Significance
Echinoderm microtubule-associated protein (EMAP)-like (EML) proteins normally function in the cytoskeleton. In some lung cancers, genetic abnormalities generate the oncogenic fusion protein EML4-anaplastic lymphoma kinase (ALK) on which the cancer cells depend for survival. We have determined the molecular structure of a conserved, tubulin-binding region of EML1 that reveals an unexpected protein fold. This region is disrupted in ∼70% of EML4-ALK fusions found in patients, causing them to be sensitive to drugs that target Hsp90, a cellular factor that stabilizes misfolded protein. Our findings will potentially enable more effective, stratified therapy of EML4-ALK nonsmall cell lung cancer and suggest that the truncation of a globular domain at the translocation breakpoint may prove generally predictive of Hsp90 inhibitor sensitivity in cancers driven by fusion oncogenes.
Keywords: structural biology, stratified medicine
Abstract
Proteins of the echinoderm microtubule-associated protein (EMAP)-like (EML) family contribute to formation of the mitotic spindle and interphase microtubule network. They contain a unique hydrophobic EML protein (HELP) motif and a variable number of WD40 repeats. Recurrent gene rearrangements in nonsmall cell lung cancer fuse EML4 to anaplastic lymphoma kinase (ALK), causing expression of several fusion oncoprotein variants. We have determined a 2.6-Å crystal structure of the representative ∼70-kDa core of EML1, revealing an intimately associated pair of β-propellers, which we term a TAPE (tandem atypical propeller in EMLs) domain. One propeller is highly atypical, having a discontinuous subdomain unrelated to a WD40 motif in place of one of its blades. This unexpected feature shows how a propeller structure can be assembled from subdomains with distinct folds. The HELP motif is not an independent domain but forms part of the hydrophobic core that joins the two β-propellers. The TAPE domain binds α/β-tubulin via its conserved, concave surface, including part of the atypical blade. Mapping the characteristic breakpoints of each EML4-ALK variant onto our structure indicates that the EML4 TAPE domain is truncated in many variants in a manner likely to make the fusion protein structurally unstable. We found that the heat shock protein 90 (Hsp90) inhibitor ganetespib induced degradation of these variants whereas others lacking a partial TAPE domain were resistant in both overexpression models and patient-derived cell lines. The Hsp90-sensitive EML4-ALK variants are exceptions to the rule that oncogenic fusion proteins involve breakpoints in disordered regions of both partners.
Rearrangements in the short arm of chromosome 2 leading to genetic fusions between EML4 [echinoderm microtubule-associated protein (EMAP)-like 4] and the gene encoding anaplastic lymphoma kinase (ALK) occur in ∼5% of cases of nonsmall cell lung cancer (NSCLC) (1). Multiple variants of the EML4-ALK fusion have been identified in NSCLC resulting from translocations at different points within the EML4 gene (2). A similar genetic fusion has also been reported between EML1 and the gene encoding another tyrosine kinase, Abelson 1 (ABL1), in T-cell acute lymphoblastic leukemia (3). These fusions display potent transforming activity due to constitutive activation of the tyrosine kinase but confer addiction to the oncogene, and inhibition of the tyrosine kinase induces apoptosis in transformed cells (4, 5). The ALK inhibitor crizotinib is a highly effective first-line therapy for NSCLC patients with EML4-ALK fusions, but because the acquirement of resistance is inevitable, often involving secondary mutations in the kinase portion of the fusion protein, additional therapeutic strategies must be identified (6, 7).
Inhibitors of the molecular chaperone heat shock protein 90 (Hsp90) are being investigated as potential cancer therapeutics because many oncoproteins are obligate Hsp90 clients and they are rapidly degraded by the proteasome when Hsp90 is inhibited (8). Hsp90 inhibitors induce the degradation of EML4-ALK variant 1 and regression in some EML4-ALK–positive tumor models (7, 9, 10). Furthermore, clinical efficacy of an Hsp90 inhibitor in EML4-ALK NSCLC has been confirmed (11), and clinical trials are ongoing. However, because neither ALK nor EML4 are native Hsp90 clients, it was proposed that Hsp90 sensitivity of EML4-ALK fusions was due to their protein-folding properties, which might expose hydrophobic residues that lead to Hsp90 recruitment (12). EML4-ALK variants incorporating various portions of EML4 differ in sensitivity to Hsp90 inhibitors, suggesting that they have different protein-folding properties (12).
In contrast to the abundance of studies on EML4-ALK fusions, the basic functions of EML proteins are relatively understudied. The archetypal EML protein was identified as the most abundant MAP in dividing echinoderm eggs and embryos, where it promotes microtubule dynamics (13, 14). Microtubule interactions of EML1-4 have been characterized in six EML proteins present in humans (15–18). More members of the family have been identified in Drosophila and Caenorhabditis elegans, which each have a single homolog (19). The EML proteins contain a variable number of WD40 repeats, and in EML1-4 these fall within an ∼70-kDa core region that begins with a conserved ∼60-amino-acid sequence that has been termed the hydrophobic EML protein (HELP) motif (15). Many EML proteins also contain a predicted coiled-coil (CC) in their N termini that is likely to mediate oligomerization.
Functional studies of EML proteins and their oncogenic fusions have had to be interpreted without knowledge of their molecular structures. For example, it has been assumed that the HELP motif forms an independent domain that perhaps mediates specific interactions with microtubules (MTs) or the plasma membrane (15, 17, 18, 20). The extent to which the folding of the WD40 repeat region is disrupted in EML4-ALK fusions is also unclear because the number, position, and arrangement of β-propellers in EML proteins are unknown. We determined the crystal structure of the ∼70-kDa core HELP/WD region of human EML1. The structure serves as an archetype for the whole family and has an unusual molecular architecture that we have termed the TAPE (tandem atypical propeller in EMLs) domain. The structure enabled us to dissect the molecular determinants of tubulin-binding and map the precise location of breakpoints in EML4-ALK patients onto the structure. We explain how the partial EML moieties that are present in each of the variants confer differences between them with respect to Hsp90 inhibitor sensitivity.
Results
Conserved Core Region of EML1 Forms a Tandem β-Propeller Structure.
We expressed fragments of human EML1–4 corresponding to an ∼70-kDa core region that is conserved across the EMAP/EML protein family, including the HELP region and WD40 repeats, in Spodoptera frugiperda Sf9 cells using the baculovirus system. Of the four proteins tested, only EML1 (amino acids 167–815) formed diffraction quality crystals, and we determined the structure using single-wavelength anomalous dispersion (SAD) phasing of a selenomethionine-labeled sample to 2.6-Å resolution (Table 1 and Fig. S1). This revealed a tandem β-propeller, which we call the TAPE domain, with one highly atypical blade built from two separate regions of the primary sequence (Fig. 1).
Table 1.
Summary of crystallographic analysis
| Data collection | |
| Space group | P1 21 1 |
| a, b, c (Å) | 91.98, 83.98, 115.52 |
| α, β, γ (°) | 90.00, 96.61, 90.00 |
| X-ray source | Diamond I03 |
| Wavelength (Å) | 0.9790 |
| Resolution range (Å) | 47.40–2.60 (2.74–2.60) |
| No. unique reflections | 53,683 (7,804) |
| Completeness (%) | 99.4 (99.6) |
| Redundancy | 3.6 (3.7) |
| Rmerge (%) | 9 (50) |
| I/σI | 10.1 (2.7) |
| Phasing (figures of merit) | |
| Before density modification | 0.26 |
| After density modification | 0.67 |
| Refinement | |
| Resolution range (Å) | 47.4–2.6 |
| Rwork/Rfree (%) | 19.95/25.08 |
| No. protein molecules; amino acids; waters; hetero molecules | 2; 1,278; 208; 25 |
| Mean B-factors (Å2) | 48.78 |
| Wilson B-factor (Å2) | 42.06 |
| r.m.s. bond lengths (Å) | 0.004 |
| r.m.s. bond angles (°) | 0.885 |
| MolProbity analysis | |
| All-atom clash-score | 25.27 |
| Poor rotamers (%) | 6.03 |
| Ramachandran outliers (%) | 0.00 |
| Ramachandran favored (%) | 93.97 |
| MolProbity score | 2.20 |
Values in parentheses are for the highest-resolution shell.
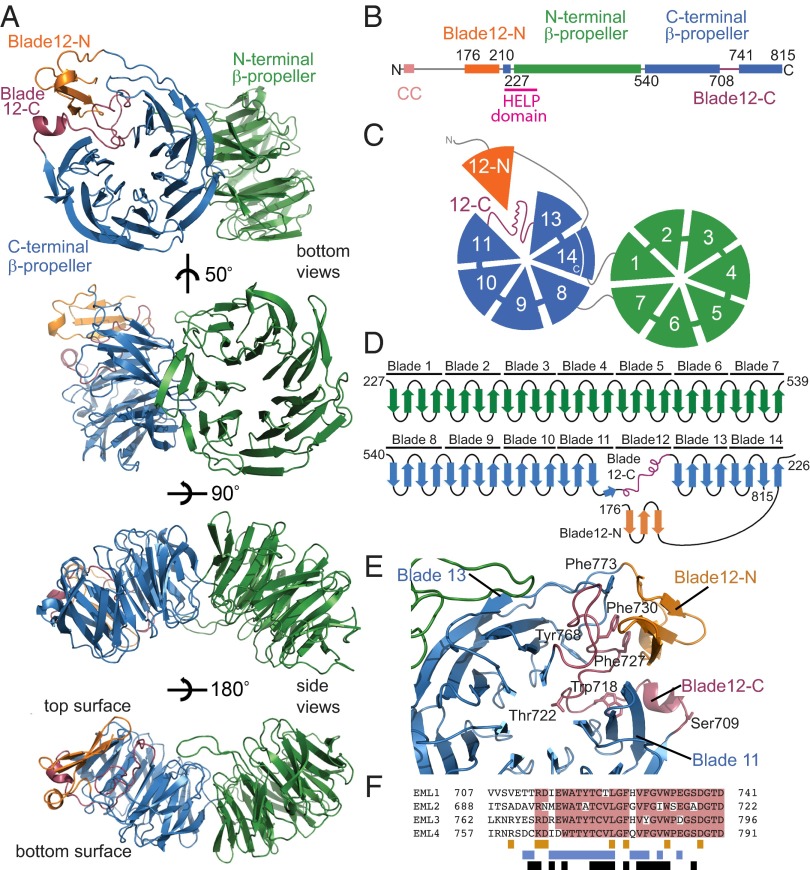
Fig. 1.
Architecture of the EML1 TAPE domain. Throughout the figure, the subdomains are colored by the following scheme: orange (blade12-N), green (N-terminal propeller), blue (C-terminal propeller), and purple (blade12-C). (A) The structure of the EML1 TAPE domain is shown as cartoon representations of bottom and side views. (B) Linear representation of the EML1 protein including residue numbering indicating domain boundaries. (C) Cartoon representation showing the bottom face of the TAPE domain with numbered blades. (D) Topology diagram showing connectivity of the secondary structure elements. (E) View of the C-terminal β-propeller from above its central channel showing how blade 12 is formed from blade12-N and blade12-C. Key residues are shown as stick representations. (F) Sequence alignment indicating sequence conservation among human EML proteins (pink) in the region corresponding to blade12-C. The colored bars under the alignment indicate residues that make contacts with blades 11 and 13 (blue), with blade12-N (orange), and with those that are buried (black).
The overall topology of the TAPE domain resembles the tandem β-propeller of actin-interacting protein 1 (AIP1) (21, 22). The two β-propellers are intimately associated with one another at an angle of ∼50°. Residues 230–540 form a seven-bladed β-propeller domain (green in Fig. 1). Each blade is a twisted, four-stranded, antiparallel β-sheet that radiates from the center of the domain and is encoded by a separate WD40 repeat. As in AIP1, but unusually for β-propeller domains, the seventh blade of this N-terminal β-propeller is self-contained rather than having its fourth strand donated by a more N-terminal part of the sequence. Residues 542–815 form the C-terminal β-propeller of EML1, which is also similar in overall size and topology to a classical seven-bladed β-propeller (blue). Its seventh blade (blade 14) is completed by a fourth strand formed by residues (220–227) immediately N-terminal to the first β-propeller. The two β-propellers communicate through contacts between the face of blade 1 and the edges of blades 14 and 13.
Blade 12 is not a canonical four-stranded β-sheet, but is composed of two subdomains, which we have termed Blade12-N and Blade12-C, that correspond to two distant stretches of primary sequence. This generates a β-propeller structure in which one of the blades is replaced by a subdomain of sequence and structure entirely unrelated to the other constituent repeats (Fig. 1 D–F). Blade12-C (amino acids 708–741) lies in the primary sequence between blades 11 and 13 and forms an unusual loop structure (purple). The fourth β-strand of blade 11 (amino acids 704–707) forms only three main-chain hydrogen bonds with the third strand before the polypeptide chain instead continues tangentially to the propeller and into the region corresponding to blade 12. Thus, in blade12-C the chain begins on the outer, bottom surface of the propeller, rather than on the top surface near the central channel as orthodox blades do (Fig. 1D). Blade12-C fills the wedge of space between blades 11 and 13 that a canonical WD40 blade would normally be expected to occupy and thereby maintains the structural integrity of the domain, but it does so without forming significant regular secondary structure (Fig. 1E). Blade12-C consists of 34 residues, fewer than a typical WD40 repeat, with a high proportion of conserved bulky aromatic residues. Blade12-N (amino acids 176–205) is N-terminal to the WD-repeat region and forms a three-stranded antiparallel β-sheet, topped by a turn of α-helix (orange). The intervening residues (amino acids 206–219) form an extended arm that reaches around blade 13 stabilized by the loop between its third and fourth strands. Blade12-N is suspended on the end of this arm and stacks against the outside of blade12-C, giving the appearance of a β-propeller blade displaced to a position outside the circle.
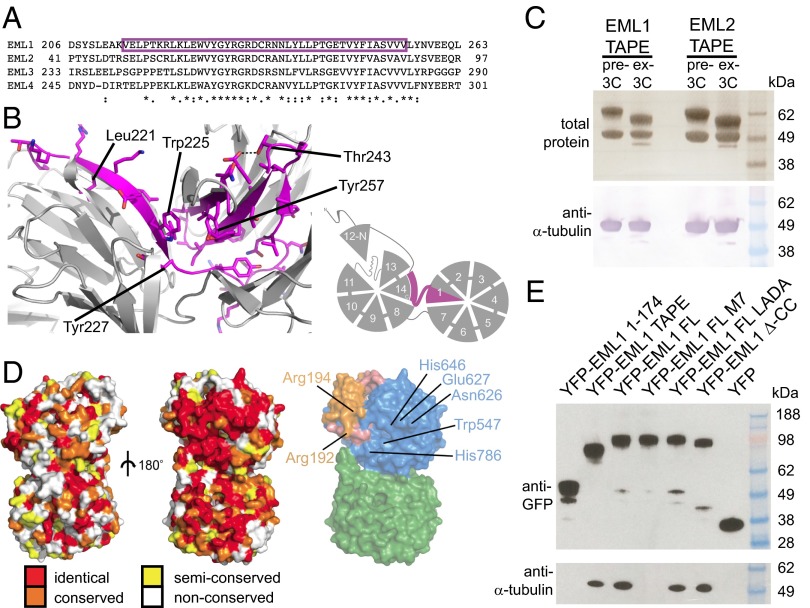
The HELP domain (amino acids 217–259 in EML1, Fig. 2A) is a characteristic and conserved component of the EML sequence (15) and has been identified previously as carrying its MT-binding activity (17, 18). The present structure shows that the HELP motif is not a distinct domain but rather forms part of the hydrophobic core of the TAPE domain and corresponds to those parts of blades 1 and 14 that are buried at the interface of the two β-propellers (Fig. 2B). Its deletion, which has been used in previous work to investigate its function, would disrupt folding of the TAPE domain. Because the HELP domain is not available for direct interaction with tubulin, we sought the molecular determinants of tubulin association in EML proteins.
Fig. 2.
A conserved patch on the concave surface of the TAPE domain binds soluble tubulin. (A) The position of the HELP motif is shown in magenta in an alignment of human EML proteins and (B) in a structural model of the interface between the β-propellers and a schematic diagram. (C) Copurification of tubulin from insect cell lysate with Strep-tagged EML1 and EML2 TAPE domains. (Upper) A Coomassie Blue-stained gel of Strep-tactin Sepharose purified proteins (before and after 3C-protease cleavage). (Lower) An anti–α-tubulin immunoblot confirming the identity of the copurified protein. (D) Surface views of the EML1 TAPE domain showing the convex (Left) and concave (Center) faces colored by sequence conservation between human EML 1–4 proteins and a surface view of the concave face of the TAPE domain colored by subdomain (Right). The positions of the conserved surface-exposed residues mutated to create M7 mutant are indicated (see also Fig. S2). (E) Copurification of tubulin from HEK293F cells with EML1 constructs under MT-depolymerizing conditions. Immunoblots of YFP-tagged proteins purified on Strep-tactin Sepharose from HEK293F cells transfected with Strep-YFP-EML1 constructs as indicated (Upper) and copurified tubulin (Lower).
TAPE Domain Binds Soluble α/β-Tubulin.
During purification of EML TAPE domains from insect cells, we noted that in each case α/β-tubulin was copurified (Fig. 2C). We mapped sequence conservation among human EML proteins onto the surface of the TAPE domain crystal structure to identify patches that might mediate binding interactions (Fig. 2D). Although there are few conserved residues on the convex surface of the domain, there is a prominent ribbon of conservation across the concave surface. We mutated seven conserved residues from the top face of the C-terminal β-propeller to disrupt this patch (the M7 mutant). We tested the ability of multiple YFP-EML1 constructs to copurify tubulin from the lysate of transfected HEK293F cells (Fig. 2E); because these experiments were carried out under MT-depolymerizing conditions, they reflect association with soluble tubulin dimers rather than MTs. The TAPE domain bound to soluble tubulin whereas the N-terminal region did not. Tubulin binding was not disrupted by a L59A, D61A mutation (LADA in Fig. 2E) of a conserved motif in the N-terminal region. Deletion of the N-terminal coiled-coil region (Δ-CC) also failed to disrupt tubulin binding, indicating that oligomerization of EML1 is not necessary for the interaction. The M7 mutation, which does not disrupt the folding of EML1 (Fig. S2), abrogated tubulin binding completely, indicating that the TAPE domain binds soluble tubulin via its concave surface.
EML4-ALK Variants Differ in Structural Viability.
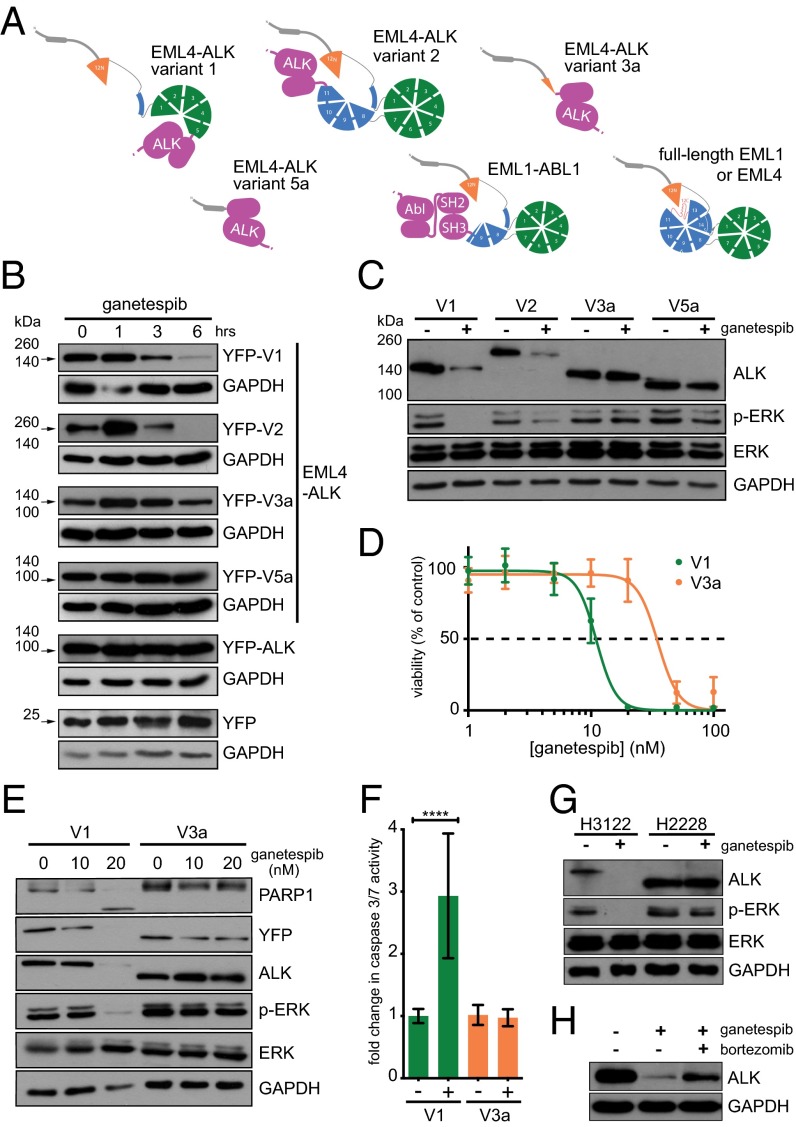
The availability of the TAPE domain crystal structure allowed us to consider the likely structural characteristics of oncogenic EML-tyrosine kinase fusion proteins that incorporate different portions of the EML structure. Fig. 3A shows several of these as schematic representations. In EML4-ALK variant 1, the breakpoint falls within the N-terminal β-propeller, and in EML4-ALK variant 2 (1) and EML1-ABL1 (3), the breakpoint falls within the C-terminal β-propeller (Fig. 3A). Fusion proteins such as these containing a partial TAPE domain would be predicted to be structurally unstable and may be dependent upon molecular chaperones to remain in solution due to exposure of an incomplete hydrophobic core. However, the portions of EML4 present in EML4-ALK variants 3a/b and 5 would not be expected to confer structural instability upon the fusion protein as they lack any core part of the TAPE domain.
Fig. 3.
The TAPE domain structure explains differential stability of EML4-ALK variants in response to Hsp90 inhibition. (A) Models of full-length EML1 and EML4 and EML1-ABL1 and EML4-ALK fusion variants based upon the EML1 TAPE domain structure are illustrated schematically. (B) MEFs were transfected with YFP, YFP-cytoplasmic ALK (amino acids 1,058–1,620: the region of ALK present in the fusion proteins), or YFP-EML4-ALK variants 1, 2, 3a, and 5a. Forty-eight hours post transfection, cells were treated with 100 nM ganetespib for a further 1, 3, and 6 h. Transient overexpression of EML4-ALK was assessed by Western blot with anti-GFP 4B10 antibody as indicated by arrows. (C) Ba/F3 clones stably expressing YFP-EML4-ALK variants 1, 2, 3a, or 5a were left untreated or treated with 20 nM ganetespib for 6 h. Expression of ALK, ERK phosphorylated at T202 and Y204 (p-ERK), and total ERK was determined by Western blot. GAPDH was used as a loading control. (D) Ba/F3 clones expressing V1 or V3a were left untreated or treated with ganetespib doses ranging from 1 to 100 nM. Cell viability was assayed after 24 h of treatment and normalized as the percentage of the untreated control for each cell line. (E) Ba/F3 clones stably expressing variants 1 or 3a were left untreated or treated with 10 or 20 nM ganetespib for 12 h. Expression of ALK, ERK phosphorylated at T202 and Y204 (p-ERK), and total ERK were determined by Western blot. Induction of apoptosis was analyzed with anti-PARP1 antibody. GAPDH was used as a loading control. (F) V1 and V3a clones were left untreated or treated with 20 nM ganetespib for 12 h and assayed for caspase 3/7 activity. (P values: V1 < 0.0001; V3a = 0.4086). (G) NCI-H3122 and NCI-H2228 (NSCLC cells endogenously expressing EML4-ALK variant 1 and 3b, respectively) were untreated or treated with 100 nM ganetespib for 6 h and analyzed by Western blot as in C. (H) NCI-H3122 cells were left untreated or treated with 100 nM ganetespib alone or in combination with 100 nM bortezomib for 6 h. ALK expression was determined by Western blot with anti-ALK.
We next compared the sensitivity of these four EML4-ALK variants to Hsp90 inhibition. Wild-type mouse embryonic fibroblasts (MEFs) were transiently transfected with different constructs expressing YFP-EML4-ALK variants 1, 2, 3a, and 5a, YFP-ALK 1054–1620 (the region of ALK present in fusions) and YFP alone. Significant degradation of ALK was observed after exposure to the Hsp90 inhibitor ganetespib (Synta Pharmaceuticals) (23) in cells transfected with EML4-ALK variants 1 or 2 but not variants 3a or 5a (Fig. 3B). To evaluate the effect of Hsp90 inhibition in cells where survival is dependent on ALK activity, we generated a system stably expressing each EML4-ALK variant using the murine hematopoietic interleukin (IL)-3–dependent Ba/F3 cell line. Once stable clones are selected and IL-3 is withdrawn, these cells become ALK-dependent. Treatment with ganetespib induced ALK degradation as observed in MEFs, with significant degradation of variants 1 and 2 only (Fig. 3C). The effect of ganetespib treatment on the extracellular signal-regulated kinase (ERK) survival pathway correlated with ALK degradation, causing significant decreases in phosphorylation of ERK on T202 and Y204 in clones expressing variants 1 and 2 but not 3a or 5a.
We next investigated whether the differential Hsp90 sensitivities of EML4-ALK variants 1 and 3 impacted on the viability of cells exposed to increasing concentrations of ganetespib, using Ba/F3 cells overexpressing variant 1 and variant 3a. Viability assays were performed for ganetespib in the two cell lines, and IC50 values were determined (V1: 11.09 nM; V3a: 35.13 nM) (Fig. 3D). Western blotting confirmed complete degradation of EML4-ALK V1 in cells treated with 20 nM ganetespib, and the consequent reduction of downstream prosurvival and proliferative signaling pathways was indicated by loss of ERK phosphorylation (Fig. 3E). We also observed PARP1 cleavage and a significant (2.9-fold) increase in caspase 3/7 activity in V1-expressing cells treated with 20 nM ganetespib, indicating the activation of apoptotic pathways, whereas similar treatment induced no such changes in cells expressing V3a (Fig. 3F).
The Hsp90 sensitivities of EML4-ALK variants were then probed in two human NSCLC cell lines that harbor endogenous EML4-ALK variant 1 (NCI-H3122) or EML4-ALK variant 3b (NCI-H2228). After 6 h exposure to ganetespib, we observed degradation of ALK in H3122 cells, accompanied by a loss of ERK phosphorylation, whereas in H2228 cells the same treatment failed to affect levels of ALK or phosphorylated ERK (Fig. 3G). We finally investigated the role of the proteasome in the degradation of EML4-ALK upon Hsp90 inhibition. H3122 cells were treated with ganetespib alone or in combination with the 26S proteasome inhibitor bortezomib (Millennium Pharmaceuticals). Cotreatment with bortezomib reduced ganetespib-induced degradation of the endogenous EML4-ALK variant 1 protein, confirming that the degradation is proteasome-dependent (Fig. 3H).
Discussion
The crystal structure of the HELP/WD40 repeat region (TAPE domain) of EML1 reveals a pair of β-propellers assembled into a single globular tubulin-binding module. The C-terminal β-propeller has a unique arrangement in which the fifth segment in the propeller is occupied by a discontinuous subdomain formed from an unusual loop structure (blade12-C) and an N-terminal portion of the domain that folds as a three-stranded β-sheet (blade12-N) (Fig. 2). This feature adds another dimension to the architectural diversity of β-propeller proteins, which have hitherto included only examples with different numbers of canonical blades, and raises the possibility that other forms of noncanonical blades may be present in β-propellers that await structural characterization.
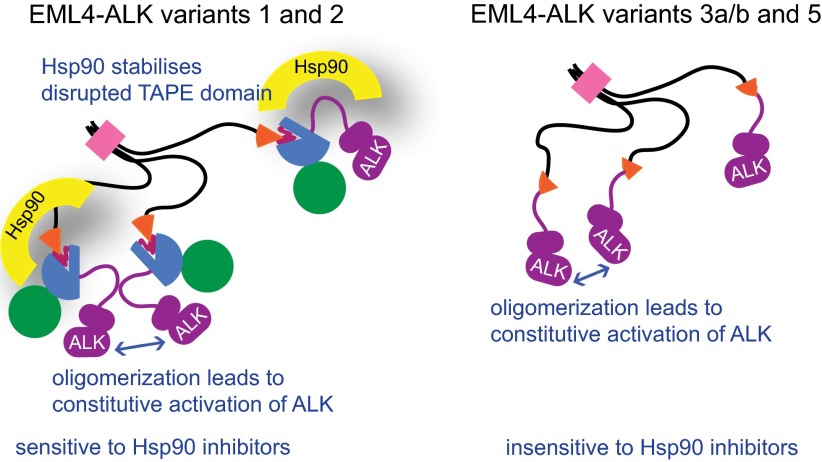
Chromosomal translocations in cancer may fuse together 5′ and 3′ fragments of two distinct genes that are then transcribed and translated as a single chimeric protein. The breakpoint in these oncogenic fusion proteins usually occurs within intrinsically disordered regions of both fusion partners rather than interrupting globular domains (24). This is not surprising because truncation of a globular domain would be expected to generate a structurally unstable protein with an exposed hydrophobic core. Analysis of the EML4-ALK breakpoints based on the crystal structure of the EML1 TAPE domain confirm that several of these variants are exceptional because they are potent oncogenes in which a globular domain is disrupted. This makes them dependent on molecular chaperones to remain stable. Indeed, EML4-ALK variant 1 has been shown to be highly unstable, and it may be more sensitive to Hsp90 inhibition than any other client protein yet observed (8, 9, 25). The sensitivity of variants 1 and 2 are due to protein mis-folding resulting in exposure of hydrophobic residues that recruit Hsp90, which is supported by the observation that this sensitivity is not found in variants 3 and 5 (12). These differences are also found in patient-derived NSCLC cells that express variant 1 or 3 endogenously (Fig. 4). Furthermore, because the HELP/WD is a single continuous domain, and not a series of independent domains, we predict that it will generally be the case that oncogenic EML fusions with breakpoints that fall within the TAPE domain, such as EML1-ABL1, will be Hsp90-dependent, whereas other fusions will be Hsp90-independent.
Fig. 4.
Models summarizing the properties of EML4-ALK variants. Constitutive activation of ALK through oligomerization via the coiled-coil region of EML4 is common to all EML4-ALK variants. However, sensitivity of variants to Hsp90 inhibitors differs depending on whether the breakpoint of the fusion protein interrupts the globular TAPE domain.
We predict that tumors harboring any EML4-ALK variants in which the TAPE domain is truncated, and indeed the EML1-ABL1 fusion where this is also the case, would be sensitive to Hsp90 inhibitors. The high response rate to Hsp90 inhibition observed clinically in EML4-ALK NSCLC (11) may be explained by the knowledge that such variants account for ∼70% of cases (2, 26). However, Hsp90 inhibitor therapy may be less efficacious in the remaining cases, in which tumors harbor variants 3a/b or 5a/b. The current standard diagnostic test for the presence of ALK rearrangements identifies fragmentation of ALK by a fluorescent in situ hybridization assay (27). Previous work (12) and our data suggest that knowledge of the specific fusion variant that is present in patient tumors may allow more effective stratification of Hsp90 therapy in EML4-ALK NSCLC. We plan to correlate variant-specific genotype with response to ganetespib treatment in an ongoing phase II clinical trial involving patients with ALK-positive NSCLC (NCT01562015). Furthermore, the truncation of a globular domain at the translocation breakpoint is likely to be generally predictive of Hsp90 inhibitor sensitivity in fusion oncogene-driven cancers.
Materials and Methods
Gene Manipulation.
Human EML cDNAs were cloned by PCR from human cDNA (Clontech). Constructs were made comprising EML1 residues 167–815 (TAPE), 1–174 (N terminus), and 80–815 (Δ-CC). The EML1 LADA mutant contained the mutations L59A and D61A. The M7 mutant contained the mutations R192S, R194S, W547A, N626T, E627S, H646A, and H786A. Mutagenesis was carried out by the Quikchange procedure (Stratagene). EML1-ABL1 and EML4-ALK variants were constructed by amplifying the appropriate regions of the ABL1 and ALK genes from human cDNA (Clontech), ligating them in tandem with EML1 or EML4 cDNAs and then fusing the genes together at the desired position by PCR.
Protein Expression and Purification.
The EML1 167–815 construct was cloned into a modified version of pFASTBAC, providing a 3C-cleavable N-terminal FLAG-Strep tag. The bacmid was made using the Bac-to-Bac system (Invitrogen) and used to produce recombinant baculovirus. To obtain selenomethionine-labeled EML1 167–815 protein, Sf9 cells were cultured in methionine-free, cysteine-free, serum-free SF-900 II media (Gibco) supplemented with cysteine and selenomethionine (Molecular Dimensions) and infected with the baculovirus. The protein was purified from clarified cell lysate on Strep-tactin Sepharose (Qiagen) in Tris-buffered saline (TBS) and eluted using 3 mM desthiobiotin (IBA). The protein was incubated with 3C protease to cleave off the tag (Fig. 2C) and then passed over a Glutathione-Sepharose column to remove the protease. Finally, the protein was separated from copurifying Sf9 tubulin by gel filtration on a Superose 6 column (GE Healthcare) in 20 mM Tris⋅HCl, pH 7.5, 80 mM NaCl, and 2 mM DTT.
Crystal Structure Determination.
Crystals of the selenomethionine-labeled EML1 TAPE domain were obtained by vapor diffusion at 18 °C in drops comprising 15 mg/mL protein solution mixed 1:1 with reservoir solution (100 mM trisodium citrate, pH 5.6, 2.0 M ammonium sulfate, 200 mM potassium/sodium tartrate, 2 mM DTT). Crystals formed within 2 d and were flash-frozen in reservoir solution with the ammonium sulfate concentration increased to 3.5 M to provide cryo-protection.
SAD data to a resolution of 2.6 Å were collected from an EML1 TAPE domain selenomethionine derivative crystal at Diamond I03. Diffraction images were indexed and scaled using iMosflm and SCALA (28). Initial phases were obtained using PHENIX (29). There were two molecules in the asymmetric unit. Some of the backbone was built automatically using BUCCANEER (28), and building was continued manually using COOT (30) until an initial model corresponding to most of the protein was obtained. Iterative rounds of manual building in COOT and refinement using REFMAC (28) and PHENIX were performed to complete the structure. Validation of the structure was carried out using MolProbity (31). Structure figures were prepared using PyMOL (www.pymol.org).
Cell Culture and Transfection.
Wild-type MEF cells were a gift from S. Oakes (University of California, San Francisco), Ba/F3 cells were provided by M. Dyer (University of Leicester, Leicester, UK), NCI-H3122 cells were obtained from the National Cancer Institute (NCI) Tumor Repository, and NCI-H2228 cells were provided by S. Gray (Institute of Molecular Medicine, Dublin). MEF, Ba/F3, H2228, and H3122 cells were cultured in RPMI medium 1640 with 2 mM Glutamax and 10% (vol/vol) FBS. Ba/F3s were supplemented with 10% (vol/vol) WEHI-3b–conditioned media (Cell Lines Service) as a source of IL-3 until after electroporation. EML4-ALK variants and ALK 1058–1620 were cloned into a version of pcDNA3.1-hygro (Invitrogen), providing an N-terminal YFP tag for transfection of MEFs and Ba/F3 cells. MEFs were transiently transfected using X-tremeGENE HP reagent (Roche). Ba/F3 cells were electroporated using the Amaxa Cell Line Nucleofector Kit V and Nucleofector II Device (Lonza). After 48 h, cells were resuspended in IL-3–free media and selected with 200 µg/mL hygromycin B (Invitrogen). HEK293F cells were grown in suspension culture in Freestyle 293 medium (Gibco) in a 5% (vol/vol) CO2 atmosphere and transfected with Strep-YFP-EML1 constructs in a modified version of pMAX (Lonza) at a density of 1 × 106 cells/mL using 2 μg/mL polyethylenimine.
Tubulin Copurification.
HEK293F cells transfected with Strep-YFP-EML1 constructs were harvested by centrifugation, resuspended in TBS containing Complete protease inhibitor mixture (Roche), and lysed by sonication. Ice-cold clarified lysates were applied to 1-mL StrepTrap columns (GE Healthcare). These were washed with ice-cold TBS and bound protein eluted with 3 mM desthiobiotin (IBA) in TBS. Eluates were analyzed by SDS/PAGE and Western blotting using anti-GFP ab6556 (Abcam) and anti–α-tubulin (Sigma) antibodies.
Cell Viability and Apoptosis Assays.
Ganetespib (STA-9090) was a gift from Synta Pharmaceuticals; bortezomib (Velcade) was from Millennium Pharmaceuticals. For cell viability assays, Ba/F3 cells were seeded in opaque 96-well plates at 10,000 cells/well and incubated for 24 h without drug or with 1–100 nM ganetespib. CellTiter-Glo assays (Promega) were performed according to manufacturer’s instructions. Data are representative of the median of three independent experiments ± SD and were fitted by nonlinear regression to derive IC50 values. Apoptosis activity was measured using the Caspase-Glo 3/7 Activity assay (Promega) after 12 h of treatment with 20 nM ganetespib and was normalized to the untreated controls for each cell line. Data are representative of the median of four independent experiments ± SD. Unpaired t test analysis was carried out for untreated vs. treated groups for each cell line. P values <0.05 were considered significant.
Western Blotting Analysis.
MEFs, Ba/F3, H3122, and H2228 cells were lysed in RIPA buffer supplemented with 50 mM NaF, 2 mM Na3VO4, and Complete protease inhibitor mixture (Roche) and clarified by centrifugation. Lysates were separated on SDS/PAGE denaturing gels and transferred to nitrocellulose membranes. Membranes were blocked in TBS containing 0.1% Tween-20 and 5% (wt/vol) milk and were probed with primary antibodies diluted in the same buffer at 4 °C overnight. Primary antibodies were anti-GFP (4B10), phospho-ERK 1/2 (Thr202/Tyr204), ERK 1/2 (L34F12), ALK (31F12) (Cell Signaling), anti-PARP1 (C2-10, Enzo Life Sciences), and GAPDH (Abcam). The secondary antibody was horseradish peroxidase conjugated anti-mouse IgG (Cell Signaling). Signal detection was performed with the Western Lightning Plus-ECL chemiluminescent system (Perkin-Elmer). All blots are representative of at least three independent experiments.
Supplementary Material
Acknowledgments
We thank Kiran Kulkarni and Ioannis Manolaridis (Institute of Cancer Research) for assistance with crystallography and Anne Straube (University of Warwick), Carolyn Moores (Birkbeck College), and Fiona Francis (University Pierre et Marie Curie, Paris) for helpful discussions. This work was supported by Royal Society Research Fellowship and Cancer Research UK Programme Grant C24461/A13231 (to R.B.) and by funding from the Association for International Cancer Research and the Wellcome Trust (A.M.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. C.D.S. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4ci8).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1322892111/-/DCSupplemental.
References
- 1.Soda M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 2.Sasaki T, Rodig SJ, Chirieac LR, Jänne PA. The biology and treatment of EML4-ALK non-small cell lung cancer. Eur J Cancer. 2010;46(10):1773–1780. doi: 10.1016/j.ejca.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Keersmaecker K, et al. Fusion of EML1 to ABL1 in T-cell acute lymphoblastic leukemia with cryptic t(9;14)(q34;q32) Blood. 2005;105(12):4849–4852. doi: 10.1182/blood-2004-12-4897. [DOI] [PubMed] [Google Scholar]
- 4.Choi YL, et al. Identification of novel isoforms of the EML4-ALK transforming gene in non-small cell lung cancer. Cancer Res. 2008;68(13):4971–4976. doi: 10.1158/0008-5472.CAN-07-6158. [DOI] [PubMed] [Google Scholar]
- 5.Takezawa K, Okamoto I, Nishio K, Jänne PA, Nakagawa K. Role of ERK-BIM and STAT3-survivin signaling pathways in ALK inhibitor-induced apoptosis in EML4-ALK-positive lung cancer. Clin Cancer Res. 2011;17(8):2140–2148. doi: 10.1158/1078-0432.CCR-10-2798. [DOI] [PubMed] [Google Scholar]
- 6.Choi YL, et al. ALK Lung Cancer Study Group EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med. 2010;363(18):1734–1739. doi: 10.1056/NEJMoa1007478. [DOI] [PubMed] [Google Scholar]
- 7.Katayama R, et al. Therapeutic strategies to overcome crizotinib resistance in non-small cell lung cancers harboring the fusion oncogene EML4-ALK. Proc Natl Acad Sci USA. 2011;108(18):7535–7540. doi: 10.1073/pnas.1019559108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neckers L, Workman P. Hsp90 molecular chaperone inhibitors: Are we there yet? Clin Cancer Res. 2012;18(1):64–76. doi: 10.1158/1078-0432.CCR-11-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Normant E, et al. The Hsp90 inhibitor IPI-504 rapidly lowers EML4-ALK levels and induces tumor regression in ALK-driven NSCLC models. Oncogene. 2011;30(22):2581–2586. doi: 10.1038/onc.2010.625. [DOI] [PubMed] [Google Scholar]
- 10.Sang J, et al. Targeted inhibition of the molecular chaperone Hsp90 overcomes ALK inhibitor resistance in non-small cell lung cancer. Cancer Discov. 2013;3(4):430–443. doi: 10.1158/2159-8290.CD-12-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sequist LV, et al. Activity of IPI-504, a novel heat-shock protein 90 inhibitor, in patients with molecularly defined non-small-cell lung cancer. J Clin Oncol. 2010;28(33):4953–4960. doi: 10.1200/JCO.2010.30.8338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heuckmann JM, et al. Differential protein stability and ALK inhibitor sensitivity of EML4-ALK fusion variants. Clin Cancer Res. 2012;18(17):4682–4690. doi: 10.1158/1078-0432.CCR-11-3260. [DOI] [PubMed] [Google Scholar]
- 13.Suprenant KA, Dean K, McKee J, Hake S. EMAP, an echinoderm microtubule-associated protein found in microtubule-ribosome complexes. J Cell Sci. 1993;104(2):445–450. doi: 10.1242/jcs.104.2.445. [DOI] [PubMed] [Google Scholar]
- 14.Hamill DR, Howell B, Cassimeris L, Suprenant KA. Purification of a WD repeat protein, EMAP, that promotes microtubule dynamics through an inhibition of rescue. J Biol Chem. 1998;273(15):9285–9291. doi: 10.1074/jbc.273.15.9285. [DOI] [PubMed] [Google Scholar]
- 15.Eichenmuller B, Everley P, Palange J, Lepley D, Suprenant KA. The human EMAP-like protein-70 (ELP70) is a microtubule destabilizer that localizes to the mitotic apparatus. J Biol Chem. 2002;277(2):1301–1309. doi: 10.1074/jbc.M106628200. [DOI] [PubMed] [Google Scholar]
- 16.Lepley DM, Palange JM, Suprenant KA. Sequence and expression patterns of a human EMAP-related protein-2 (HuEMAP-2) Gene. 1999;237(2):343–349. doi: 10.1016/s0378-1119(99)00335-2. [DOI] [PubMed] [Google Scholar]
- 17.Tegha-Dunghu J, et al. EML3 is a nuclear microtubule-binding protein required for the correct alignment of chromosomes in metaphase. J Cell Sci. 2008;121(Pt 10):1718–1726. doi: 10.1242/jcs.019174. [DOI] [PubMed] [Google Scholar]
- 18.Pollmann M, et al. Human EML4, a novel member of the EMAP family, is essential for microtubule formation. Exp Cell Res. 2006;312(17):3241–3251. doi: 10.1016/j.yexcr.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 19.Suprenant KA, et al. Conservation of the WD-repeat, microtubule-binding protein, EMAP, in sea urchins, humans, and the nematode C. elegans. Dev Genes Evol. 2000;210(1):2–10. doi: 10.1007/pl00008183. [DOI] [PubMed] [Google Scholar]
- 20.Hueston JL, et al. The C. elegans EMAP-like protein, ELP-1 is required for touch sensation and associates with microtubules and adhesion complexes. BMC Dev Biol. 2008;8(110):110. doi: 10.1186/1471-213X-8-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohri K, Vorobiev S, Fedorov AA, Almo SC, Ono S. Identification of functional residues on Caenorhabditis elegans actin-interacting protein 1 (UNC-78) for disassembly of actin depolymerizing factor/cofilin-bound actin filaments. J Biol Chem. 2004;279(30):31697–31707. doi: 10.1074/jbc.M403351200. [DOI] [PubMed] [Google Scholar]
- 22.Voegtli WC, Madrona AY, Wilson DK. The structure of Aip1p, a WD repeat protein that regulates Cofilin-mediated actin depolymerization. J Biol Chem. 2003;278(36):34373–34379. doi: 10.1074/jbc.M302773200. [DOI] [PubMed] [Google Scholar]
- 23.Ying W, et al. Ganetespib, a unique triazolone-containing Hsp90 inhibitor, exhibits potent antitumor activity and a superior safety profile for cancer therapy. Mol Cancer Ther. 2012;11(2):475–484. doi: 10.1158/1535-7163.MCT-11-0755. [DOI] [PubMed] [Google Scholar]
- 24.Hegyi H, Buday L, Tompa P. Intrinsic structural disorder confers cellular viability on oncogenic fusion proteins. PLOS Comput Biol. 2009;5(10):e1000552. doi: 10.1371/journal.pcbi.1000552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Z, et al. Inhibition of ALK, PI3K/MEK, and HSP90 in murine lung adenocarcinoma induced by EML4-ALK fusion oncogene. Cancer Res. 2010;70(23):9827–9836. doi: 10.1158/0008-5472.CAN-10-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, et al. Fusion of EML4 and ALK is associated with development of lung adenocarcinomas lacking EGFR and KRAS mutations and is correlated with ALK expression. Mol Cancer. 2010;9:188. doi: 10.1186/1476-4598-9-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thunnissen E, et al. EML4-ALK testing in non-small cell carcinomas of the lung: A review with recommendations. Virchows Arch. 2012;461(3):245–257. doi: 10.1007/s00428-012-1281-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 29.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 11):1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 30.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.