Significance
Auxin is a critical plant hormone that regulates every aspect of plant growth and development. AUXIN RESPONSE FACTOR (ARF) transcription factors control auxin-regulated gene transcription, and their activity is regulated by AUXIN/INDOLE 3-ACETIC ACID repressor proteins. This work identifies that dimerization of the repressor with the transcription factor is insufficient to repress activity, suggesting that multimerization is the mechanism of repressing ARF transcriptional activity and further raising the possibility that multimerization in other systems may play roles in transcriptional repression.
Abstract
In plants, the AUXIN RESPONSE FACTOR (ARF) transcription factor family regulates gene expression in response to auxin. In the absence of auxin, ARF transcription factors are repressed by interaction with AUXIN/INDOLE 3-ACETIC ACID (Aux/IAA) proteins. Although the C termini of ARF and Aux/IAA proteins facilitate their homo- and heterooligomerization, the molecular basis for this interaction remained undefined. The crystal structure of the C-terminal interaction domain of Arabidopsis ARF7 reveals a Phox and Bem1p (PB1) domain that provides both positive and negative electrostatic interfaces for directional protein interaction. Mutation of interface residues in the ARF7 PB1 domain yields monomeric protein and abolishes interaction with both itself and IAA17. Expression of a stabilized Aux/IAA protein (i.e., IAA16) bearing PB1 mutations in Arabidopsis suggests a multimerization requirement for ARF protein repression, leading to a refined auxin-signaling model.
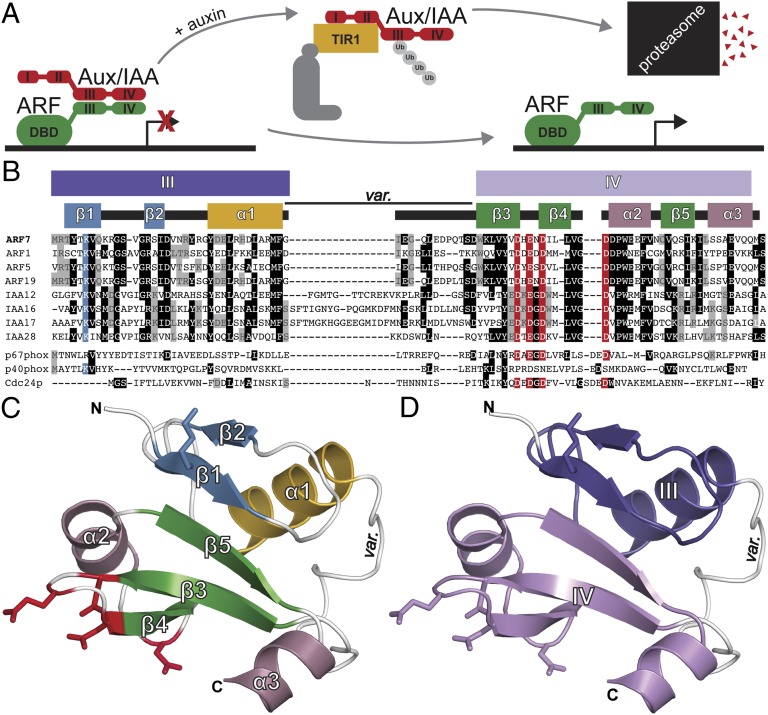
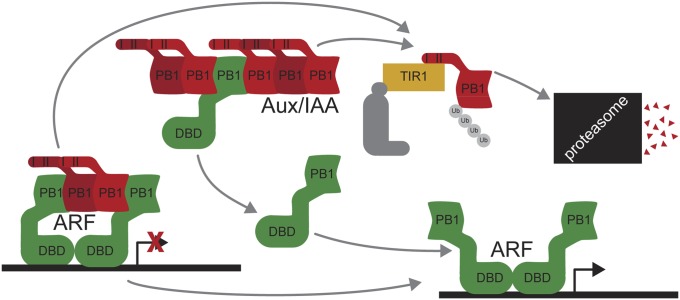
The phytohormone auxin regulates cell division and elongation to drive plant growth and development (1). Auxin perception and auxin-regulated gene expression control requires three protein families: auxin-perceiving F-box proteins, Auxin/INDOLE 3-ACETIC ACID (Aux/IAA) repressor proteins, and AUXIN RESPONSE FACTOR (ARF) transcription factors (Fig. 1A). In Arabidopsis thaliana, a repression–derepression mechanism regulates auxin signaling. Under low auxin concentrations, Aux/IAA proteins repress ARF transcription factors via direct interaction (2, 3). When auxin concentrations are high, a coreceptor complex, comprised of an F-box protein from the TRANSPORT INHIBITOR REPONSE1 (TIR1)/AUXIN SIGNALING F-BOX PROTEIN (AFB) family and an Aux/IAA protein, directly binds auxin (4–6). The F-box protein participates in a Skp1–Cullin–F-box (SCF) E3 ubiquitin ligase (5–7), which polyubiquitylates and targets the Aux/IAA protein for degradation (8). This degradation event relieves ARF transcription factor repression, thereby allowing auxin-regulated gene transcription (3).
Fig. 1.
ARF7 contains a C-terminal type I/II PB1 domain. (A) In the current auxin-signaling model, Aux/IAAs dimerize with and repress ARF transcription factors in the absence of auxin. In the presence of auxin, Aux/IAAs interact with SCFTIR1 resulting in repressor degradation, freeing ARFs for auxin-responsive gene transcription. (B) Sequence alignment of ARF and Aux/IAA proteins identify canonical PB1 domain features, including a conserved lysine (blue) and the OPCA-like motif (red) that align with type I (p67phox and Cdc24p) and type II (p40phox) PB1 domains. The ARF and Aux/IAA PB1 domain variable region (var.) is also indicated. (C) The ARF7PB1 crystal structure reveals that the ARF7 C terminus is a PB1 domain. Secondary structure features are labeled and colored as in B with modeled lysine (blue) and OPCA (red) residues shown as stick representations. (D) ARF7PB1 structural features define ARF and Aux/IAA sequence motifs III and IV, which are colored as in B.
The Arabidopsis genome encodes 29 Aux/IAA proteins that contain four conserved sequence motifs: (i) an amino-terminal repression domain, region I; (ii) an SCFTIR1/AFB recognition sequence, region II; (iii) a region containing a predicted βαα motif, region III; and (iv) an acidic region, region IV (Fig. 1A) (9, 10). Regions III and IV of Aux/IAA proteins enable interaction with other Aux/IAA (11) and ARF (12) proteins. Additionally, Arabidopsis has 22 ARF-encoding genes (2) which contain three conserved regions of homology: an amino-terminal B3-type DNA-binding domain (DBD) and two C-terminal regions that share homology with Aux/IAA protein domains III and IV; the N-terminal DBD and C-terminal III and IV domains are connected by a variable middle domain conferring either activation or repression properties (Fig. 1A) (2, 13). Auxin-responsive gene expression may be attenuated by Aux/IAA and ARF pairing (14) and ARF–ARF, ARF–Aux/IAA, and Aux/IAA–Aux/IAA interactions display specificity (15). A lack of structural data has precluded understanding ARF–ARF and ARF–Aux/IAA interactions.
ARF and Aux/IAA protein bioinformatic analysis (16) suggests that the III–IV region may form a type I/II Phox and Bem1p (PB1) protein–protein interaction domain (17–19). PB1 domains adopt a β-grasp fold (19) and may display an acidic surface (type I), a basic surface (type II), or both surfaces (type I/II) on opposite faces of the domain structure to allow for front-to-back orientation of multiple PB1 domains (17, 19). ARF and Aux/IAA putative PB1 domains contain both an invariant lysine typical of type II PB1 domains and a type I PB1 series of acidic residues (D-x-D/E-x-D-xn-D/E; known as the octicosapeptide repeat, p40phox and budding yeast Cdc24p,atypical PKC-interaction domain (OPCA) motif; Fig. 1B) (16), consistent with the possibility that ARF and Aux/IAA C termini may be type I/II PB1 domains.
Here, we provide crystallographic evidence that the C-terminal region of Arabidopsis ARF7 adopts a PB1 fold to mediate interaction with both ARF7 and IAA17 in vivo. In planta analysis demonstrates that Aux/IAA activity by dimerization is insufficient to repress auxin responses, suggesting that multimerization through the PB1 domain may be required for ARF repression. Identification of multimerization as the potential molecular basis for ARF and Aux/IAA interaction refines our understanding of the control of auxin response repression.
Results
Crystal Structure of Arabidopsis ARF7 PB1 Domain.
To investigate the structural basis of ARF and Aux/IAA interactions, we focused on Arabidopsis ARF7, a well-studied activating ARF with roles in light responses and root and hypocotyl growth (20). For crystallographic studies, a truncated version of ARF7 (ARF7PB1K1042A, opca) consisting of amino acids M1037-S1127 with K1042A, D1092A, and D1096A mutations to reduce protein aggregation was generated based on output from the Phyre2 protein-fold recognition server (21).
The ARF7PB1K1042, opca 3D structure was determined to 2.4-Å resolution using single-wavelength anomalous dispersion phasing from selenomethionine-substituted protein (Fig. 1C and Table S1). The ARF7PB1 monomer adopts a canonical PB1 fold with slight modifications (Fig. 1C). The overall structure reveals two subdomains corresponding to sequence regions III and IV, as originally noted for the ARF and Aux/IAA proteins (Fig. 1D). The N-terminal III region consists of an antiparallel β-sheet (β1–β2) and α1. The position of the invariant lysine (K1042) is on the surface-exposed face of β1. The C-terminal IV region contains a second antiparallel β-sheet (β3–β5) and two α-helices (α2 and α3). The cluster of acidic residues forming the OPCA-like motif is located on the loops flanking β4. The two halves are joined by the α1–β3 loop, which is variable in length and sequence across ARF and Aux/IAA proteins (Fig. 1B and Fig. S1). This PB1 scaffold places the lysine and OPCA motif residues on opposite faces of the structure.
The secondary structural elements of ARF7PB1 adopt a β-grasp fold. A Dali database search of the Protein Data Bank (22) identified >300 similar structures (Z-score > 5), including other PB1 domains, PB1 domain-containing proteins, and ubiquitin moieties. ARF7PB1 contains more disordered regions than the PB1 domains of protein kinase C iota and partitioning defective-6 homolog alpha (PDB: 1WMH) (23), the structures that share highest structural homology to ARF7PB1. These data confirm the presence of a modified PB1 domain in the C terminus of ARF7.
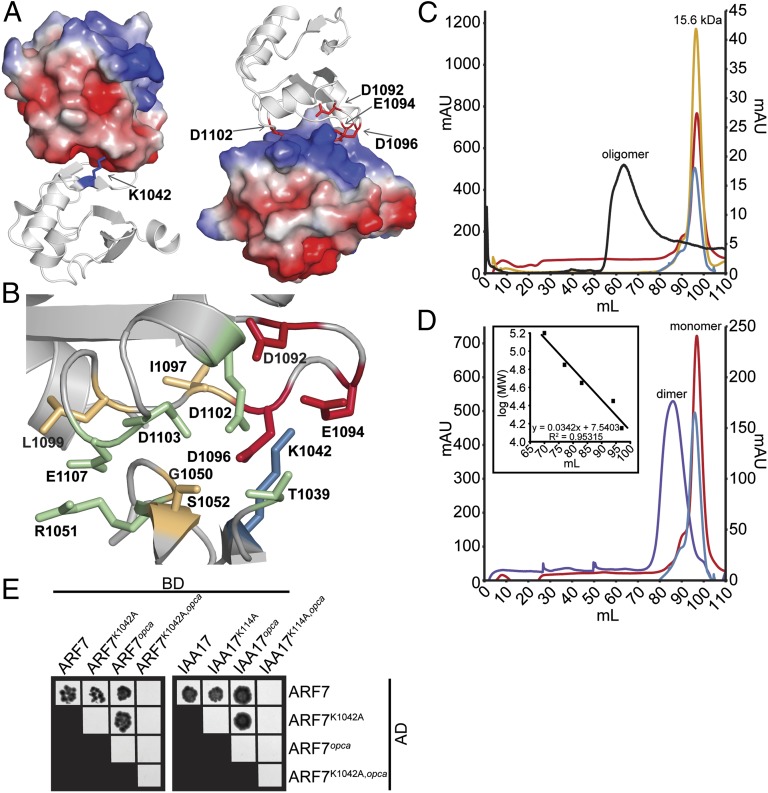
The electrostatic surface potential of monomeric ARF7PB1 with K1042A, D1092A, and D1096A remutated to wild-type residues in silico reveals distinct charged patches on opposite faces of the protein (Fig. 2A). These basic and acidic regions contain the invariant lysine and OPCA motif, respectively. The ARF7PB1 domain crystallized with 16 molecules in the asymmetric unit and packing of monomers suggests how PB1 domains may orient to facilitate protein–protein interaction. Charge complementarity between these PB1 domain faces contributed to an in-crystal protein–protein interaction interface that orients two molecules in a front-to-back fashion. Protein–protein interface analysis between chains A and P using Proteins, Interfaces, Structures and Assemblies (PISA) database (24) reveals a 497-Å2 interaction surface that encompasses the area surrounding the modeled side-chains of K1042 (invariant Lys) and D1092, E1094, D1096, and D1102 (OPCA motif) (Fig. 2B). Of the 27 predicted interface residues, nine—five on chain A (I1097, L1099, D1102, D1103, and E1107) and four on chain P (T1039, G1050, R1051, and S1052)—participate in charge–charge interactions and/or hydrogen bonding. Whereas some of the residues involved in this dimer interface—e.g., R1051 and L1099—are highly conserved in all ARF and Aux/IAA proteins, others—e.g., T1039, G1050, S1052, and I1097—vary considerably across the ARF and Aux/IAA proteins of Arabidopsis (Fig. S1). Interestingly, residues conserved in ARF proteins, but not in most Aux/IAA proteins—e.g., D1103 and E1107—may lend clues toward understanding protein–protein interaction affinities among ARF and Aux/IAA family members.
Fig. 2.
ARF and Aux/IAA PB1 domain interactions are driven by charge–charge interactions. (A) The modeled ARF7PB1 electrostatic surface potential reveals positive (blue) and negative (red) interaction interface, containing the invariant lysine and OPCA motif, respectively. (B) PISA predictions of residue contributions to interactions identified the ARF7PB1 interaction interface containing the invariant lysine (blue), OPCA motif (red), and residues important for hydrogen bonding (orange) and charge–charge interactions (green). (C) Size-exclusion chromatography reveals that wild-type ARF7Cterm exists as soluble aggregate in solution (black), whereas mutations in the invariant lysine (blue), the OPCA motif (red), or both (yellow) afford monomeric protein. (D) Size-exclusion chromatography reveals that mixing ARF7CtermK1042A (blue) and ARF7Ctermopca (red), which exist as monomeric protein in solution, results in the formation of a dimer (ARF7CtermK1042A and ARF7Ctermopca mixed in a 1:1 ratio, purple). Inset, protein molecular weight standards. (E) Yeast two-hybrid assays reveal that introduction of single unlike PB1 mutations in ARF7 and IAA17 does not affect ARF–ARF or ARF–Aux/IAA interactions. Combining both PB1 mutations (lysine and opca) or like mutations—e.g., both proteins contain lysine or OPCA mutations—abrogates protein–protein interactions. BD, binding domain; AD, activation domain.
PB1 Mutations Disrupt ARF7 Protein Interactions.
Mutation of either the invariant lysine or the aspartates in the OPCA motif abolish PB1 protein–protein interactions (19). We reasoned that if ARF7 self-interacts through canonical PB1 domain interactions, mutation of these residues would ameliorate multimerization. ARF7 K1042 corresponds to the PB1 invariant lysine, and D1092, E1094, D1096, and D1102 form the PB1 OPCA-like motif (Fig. 1B). We examined the in-solution oligomerization of wild-type ARF7PB1, ARF7PB1K1042A, ARF7PB1opca (ARF7PB1D1092A, D1095A), and ARF7PB1K1042A, opca using size-exclusion chromatography. Wild-type ARF7PB1 formed high molecular weight aggregates; however, the ARF7PB1K1042A, ARF7PB1opca, and ARF7PB1K1042A, opca variants migrated as monomeric proteins (Fig. 2C). This result indicates that mutation of key interaction residues on either one or both faces of the ARF7 PB1 domain disrupts oligomerization. We further found that mixing ARF7PB1K1042A and ARF7PB1opca, each of which retain a single compatible electrostatic face, resulted in formation of a dimer (Fig. 2D), suggesting that ARF7 PB1 domain electrostatic potential drives these interactions.
To examine in vivo interaction roles for ARF7 K1042A and the aspartates in the OPCA motif, we performed yeast two-hybrid assays using wild-type and mutant versions of ARF7Cterm and an Aux/IAA interaction partner, IAA17 (Fig. 2E and Fig. S2). Expression of the ARF7Cterm and IAA17 in yeast was confirmed by immunoblot analysis (Fig. S2A). As previously described (15), we found that ARF7Cterm interacted with itself and IAA17 (Fig. S2B). Additionally, ARF7Cterm interacted with ARF7CtermK1042A, ARF7Ctermopca, IAA17K114A, and IAA17opca (Fig. 2E), suggesting that a single functional electrostatic interaction domain is sufficient for protein–protein interaction. However, ARF7Cterm failed to interact with ARF7CtermK1042A, opca or IAA17K114A, opca, indicating that disruption of both electrostatic faces abolishes protein interaction. Intriguingly, like mutant proteins—e.g., Lys mutant with Lys mutant and opca mutant with opca mutant—failed to interact, consistent with the in vitro ARF7Cterm size-exclusion data (Fig. 2C) and suggesting that the presence of only a positive or negative face is insufficient to drive protein–protein interaction in vivo. Additionally, IAA17K114A, opca failed to interact with ARF7Cterm (Fig. 2E), suggesting IAA17 also contains a functional PB1 domain.
Multimerization of ARF7 PB1.
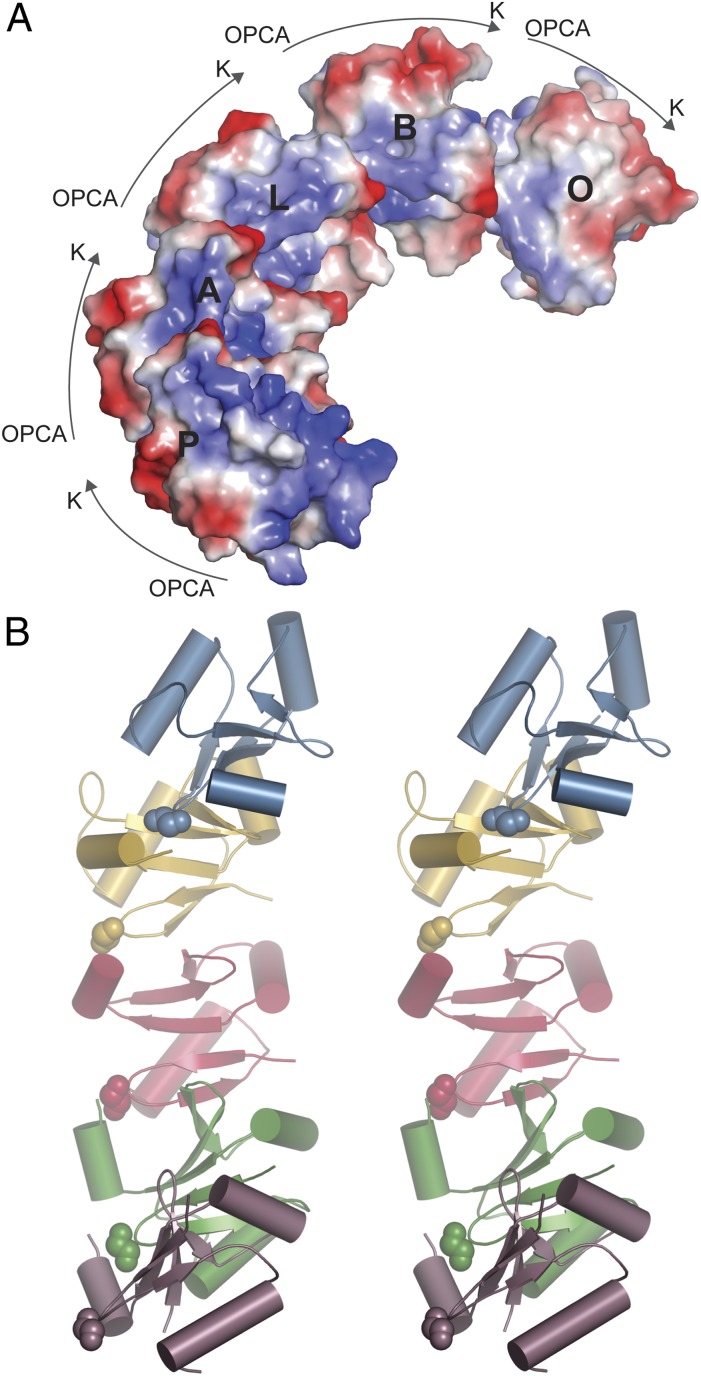
The ARF7PB1 crystal structure contained 16 noncrystallographic symmetry-related molecules. Each chain, which is a complete ARF7PB1 domain, is labeled A to P. Although ARF7PB1K1042A, opca is monomeric in solution, chains A, B, L, O, and P packed to form an extended directional ARF7PB1 pentamer within the crystal (Fig. 3). In this arrangement, the invariant ARF7PB1 lysine orients toward the OPCA motif of the next PB1 domain, as described above for the A–P chain interaction, to form a curved helix topology. Overall, each protein–protein interface in the ARF7PB1 pentamer retains a similar overall orientation and set of interactions observed with the association of PB1 domains from other proteins (17, 19). This pentameric arrangement of ARF7PB1 molecules presents negative charge toward the outer side of the curve and positive charge along the inner face and places the PB1 domain N-terminal side staggered along the curved multimer outer face. In contrast to the original model of dimeric homo- and heterooligomerization of ARF and Aux/IAA proteins, this observation suggests the possible formation of higher-order ARF and Aux/IAA type I/II PB1 domain multimers, which may have a functional role in modulating auxin responses.
Fig. 3.
ARF7 PB1 domains formed an extended directional pentamer within the crystal. (A) The ARF7PB1 crystal contains an oriented ARF7PB1 domain pentamer of chains A, B, L, O, and P. (B) Stereo view of ARF7PB1 pentamer topology suggests formation of a curved helix multimer. The N-terminal residue of each chain is shown as a space-filling model.
Aux/IAA PB1 Domain Mutation Effects on ARF Repression in Planta.
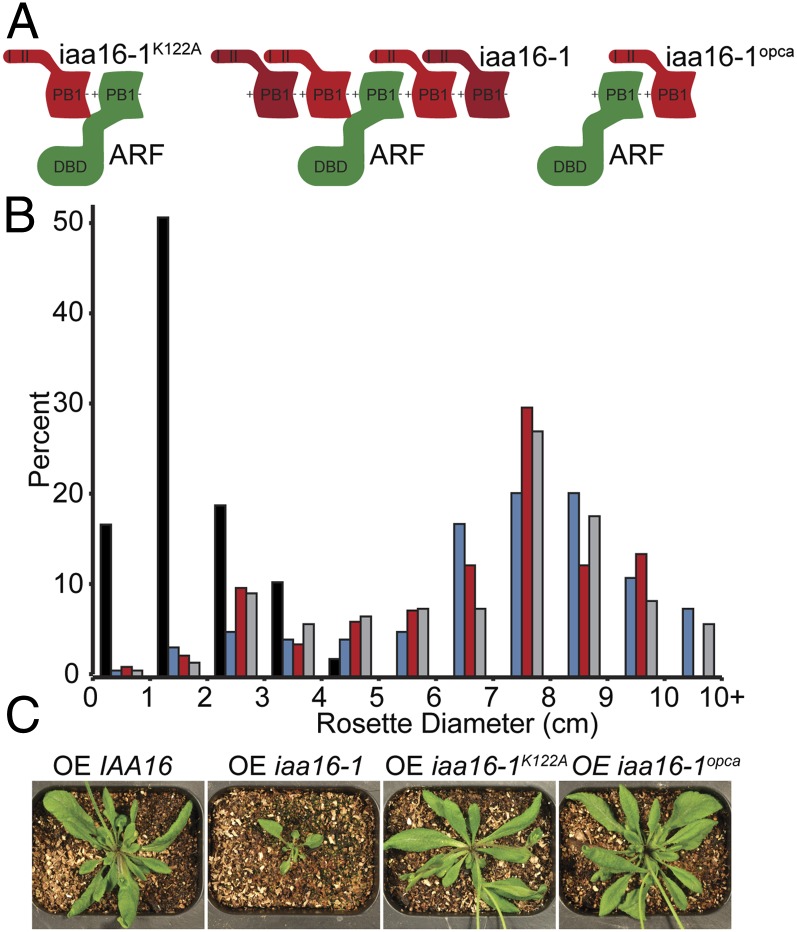
The current auxin-signaling model suggests that Aux/IAA protein dimerization with ARF proteins drives transcription factor repression (Fig. 1A). However, the structure of ARF7PB1 (Figs. 1B and 2A) and the in vitro and in vivo interaction data (Figs. 2 C–E and 3) suggest the possibility that ARF and Aux/IAA proteins multimerize. To understand the potential role of Aux/IAA protein multimerization in regulating auxin response, we combined an Aux/IAA protein gain-of-function mutation, which results in repressor hyperaccumulation and constitutive ARF repression (10), with PB1 domain mutations in planta. As previously described (25), overexpression of the iaa16-1 gain-of-function mutation in wild-type Arabidopsis resulted in stunted growth, decreased rosette diameter, infertility, and lack of apical dominance. However, overexpressing iaa16-1K122A or iaa16-1opca resulted in wild-type phenotypes (Fig. 4). Because iaa16-1K122A and iaa16-1opca each retain one of the two PB1 interaction interfaces and because only a single electrostatic face is necessary for dimerization (Fig. 2D), these proteins are capable of interaction with a single face of a target ARF. Loss of only a single-interaction interface abolished iaa16-1 repressive activity, consistent with the possibility that IAA16 multimerization, not dimerization, with target ARF proteins is necessary for repressor activity.
Fig. 4.
Aux/IAA PB1 domain dimerization is insufficient for ARF repression in Arabidopsis. (A) Models representing stabilized iaa16-1 with a single-interaction interface. (B) Histograms and (C) representative photographs of rosette diameters from wild-type plants overexpressing IAA16 (gray; n = 117), iaa16-1 (black; n = 47), iaa16-1K122A (blue; n = 117), or iaa16-1opca (red; n = 80) reveal that mutation of the PB1 invariant lysine or OPCA motif abrogates iaa16-1 low-auxin phenotypes.
Discussion
Over the past 15 y, identification both of TIR1/AFB family as auxin receptors (5, 6) and their “molecular glue” mechanism of action (4) filled in key molecular details of auxin perception. However, molecular understanding of ARF and Aux/IAA auxin-signaling components has been hampered by complexity and redundancy of these protein families in planta and solubility, expression, and protein behavior issues in vitro. Recent bioinformatic-based predictions (16) have facilitated ARF and Aux/IAA protein molecular analysis by allowing us to make mutations abrogating ARF and Aux/IAA protein aggregation. Crystallographic studies reveal that the ARF7 C terminus folds into a PB1 domain that defines the III/IV sequence motif of ARF and Aux/IAA proteins responsible for protein–protein interaction (Figs. 1 and 2). Based on sequence homology, we hypothesize that additional ARF proteins also contain PB1 domains (Fig. S1). Further, sequence analysis and in vivo and in planta data reveal that Aux/IAA proteins also likely contain PB1 domains necessary for auxin signal regulation (Fig. 1B and Fig. S1).
The traditional auxin-signaling model depicts a single Aux/IAA protein interacting with a single ARF protein in the absence of auxin (Fig. 1A). Auxin promotes degradation of Aux/IAA repressors releasing ARF proteins, either as monomers or dimers, to promote gene transcription (2). Interaction of PB1 domains requires both acidic and basic surfaces on opposite faces of the domain structure to allow for front-to-back orientation of multiple PB1 domains (17, 19). Here, our data suggest Aux/IAA repression of ARF proteins may require protein multimerization. Wild-type ARF7Cterm forms a high-molecular-weight species in solution, but mutation of the invariant lysine, the OPCA motif, or both results in a stable monomeric form (Fig. 2C). Moreover, mixing of ARF7PB1K1042A and ARF7PB1opca, which retain charge-complementarity, results in dimer formation (Fig. 2D). Additionally, yeast two-hybrid assays (Fig. 2E) indicate that both an acidic face and a basic face of the PB1 domain are needed for ARF7 PB1 domain interaction. A plausible model for formation ARF7PB1 domain interaction involves positioning of the invariant lysine and OPCA motif to yield a front-to-back orientation of domains. Although the ARF7PB1 crystal structure contains 16 molecules in the asymmetric unit with a variety of packing orientations, it is intriguing that five molecules associate with the predicted front-to-back orientation (Fig. 3).
Similarly, overexpression in Arabidopsis of a stabilized Aux/IAA protein capable of interaction with a single face of a target ARF fails to result in low-auxin phenotypes (Fig. 4), suggesting that dimerization at either face of the ARF protein is insufficient to repress activity. In addition, a recent study identified an intragenic domain IV mutation suppressor of the rice gain-of-function Osiaa23 mutant (26). This Osiaa23-R5 point mutation, predicted to be oriented on the acidic face of the PB1 domain of the stabilized Osiaa23 Aux/IAA protein, resulted in phenotypes consistent with restored auxin responsiveness (26). This study with a rice Aux/IAA protein combined with our Arabidopsis IAA16 data (Fig. 4) suggests Aux/IAA protein dimerization with ARF proteins is insufficient to inhibit ARF activity across multiple Aux/IAA proteins and in diverse species.
These in planta results lead to two major possible conclusions: (i) ARF protein repression depends upon Aux/IAA protein oligomerization and (ii) ARF protein repression requires a minimum number of Aux/IAA proteins. We speculate that generating larger homogeneous or heterogeneous Aux/IAA protein repression chains may ensure auxin signal specificity and fidelity (Fig. 5)—an idea supported by the assorted interactions between ARF and Aux/IAA proteins (15). Further, because ARF C termini are unnecessary for ARF transcriptional activity (27), our results provide a specific function for ARF and Aux/IAA PB1 domains.
Fig. 5.
A refined auxin-signaling model. Higher-order oligomerization of Aux/IAA proteins may be required for ARF repression. In the presence of auxin, Aux/IAA proteins are targeted for degradation by the SCFTIR1 complex, freeing ARF proteins for auxin-responsive gene transcription. ARF–ARF interactions can occur through the DBD (28) and/or through the PB1 domain.
Analyzing alterations within the PB1 domain between different ARF and Aux/IAA isoforms may provide insight into dissecting the complex web of auxin signaling. Examining variance within the interface interaction residues and the inserts and modifications within the variable α1–β3 loop could enable interaction partner predictions when coupled with developmental and tissue-specific expression data. In addition, recent crystallographic analysis of the N-terminal B3 DNA-binding domains of Arabidopsis ARF1 and ARF5 reveals how the spacing of these domains acts as molecular calipers for interaction with auxin response elements, which provides insight on ARF transcription factor preferences for target sites (28). Intriguingly, ARF dimerize at their DNA-binding domain (28) and interact with other ARF or Aux/IAA proteins via their PB1 domain (domains III and IV) with specificity (15).
In summary, ARF and Aux/IAA protein PB1 domains are necessary for auxin signal attenuation. In the future, analysis of the ARF and Aux/IAA protein PB1 interface may provide insight into the in planta specificities of ARF and Aux/IAA protein complexes. These findings provide an important step toward further dissecting the complex auxin-signaling web.
Materials and Methods
Vector Construction.
For all vectors, ARF7 C-terminal truncations, IAA16, and IAA17 were PCR-amplified from cDNA and cloned into pENTR/d-TOPO (Life Technologies). Site-directed mutagenesis was performed using the QuikChange Lightning Multi Site-Directed Mutagenesis Kit (Agilent). Targeted mutations were generated based on previously described mutations: iaa16-1 (25) and PB1 mutations (reviewed in ref. 19).
All other constructs were subcloned from the pENTR/d-TOPO constructs. For protein expression, truncations of ARF7 (Cterm or PB1) were cloned into pET-28a (Invitrogen) using NdeI and XhoI restriction sites. For yeast two-hybrid assays, ARF7Cterm and IAA17 were cloned into the pBI-770 “bait” vector, which contains the GAL4 DNA-binding domain, and the pBI-771 “prey” vector, which contains the GAL4 activation domain (29) using SalI and NotI restriction sites. For expression in plants, IAA16 was cloned into pEarleyGate100 (30) using Gateway LR Clonase II (Life Technologies) to create plant expression constructs of genes driven behind the strong 35S promoter.
Yeast Two-Hybrid Assays.
Plasmids were transformed (31) into the Saccharomyces cerevisiae strain YPB2 (MATa ura3-52 his3-200 ade2-101 lys2-801 trp1-901 leu2-3,112 canR gal4-542 Gal80-538 LYS2::GALIUAS-LEU2TATA-HIS3 URA3::GAL417mers(×3)-CyClTATA-lacZ) (32) with the pBI770 or pBI771 yeast two-hybrid constructs containing either ARF7Cterm or IAA17 tethered to the DNA-binding domain (DBD) or activation domain (AD) of GAL4. Transformants were selected on synthetic complete (SC) growth media lacking leucine and tryptophan (SC Leu− Trp−). Individual colonies from the initial transformation were streaked onto SC Leu− Trp− plates for secondary selection. Individual colonies from these plates were resuspended in 60 µL dH2O in a 96-well plate and plated onto either SC, Leu−, Trp− plates or SC plates lacking leucine, tryptophan, and histidine supplemented with 3-amino-1,4,5-triazole (3-AT) using an inoculation frogger. Plates were incubated at 30 °C and photographed after 5 d of growth.
Protein Expression and Purification.
ARF7Cterm and ARF7 PB1 were expressed in Escherichia coli (DE3) Rosetta (Invitrogen) as N-terminal His-tagged proteins. Bacterial cultures were grown at 37 °C to an A600nm = ∼0.5. Protein expression was induced with a final concentration of 1 mM isopropyl β-D-1-thiogalactopyranoside, then grown for 18 h at 18 °C. Bacterial cells were pelleted and resuspended in lysis buffer [50 mM Tris pH 8.0, 20 mM imidazole, 500 mM NaCl, 10% (vol/vol) glycerol, 1% Tween-20]. Resuspended cells were lysed by sonication, and cell debris was pelleted by centrifugation. The soluble cell lysate was passed over a Ni2+-nitrilotriacetic acid (NTA) chromatography column. The column was washed with wash buffer [50 mM Tris pH 8.0, 20 mM imidazole, 500 mM NaCl, 10% (vol/vol) glycerol] and bound protein eluted with elution buffer [50 mM Tris pH 8.0, 250 mM imidazole, 500 mM NaCl, 10% (vol/vol) glycerol]. The His tag was then removed using overnight thrombin digest (1 U thrombin per milligram of protein; 4 °C) in dialysis against wash buffer. Thrombin was removed using a benzamidine–Sepharose column, and protein lacking the His tag was collected as the flow-through of a second Ni2+–NTA column. Size-exclusion chromatography was performed using either a HiLoad Superdex-75 or -200 prep grade FPLC column (GE Healthcare) equilibrated in 25 mM Tris pH 9.0, 100 mM NaCl buffer. All protein quantification was carried out using Bradford protein assay reagents (BioRad) with BSA as a standard. SeMet-substituted protein was generated by growth in M9 minimal media supplemented with SeMet (33).
Protein Crystallography.
Native ARF7PB1K1042A, opca crystals were obtained in 25% (wt/vol) PEG-6000, 0.1 M Pipes, pH 6.5 using the hanging-drop vapor diffusion method at 4 °C in 2-µL drops containing a 1:1 ratio of protein (8–10 mg/mL):crystallization condition. Using a similar crystallization approach, SeMet-substituted ARF7PB1K1042A, opca crystals were obtained in 20% (vol/vol) PEG-3350, 0.1 M Pipes, pH 6.5. Crystals were stabilized in cryoprotectant [crystallization buffer supplemented with 25–30% (vol/vol) glycerol] for data collection at 100 K. All diffractions were collected at beamline 19ID of the Argonne National Lab Advanced Photon Source. The images were indexed, integrated, and scaled using HKL-3000 (Table S1) (34). For single-wavelength anomalous diffraction phasing, selenium sites were calculated using HySS (35) with density modification performed in RESOLVE (36) integrated into the PHENIX AutoSol wizard (37). AutoSol built the N-terminal 50 amino acids of 12 out of 16 chains in the asymmetric unit. Manual building of the remaining residues and chains was performed in COOT (38). Iterative rounds of model building and refinement in PHENIX (39) led to an initial model of the SeMet-substituted protein. This solution was then used as a search model in PHASER (40) to determine phases for the native data set.
Plant Transformation, Growth Conditions, and Phenotypic Assays.
The Columbia (Col-0) ecotype of A. thaliana was used as wild type in the described experiment. Col-0 plants were transformed via floral dip transformation (41) with Agrobacterium tumefaciens strain GV3101 (42) carrying the plant transformation constructs. To acquire transformed plants overexpressing stabilized Aux/IAA proteins, T1 seeds were surface-sterilized and germinated on plates containing plant nutrient media [PN (43)], solidified with 0.6% agar, and supplemented with Basta (10 µg/mL) and timentin (20 ng/mL). Plants were grown on PN plates at 24 °C with 24-h days. Putative transformants were selected based on survival on Basta. Transformants were transferred to soil and grown under the same conditions. After 7 d in soil, all transformants were sprayed with 1.2 mg/mL Basta (Finale, Bayer) on days 7 and 9 after transplanting. On day 32 (after initial germination on plates), plants were photographed, and rosette diameter was measured using ImageJ (44).
Supplementary Material
Acknowledgments
This research was supported by the National Science Foundation (NSF) Graduate Research Fellowship Program (2011101911 to D.A.K.), the National Institutes of Health (R00 GM089987-03 to L.C.S.), the NSF (MCB-1157771 to J.M.J.), and the US Department of Agriculture–National Institute of Food and Agriculture Fellowship Program (MOW-2010-05240 to C.S.W.). Portions of this research were carried out at the Argonne National Laboratory Structural Biology Center of the Advanced Photon Source, a national user facility operated by the University of Chicago for the Department of Energy Office of Biological and Environmental Research (DE-AC02-06CH11357).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4NJ6 and 4NJ7).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1400074111/-/DCSupplemental.
References
- 1.Woodward AW, Bartel B. Auxin: Regulation, action, and interaction. Ann Bot (Lond) 2005;95(5):707–735. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guilfoyle TJ, Hagen G. Auxin response factors. Curr Opin Plant Biol. 2007;10(5):453–460. doi: 10.1016/j.pbi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 3.Tiwari SB, Wang X-J, Hagen G, Guilfoyle TJ. AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell. 2001;13(12):2809–2822. doi: 10.1105/tpc.010289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan X, et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446(7136):640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 5.Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435(7041):446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 6.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435(7041):441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 7.Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature. 2001;414(6861):271–276. doi: 10.1038/35104500. [DOI] [PubMed] [Google Scholar]
- 8.Maraschin FdS, Memelink J, Offringa R. Auxin-induced, SCF(TIR1)-mediated poly-ubiquitination marks AUX/IAA proteins for degradation. Plant J. 2009;59(1):100–109. doi: 10.1111/j.1365-313X.2009.03854.x. [DOI] [PubMed] [Google Scholar]
- 9.Hagen G, Guilfoyle T. Auxin-responsive gene expression: Genes, promoters and regulatory factors. Plant Mol Biol. 2002;49(3-4):373–385. [PubMed] [Google Scholar]
- 10.Liscum E, Reed JW. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol. 2002;49(3-4):387–400. [PubMed] [Google Scholar]
- 11.Kim J, Harter K, Theologis A. Protein-protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci USA. 1997;94(22):11786–11791. doi: 10.1073/pnas.94.22.11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9(11):1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tiwari SB, Hagen G, Guilfoyle T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell. 2003;15(2):533–543. doi: 10.1105/tpc.008417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weijers D, Jürgens G. Funneling auxin action: specificity in signal transduction. Curr Opin Plant Biol. 2004;7(6):687–693. doi: 10.1016/j.pbi.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 15.Vernoux T, et al. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol Syst Biol. 2011;7:508. doi: 10.1038/msb.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guilfoyle TJ, Hagen G. Getting a grasp on domain III/IV responsible for Auxin Response Factor-IAA protein interactions. Plant Sci. 2012;190:82–88. doi: 10.1016/j.plantsci.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 17.Noda Y, et al. Molecular recognition in dimerization between PB1 domains. J Biol Chem. 2003;278(44):43516–43524. doi: 10.1074/jbc.M306330200. [DOI] [PubMed] [Google Scholar]
- 18.Ito T, Matsui Y, Ago T, Ota K, Sumimoto H. Novel modular domain PB1 recognizes PC motif to mediate functional protein-protein interactions. EMBO J. 2001;20(15):3938–3946. doi: 10.1093/emboj/20.15.3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sumimoto H, Kamakura S, Ito T. Structure and function of the PB1 domain, a protein interaction module conserved in animals, fungi, amoebas, and plants. Sci STKE. 2007;2007(401):re6. doi: 10.1126/stke.4012007re6. [DOI] [PubMed] [Google Scholar]
- 20.Okushima Y, et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell. 2005;17(2):444–463. doi: 10.1105/tpc.104.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: A case study using the Phyre server. Nat Protoc. 2009;4(3):363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 22.Holm L, Rosenström P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Suppl 2):W545-9. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirano Y, et al. Structure of a cell polarity regulator, a complex between atypical PKC and Par6 PB1 domains. J Biol Chem. 2005;280(10):9653–9661. doi: 10.1074/jbc.M409823200. [DOI] [PubMed] [Google Scholar]
- 24.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 25.Rinaldi MA, Liu J, Enders TA, Bartel B, Strader LC. A gain-of-function mutation in IAA16 confers reduced responses to auxin and abscisic acid and impedes plant growth and fertility. Plant Mol Biol. 2012;79(4-5):359–373. doi: 10.1007/s11103-012-9917-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ni J, et al. Intragenic suppressor of osiaa23 revealed a conserved tryptophan residue crucial for protein-protein interactions. PLoS ONE. 2014;9(1):e85358. doi: 10.1371/journal.pone.0085358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang S, Hagen G, Guilfoyle TJ. ARF-Aux/IAA interactions through domain III/IV are not strictly required for auxin-responsive gene expression. Plant Signal Behav. 2013;8(6):e24526. doi: 10.4161/psb.24526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boer DR, et al. Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell. 2014;156(3):577–589. doi: 10.1016/j.cell.2013.12.027. [DOI] [PubMed] [Google Scholar]
- 29.Kohalmi S, Nowak J, Crosby WL. cDNA Library Screening with the Yeast Two-Hybrid System. Saskatoon, SK, Canada: Plant Biotechnol Inst; 1995. [Google Scholar]
- 30.Earley KW, et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45(4):616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 31.Gietz R, Schiestl R. Transforming yeast with DNA. Methods Mol Cell Biol. 1995;5:255–269. [Google Scholar]
- 32.Bartel PL, Chien CT, Sternglanz R, Fields S. Cellular Interactions in Development: A Practical Approach. Oxford: Oxford Univ Press; 1993. pp. 153–179. [Google Scholar]
- 33.Doublié S. Production of selenomethionyl proteins in prokaryotic and eukaryotic expression systems. Methods Mol Biol. 2007;363:91–108. doi: 10.1007/978-1-59745-209-0_5. [DOI] [PubMed] [Google Scholar]
- 34.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: The integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 8):859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 35.Grosse-Kunstleve RW, Adams PD. Substructure search procedures for macromolecular structures. Acta Crystallogr D Biol Crystallogr. 2003;59(Pt 11):1966–1973. doi: 10.1107/s0907444903018043. [DOI] [PubMed] [Google Scholar]
- 36.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr. 2000;56(Pt 8):965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Terwilliger TC, et al. Decision-making in structure solution using Bayesian estimates of map quality: The PHENIX AutoSol wizard. Acta Crystallogr D Biol Crystallogr. 2009;65(Pt 6):582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 39.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fortelle E, Bricogne G. Maximum-likelihood heavy-atom parameter refinement for the multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 41.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 42.Koncz C, Schell J. The promoter of the TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet. 1986;204:383–396. [Google Scholar]
- 43.Haughn GW, Somerville C. Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol Gen Genet. 1986;204:430–434. [Google Scholar]
- 44.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.