK266-22B-Fluc. No significant difference between the transfected and the parental cells was observed in terms of proliferation or tumorigenicity.
Western Blot Analysis.
cTK266-22B-Fluc cells were treated with different concentrations of Dox to induce apoptosis. Twenty-four hours later, the cells were lysed in a lysis buffer [20 mM Hepes, 20% (vol/vol) glycerol, 500 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 1 mM DTT, 0.1% Triton X-100] and a protease inhibitor mixture (1:100, Invitrogen) at 4 °C for 20 min. An equal amount of protein (300 μg) was loaded and resolved on 4–12% SDS/PAGE gels and electroblotted onto a nitrocellulose membrane (0.45 μm, Invitrogen). A goat polyclonal anti–HSV1-TK antibody (1:100) was used as the primary antibody to detect HSV1-TK; the immunoreactivity was evaluated using an HRP-conjugated donkey anti-goat IgG antibody (1:2,500). The HRP activities were then visualized by using an ECL advanced Western blotting detection kit (Thermo Scientific) followed by exposure to ECL X-ray film (Amersham Biosciences).
Caspase-3 Activity Assay.
The activity of caspase-3 was determined by a caspase colorimetric assay kit (Sigma-Aldrich) according to the manufacturer’s protocol. Briefly, the cells treated with different doses of Dox were collected, washed with PBS, and lysed in a lysis buffer. Subsequently, the caspase-3 substrate (Ac-DEVD-pNA) was characterized with a spectrophotometer (Synergy II, BioTek) at a wavelength of 405 nm, and the pNA standard was used for calibration of caspase-3 activity in samples.
18F-FHBG Cell Uptake Studies.
After Dox treatment, uptake of 18F-FHBG was assessed in transiently transfected 22B cells or cTK266-22B-Fluc cells. In the case of IDN6556 pan-caspase inhibitor, 20 µM IDN6556 was used in combination with Dox for cell treatment. The cells were incubated with 5 μCi/mL of 18F-FHBG at 37 °C for 2 h, and then the cells were washed twice with PBS (pH 7.2). Then the cells in each well were harvested with 0.1 N NaOH and the cell-associated radioactivity was determined with a γ-counter (1480 Wizard 3, Perkin–Elmer). Triplicate samples were measured for all uptake studies. A parallel set of plates was used to determine relative bioluminescence signal and total protein content.
Animal Model.
All animal experiments were approved by the Institutional Animal Care and Use Committee of Clinical Center at the National Institutes of Health. Six-week-old female athymic mice were obtained from Harlan Laboratories and maintained on a standard diet at room temperature. Xenografted tumor models were prepared by s.c. injection of 5 × 106 cTK266-22B-Fluc cells suspended in 100 μL PBS into nude mice. Tumor size was monitored with a digital caliper, and tumor volume was calculated as ab2/2, where a represents the longest diameter and b represents the shortest diameter. When tumors reached a diameter of about a = 6 mm, mice were randomized into two groups (n = 6 per group). One group of mice received one dose of 10 mg/kg of Dox through i.v. administration, whereas the control group of mice received one dose of PBS only. Doses were selected to mimic high-dose strategies used in clinical chemotherapy regimens. Animal status was monitored closely throughout the experiment.
In Vivo BLI.
Before imaging, mice were anesthetized with a 2% (vol/vol) isoflurane–air mixture and given a single i.p. injection of 150 mg/kg d-luciferin in normal saline. BLI was accomplished using a Xenogen Lumina II system (Caliper Life Sciences) between 10 and 20 min postluciferin administration. During image acquisition, isoflurane anesthesia was maintained using a nose delivery system, and animal body temperature was maintained using a temperature-controlled bed. Signal intensity was quantified as the sum of all detected photon counts within a region of interest (ROI) over the tumor site. Images were analyzed with Living Image v3.1 software (Caliper Life Sciences). Bioluminescence signal was recorded as average radiance with the unit of photons/s/cm2/sr.
Micro-PET Imaging in Living Mice.
After BLI, the same group of mice (n = 6), under isoflurane anesthesia, were injected via a tail vein with 18F-FHBG (300 μCi per mouse). Ten-minute static scans were acquired at 1 and 2 h after injection with a dedicated small animal Inveon PET scanner (Preclinical Solution, Siemens). The images were reconstructed using a 2D ordered-subset expectation maximization algorithm without correction for attenuation or scattering. For each scan, ROIs were drawn over the tumor region using the vendor’s software (ASI Pro-5.2.4.0) on decay-corrected whole-body coronal images. The radioactivity concentrations (accumulation) within the tumors were obtained from mean pixel values within the multiple ROI volume and then converted to megabecquerel per milliliter per minute using the calibration factor determined for the Inveon PET system. These values were then divided by the administered activity to obtain (assuming a tissue density of 1 g/mL) an image-ROI–derived percent injected dose per gram (%ID/g).
Statistical Analysis.
Quantitative data were expressed as mean ± SD. Means were compared using Student’s unpaired test. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported by the National Basic Research and Development Program of China (973 Program) (2011CB707702, 2013CB733802, and 2014CB744503), National Natural Science Foundation of China (81301214, 81090272, 81227901, and 81371596), and the Intramural Research Program, National Institute of Biomedical Imaging and Bioengineering, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321374111/-/DCSupplemental.
References
- 1.Cotter TG. Apoptosis and cancer: The genesis of a research field. Nat Rev Cancer. 2009;9(7):501–507. doi: 10.1038/nrc2663. [DOI] [PubMed] [Google Scholar]
- 2.Danial NN, Korsmeyer SJ. Cell death: Critical control points. Cell. 2004;116(2):205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 3.Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267(5203):1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 4.Rupnow BA, Knox SJ. The role of radiation-induced apoptosis as a determinant of tumor responses to radiation therapy. Apoptosis. 1999;4(2):115–143. doi: 10.1023/a:1009675028784. [DOI] [PubMed] [Google Scholar]
- 5.Dive C, Evans CA, Whetton AD. Induction of apoptosis—New targets for cancer chemotherapy. Semin Cancer Biol. 1992;3(6):417–427. [PubMed] [Google Scholar]
- 6.Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411(6835):342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- 7.Therasse P, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92(3):205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 8.Green AM, Steinmetz ND. Monitoring apoptosis in real time. Cancer J. 2002;8(2):82–92. doi: 10.1097/00130404-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 9.Blankenberg FG, Strauss HW. Recent advances in the molecular imaging of programmed cell death: Part II—Non-probe-based MRI, ultrasound, and optical clinical imaging techniques. J Nucl Med. 2013;54(1):1–4. doi: 10.2967/jnumed.112.111740. [DOI] [PubMed] [Google Scholar]
- 10.Blankenberg FG. In vivo detection of apoptosis. J Nucl Med. 2008;49(Suppl 2):81S–95S. doi: 10.2967/jnumed.107.045898. [DOI] [PubMed] [Google Scholar]
- 11.Blankenberg FG. Imaging the molecular signatures of apoptosis and injury with radiolabeled annexin V. Proc Am Thorac Soc. 2009;6(5):469–476. doi: 10.1513/pats.200901-001AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blankenberg FG, et al. In vivo detection and imaging of phosphatidylserine expression during programmed cell death. Proc Natl Acad Sci USA. 1998;95(11):6349–6354. doi: 10.1073/pnas.95.11.6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krysko O, De Ridder L, Cornelissen M. Phosphatidylserine exposure during early primary necrosis (oncosis) in JB6 cells as evidenced by immunogold labeling technique. Apoptosis. 2004;9(4):495–500. doi: 10.1023/B:APPT.0000031452.75162.75. [DOI] [PubMed] [Google Scholar]
- 14.Riedl SJ, Shi Y. Molecular mechanisms of caspase regulation during apoptosis. Nat Rev Mol Cell Biol. 2004;5(11):897–907. doi: 10.1038/nrm1496. [DOI] [PubMed] [Google Scholar]
- 15.Shi Y. Mechanisms of caspase activation and inhibition during apoptosis. Mol Cell. 2002;9(3):459–470. doi: 10.1016/s1097-2765(02)00482-3. [DOI] [PubMed] [Google Scholar]
- 16.Green DR. Apoptotic pathways: The roads to ruin. Cell. 1998;94(6):695–698. doi: 10.1016/s0092-8674(00)81728-6. [DOI] [PubMed] [Google Scholar]
- 17.Edgington LE, et al. Noninvasive optical imaging of apoptosis by caspase-targeted activity-based probes. Nat Med. 2009;15(8):967–973. doi: 10.1038/nm.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Saint-Hubert M, Prinsen K, Mortelmans L, Verbruggen A, Mottaghy FM. Molecular imaging of cell death. Methods. 2009;48(2):178–187. doi: 10.1016/j.ymeth.2009.03.022. [DOI] [PubMed] [Google Scholar]
- 19.Bullok KE, et al. Biochemical and in vivo characterization of a small, membrane-permeant, caspase-activatable far-red fluorescent peptide for imaging apoptosis. Biochemistry. 2007;46(13):4055–4065. doi: 10.1021/bi061959n. [DOI] [PubMed] [Google Scholar]
- 20.Kim K, et al. Cell-permeable and biocompatible polymeric nanoparticles for apoptosis imaging. J Am Chem Soc. 2006;128(11):3490–3491. doi: 10.1021/ja057712f. [DOI] [PubMed] [Google Scholar]
- 21.Quinti L, Weissleder R, Tung CH. A fluorescent nanosensor for apoptotic cells. Nano Lett. 2006;6(3):488–490. doi: 10.1021/nl0524694. [DOI] [PubMed] [Google Scholar]
- 22.Barnett EM, Zhang X, Maxwell D, Chang Q, Piwnica-Worms D. Single-cell imaging of retinal ganglion cell apoptosis with a cell-penetrating, activatable peptide probe in an in vivo glaucoma model. Proc Natl Acad Sci USA. 2009;106(23):9391–9396. doi: 10.1073/pnas.0812884106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laxman B, et al. Noninvasive real-time imaging of apoptosis. Proc Natl Acad Sci USA. 2002;99(26):16551–16555. doi: 10.1073/pnas.252644499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galbán S, et al. Imaging proteolytic activity in live cells and animal models. PLoS ONE. 2013;8(6):e66248. doi: 10.1371/journal.pone.0066248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coppola JM, Ross BD, Rehemtulla A. Noninvasive imaging of apoptosis and its application in cancer therapeutics. Clin Cancer Res. 2008;14(8):2492–2501. doi: 10.1158/1078-0432.CCR-07-0782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanno A, Yamanaka Y, Hirano H, Umezawa Y, Ozawa T. Cyclic luciferase for real-time sensing of caspase-3 activities in living mammals. Angew Chem Int Ed Engl. 2007;46(40):7595–7599. doi: 10.1002/anie.200700538. [DOI] [PubMed] [Google Scholar]
- 27.Ray P, De A, Patel M, Gambhir SS. Monitoring caspase-3 activation with a multimodality imaging sensor in living subjects. Clin Cancer Res. 2008;14(18):5801–5809. doi: 10.1158/1078-0432.CCR-07-5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niers JM, Kerami M, Pike L, Lewandrowski G, Tannous BA. Multimodal in vivo imaging and blood monitoring of intrinsic and extrinsic apoptosis. Mol Ther. 2011;19(6):1090–1096. doi: 10.1038/mt.2011.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones T. The imaging science of positron emission tomography. Eur J Nucl Med. 1996;23(7):807–813. doi: 10.1007/BF00843711. [DOI] [PubMed] [Google Scholar]
- 30.Dong D, et al. Spontaneous and controllable activation of suicide gene expression driven by the stress-inducible grp78 promoter resulting in eradication of sizable human tumors. Hum Gene Ther. 2004;15(6):553–561. doi: 10.1089/104303404323142006. [DOI] [PubMed] [Google Scholar]
- 31.Tjuvajev JG, et al. Noninvasive imaging of herpes virus thymidine kinase gene transfer and expression: A potential method for monitoring clinical gene therapy. Cancer Res. 1996;56(18):4087–4095. [PubMed] [Google Scholar]
- 32.Cowsill C, et al. Central nervous system toxicity of two adenoviral vectors encoding variants of the herpes simplex virus type 1 thymidine kinase: Reduced cytotoxicity of a truncated HSV1-TK. Gene Ther. 2000;7(8):679–685. doi: 10.1038/sj.gt.3301147. [DOI] [PubMed] [Google Scholar]
- 33.Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: The PEST hypothesis. Science. 1986;234(4774):364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- 34.Gardberg A, Shuvalova L, Monnerjahn C, Konrad M, Lavie A. Structural basis for the dual thymidine and thymidylate kinase activity of herpes thymidine kinases. Structure. 2003;11(10):1265–1277. doi: 10.1016/j.str.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 35.Wild K, Bohner T, Folkers G, Schulz GE. The structures of thymidine kinase from herpes simplex virus type 1 in complex with substrates and a substrate analogue. Protein Sci. 1997;6(10):2097–2106. doi: 10.1002/pro.5560061005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Z, Chui WK, Ho PC. Design of a multifunctional PLGA nanoparticulate drug delivery system: Evaluation of its physicochemical properties and anticancer activity to malignant cancer cells. Pharm Res. 2009;26(5):1162–1171. doi: 10.1007/s11095-009-9837-y. [DOI] [PubMed] [Google Scholar]
- 37.Wang Z, Chui WK, Ho PC. Integrin targeted drug and gene delivery. Expert Opin Drug Deliv. 2010;7(2):159–171. doi: 10.1517/17425240903468696. [DOI] [PubMed] [Google Scholar]
- 38.Jänicke RU, Sprengart ML, Wati MR, Porter AG. Caspase-3 is required for DNA fragmentation and morphological changes associated with apoptosis. J Biol Chem. 1998;273(16):9357–9360. doi: 10.1074/jbc.273.16.9357. [DOI] [PubMed] [Google Scholar]
- 39.Niu G, Chen X. Apoptosis imaging: Beyond annexin V. J Nucl Med. 2010;51(11):1659–1662. doi: 10.2967/jnumed.110.078584. [DOI] [PubMed] [Google Scholar]
- 40.Paulmurugan R, Umezawa Y, Gambhir SS. Noninvasive imaging of protein-protein interactions in living subjects by using reporter protein complementation and reconstitution strategies. Proc Natl Acad Sci USA. 2002;99(24):15608–15613. doi: 10.1073/pnas.242594299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michnick SW, Remy I, Campbell-Valois FX, Vallée-Bélisle A, Pelletier JN. Detection of protein-protein interactions by protein fragment complementation strategies. Methods Enzymol. 2000;328:208–230. doi: 10.1016/s0076-6879(00)28399-7. [DOI] [PubMed] [Google Scholar]
- 42.Massoud TF, Paulmurugan R, Gambhir SS. A molecularly engineered split reporter for imaging protein-protein interactions with positron emission tomography. Nat Med. 2010;16(8):921–926. doi: 10.1038/nm.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scott CP, Abel-Santos E, Wall M, Wahnon DC, Benkovic SJ. Production of cyclic peptides and proteins in vivo. Proc Natl Acad Sci USA. 1999;96(24):13638–13643. doi: 10.1073/pnas.96.24.13638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iwai H, Lingel A, Pluckthun A. Cyclic green fluorescent protein produced in vivo using an artificially split PI-PfuI intein from Pyrococcus furiosus. J Biol Chem. 2001;276(19):16548–16554. doi: 10.1074/jbc.M011639200. [DOI] [PubMed] [Google Scholar]
- 45.Wu H, Hu Z, Liu XQ. Protein trans-splicing by a split intein encoded in a split DnaE gene of Synechocystis sp. PCC6803. Proc Natl Acad Sci USA. 1998;95(16):9226–9231. doi: 10.1073/pnas.95.16.9226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.