Significance
Deafness is caused largely by the death of sensory hair cells in the inner ear. In contrast to nonmammalian vertebrates, human hair cells do not regenerate. Understanding the mechanisms that regulate hair cell regeneration in zebrafish may shed light on the factors that prevent hair cell regeneration in mammals. RNA-Seq analysis of regenerating sensory organs uncovered dynamic changes in the expression of signaling pathways during zebrafish hair cell regeneration. Unexpectedly, the Wnt/β-catenin, Notch, and Fgf pathways are downregulated following hair cell death, whereas the Jak1/Stat3 pathway and cell cycle are activated. We propose that mimicking the zebrafish activation status of a combination of pathways at the correct time points in mammals may improve the chances of triggering regeneration of functional hair cells.
Keywords: signaling pathway analysis, RNA sequencing, neuromast, Jak/Stat3, cdkn1b
Abstract
Deafness caused by the terminal loss of inner ear hair cells is one of the most common sensory diseases. However, nonmammalian animals (e.g., birds, amphibians, and fish) regenerate damaged hair cells. To understand better the reasons underpinning such disparities in regeneration among vertebrates, we set out to define at high resolution the changes in gene expression associated with the regeneration of hair cells in the zebrafish lateral line. We performed RNA-Seq analyses on regenerating support cells purified by FACS. The resulting expression data were subjected to pathway enrichment analyses, and the differentially expressed genes were validated in vivo via whole-mount in situ hybridizations. We discovered that cell cycle regulators are expressed hours before the activation of Wnt/β-catenin signaling following hair cell death. We propose that Wnt/β-catenin signaling is not involved in regulating the onset of proliferation but governs proliferation at later stages of regeneration. In addition, and in marked contrast to mammals, our data clearly indicate that the Notch pathway is significantly down-regulated shortly after injury, thus uncovering a key difference between the zebrafish and mammalian responses to hair cell injury. Taken together, our findings lay the foundation for identifying differences in signaling pathway regulation that could be exploited as potential therapeutic targets to promote either sensory epithelium or hair cell regeneration in mammals.
Hearing disorders are the most common sensory disabilities in humans and can be either inherited or acquired. Degeneration and death of sensory hair cells in the inner ear is the underlying cause of age-related, noise-, antibiotic-, or chemotherapy drug-induced hearing loss (www.nidcd.nih.gov). Such loss is permanent, because mammalian inner ear hair cells do not regenerate. The inability of mammals to restore sensory hair cells after injury is intriguing given that (i) mammalian vestibular support cells exhibit low levels of proliferation after injury; (ii) cochlear support cells of newborn mice are able to proliferate subsequent to hair cell death in cell culture (1); and (iii) other vertebrates such as birds, amphibians, and fish replace their sensory hair cells continuously and, importantly, are capable of regenerating hair cells after damage (2–10).
Work in birds has shed light on the mechanisms regulating sensory hair cell regeneration in vertebrates (2, 4, 11–18). For instance, during sensory hair cell regeneration in the chick, the majority of hair cells arise by proliferation of neighboring support cells (19). Given that mammalian support cells do not respond to hair cell death by proliferating, molecular studies in chick have focused on identifying the mitogenic signals as well as the target genes that induce re-entering of the cell cycle in support cells (11, 20–22). For example, the transcription factors Atoh1 and Sox2 and the Wnt/β-catenin and Notch pathways have been shown to play crucial roles (23), and gene-expression analyses using microarrays have identified hundreds of genes involved in chick hair cell regeneration (18, 24).
The Wnt/β-catenin pathway is used repeatedly during hair cell development, and recent studies have emphasized the importance of Wnt/β-catenin signaling in modulating the proliferation of support cells. For example, activated β-catenin causes proliferation in dissociated chicken utricular epithelia and ectopic expression of hair cell differentiation in the chicken basilar papilla (17, 25). Likewise, during mouse cochlear development Wnt/β-catenin signaling activates proliferation of support cells and is required subsequently for their differentiation into hair cells (3, 26–29).
Perturbing the signaling pathways known to be activated during chick inner ear regeneration therefore may provide an opportunity to modulate regenerative capacities in the mammalian inner ear. For instance, it has been shown recently that down-regulation of Notch signaling in adult mice after injury induces the transdifferentiation of support cells into hair cells and can partially restore hearing (30). However, this manipulation causes the loss of support cells, and the implications of this loss for long-term hearing restoration are unknown.
Significant progress has been made in the study of sensory hair regeneration. However, in part because of the complexity of spatial and temporal gene interactions and signaling dynamics, our knowledge still is too incomplete to enable us to promote the generation of a functional epithelium after injury in mammals. The zebrafish lateral line provides an experimentally accessible system to define the complex signaling events triggered by injury and regeneration, because these cells can be killed acutely and then regenerate rapidly (31). These characteristics allow the detection of changes in gene expression at defined time points following hair cell death.
Zebrafish and the majority of all other aquatic vertebrates possess hair cells not only in their vestibular system in the ear but also in their skin. These hair cells are part of the sensory lateral line system. The lateral line system senses water motion and initiates the appropriate behavioral response for capturing prey, avoiding predators, and schooling. The morphology and function of sensory hair cells are evolutionarily conserved from fish to mammals (32, 33). Lateral line hair cells are located in the center of a mechanosensory organ known as the neuromast and are surrounded by inner support cells and an outer ring of mantle cells. Importantly, despite the unusual location of the hair cells on the trunk, lateral line and ear hair cells develop and differentiate by similar developmental mechanisms. For example, mutations in genes causing deafness in humans also disrupt hair cell function in the zebrafish lateral line and vestibular system (34). Because of its genetic tractability and the accessibility of sensory organs to experimental manipulation and visualization, the zebrafish recently has been recognized as an excellent model for discovering and functionally characterizing genes crucial for hair cell development and regeneration (16, 19, 35, 36).
In zebrafish hair cell regeneration occurs via the proliferation and differentiation of inner support cells (5, 6, 9, 37, 38), but detailed lineage analyses have not yet been performed, and the stem cell population has not been identified. Mantle cells are good stem cell candidates, because they migrate to form neuromasts on regenerating tail tips and also form postembryonic neuromasts by a process called “stitching.” However, after chemical ablation of hair cells by antibiotic treatment, mantle cells proliferate much less frequently than inner support cells (9). Possibly, mantle cells replenish inner support cells that then differentiate into hair cells. Mantle cells have not been shown to differentiate directly into hair cells (37, 38). Therefore, both mantle and inner support cells seem to be involved in reestablishing proper neuromast morphology after hair cell death.
To identify the activation status of signaling pathways during the first few hours of regeneration, we performed RNA-Seq gene expression analyses of zebrafish mantle and inner support cells isolated by FACS. We discovered that the Notch pathway is transiently down-regulated immediately after hair cell death; to our knowledge, this down-regulation has not been previously reported in other studies of regenerating species. Our studies also demonstrate that Wnt/β-catenin signaling is not among the earliest pathways to respond to hair cell death but instead is up-regulated relatively late during the regenerative response. These results suggest that Wnt/β-catenin signaling does not trigger proliferation by down-regulating cyclin-dependent kinase inhibitors immediately after hair cell death but rather regulates proliferation at later stages of regeneration. Our findings have defined the temporal regulation of key signaling pathways associated with the early stages of hair cell regeneration and suggest that the successful initiation of mammalian hair cell regeneration, either in vivo or in vitro, might require precise temporal manipulation of these pathways.
Results
Isolation of Mantle and Inner Support Cells by FACS.
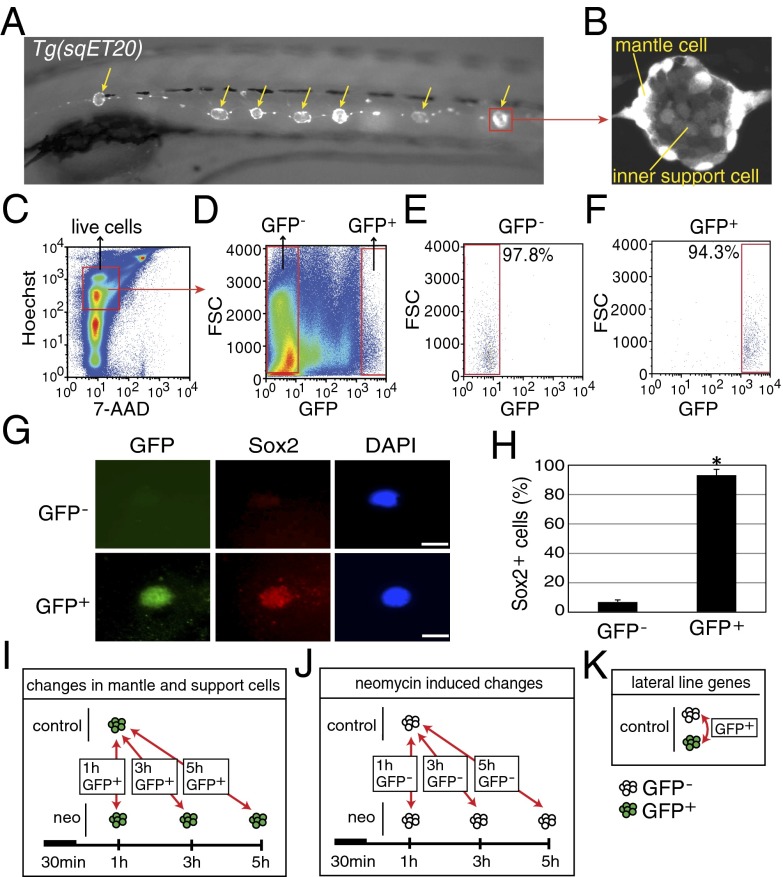
To identify changes in gene expression in mantle and inner support cells after hair cell death, we isolated GFP+ cells from regenerating and control Tg(sqET20) transgenic zebrafish via FACS (Fig. 1 A and B) (39). Tg(sqET20) larvae express GFP strongly in mantle cells and to a lesser degree in inner support cells (Fig. 1B). Hair cells are GFP− and therefore are excluded from the GFP+ population. We isolated GFP− and GFP+ cells from Tg(sqET20) control larvae at 5 days postfertilization (dpf) and from neomycin-treated larvae at 1, 3, and 5 h after neomycin treatment. Larvae were dissociated into a single-cell suspension in a trypsin solution and stained with 7-aminoactinomycin D (7-AAD) and Hoechst 33342 to exclude dead cells and cell debris (Fig. 1C). Live GFP+ cells (7-AAD−, Hoechst+, GFP+ cells) and control GFP− cells (7-AAD−, Hoechst+, GFP− cells) were sorted by FACS (Fig. 1D). The sorting yielded highly GFP+-enriched cell populations, as demonstrated by a postsort reanalysis of the collected cells (Fig. 1 E and F). GFP+-sorted cells showed 94.4 ± 1.3% enrichment (n = 7). To confirm that GFP+ cells are lateral line cells, we performed immunostaining with an antibody against the support cell marker Sox2 that labels inner support cells as well as some mantle cells (Fig. 1G and Fig. S1) (40). Counts of Sox2+ FACS-purified cells revealed that 95% of GFP+ cells but only 6.6% of GFP− cells were positive for Sox2 (Fig. 1H). Additionally, we performed RT-PCR for genes highly expressed in neuromast cell types and with limited expression in other tissues. RT-PCR for gfp, eya1, klf4b, and cldnb showed that these genes are expressed at significantly higher levels in GFP+ than in GFP− cells (Fig. S2 A–D). In contrast, genes expressed in tissues outside the lateral line system, such as rhodopsin (rho, eye), villin1 (vil1, gut), and myod1 (muscle), are expressed at much lower levels in GFP+ cells (Fig. S2 E–G). In Tg(sqET20) larvae, mantle cells express GFP most strongly, but inner support cells respond more robustly to hair cell death by proliferating (5, 6). Because Tg(sqET20) larvae show weaker GFP expression in inner support cells, we were concerned that our cell sorts would exclude inner support cells. However, RT-PCR with inner support cell markers such as klf4b showed that inner support cells were included in the GFP+ cell populations (Fig. S2C). Combined, these studies demonstrated that our FACS protocol yielded a highly enriched pool of lateral line cells containing mantle and inner support cells but not hair cells.
Fig. 1.
Purification of mantle and inner support cells from Tg(sqET20). (A) In 5-dpf Tg(sqET20) larvae GFP is expressed in neuromasts (arrows) and in the interneuromast cells that connect adjacent neuromasts. (B) Magnification of a neuromast shows that mantle cells are strongly labeled by GFP, but inner support cells are weakly labeled. (C and D) Dissociated cells of Tg(sqET20) larvae at 5 dpf were FACS sorted using a two-gate sorting strategy. Live cells were gated by 7-AAD− and Hoechst+ (C) and subsequently were gated by forward scatter (FSC) and GFP intensity to isolate GFP− and GFP+ cells (D). (E and F) A postsorting experiment shows that GFP− cells are 97.8% pure (E) and GFP+ cells are 94.3% pure (F). (G) Sorted GFP− and GFP+ cells were stained with Sox2 antibody and DAPI. (Upper) A GFP− cell is Sox2−. (Lower) A GFP+ cell expresses Sox2. (Scale bars: 5 μm.) (H) Quantification of Sox2+ cells in each population as mean ± SD; *P < 0.01 by t test. (I) Regeneration-induced mantle and inner support cell genes were identified at each time point by comparing GFP+ neomycin-treated cells with GFP+ untreated samples. (J) Neomycin-induced genes were identified by comparing GFP− neomycin-treated cells at each time point with GFP− untreated samples. Neomycin-induced genes subsequently were subtracted from the gene lists obtained with GFP+ cells to select for regeneration-specific changes in gene expression in inner support and mantle cells. (K) Comparisons between GFP+ and GFP− cells of untreated larvae generated a list of genes enriched in lateral line cells.
To investigate the temporal activation or repression of individual genes and signaling pathways in GFP+ mantle and inner support cells during the first few hours of regeneration, we examined GFP+ and GFP− cells at 1, 3, and 5 h after a 30-min treatment with neomycin (Fig. 1I). Sorts for each time point were performed in triplicate and subjected to RNA-Seq analysis. Bactericidal antibiotics cause oxidative stress in mammalian cells, leading us to speculate that sublethal concentrations of neomycin may cause changes in gene expression unrelated to hair cell death (41). To control for this nonspecific effect, we examined gene expression in GFP− cells in the presence or absence of neomycin (Fig. 1J). Although this population includes all larval nonlateral cells, it allows us to control for a nonspecific neomycin effect when comparing GFP+ cells from treated and control larvae.
Analysis of the RNA-Seq Results.
Sequencing libraries were prepared from mRNAs of collected cells and were quality checked on a Bioanalyzer. We performed 50-bp single-end RNA-Seq using the Illumina platform. To test the enrichment of lateral line genes further, we examined the fragments per kilobase of transcript per million mapped reads (FPKM) values for known lateral line genes and for genes expressed in other tissues (Fig. S3). The results confirmed the RT-PCR analysis described above. Pan lateral line, as well as inner support cell, markers show much higher FPKM values in FACS-sorted GFP+ cells than in GFP− cells. As suggested by the RT-PCR results, the FPKM values for the inner support cell markers pea3 and klf4b are significantly higher in the GFP+ than in the GFP− cells, confirming that inner support cells were included in our FACS sorts. This enrichment of lateral line genes is supported by the information in Dataset S1, in which neuromast-specific genes were identified by comparing the expression profiles of untreated GFP+ and GFP− cells. In addition to known lateral line genes, the resulting lists of differentially expressed genes provide a valuable resource of as yet uncharacterized genes that potentially could play important roles in hair cell development and/or regeneration (Fig. 1K and Dataset S1). The number of genes enriched in GFP+ cells relative to GFP− controls at an adjusted p-value ≤0.05 is 1,670 (SI Materials and Methods and Dataset S1). This dataset also contains many of the genes reported in the dataset of Steiner et al. (42), who identified mantle cell-specific genes using a different transgenic line (see below).
Gene Identification for Each Time Point.
To identify genes from transcripts specifically enriched or depleted in GFP+ mantle and inner support cells after hair cell death, we created several comparisons between the RNA-Seq datasets. Ratios of gene expression were created between the neomycin-treated GFP+ cells at 1, 3, and 5 h and the nontreated GFP+ cells at 1 h to identify genes responding to hair cell death (Fig. 1I). To control for a nonspecific neomycin response, a similar set of ratios was created between the neomycin-treated GFP− cells at each time point and the untreated GFP− cells at 1 h (Fig. 1J). The neomycin-induced genes were subtracted from all gene lists generated with GFP+ cells. We then used ratios and P values between datasets to select genes of interest at any given time point (SI Materials and Methods and Dataset S2). Genes identified by these criteria are marked with a numeric flag with positive numbers indicating up-regulated and negative numbers indicating down-regulated genes. The numeric value indicates the time point at which a gene is up- or down-regulated (Dataset S2, flagged column). A principal component analysis of the biological replicates of GFP− and GFP+ cell populations at the three different time points demonstrates that GFP+ cells are very different from GFP− cells. In addition, cell sorts performed in triplicate for each time point are highly reproducible (Fig. S4).
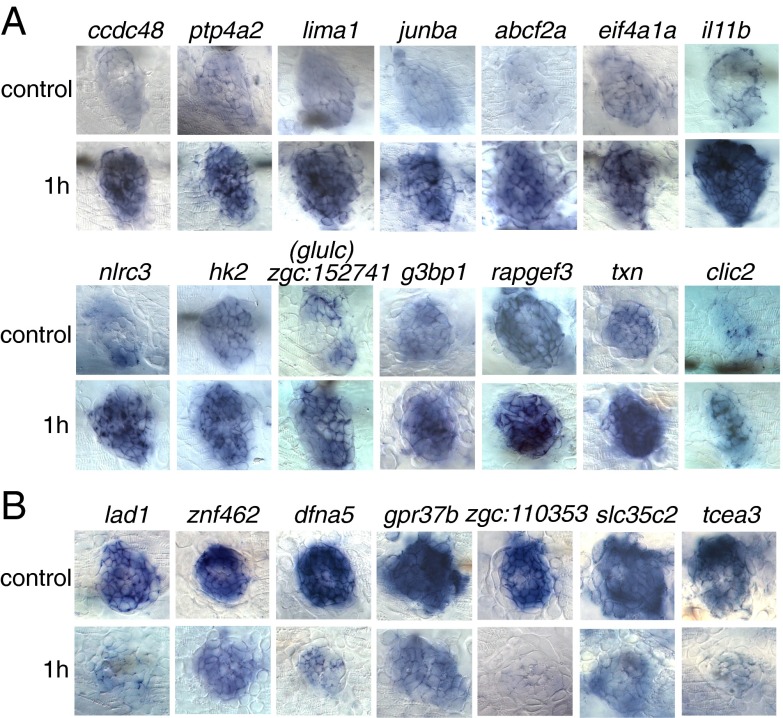
To define a set of the top 100 up- and down-regulated genes to use as candidates for validation, we ranked 193 up-regulated and 200 down-regulated significant (flagged) genes from the 1-h dataset as a function of the ratio and general abundance (SI Materials and Methods and Table S1). We validated the RNA-Seq results by performing in situ hybridizations with 28 up-regulated and 21 down-regulated genes selected from the top 100 gene list in larvae 1 h after neomycin treatment (Table S2). All 28 up-regulated genes are expressed in the lateral line, and 20 of these genes show up-regulation by in situ hybridization after neomycin treatment. Of the 21 down-regulated genes, 19 are expressed in the lateral line, and 12 genes are detectably down-regulated by in situ hybridization (Fig. 2 and Table S2). These experiments demonstrated that the FACS sorting followed by RNA-Seq analysis produced high-quality results that enable us to study hair cell regeneration in zebrafish in detail.
Fig. 2.
Validation by in situ hybridization of a selection of 14 genes up-regulated (A) and seven genes down-regulated (B) at 1 h after neomycin treatment. (Upper Rows) Untreated larvae (control) at 5 dpf. (Lower Rows) Treated larvae.
Pathway Analysis at 1, 3, and 5 h After Hair Cell Death.
The Wnt/β-catenin, Notch, Jak/Stat, Bmp, and Fgf pathways are important for hair cell development as well as for the regeneration of hair cells in fish and chick epithelia (15, 23, 43). Our understanding of which pathways act upstream and how these pathways affect each other is still very limited. We performed a temporal expression analysis at 1, 3, and 5 h after hair cell death to determine when particular pathways are activated or inhibited; this temporal information provides clues into how they might interact with each other. We also aimed to identify other signaling pathways and record any dynamic changes in their behavior that could indicate how they integrate with the Notch and Wnt/β-catenin signaling pathways. To identify enriched pathways in our datasets, we used the signaling pathway impact analysis (SPIA) package in R (44). This software tool not only takes into account how many genes belong to a pathway but also focuses on the importance of a gene by analyzing the perturbation of the pathway if the gene expression changes. Therefore, even pathways that are represented by only a small number of genes can be classified as enriched if these genes are important regulators of the pathway. For the SPIA analysis we included all flagged genes (flagged column) listed in Dataset S2. For each time point the up- and down-regulated genes were combined and analyzed against a panel of pathways from zebrafish defined in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (45). The 1-h postneomycin treatment time point contained 509 genes from the National Center for Biotechnology Information Entrez database. The 3-h postneomycin treatment time point contained 401 Entrez genes, and the 5-h postneomycin treatment time point contained 418 genes.
The results of the SPIA analysis are shown in Table S3. Automated pathway annotation is powerful but subject to caveats, because genes can be annotated to function in several pathways, some of which might not be active in the lateral line. In addition, in the KEGG database, the Wnt pathway encompasses the canonical Wnt/β-catenin, the PCP, and the Wnt- regulated calcium pathways, potentially providing erroneous status assignment of the Wnt/β-catenin pathway. We therefore tested the SPIA analysis by in situ hybridization experiments with pathway members at various time points of regeneration (see below and Figs. 3–5). For improved visualization we generated heat maps of log2 ratio values of KEGG pathway members. We show only the genes that were flagged in our analysis and reported to be expressed in the lateral line by the zebrafish information network (www.zfin.org) or prior reports (46, 47).
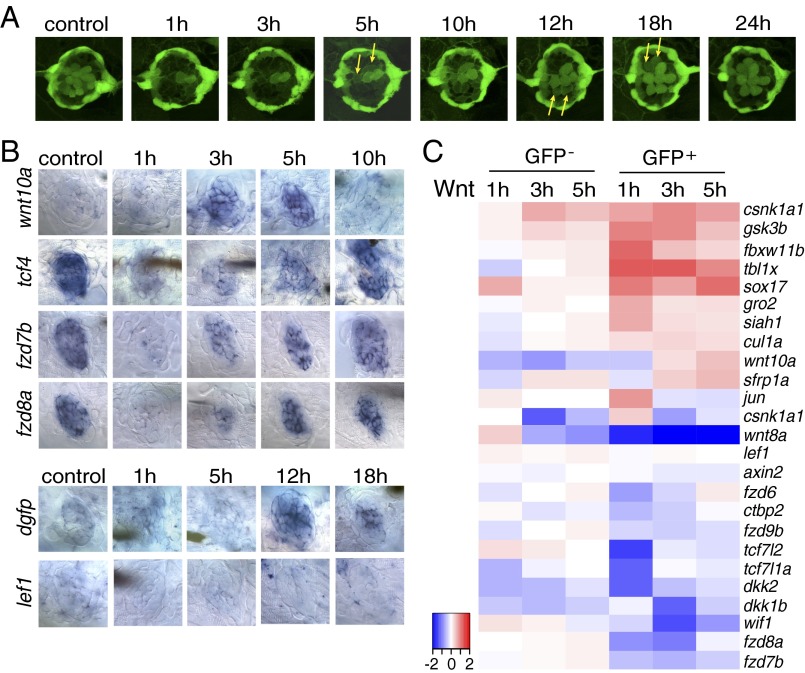
Fig. 3.
The Wnt/β-catenin pathway is inactive during the early stages of hair cell regeneration. (A) Still images of a Tg(sqET20;sqET4) larval neuromast in the process of neomycin-induced regeneration. All hair cells, except for two immature hair cells, were killed by neomycin by 1 h after neomycin treatment. Two newly formed hair cells (arrows) start to express GFP at 5 h, and other pairs of hair cells (arrows) appear at 12 and 18 h. (B) In situ hybridization of Wnt/β-catenin pathway genes in 5-dpf untreated (control) larvae and in larvae at different time points after neomycin treatment. Expression of wnt10a is increased at 3 and 5 h. Expression of tcf7l2 (tcf4), fzd7b, and fzd8a is largely decreased at 1 h after neomycin. The Wnt/β-catenin reporter line Tg(Tcf/Lef-miniP:dGFP) demonstrates activation starting at 12 h as shown by dgfp expression, but lef1 is not induced in regenerating neuromasts. (C) Heat map of selected Wnt/β-catenin pathway genes based on RNA-Seq results. Log2 ratios of GFP− and GFP+ cells at 1, 3, and 5 h after neomycin treatment are color coded (red: up-regulation, blue: down-regulation). Genes that are modulated in GFP− cells are neomycin-induced genes.
Fig. 5.
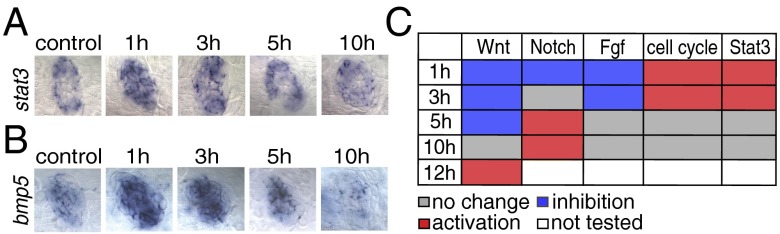
The Jak1/Stat3 pathway is activated during the early stages of hair cell regeneration. (A) In situ hybridization shows that stat3 is increased at 1 and 3 h after neomycin treatment. (B) bmp5 is up-regulated at 1 h after neomycin treatment in situ. (C) Table depicting the status of signaling pathways at different time points after hair cell death. All comparisons (no change, inhibition, or activation) were evaluated relative to untreated control samples. The Wnt, Notch, and Fgf pathways are inhibited immediately after neomycin treatment, whereas the cell cycle and Jak1/Stat3 pathways are activated during the early stages of regeneration. The Wnt and Notch pathways are reactivated at 12 h and 5 h after neomycin treatment, respectively.
The Wnt/β-Catenin Pathway Is Not Activated During Early Stages of Lateral Line Hair Cell Regeneration.
The analysis of hair cell regeneration in a Tg(sqEt20;sqEt4) double-transgenic larva allows us to draw comparisons between gene expression and cellular changes at particular time points (Fig. 3A) (39). At 1 h after neomycin treatment almost all hair cells have been eliminated. Two hair cells remain that were too immature to be killed by neomycin. Proliferation of inner support cells begins within the first 2 h of regeneration (37). At 5 h two newly differentiating hair cells start to express GFP (Fig. 3A; arrows), and by 12 h after neomycin treatment six hair cells are present.
Wnt/β-catenin signaling is required for proliferation in developing and regenerating neuromasts (26, 48). Unexpectedly, the SPIA pathway analysis suggested that the Wnt/β-catenin pathway is inactive at 1 h after hair cell death (Table S3). To investigate this finding in more detail, we performed in situ hybridization experiments with Wnt/β-catenin pathway genes in untreated and treated larvae at various time points after hair cell death. Our analysis showed that Wnt/β-catenin signaling is not active in 5-dpf untreated control neuromasts. The ligand wnt10a is not expressed in 5-dpf control neuromasts, and we failed to detect any expression of dgfp in the Tg(Tcf/Lef-miniP:dGFP) Wnt/β-catenin reporter line (Fig. 3B) (49). Even though Wnt/β-catenin signaling is inactive, the β-catenin cotranscription factor tcf7l2 (tcf4) is expressed. Likewise, the Wnt receptors fzd7b and 8a are present, although whether they act as receptors for canonical or noncanonical Wnt signaling in neuromasts is unknown (Fig. 3B) (50, 51).
At 1 h after neomycin treatment tcf7l2 (tcf4), frz7b, and fzd8a are drastically down-regulated, and, along with wnt10a, their expression in the Wnt/β-catenin Tg(Tcf/Lef-miniP:dGFP) reporter line remains low or undetectable. At 3 h after neomycin treatment wnt10a, tcf7l2 (tcf4) and fzd7b, and fzd8a are up-regulated, leading to Wnt/β-catenin pathway activation at 12 h as demonstrated by dgfp expression in the Tg(Tcf/Lef-miniP:dGFP) reporter line (Fig. 3B). The activation status of other Wnt/β-catenin pathway genes at the different time points is shown in Fig. 3C. Genes that change expression in the GFP− cells represent neomycin-induced genes. Our analyses reveal that Wnt/β-catenin signaling is a late-responding pathway and does not play a role in the initiation of regeneration.
The Notch Pathway Is Inhibited 1 h After Neomycin Treatment.
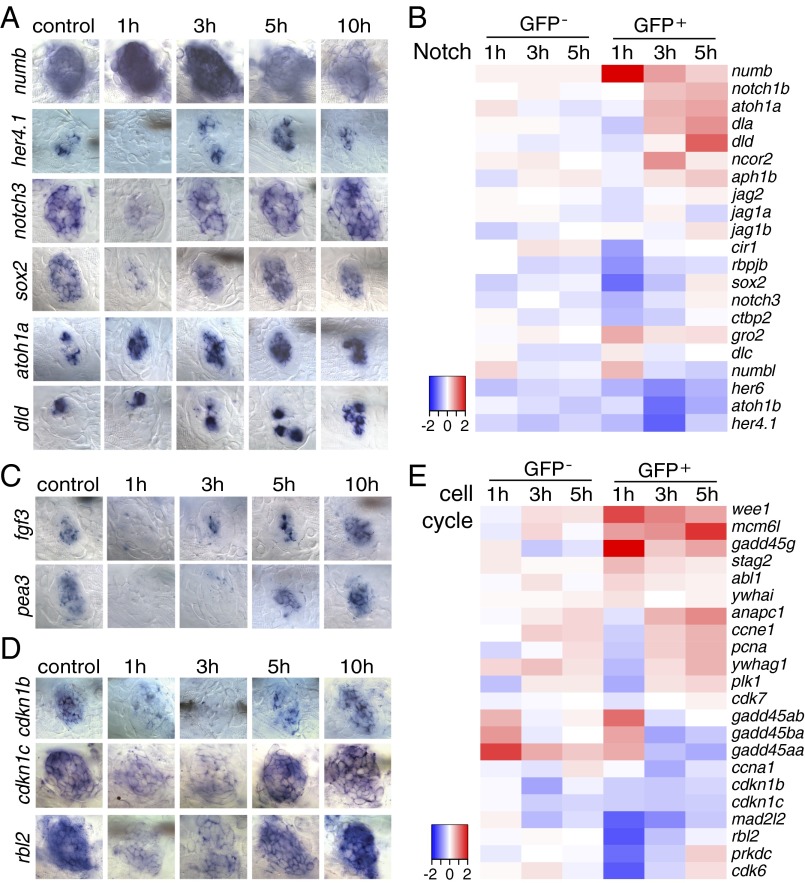
The Notch pathway is of particular interest because experimental down-regulation of Notch induces transdifferentiation of mammalian support cells into hair cells (30, 52–57). In contrast to previous reports in zebrafish and chicken (9, 13), our SPIA pathway analysis suggested that the Notch pathway is inhibited immediately after hair cell death because of simultaneous up-regulation of numb (Fig. 4 A and B). Numb inhibits Notch activity by regulating postendocytic sorting events and degradation of the Notch receptor and thereby regulates neural stem cell homeostasis (58, 59). However, because Numb is a multifunctional membrane-associated protein with additional functions independent of the regulation of Notch signaling, we tested if the Notch pathway is indeed down-regulated by in situ hybridization of Notch pathway members. Our analysis of the Notch target genes her4.1 and notch3 confirms that Notch signaling is attenuated immediately 1 h after neomycin treatment and is accompanied by down-regulation of sox2 (Fig. 4 A and B) and up-regulation of atoh1a (Fig. 4 A and B). The atoh1a target deltaD (dld) is up-regulated between 3–5 h after neomycin treatment, reflecting the appearance of hair cell precursors (Fig. 4 A and B) (60). Notch signaling must be reinitiated for proper fate specification into hair and support cells and proper organ size control (9, 60). Accordingly, between 3 and 5 h after neomycin treatment (Fig. 4 A and B), we observed renewed activation of the Notch target genes her4.1, notch3, and sox2 that likely is caused by the up-regulation of dld (60).
Fig. 4.
Inhibition of Notch and Fgf pathways is accompanied with cell cycle reentry immediately after hair cell ablation. (A, C, and D) In situ hybridization of Notch pathway, Fgf pathway, and cell cycle genes in 5-dpf untreated and neomycin-treated larvae. (A) The Notch pathway inhibitor numb is increased at 1 h after neomycin. The Notch target genes her4.1, notch3, and sox2 are decreased at 1 h and up-regulated at 3 h after neomycin treatment. atoh1a is up-regulated starting at 1 h after neomycin, and its target dld is increased at 5 and 10 h. (C) fgf3 and its target pea3 are down-regulated at 1 and 3 h after neomycin treatment. (D) Cell cycle inhibitory genes cdkn1b, cdkn1c, and rbl2 are decreased at 1 and 3 h after neomycin compared with control. (B and E) Heat maps of selected Notch pathway genes and cell cycle genes.
We postulate that down-regulation of Notch signaling is a characteristic event during hair cell regeneration in zebrafish. In contrast to zebrafish, the Notch target genes are decreased only modestly, and Notch signaling is still active, in hair cell-depleted neonatal mouse cochleae and 3- to 4-wk-old mouse utricles (55, 56, 61). Therefore we hypothesize that the failure of transient Notch down-regulation in mammals could be one of the reasons that mammalian hair cells do not regenerate spontaneously after injury.
Fgf Signaling Is Down-Regulated Immediately After Hair Cell Death.
The Fgf pathway initiates dld and subsequently Notch signaling in the mouse cochlea, chicken inner ear, and zebrafish lateral line neuromasts (23, 60). Both pathways are active in 5-dpf untreated neuromasts (Fig. 4 A and C and Fig. S5D). Identically to Notch pathway members, fgf3 and its transcriptional target pea3 are turned off 1 h after neomycin treatment. However, the Notch target her4.1 is reactivated at 3 h, whereas pea3 is not transcribed until 5 h (Fig. 4 A and C). Therefore, it is possible that Notch signaling does not depend on Fgf signaling during regeneration; however, this hypothesis needs to be tested. The rapid down-regulation of Fgf signaling is in accordance with findings in chicken sensory epithelia, in which Fgfr3 is highly expressed in support cells but is down-regulated rapidly after hair cell death (62). Additionally, in the chicken down-regulation of Fgf signaling is required for regeneration to occur, because the addition of Fgf inhibits support cell division after damage (63).
The Cell Cycle Exit Genes cdkn1b, cdkn1c, and rbl2 Are Down-Regulated at 1 h After Hair Cell Death.
The failure to down-regulate cell cycle exit regulators is one of the reasons mammalian support cells do not proliferate in response to hair cell death (10, 12, 64, 65). Determining the earliest changes in the expression of cell cycle regulators and mapping correlating changes in signaling pathway activations will aid in identifying candidate pathways involved in cell cycle reentry. We detected the earliest changes in cell cycle genes at 1 h after neomycin treatment. The cyclin-dependent kinase (cdk) inhibitors cdkn1b (p27/kip1) and cdkn1c (p57/kip2), as well as the pocket domain-containing rbl2, are down-regulated 1 h after neomycin treatment and are not up-regulated again until 5–10 h after neomycin treatment (Fig. 4 D and E). Time-lapse analyses revealed that support cells enter the cell cycle 1–5 h after neomycin treatment (37), likely as a consequence of the down-regulation of the cdk inhibitors at 1 h after neomycin treatment. Because we did not observe any signs of Wnt/β-catenin signaling activation, we conclude that Wnt/β-catenin signaling is not required for the reentry of support cells into the cell cycle immediately after hair cell death. In contrast, down-regulation of Notch and Fgf signaling correlates with the down-regulation of cdk inhibitors. It is possible that active Notch or Fgf signaling induces cdk inhibitors, causing cells to exit the cell cycle or become quiescent (66). However, manipulation of Notch signaling does not affect the cell cycle in chick or murine inner ears (13, 30), suggesting that signaling pathways other than Wnt/β-catenin and Notch are involved in triggering or inhibiting proliferation.
The Jak1/Stat3 Pathway Is Up-Regulated Immediately After Neomycin Treatment.
The Jak1/Stat3 pathway regulates a plethora of cellular processes, such as cell proliferation, cell differentiation, and cell survival (67, 68). The Jak1/Stat3 pathway is up-regulated during zebrafish inner ear regeneration and is required for proliferation of cardiomyocytes during zebrafish heart regeneration (36, 69). Likewise, in zebrafish lateral line regeneration stat3 is up-regulated immediately, 1 h after neomycin treatment, as evidenced by in situ hybridization (Fig. 5A and Fig. S5A). Other Jak1/Stat3 pathway members that are highly up-regulated are jak1, interleukin 6 signal transducer (il6st), the stat inhibitor and target gene suppressor of cytokine signaling (socs3b), and the ligand interleukin 11b (il11b) (Fig. 2A and Fig. S5A). In zebrafish heart regeneration, the proliferation of cardiomyocytes is regulated by the Jak1/Stat3 downstream target relaxin 3a (rln3a). rln3a also is highly up-regulated in GFP+ cells 3 h after neomycin treatment (Fig. S5A) (69). Possibly, Jak1/Stat3 signaling regulates proliferation in support cells in the first few hours after hair cell death, whereas Wnt/β-catenin signaling is required for proliferation during later stages of regeneration.
Another signaling pathway that is modulated during zebrafish hair cell regeneration and that might interact with the pathways described above is the Bmp pathway. Bmp signaling influences hair cell differentiation during chicken and mouse inner ear development (70–73). During zebrafish hair cell regeneration, bmp5 is up-regulated between 1 and 5 h after hair cell death (Fig. 5B and Fig. S5B), whereas the BMP inhibitor noggin 3 (nog3) is not down-regulated until 5 h after hair cell death (Fig. S5B). Because ligand expression by itself is not a reliable indicator for signaling pathway activation, future studies will need to determine how the activation status of Bmp signaling is affected by bmp5 and nog3.
Other pathways that respond to zebrafish lateral line hair cell death are the Mapk (Fig. S5 C and D), TNF-α, (Fig. S5E), insulin (Fig. S5F), and apoptosis signaling (Fig. S5G) pathways and nitric oxide (NO) and reactive oxygen species (ROS) (Fig. S5H), some of which increase proliferation in undamaged chicken vestibular support cells (74). For example, we observed up-regulation of nox1, noxo1a, and noxa1 (Fig. S5H and Dataset S2). NO is a radical that regulates a wide variety of physiological processes and is important during liver regeneration (75).
We summarized the activation status of some of the major signaling pathways based on the in situ hybridization patterns of pathway members in Fig. 5C. The Wnt/β-catenin pathway is not active in homeostatic or newly regenerating neuromasts, and the Notch and Fgf pathways are down-regulated during the first few hours of regeneration. The Wnt/β-catenin and Notch pathways seem to play a role only during later stages of regeneration. The cell cycle is activated immediately, possibly via cytokine stimulation of the Jak1/Stat3 pathway.
Independent Microarray Studies Support the Differential Expression of Genes Not Associated with Key Signaling Pathways.
An independent expression analysis performed by Steiner et al. and reported in this issue (42), used a transgenic line with an alkaline phosphatase enhancer [Tg(-4.7alp:mCherry)] instead of the Tg(sqET20) line we used to isolate mantle cells by FACS analysis. We compared our respective datasets to determine how similar the data are. We first analyzed the overlap of genes enriched in untreated mantle/lateral line cells in both studies. The obtained results are a clear indicator of how similar the sorted cell populations in the two studies are. We chose the top 1,000 up-regulated genes from our Dataset S1 by p-value and then examined how many of these genes also are among the top 1,000 genes identified by Steiner et al. (dataset S1 in ref. 42). If the datasets were independent and unrelated, we would expect 53 genes to overlap by chance. We identified 201 common genes with a p-value of 3.9e-107, demonstrating that our two datasets are significantly related. All six mantle cell-specific genes identified by Steiner et al. (42) also show enrichment in our untreated GFP+ cell population (Table S4A, blue rows). Ten of the 26 top up- and down-regulated genes that we validated by in situ hybridization after hair cell loss (Table S2) and for which we found a match in the Steiner dataset also show significant changes in the Steiner microarray dataset (Table S4B, blue rows) (42). Several of the genes that were not present in the Steiner dataset are expressed more highly by in situ hybridization in inner support cells than in mantle cells, reflecting the nature of the transgenic lines used and the type of cells purified. Nine of the 14 genes that Steiner et al. identified by microarray analysis, by RT-PCR, or by in situ hybridization (42) also showed the same trend in expression changes in our RNA-Seq analysis (Table S4C, blue rows).
The overlap between a subset of our data and that of Steiner et al. (42), combined with the identification and corresponding validation of genes in our study, strongly indicates that the two lines of investigation are complementary, with most of the targets identified by Steiner et al. (42) found in the mantle cells and the majority of genes identified in our study expressed primarily in the inner support cells. As such, our respective studies both validate each other and help resolve the cell-type specificity of the revealed differential gene expressions.
Discussion
Several studies have aimed to induce mammalian hair cell regeneration by inactivating or activating candidate genes and pathways. Knockdown of the cell cycle exit regulators Cdkn1b (p27/Kip1), p19/Ink4d, and pRb led to the initiation of cell cycle reentry and the differentiation of hair cells in injured epithelia in mice (20, 21, 64, 76, 77). Unfortunately, these ectopic hair cells cannot be maintained and eventually apoptose. Because direct manipulation of cell cycle regulators has not been successful in reconstituting a fully functional sensory epithelium in mammals, it is important to identify the mitogens that activate cell cycle entry in regenerating organisms. It is plausible that manipulations of upstream signaling events are a more promising avenue to regenerate mammalian hair cells.
More recently, in mice the down-regulation of Notch signaling caused transdifferentiation of support cells into functional hair cells without inducing proliferation (30). However, the number of regenerated hair cells was modest, and it is unclear if the accompanied depletion of support cells affects the function of the partially regenerated sensory epithelium in the long term. Although these efforts have not resulted in the regeneration of a fully functional, morphologically normal epithelium, they clearly demonstrate that the mammalian cells of the inner ear can be manipulated and coaxed away from their terminally differentiated state.
Knowledge gained from studying the development of hair cells has demonstrated that the underlying molecular mechanisms are complex, with a multitude of interacting signaling pathways and feedback loops that ensure that an unspecified progenitor cell transitions to a prosensory cell that ultimately differentiates into a functional sensory cell. We also have learned that signaling pathways are activated and inhibited repeatedly as hair cell progenitors are specified and differentiate, further complicating their experimental manipulation. Hence, a global up- or down-regulation of a particular pathway is unlikely to lead to fully functional sensory epithelia.
We hypothesize that the regeneration of a complete sensory epithelium will require the manipulation of several pathways in the correct order. However, the model systems currently used to study the regeneration of sensory epithelia present unique obstacles. For example, in chicken, antibiotic-induced hair cell death occurs over a 24-h period, and the proliferation of support cells and differentiation of hair cells occurs 1–4 wk after injury (19). This long time frame makes it difficult to determine the precise time point when pathways are turned on and off, because support cells will be in different stages of regeneration. Likewise, a previous expression study of the regenerating zebrafish ear was performed 24 h after the larvae were exposed to noise that does not cause death simultaneously in all hair cells (36). Further adding to the complexity, genes are used repeatedly during hair cell development and can turn from an activator into a repressor (23). We have selected the zebrafish lateral line tissue to elucidate the gene-expression patterns triggered by hair cell death. The zebrafish lateral line is particularly well suited for these analyses, because all hair cells can be killed acutely within 30 min, and regeneration occurs within 24 h (Fig. 3A), allowing us to determine the earliest signals and responses in support cells after hair cell death.
Wnt/β-Catenin Signaling Is Not Detected Early in Lateral Line Hair Cell Regeneration.
Our analyses focused on pathways involved in hair cell development as they might be reused during regeneration (15, 23, 43, 78, 79). For example, Wnt/β-catenin signaling is crucial for the development of many organs across the animal kingdom (80). During sensory development, Wnt/β-catenin signaling governs the proliferation of unspecified progenitor cells, renders them competent to acquire a neural cell fate, and subsequently is involved in the differentiation of these postmitotic prosensory cells (3, 23, 81). Wnt/β-catenin signaling also is involved in regeneration. Wnt/β-catenin signaling controls stem cell maintenance, quiescence, and proliferation during mammalian epithelial hair cell, zebrafish fin, mammalian intestine, hydra head, and planarian regeneration (82–86). Importantly, Wnt/β-catenin also is crucial for regulating proliferation in developing and regenerating zebrafish neuromasts (26, 48).
In the mouse cochlea, ectopic Wnt/β-catenin signaling promotes proliferation of Lgr5+ support cells via the down-regulation of Cdkn1c (p27/Kip1); however, this effect diminishes as the animals mature (3, 20, 28, 81, 87). Because Wnt/β-catenin signaling is down-regulated in the mammalian cochlea after birth, it was hypothesized that the absence of Wnt/β-catenin signaling is responsible for the inability of support cells to proliferate in response to hair cell death (3, 28, 29, 65). Therefore, recent studies have focused on up-regulating the Wnt/β-catenin pathway in uninjured cochleae with the expectation of coaxing mammalian support cells into proliferating and producing hair cells (3, 17, 28, 29, 81). These experiments indeed yielded additional hair cells, but the epithelium was disorganized.
Surprisingly, our data demonstrate that in zebrafish, as in adult mice cochleae, Wnt/β-catenin signaling is inactive in homeostatic neuromasts and is up-regulated only at 12 h after neomycin treatment. Therefore Wnt/β-catenin signaling is unlikely to play a role in the initial triggering of the regenerative response in zebrafish, even though it is sufficient and necessary to induce proliferation during later stages of regeneration, for example by inhibiting Cdkn1b (p27/Kip1) and regulating dyclinD1 (3, 27, 87).
Notch Signaling Is Down-Regulated Early in Lateral Line Hair Cell Regeneration.
A growing number of studies have explored how the manipulation of Notch signaling could aid in regenerating mammalian hair cells. It is well established that the inhibition of Notch signaling causes an increase in the number of hair cells in uninjured and injured chicken, guinea pig, and mouse utricles and cochleae and in injured zebrafish ears and lateral line neuromasts (52, 53, 55–57, 88). The presence of Notch is detrimental to hair cell differentiation, because it blocks the proneural gene atoh1a and thereby maintains support cells (9, 13, 65, 89, 90). The ability of support cells to transdifferentiate into hair cells after down-regulation of Notch signaling was exploited recently to restore a modest amount of hair cells in the injured adult mouse cochlea that led to some promising functional recovery of hearing (30). This experimental down-regulation of Notch signaling recapitulates our observations during zebrafish lateral line hair cell regeneration. In zebrafish, Notch signaling is transiently down-regulated 1 h after hair cell death, correlating with the attenuation of sox2 and up-regulation of atoh1a. The interactions between Notch signaling, sox2, and atoh1a have not yet been tested in the zebrafish lateral line; however, because of their behavior during regeneration, it is very likely that their regulation is similar to that occuring during mouse, Xenopus, and zebrafish central nervous system or sensory organ development (23, 52, 61, 91–93). During murine ear development Sox2 expression depends on Notch signaling and is required for the acquisition of neural potential in the prosensory epithelium (52). Subsequently, for neurogenesis to proceed, Sox2 is shut off via the accumulation of proneural genes, such as Atoh (93–97). In contrast, Sox2 is maintained in Notch+ support cells, where it inhibits hair cell differentiation by inhibiting the proneural basic helix-loop-helix gene Atoh1a (91).
It is plausible that the down-regulation of Notch signaling in neuromasts is required during the earliest stages of hair cell regeneration to down-regulate sox2 in support cells, allowing these cells to acquire a proneural character via the up-regulation of atoh1a. Because Notch signaling needs to be reinitiated for fate specification of hair and support cells and control of organ size (9, 60), the Notch target genes her4.1, notch3, and sox2 are again up-regulated between 3 and 5 h after neomycin treatment (Fig. 4 A and B). These genes likely are activated by the up-regulation of dld (60). Therefore, we hypothesize that the orchestration of genes downstream of Notch signaling appears to be the same during zebrafish hair cell development and regeneration.
It has not been appreciated previously that the down-regulation of Notch signaling could be a hallmark of spontaneously regenerating sensory epithelia, because it was reported that the Notch signaling pathway is up-regulated in response to hair cell death in chicken, guinea pigs, and zebrafish (9, 88, 98). However, these expression analyses were performed at later time points during the regeneration process, when Notch signaling is required once again for hair cell versus support cell specification to occur. We also observed that zebrafish Notch target genes are reexpressed at 3 h after hair cell death (Fig. 4A).
Because the zebrafish lateral line system allows us to kill hair cells very acutely, causing a relatively synchronized response in support cells, we were able to detect the immediate, transient down-regulation of Notch signaling after hair cell death. In the injured mammalian cochlea or utricle such drastic down-regulation of Notch target genes, such as Sox2 and several Hes and Hey genes, does not occur (55, 56); the absence of such down-regulation could be one of the reasons mammalian hair cells do not regenerate.
The Regulation of Notch Signaling in Combination with Mitogenic Signals May Offer Novel Therapeutic Strategies to Promote Hair Cell Regeneration in Mammals.
Our data suggest that identifying the molecules that down-regulate Notch signaling in regenerating species will be important in efforts to understand why mammals fail to regenerate injured hair cells. Our RNA-Seq analyses identified the transmembrane protein Numb as a potential candidate for inhibiting Notch signaling. Future studies should focus on the sufficiency of Numb to down-regulate Notch signaling in zebrafish and understanding Numb's temporal expression patterns during homeostasis and after injury in other regenerating species, as well as in mice. Numb could open a new therapeutic avenue for manipulating Notch signaling in mammals and facilitating successful regeneration of hair cells in mammals.
Even though down-regulation of Notch signaling induced differentiation of new hair cells in mammals, these hair cells developed via transdifferentiation of support cells into hair cells, leading to the depletion of support cells (30). The down-regulation of Notch was not sufficient to induce support cell proliferation as observed in spontaneously regenerating species. Zebrafish regenerate hair cells mainly or exclusively by proliferation of support cells, and regenerating chicken hair cells arise via transdifferentiation, as well as proliferation (19). In zebrafish we observe that the down-regulation of Notch signaling at 1 h after neomycin treatment correlates with the down-regulation of cyclin-dependent kinase inhibitors cdkn1b (p27/kip1) and cdkn1c (p57/kip2) (Fig. 4 A and D). This result suggests that Notch could be an activator of cdkn1b (p27/kip1) that keeps cells out of the cell cycle and therefore that the down-regulation of Notch signaling could be a crucial event for the onset of regeneration. Indeed in the murine inner ear, the Notch target Sox2 regulates Cdkn1b (p27/Kip1), similar to activity described in the epidermis (99, 100) but in contrast to the activity in the intestinal crypt and inner ear, where Notch inhibits Cdkn1b (p27/Kip1) (101, 102). In zebrafish the attenuation of Notch signaling has no effect on proliferation in uninjured sensory organs and has only a proproliferative effect on support cells after injury (9). In chicken cochleae the proliferation rate is not increased after injury, nor does inhibition of Notch cause cell cycle reentry in mice cochleae (13, 19, 30). Based on these findings it was concluded that Notch signaling is not required to initiate proliferation during regeneration (13). Therefore, it is possible that Cdk inhibitors are regulated by as yet unidentified, Notch-independent signals. Possibly, Fgf signaling inhibits proliferation, as demonstrated in the chicken inner ear (63).
Because complete regeneration of sensory epithelia in mammals requires coaxing support cells into hair cell differentiation as well as stimulating the support cells to re-enter the cell cycle and self-renew, manipulation of Notch signaling is not sufficient and needs to be married with manipulations of mitogenic signals. Furthermore, similar to our observations during zebrafish hair cell regeneration, Notch signaling must be reinitiated for the inhibition of proneural genes and specification of support cells (12, 23, 91). A temporally restricted Notch down-regulation or a combination of Notch down-regulation with subsequent ectopic expression of support cell inducers, such as Sox2, could be fruitful.
Our efforts presented here have characterized the pattern of temporal activation of signaling pathways during zebrafish sensory hair cell regeneration. These results will aid in the design and interpretation of functional studies aimed at elucidating the gene-regulatory network underlying zebrafish hair cell regeneration and can guide experiments to trigger hair cell regeneration in mammals.
Materials and Methods
Fish Strains and Neomycin Treatment.
Fish lines Tg(sqET20) and Tg(sqET4) were gifts from Vladimir Korzh (Institute of Molecular and Cell Biology, Singapore) (39). Tg(Tcf/Lef-miniP:dGFP) was a gift from T. Ishitani (Kyushu University, Fukuoka, Japan) (49). Larvae were generated by paired matings and raised in 0.5× E2 medium at 28.5 °C. GFP+ larvae were sorted from wild-type larvae at 48 h after fertilization under fluorescent stereoscope. For neomycin treatment, 5-dpf larvae were treated for 30 min with 300 μM neomycin (Fisher BioReagents) diluted in 0.5× E2 medium, rinsed three times, and recovered in 0.5× E2 medium at 28.5 °C for the indicated time.
FACS.
Two hundred 5-dpf untreated (control) or neomycin-treated Tg(sqET20) larvae were anesthetized and collected in a 2-mL tube. Then 1.5 mL cold trypsin solution (0.5 g/L; T3924; Sigma) was added and gently triturated with a 1,000-μL pipette tip for 20 min on ice. Cells then were filtered with a 70-μm cell strainer (BD Transduction Laboratories) to remove the undissociated tissue and were washed twice with cold PBS. Cells were resuspended in 2.5 mL PBS and stained with 5 μg/mL 7-AAD (BD) and 5 μg/mL Hoechst33342 (BD) on ice for 30 min. High-speed FACS was performed using a Legacy MoFlo (Beckman Coulter) equipped with 488-nm and 350-nm water-cooled lasers (Coherent). A two-gate FACS strategy was used to select only living target cells from excesses of dead cells and cellular debris of the larval cell suspensions described above. The first gate of the two-gate strategy was created on a 7-AAD versus Hoechst dot plot. It encompassed events that were negative for the membrane-impermeable 7-AAD DNA dye (7-AAD−) and brightly positive for the membrane-permeable Hoechst DNA dye (HO+) (Fig. 1C). Only events meeting this 7-AAD−/HO+ criterion were passed through to a GFP versus forward scatter dot plot. From this second dot plot, GFP− and GFP+ events were gated for FACS isolation. As such, live neuromast mantle and inner support cells from 5-dpf Tg(sqET20) larvae were defined as 7-AAD−/HO+/GFP+ events and were FACS isolated and collected directly into TRIzol. This GFP+ population comprised 0.42 ± 0.21% (n = 12) of the live cell fraction (7-AAD−/HO+). Live 7-AAD−/HO+/GFP− somatic cells were collected as a control population from the same larval cell suspensions. The target populations also were collected into PBS for postsorting purity verifications and Sox2 antibody staining.
RNA Extraction, Library Construction and RNA-Seq.
About 30,000 GFP− or GFP+ cells were used for total RNA extraction following the manufacturer’s manual. RNA samples were quality checked with a Bioanalyzer (Agilent Technologies). Short fragment libraries for RNA sequencing were made with poly-A–selected mRNA using the Illumina TruSeq RNA library construction kits v2 (Illumina). The resulting libraries were purified using Agencourt AMPure XP system (Backman Coulter) and then were quantified using a Bioanalyzer or Qubit Fluorometer (Life Technologies). Libraries were pooled, requantified, and run as 50-bp single-end lanes on an Illumina HiSEq 2000 instrument, using HiSeq Control Software 1.5.15.1 and Real-Time Analysis (RTA) version 1.13.48.0. Secondary analysis version CASAVA-1.8.2 was run to demultiplex reads and generate FASTQ files.
Real-Time PCR and Statistical Analysis.
Real-time PCR was performed using the Qiagen OneStep RT-PCR kit in an ABI7900 PCR machine (Applied Biosystems). Data were expressed as mean ± SD. Pairwise comparisons were performed using the Student t test. See SI Materials and Methods for additional details.
Supplementary Material
Acknowledgments
We thank Dr. V. Korzh (Institute of Molecular and Cell Biology) and T. Ishitani (Kyushu University) for the Tg(sqEt20), Tg(sqEt4) and Tg(Tcf/Lef-miniP:dGFP) fish lines; Ariel Paulson for bioinformatics advice; Marina Venero-Galanternik and Alejandro Sánchez Alvarado for critically reading the manuscript; the Stowers zebrafish facility for excellent fish care; and the Stowers Molecular Biology Core for Illumina sequencing. This study was supported by funding from the Deafness Research Foundation (now Hearing Health Foundation), the National Organization for Hearing Research Foundation, and the Innisfree Foundation and by National Institute on Deafness and Other Communication Disorders American Recovery and Reinvestment Act Grant 1RC1DC010631 (to T.P.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE56176).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1402898111/-/DCSupplemental.
References
- 1.White PM, Doetzlhofer A, Lee YS, Groves AK, Segil N. Mammalian cochlear supporting cells can divide and trans-differentiate into hair cells. Nature. 2006;441(7096):984–987. doi: 10.1038/nature04849. [DOI] [PubMed] [Google Scholar]
- 2.Corwin JT, Oberholtzer JC. Fish n’ chicks: Model recipes for hair-cell regeneration? Neuron. 1997;19(5):951–954. doi: 10.1016/s0896-6273(00)80386-4. [DOI] [PubMed] [Google Scholar]
- 3.Jacques BE, et al. A dual function for canonical Wnt/β-catenin signaling in the developing mammalian cochlea. Development. 2012;139(23):4395–4404. doi: 10.1242/dev.080358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bermingham-McDonogh O, Rubel EW. Hair cell regeneration: Winging our way towards a sound future. Curr Opin Neurobiol. 2003;13(1):119–126. doi: 10.1016/s0959-4388(03)00018-7. [DOI] [PubMed] [Google Scholar]
- 5.Williams JA, Holder N. Cell turnover in neuromasts of zebrafish larvae. Hear Res. 2000;143(1-2):171–181. doi: 10.1016/s0378-5955(00)00039-3. [DOI] [PubMed] [Google Scholar]
- 6.Harris JA, et al. Neomycin-induced hair cell death and rapid regeneration in the lateral line of zebrafish (Danio rerio) J Assoc Res Otolaryngol. 2003;4(2):219–234. doi: 10.1007/s10162-002-3022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hernández PP, Moreno V, Olivari FA, Allende ML. Sub-lethal concentrations of waterborne copper are toxic to lateral line neuromasts in zebrafish (Danio rerio) Hear Res. 2006;213(1-2):1–10. doi: 10.1016/j.heares.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 8.López-Schier H, Hudspeth AJ. A two-step mechanism underlies the planar polarization of regenerating sensory hair cells. Proc Natl Acad Sci USA. 2006;103(49):18615–18620. doi: 10.1073/pnas.0608536103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma EY, Rubel EW, Raible DW. Notch signaling regulates the extent of hair cell regeneration in the zebrafish lateral line. J Neurosci. 2008;28(9):2261–2273. doi: 10.1523/JNEUROSCI.4372-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Groves AK. The challenge of hair cell regeneration. Exp Biol Med (Maywood) 2010;235(4):434–446. doi: 10.1258/ebm.2009.009281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Torchinsky C, Messana EP, Arsura M, Cotanche DA. Regulation of p27Kip1 during gentamicin mediated hair cell death. J Neurocytol. 1999;28(10-11):913–924. doi: 10.1023/a:1007082424477. [DOI] [PubMed] [Google Scholar]
- 12.Stone JS, Cotanche DA. Hair cell regeneration in the avian auditory epithelium. Int J Dev Biol. 2007;51(6-7):633–647. doi: 10.1387/ijdb.072408js. [DOI] [PubMed] [Google Scholar]
- 13.Daudet N, et al. Notch regulation of progenitor cell behavior in quiescent and regenerating auditory epithelium of mature birds. Dev Biol. 2009;326(1):86–100. doi: 10.1016/j.ydbio.2008.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ryals BM, Rubel EW. Hair cell regeneration after acoustic trauma in adult Coturnix quail. Science. 1988;240(4860):1774–1776. doi: 10.1126/science.3381101. [DOI] [PubMed] [Google Scholar]
- 15.Cotanche DA, Kaiser CL. Hair cell fate decisions in cochlear development and regeneration. Hear Res. 2010;266(1-2):18–25. doi: 10.1016/j.heares.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rubel EW, Furrer SA, Stone JS. A brief history of hair cell regeneration research and speculations on the future. Hear Res. 2013;297:42–51. doi: 10.1016/j.heares.2012.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alvarado DM, et al. An RNA interference-based screen of transcription factor genes identifies pathways necessary for sensory regeneration in the avian inner ear. J Neurosci. 2011;31(12):4535–4543. doi: 10.1523/JNEUROSCI.5456-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hawkins RD, et al. Large scale gene expression profiles of regenerating inner ear sensory epithelia. PLoS ONE. 2007;2(6):e525. doi: 10.1371/journal.pone.0000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brignull HR, Raible DW, Stone JS. Feathers and fins: Non-mammalian models for hair cell regeneration. Brain Res. 2009;1277:12–23. doi: 10.1016/j.brainres.2009.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen P, Segil N. p27(Kip1) links cell proliferation to morphogenesis in the developing organ of Corti. Development. 1999;126(8):1581–1590. doi: 10.1242/dev.126.8.1581. [DOI] [PubMed] [Google Scholar]
- 21.Löwenheim H, et al. Gene disruption of p27(Kip1) allows cell proliferation in the postnatal and adult organ of corti. Proc Natl Acad Sci USA. 1999;96(7):4084–4088. doi: 10.1073/pnas.96.7.4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weber T, et al. Rapid cell-cycle reentry and cell death after acute inactivation of the retinoblastoma gene product in postnatal cochlear hair cells. Proc Natl Acad Sci USA. 2008;105(2):781–785. doi: 10.1073/pnas.0708061105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munnamalai V, Fekete DM. Wnt signaling during cochlear development. Semin Cell Dev Biol. 2013;24(5):480–489. doi: 10.1016/j.semcdb.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hawkins RD, et al. Gene expression differences in quiescent versus regenerating hair cells of avian sensory epithelia: Implications for human hearing and balance disorders. Hum Mol Genet. 2003;12(11):1261–1272. doi: 10.1093/hmg/ddg150. [DOI] [PubMed] [Google Scholar]
- 25.Stevens CB, Davies AL, Battista S, Lewis JH, Fekete DM. Forced activation of Wnt signaling alters morphogenesis and sensory organ identity in the chicken inner ear. Dev Biol. 2003;261(1):149–164. doi: 10.1016/s0012-1606(03)00297-5. [DOI] [PubMed] [Google Scholar]
- 26.Head JR, Gacioch L, Pennisi M, Meyers JR. Activation of canonical Wnt/β-catenin signaling stimulates proliferation in neuromasts in the zebrafish posterior lateral line. Dev Dyn. 2013;242(7):832–846. doi: 10.1002/dvdy.23973. [DOI] [PubMed] [Google Scholar]
- 27.Wada H, et al. Wnt/Dkk negative feedback regulates sensory organ size in zebrafish. Curr Biol. 2013;23(16):1559–1565. doi: 10.1016/j.cub.2013.06.035. [DOI] [PubMed] [Google Scholar]
- 28.Shi F, Hu L, Edge AS. Generation of hair cells in neonatal mice by β-catenin overexpression in Lgr5-positive cochlear progenitors. Proc Natl Acad Sci USA. 2013;110(34):13851–13856. doi: 10.1073/pnas.1219952110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chai R, et al. Wnt signaling induces proliferation of sensory precursors in the postnatal mouse cochlea. Proc Natl Acad Sci USA. 2012;109(21):8167–8172. doi: 10.1073/pnas.1202774109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mizutari K, et al. Notch inhibition induces cochlear hair cell regeneration and recovery of hearing after acoustic trauma. Neuron. 2013;77(1):58–69. doi: 10.1016/j.neuron.2012.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Owens KN, et al. Ultrastructural analysis of aminoglycoside-induced hair cell death in the zebrafish lateral line reveals an early mitochondrial response. J Comp Neurol. 2007;502(4):522–543. doi: 10.1002/cne.21345. [DOI] [PubMed] [Google Scholar]
- 32.Duncan JS, Fritzsch B. Evolution of sound and balance perception: Innovations that aggregate single hair cells into the ear and transform a gravistatic sensor into the organ of corti. Anat Rec (Hoboken) 2012;295(11):1760–1774. doi: 10.1002/ar.22573. [DOI] [PubMed] [Google Scholar]
- 33.Whitfield TT. Zebrafish as a model for hearing and deafness. J Neurobiol. 2002;53(2):157–171. doi: 10.1002/neu.10123. [DOI] [PubMed] [Google Scholar]
- 34.Nicolson T. The genetics of hearing and balance in zebrafish. Annu Rev Genet. 2005;39:9–22. doi: 10.1146/annurev.genet.39.073003.105049. [DOI] [PubMed] [Google Scholar]
- 35.Behra M, et al. Phoenix is required for mechanosensory hair cell regeneration in the zebrafish lateral line. PLoS Genet. 2009;5(4):e1000455. doi: 10.1371/journal.pgen.1000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang J, et al. The stat3/socs3a pathway is a key regulator of hair cell regeneration in zebrafish. [corrected] J Neurosci. 2012;32(31):10662–10673. doi: 10.1523/JNEUROSCI.5785-10.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wibowo I, Pinto-Teixeira F, Satou C, Higashijima S-I, López-Schier H. Compartmentalized Notch signaling sustains epithelial mirror symmetry. Development. 2011;138(6):1143–1152. doi: 10.1242/dev.060566. [DOI] [PubMed] [Google Scholar]
- 38.Mirkovic I, Pylawka S, Hudspeth AJ. Rearrangements between differentiating hair cells coordinate planar polarity and the establishment of mirror symmetry in lateral-line neuromasts. Biol Open. 2012;1(5):498–505. doi: 10.1242/bio.2012570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parinov S, Kondrichin I, Korzh V, Emelyanov A. Tol2 transposon-mediated enhancer trap to identify developmentally regulated zebrafish genes in vivo. Dev Dyn. 2004;231(2):449–459. doi: 10.1002/dvdy.20157. [DOI] [PubMed] [Google Scholar]
- 40.Hernández PP, Olivari FA, Sarrazin AF, Sandoval PC, Allende ML. Regeneration in zebrafish lateral line neuromasts: Expression of the neural progenitor cell marker sox2 and proliferation-dependent and-independent mechanisms of hair cell renewal. Dev Neurobiol. 2007;67(5):637–654. doi: 10.1002/dneu.20386. [DOI] [PubMed] [Google Scholar]
- 41.Kalghatgi S, et al. Bactericidal antibiotics induce mitochondrial dysfunction and oxidative damage in Mammalian cells. Sci Transl Med. 2013;5(192):92ra85. doi: 10.1126/scitranslmed.3006055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steiner AB, Kim T, Cabot V, Hudspeth AJ. Dynamic gene expression by putative hair-cell progenitors during regeneration in the zebrafish lateral line. Proc Natl Acad Sci USA. 2014;111:E1393–E1401. doi: 10.1073/pnas.1318692111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fritzsch B, et al. Dissecting the molecular basis of organ of Corti development: Where are we now? Hear Res. 2011;276(1-2):16–26. doi: 10.1016/j.heares.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tarca AL, et al. A novel signaling pathway impact analysis. Bioinformatics. 2009;25(1):75–82. doi: 10.1093/bioinformatics/btn577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ogata H, et al. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27(1):29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Behra M, et al. Transcriptional signature of accessory cells in the lateral line, using the Tnk1bp1:EGFP transgenic zebrafish line. BMC Dev Biol. 2012;12:6. doi: 10.1186/1471-213X-12-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gallardo VE, et al. Molecular dissection of the migrating posterior lateral line primordium during early development in zebrafish. BMC Dev Biol. 2010;10:120. doi: 10.1186/1471-213X-10-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jacques BE, et al. The role of Wnt/β-catenin signaling in proliferation and regeneration of the developing basilar papilla and lateral line. Dev Neurobiol. 2013 doi: 10.1002/dneu.22134. [DOI] [PubMed] [Google Scholar]
- 49.Shimizu N, Kawakami K, Ishitani T. Visualization and exploration of Tcf/Lef function using a highly responsive Wnt/β-catenin signaling-reporter transgenic zebrafish. Dev Biol. 2012;370(1):71–85. doi: 10.1016/j.ydbio.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 50.Medina A, Reintsch W, Steinbeisser H. Xenopus frizzled 7 can act in canonical and non-canonical Wnt signaling pathways: Implications on early patterning and morphogenesis. Mech Dev. 2000;92(2):227–237. doi: 10.1016/s0925-4773(00)00240-9. [DOI] [PubMed] [Google Scholar]
- 51.Grumolato L, et al. Canonical and noncanonical Wnts use a common mechanism to activate completely unrelated coreceptors. Genes Dev. 2010;24(22):2517–2530. doi: 10.1101/gad.1957710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dabdoub A, et al. Sox2 signaling in prosensory domain specification and subsequent hair cell differentiation in the developing cochlea. Proc Natl Acad Sci USA. 2008;105(47):18396–18401. doi: 10.1073/pnas.0808175105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takebayashi S, et al. Multiple roles of Notch signaling in cochlear development. Dev Biol. 2007;307(1):165–178. doi: 10.1016/j.ydbio.2007.04.035. [DOI] [PubMed] [Google Scholar]
- 54.Collado MS, et al. The postnatal accumulation of junctional E-cadherin is inversely correlated with the capacity for supporting cells to convert directly into sensory hair cells in mammalian balance organs. J Neurosci. 2011;31(33):11855–11866. doi: 10.1523/JNEUROSCI.2525-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jung JY, et al. siRNA targeting Hes5 augments hair cell regeneration in aminoglycoside-damaged mouse utricle. Mol Ther. 2013;21(4):834–841. doi: 10.1038/mt.2013.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Korrapati S, Roux I, Glowatzki E, Doetzlhofer A. Notch signaling limits supporting cell plasticity in the hair cell-damaged early postnatal murine cochlea. PLoS ONE. 2013;8(8):e73276. doi: 10.1371/journal.pone.0073276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin V, et al. Inhibition of Notch activity promotes nonmitotic regeneration of hair cells in the adult mouse utricles. J Neurosci. 2011;31(43):15329–15339. doi: 10.1523/JNEUROSCI.2057-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McGill MA, Dho SE, Weinmaster G, McGlade CJ. Numb regulates post-endocytic trafficking and degradation of Notch1. J Biol Chem. 2009;284(39):26427–26438. doi: 10.1074/jbc.M109.014845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Song Y, Lu B. Interaction of Notch signaling modulator Numb with α-Adaptin regulates endocytosis of Notch pathway components and cell fate determination of neural stem cells. J Biol Chem. 2012;287(21):17716–17728. doi: 10.1074/jbc.M112.360719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matsuda M, Chitnis AB. Atoh1a expression must be restricted by Notch signaling for effective morphogenesis of the posterior lateral line primordium in zebrafish. Development. 2010;137(20):3477–3487. doi: 10.1242/dev.052761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang G-P, et al. Notch signaling and Atoh1 expression during hair cell regeneration in the mouse utricle. Hear Res. 2010;267(1-2):61–70. doi: 10.1016/j.heares.2010.03.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bermingham-McDonogh O, Stone JS, Reh TA, Rubel EW. FGFR3 expression during development and regeneration of the chick inner ear sensory epithelia. Dev Biol. 2001;238(2):247–259. doi: 10.1006/dbio.2001.0412. [DOI] [PubMed] [Google Scholar]
- 63.Oesterle EC, Bhave SA, Coltrera MD. Basic fibroblast growth factor inhibits cell proliferation in cultured avian inner ear sensory epithelia. J Comp Neurol. 2000;424(2):307–326. doi: 10.1002/1096-9861(20000821)424:2<307::aid-cne9>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 64.Chen P, et al. Progressive hearing loss in mice lacking the cyclin-dependent kinase inhibitor Ink4d. Nat Cell Biol. 2003;5(5):422–426. doi: 10.1038/ncb976. [DOI] [PubMed] [Google Scholar]
- 65.Burns JC, Corwin JT. A historical to present-day account of efforts to answer the question: “What puts the brakes on mammalian hair cell regeneration?”. Hear Res. 2013;297:52–67. doi: 10.1016/j.heares.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu Z, et al. Regulation of p27Kip1 by Sox2 maintains quiescence of inner pillar cells in the murine auditory sensory epithelium. J Neurosci. 2012;32(31):10530–10540. doi: 10.1523/JNEUROSCI.0686-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hirano T, Ishihara K, Hibi M. Roles of STAT3 in mediating the cell growth, differentiation and survival signals relayed through the IL-6 family of cytokine receptors. Oncogene. 2000;19(21):2548–2556. doi: 10.1038/sj.onc.1203551. [DOI] [PubMed] [Google Scholar]
- 68.Zhang SS, et al. STAT3 activation in response to growth factors or cytokines participates in retina precursor proliferation. Exp Eye Res. 2005;81(1):103–115. doi: 10.1016/j.exer.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 69.Fang Y, et al. Translational profiling of cardiomyocytes identifies an early Jak1/Stat3 injury response required for zebrafish heart regeneration. Proc Natl Acad Sci USA. 2013;110(33):13416–13421. doi: 10.1073/pnas.1309810110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kamaid A, Neves J, Giráldez F. Id gene regulation and function in the prosensory domains of the chicken inner ear: A link between Bmp signaling and Atoh1. J Neurosci. 2010;30(34):11426–11434. doi: 10.1523/JNEUROSCI.2570-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pujades C, Kamaid A, Alsina B, Giraldez F. BMP-signaling regulates the generation of hair-cells. Dev Biol. 2006;292(1):55–67. doi: 10.1016/j.ydbio.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 72.Hwang CH, et al. Role of bone morphogenetic proteins on cochlear hair cell formation: Analyses of Noggin and Bmp2 mutant mice. Dev Dyn. 2010;239(2):505–513. doi: 10.1002/dvdy.22200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ohyama T, et al. BMP signaling is necessary for patterning the sensory and nonsensory regions of the developing mammalian cochlea. J Neurosci. 2010;30(45):15044–15051. doi: 10.1523/JNEUROSCI.3547-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Oesterle EC, Tsue TT, Rubel EW. Induction of cell proliferation in avian inner ear sensory epithelia by insulin-like growth factor-I and insulin. J Comp Neurol. 1997;380(2):262–274. doi: 10.1002/(sici)1096-9861(19970407)380:2<262::aid-cne8>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 75.Carnovale CE, Ronco MT. Role of nitric oxide in liver regeneration. Ann Hepatol. 2012;11(5):636–647. [PubMed] [Google Scholar]
- 76.Sage C, et al. Proliferation of functional hair cells in vivo in the absence of the retinoblastoma protein. Science. 2005;307(5712):1114–1118. doi: 10.1126/science.1106642. [DOI] [PubMed] [Google Scholar]
- 77.Sage C, et al. Essential role of retinoblastoma protein in mammalian hair cell development and hearing. Proc Natl Acad Sci USA. 2006;103(19):7345–7350. doi: 10.1073/pnas.0510631103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kelley MW. Regulation of cell fate in the sensory epithelia of the inner ear. Nat Rev Neurosci. 2006;7(11):837–849. doi: 10.1038/nrn1987. [DOI] [PubMed] [Google Scholar]
- 79.Sienknecht UJ, Fekete DM. Mapping of Wnt, frizzled, and Wnt inhibitor gene expression domains in the avian otic primordium. J Comp Neurol. 2009;517(6):751–764. doi: 10.1002/cne.22169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.van Amerongen R, Nusse R. Towards an integrated view of Wnt signaling in development. Development. 2009;136(19):3205–3214. doi: 10.1242/dev.033910. [DOI] [PubMed] [Google Scholar]
- 81.Shi F, Kempfle JS, Edge AS. Wnt-responsive Lgr5-expressing stem cells are hair cell progenitors in the cochlea. J Neurosci. 2012;32(28):9639–9648. doi: 10.1523/JNEUROSCI.1064-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Galliot B, Chera S. The Hydra model: Disclosing an apoptosis-driven generator of Wnt-based regeneration. Trends Cell Biol. 2010;20(9):514–523. doi: 10.1016/j.tcb.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 83.Fleming HE, et al. Wnt signaling in the niche enforces hematopoietic stem cell quiescence and is necessary to preserve self-renewal in vivo. Cell Stem Cell. 2008;2(3):274–283. doi: 10.1016/j.stem.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li L, Bhatia R. Stem cell quiescence. Clin Cancer Res. 2011;17(15):4936–4941. doi: 10.1158/1078-0432.CCR-10-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bergmann A, Steller H. Apoptosis, stem cells, and tissue regeneration. Sci Signal. 2010;3(145):re8. doi: 10.1126/scisignal.3145re8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Blanpain C, Horsley V, Fuchs E. Epithelial stem cells: Turning over new leaves. Cell. 2007;128(3):445–458. doi: 10.1016/j.cell.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Oesterle EC, Chien WM, Campbell S, Nellimarla P, Fero ML. p27(Kip1) is required to maintain proliferative quiescence in the adult cochlea and pituitary. Cell Cycle. 2011;10(8):1237–1248. doi: 10.4161/cc.10.8.15301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hori R, et al. Pharmacological inhibition of Notch signaling in the mature guinea pig cochlea. Neuroreport. 2007;18(18):1911–1914. doi: 10.1097/WNR.0b013e3282f213e0. [DOI] [PubMed] [Google Scholar]
- 89.Cafaro J, Lee GS, Stone JS. Atoh1 expression defines activated progenitors and differentiating hair cells during avian hair cell regeneration. Dev Dyn. 2007;236(1):156–170. doi: 10.1002/dvdy.21023. [DOI] [PubMed] [Google Scholar]
- 90.Groves AK, Zhang KD, Fekete DM. The genetics of hair cell development and regeneration. Annu Rev Neurosci. 2013;36:361–381. doi: 10.1146/annurev-neuro-062012-170309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Agathocleous M, et al. A directional Wnt/beta-catenin-Sox2-proneural pathway regulates the transition from proliferation to differentiation in the Xenopus retina. Development. 2009;136(19):3289–3299. doi: 10.1242/dev.040451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sweet EM, Vemaraju S, Riley BB. Sox2 and Fgf interact with Atoh1 to promote sensory competence throughout the zebrafish inner ear. Dev Biol. 2011;358(1):113–121. doi: 10.1016/j.ydbio.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Van Raay TJ, et al. Frizzled 5 signaling governs the neural potential of progenitors in the developing Xenopus retina. Neuron. 2005;46(1):23–36. doi: 10.1016/j.neuron.2005.02.023. [DOI] [PubMed] [Google Scholar]
- 94.Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39(5):749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- 95.Kan L, et al. Sox1 acts through multiple independent pathways to promote neurogenesis. Dev Biol. 2004;269(2):580–594. doi: 10.1016/j.ydbio.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 96.Wang X, et al. Wnt signaling regulates postembryonic hypothalamic progenitor differentiation. Dev Cell. 2012;23(3):624–636. doi: 10.1016/j.devcel.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Puligilla C, Dabdoub A, Brenowitz SD, Kelley MW. Sox2 induces neuronal formation in the developing mammalian cochlea. J Neurosci. 2010;30(2):714–722. doi: 10.1523/JNEUROSCI.3852-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stone JS, Rubel EW. Delta1 expression during avian hair cell regeneration. Development. 1999;126(5):961–973. doi: 10.1242/dev.126.5.961. [DOI] [PubMed] [Google Scholar]
- 99.Liu Z, et al. Regulation of p27Kip1 by Sox2 maintains quiescence of inner pillar cells in the murine auditory sensory epithelium. J Neurosci. 2012;32(31):10530–10540. doi: 10.1523/JNEUROSCI.0686-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rangarajan A, et al. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 2001;20(13):3427–3436. doi: 10.1093/emboj/20.13.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Riccio O, et al. Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27Kip1 and p57Kip2. EMBO Rep. 2008;9(4):377–383. doi: 10.1038/embor.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Murata J, et al. Notch-Hes1 pathway contributes to the cochlear prosensory formation potentially through the transcriptional down-regulation of p27Kip1. J Neurosci Res. 2009;87(16):3521–3534. doi: 10.1002/jnr.22169. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.