Significance
Serum amyloid A (SAA) is a major serum acute-phase protein and a cause of secondary amyloidosis, which impacts ∼1% of patients with chronic inflammation such as rheumatoid arthritis and neoplastic diseases. The lack of structural information has hampered our understanding of SAA-mediated amyloidosis and the development of effective therapies. Here we report a crystal structure of human SAA1.1 as a prototypic member of the family. SAA1.1 exists as a hexamer with subunits displaying a unique four-helix bundle fold. We further defined binding sites for heparin and high-density lipoprotein, identified major amyloidogenic epitopes, and visualized SAA-mediated protofibril formation using electron microscopy. These studies provide mechanistic insights into amyloidogenic conformational transition of SAA.
Keywords: heparan sulfate binding site, HDL binding site
Abstract
Serum amyloid A (SAA) represents an evolutionarily conserved family of inflammatory acute-phase proteins. It is also a major constituent of secondary amyloidosis. To understand its function and structural transition to amyloid, we determined a structure of human SAA1.1 in two crystal forms, representing a prototypic member of the family. Native SAA1.1 exists as a hexamer, with subunits displaying a unique four-helix bundle fold stabilized by its long C-terminal tail. Structure-based mutational studies revealed two positive-charge clusters, near the center and apex of the hexamer, that are involved in SAA association with heparin. The binding of high-density lipoprotein involves only the apex region of SAA and can be inhibited by heparin. Peptide amyloid formation assays identified the N-terminal helices 1 and 3 as amyloidogenic peptides of SAA1.1. Both peptides are secluded in the hexameric structure of SAA1.1, suggesting that the native SAA is nonpathogenic. Furthermore, dissociation of the SAA hexamer appears insufficient to initiate amyloidogenic transition, and proteolytic cleavage or removal of the C-terminal tail of SAA resulted in formation of various-sized structural aggregates containing ∼5-nm regular repeating protofibril-like units. The combined structural and functional studies provide mechanistic insights into the pathogenic contribution of glycosaminoglycan in SAA1.1-mediated AA amyloid formation.
Secondary amyloidosis develops in patients with chronic inflammatory, infectious, and neoplastic diseases such as rheumatoid arthritis, inflammatory bowel disease, tuberculosis, and renal cell carcinoma (1). The overall incidence of systemic secondary amyloidosis in Western nations ranges from 0.5 to 0.86%, but increases significantly in certain patients (2–4). Deposition of the amyloid fibril in secondary amyloidosis (AA amyloid) can lead to progressive loss of splenic and renal functions. The main constituent of AA amyloid is the serum amyloid A protein (SAA), a major acute-phase protein and serum marker for inflammatory diseases (5–9). Amyloid fibrils are protein aggregates with highly ordered characteristic β-sheet structures (10). Current therapy for secondary amyloidosis is to use anti-inflammatory/immunosuppressive drugs or colchicine to reduce SAA levels (1). However, adequate suppression of SAA production is often unsuccessful. New approaches are needed to target the process of amyloid formation (11). SAA proteins are highly conserved throughout evolution. They are found in mammals and vertebrates, and even the more ancient sea cucumber (9). However, there is no 3D structure available for the SAA family of proteins. Although the proteolytic fragments of SAA are the main constituents of AA amyloid fibrils (12), SAA is normally complexed with high-density lipoprotein (HDL) in the serum, and the dissociation of SAA from HDL in the pathogenesis of AA amyloid formation remains unclear. To understand the structural and functional aspects of SAA-mediated pathogenic amyloid formation, we determined the crystal structure of a major amyloid-susceptible human SAA, SAA1.1, characterized the glycosaminoglycan and HDL binding sites, and investigated the structural mechanism of AA amyloidogenesis. These structural insights help to understand the role of heparan sulfate in SAA release from HDL and to develop novel therapeutic compounds for the treatment of AA amyloid.
Results
The Structure of SAA1.1 Displays a Unique Antiparallel Four-Helix Bundle Fold.
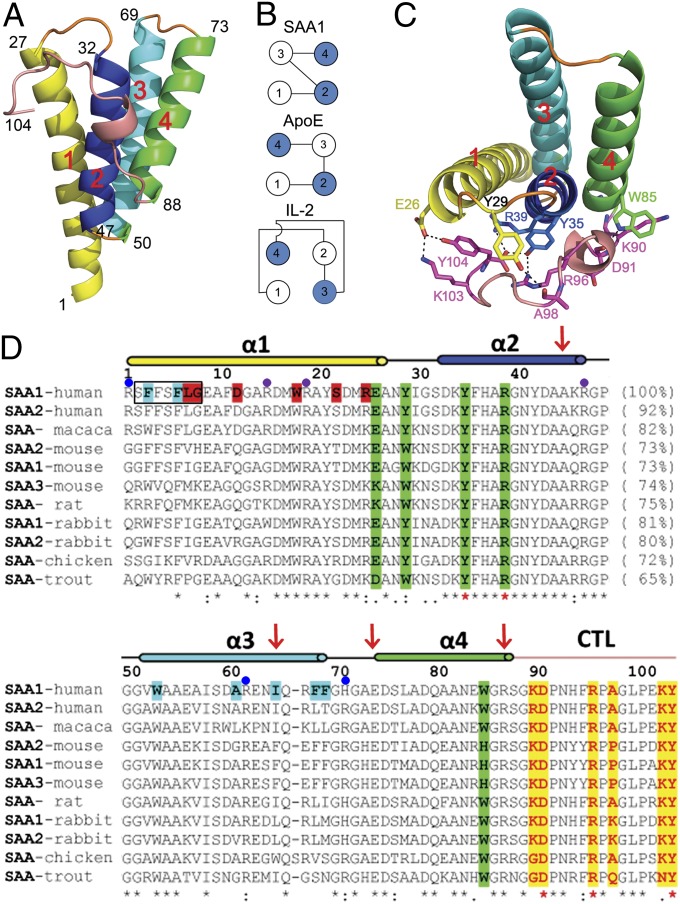
We expressed human SAA1.1 in Escherichia coli and purified the protein as previously described (13). The refolded SAA1.1 was crystallized in the orthorhombic space group P21212 in the presence of 2 M (NH4)2SO4, and its structure was solved by multiwavelength anomalous dispersion using a selenomethionine derivative to 2.2-Å resolution with refined Rwork and Rfree values of 0.204 and 0.256, respectively (Fig. S1A and Table S1). The SAA1.1 monomer forms an up-down-up-down four-helix bundle structure distinct from that of cytokines but similar to that of the apolipoprotein E (ApoE) N-terminal domain, except that the positions of helices 3 and 4 are switched (Fig. 1 and Fig. S1B) (14–16). Helices 1, 2, 3, and 4 are composed of residues 1–27, 32–47, 50–69, and 73–88, respectively. However, unlike other helix bundles, the SAA1.1 helix bundle is cone-shaped, with the N termini of helices 1 and 3 and the C termini of helices 2 and 4 packing closely together (Fig. 1A). The C-terminal tail, residues 89–104, is well-ordered and wraps around one face of the bundle, forming multiple salt bridges and hydrogen bonds with three of the four α-helices, including Glu-26 and Tyr-29 of helix 1, Tyr-35 and Arg-39 of helix 2, and Trp-85 and Gly-86 of helix 4 (Fig. 1C). Notably, the C-terminal carboxylate of Tyr-104 forms part of a four-residue charge cluster involving Tyr-35, Arg-39, and Arg-96 (Fig. 1C). In this charge quartet, both the C-terminal carboxyl groups and the OH of Tyr-35 form bifurcated salt bridges with Arg-39 and Arg-96, effectively locking up the carboxylate termini. Indeed, this charge quartet is found invariant in SAA from all species (Fig. 1D). In addition to the charge quartet, the C-terminal tail also contains three invariant proline residues at 92, 97, and 101, further restricting the conformation of the loop. The extensive conserved interactions between the C-terminal tail and the SAA1.1 helices suggest that the tail function stabilizes the SAA1.1 helix bundle structure. Overall, SAA proteins are highly conserved and share 61–80% in sequence identities from sea cucumber to rabbit to human, suggesting that all SAA family members share a common structural fold.
Fig. 1.
Monomeric structure of SAA1.1. (A) Ribbon representation of the crystal structure of human SAA1.1 with α-helices 1 (residues 1–27), 2 (residues 32–47), 3 (residues 50–69), and 4 (residues 73–88) and the C-terminal tail (residues 89–104) colored in yellow, blue, cyan, green, and sand, respectively. (B) Schematic illustration of observed SAA, ApoE, and cytokine four–α-helix bundles. Open and filled circles represent helices pointing up and down, respectively, from the plane of the paper. (C) The packing between the four-helix bundle and the C-terminal tail of SAA1.1, with hydrogen bonds and salt bridges shown as dotted lines. (D) Sequence alignment of SAA proteins. The secondary-structure elements (lines indicate loops; cylinders indicate helices) are indicated above the sequence. The lengths of representative SAA1 peptides identified in AA amyloid fibrils are indicated by red arrows. The four-helix bundle and the C-terminal tail (CTL) interface residues of SAA1 are shaded in yellow and green, respectively. The predicted amyloidogenic residues in human SAA1 are highlighted in black boxes. The dimerization and trimerization residues in the SAA1 hexamer are shaded in red and cyan, respectively. The cluster mutation sites at the center pore and apex of the SAA1 hexamer are indicated by purple and blue dots, respectively. The semiinvariant and invariant residues are indicated by dots and asterisks, respectively, under their sequences.
Hexameric Structure of SAA1.1.
Although there are four SAA1.1 monomers in the asymmetric unit of the orthorhombic SAA crystals, they do not form any sensible oligomeric assembly (Fig. S1D), in contrast to published oligomers of SAA (17, 18). This prompted us to question whether the presence of 4 M guanidine hydrochloride (Gn⋅HCl) during SAA purification disrupted its native oligomerization state, and led us to construct an N-terminal six-histidine–tagged SAA1.1 (his6SAA1.1) that can be expressed and purified in the absence of denaturants. The N-terminal histidine-tagged SAA1.1 was crystallized in PEG 2000 monomethyl ether in the space group R32, and its structure was solved to 2.5 Å by a molecular replacement method using monomeric SAA as the search model and refined to final Rwork and Rfree values of 0.249 and 0.258, respectively (Table S1). Unlike the orthorhombic crystals containing only the monomeric structure, SAA1.1 associates into a hexameric structure in the trigonal crystal through noncrystallographic twofold and crystallographic threefold axes (Fig. 2 and Fig. S1C). The subunit structures of SAA in this dimer of trimers are identical to each other and to that of orthorhombic SAA, with a root-mean-square deviation of less than 0.2 Å among all Cα atoms. Thus, the monomeric structure of SAA1 is stable in solution.
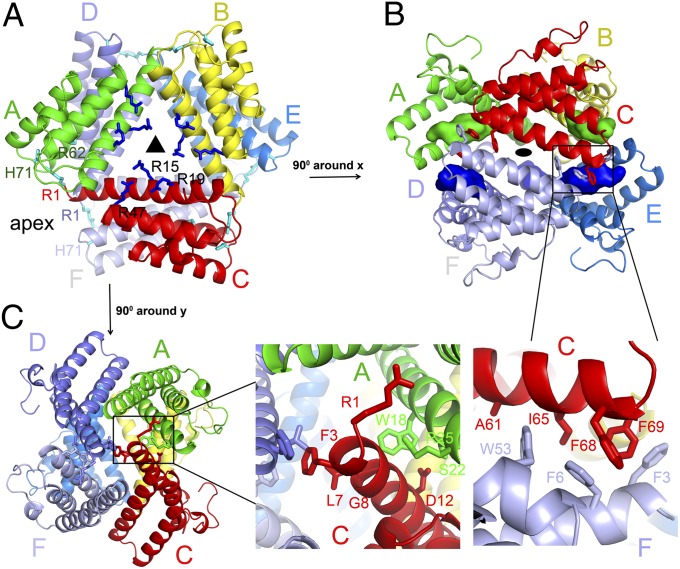
Fig. 2.
Hexameric structure of SAA1. (A) Two orthogonal views of the hexameric SAA1 structure in space group R32. The SAA1 hexamer was centered on the crystallographic threefold axis with a twofold axis-related dimer in the asymmetric unit. (B and C) Trimer interface (B) and dimer interface (C) residues in the SAA1 hexamer. The key contact residues are indicated by sticks in the enlarged views. The buried N-terminal helix 1 is shown as a colored surface.
The trimeric SAA is formed by subunits A, B, and C and involves residues from the first α-helix through a circular head-to-tail packing between the monomers (Fig. 2A). Each trimeric interface consists of a combination of hydrogen bonds, salt bridges, and hydrophobic interactions, burying ∼1,580 Å2 of solvent-accessible area. For example, whereas the N-terminal helix 1 of subunit A packs against the C-terminal helix 1 of subunit B, the C-terminal helix 1 of A forms a set of identical interactions with the N-terminal helix 1 of C. Specifically, a network of hydrogen bonds and salt bridges is formed between Arg-1 of subunit A and Asp-33 of subunit B, as well as between Gly-8 and Asp-12 of subunit A and Trp-18, Ser-22, and Arg-25 of subunit B (Fig. 2C). In addition, Phe-4 from subunit A forms hydrophobic interactions with Ile-65 from subunit B. The dimers are formed between trimers ABC and DEF with a largely hydrophobic interface (Fig. 2B). For example, Phe-3 of subunit A contacts Phe-3 and Phe-4 from subunit E, whereas Phe-6, Leu-7, and Trp-53 of subunit A interact with Ala-61, Ile-65, Phe-68, and Phe-69 from subunit D. Collectively, the dimer interface between the two SAA1 trimers buries a surface area of ∼3,180 Å2. Interestingly, this primarily hydrophobic dimeric interface buries the hydrophobic face of each trimer, leaving the hydrophilic faces exposed to the solvent (Fig. S2A). The replacement of Trp-53, Ile-65, Phe-68, and Phe-69 with alanine residues at the dimer interface resulted in a severe aggregation of SAA1 proteins, indicating their role in the oligomerization of SAA1. However, an alanine substitution of the trimer interface residues Trp-18, Ser-22, and Arg-25 did not disrupt the oligomeric state of SAA1 (Fig. S2B). Unlike the well-conserved interacting residues between the C-terminal tail and the helix bundle in each subunit, the dimer and trimer interface residues of the SAA1 hexamer are less conserved among different species (Fig. 1D), arguing that other SAA proteins may form different oligomers (17). It is worthwhile to mention that not all SAA proteins form amyloid fibrils. The oligomeric state and its intrinsic stability of different SAA proteins may influence their propensities for amyloid formation.
Oligomeric State of Human SAA1 in Solution.
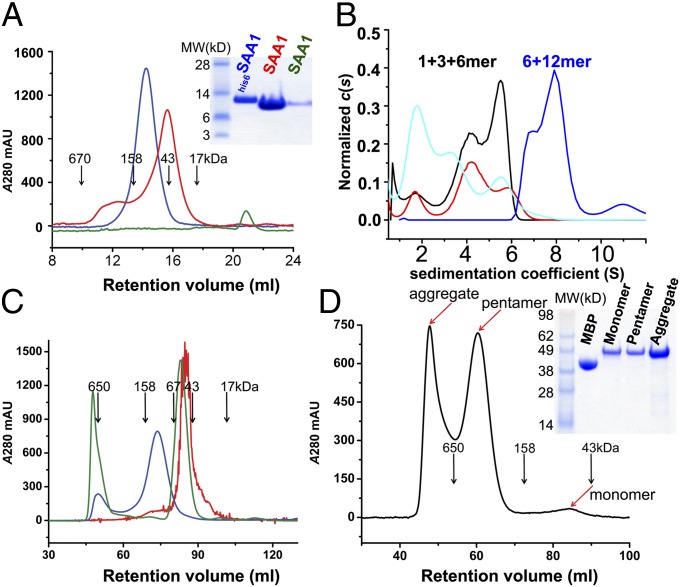
Biochemical characterizations of murine SAA2.2 showed the presence of various oligomeric forms such as hexamers, octamers, and monomers (17–19). Because mouse SAA2.2 is amyloid-resistant, it is not clear whether the major amyloid-prone human SAA1.1 adopts similar oligomeric forms. The refolded SAA1.1 has an apparent molecular mass of 43 kDa by size-exclusion chromatography (SEC), consistent with it being a trimer or tetramer (Fig. 3A) (20). Further analytical ultracentrifugation (AUC) experiments showed a sedimentation velocity (SV) profile of the refolded SAA1.1 consisting of monomers, trimers, and hexamers, with the majority being trimers at a concentration of ∼0.36 mg/mL but becoming monomers at lower concentrations (Fig. 3B). Consistent with the SV/AUC data, the equilibrium sedimentation curves of the refolded SAA1.1 can be fit with three apparent molecular mass species of 11.9, 35.9, and 74.7 kDa (Fig. S2C). Under the crystallization condition of a high concentration of (NH4)2SO4, refolded SAA1.1 existed only as monomers (Fig. 3A). To confirm whether native SAA1.1 forms hexamers in solution, the oligomeric state of his6SAA1.1 was examined using SEC and AUC experiments. Both gel-filtration and SV/AUC analyses showed a larger apparent molecular mass (∼125 kDa) of his6SAA1.1 (Fig. 3 A and B), and further equilibrium sedimentation analysis showed that a purified his6SAA1.1 sample contained a mixture of hexamers (∼30%) and dodecamers (∼70%) (Fig. S2D). Treatment of this his6SAA1.1 with 4 M Gn⋅HCl or mild detergent converted it to a trimeric form similar to that of the refolded SAA1.1 (Fig. 3C and Fig. S3A), indicating that the native SAA1 oligomerizes as a dimer of trimers.
Fig. 3.
Oligomeric state of SAA1.1. (A) Analytical size-exclusion profiles of his6SAA1.1 (blue), refolded SAA1.1 (red), and ammonium sulfate-treated SAA1.1 (green) on a Superdex 200 10/30 column. (B) Sedimentation coefficient distribution for his6SAA1.1 (blue) and the refolded SAA1 at 0.36 mg/mL (black), 0.18 mg/mL (red), and 0.06 mg/mL (cyan). (C) Size-exclusion chromatography comparison of native (blue) and Gn⋅HCl-treated his6SAA1.1 (green) with refolded SAA1.1 (red) on a Superdex 200 16/60 column. (D) Size-exclusion profile of MBP-SAA189 protein. (A and D) (Insets) SDS gels of the corresponding samples. mAU, milliabsorption unit.
Glycosaminoglycan Binding Sites on SAA1.1.
Glycosaminoglycans (GAGs), in particular heparin/heparan sulfate (HS), are known to associate with AA amyloidosis (21, 22). Previous studies showed that the binding of mouse SAA to HS depends primarily on charge interactions, and that the association promotes the HDL release of SAA and facilitates their aggregation (23–25). A number of basic residues in the C-terminal region of mouse SAA including Arg-83, His-84, Arg-86, Lys-89, Arg-95, and Lys-102 as well as His-36 have been implicated through studies of SAA peptide fragments and pH-dependent binding to HS. However, Arg-83 and His-84 of mouse SAA1 are not conserved in human SAA1.1. Arg-96 and Lys-103 (the equivalent of mouse Arg-95 and Lys-102) are involved in the C-terminal salt bridge clusters, and thus are partially buried and charge-neutral (Fig. 1C). The hexameric association of SAA revealed two positively charged patches on the surface of each monomer. One patch is near the hexameric center of SAA1 and consists of Arg-15, Arg-19, and Arg-47 from each monomer forming a nine-arginine charge patch around the center pore of SAA on each trimer face (Fig. 4). The other resides at the outer trimeric apex near its dimer interface and consists of Arg-1, Arg-62, and His-71 from the twofold axis-related monomer to form a six-arginine charge patch at each trimeric apex. To address the involvement of these two patches in heparan sulfate binding, we generated two alanine-cluster mutations: a triple mutation of R15A/R19A/R47A at the center pore and a triple mutation of R1A/R62A/H71A at the trimer apex. Both triple-alanine mutants exhibited apparent wild type-like hexamer association by SEC (Fig. S3B), suggesting no gross conformational disruptions due to the mutations. Their binding to heparin was characterized using ELISA as well as surface plasmon resonance by Biacore (Fig. 4D and Fig. S4). The heparin binding affinities measured by Biacore were 230 ± 27 nM for the wild type, 1,009 ± 163 nM for the pore charge mutant, and 1,712 ± 601 nM for the trimer apex mutant of SAA. Thus, the two charge mutations resulted in four- and sevenfold weaker affinities to heparin binding, suggesting that both regions are involved in GAG binding. These structure-based mutational analyses support the involvement of positively charged residues in GAG binding and define at least two HS binding sites on SAA.
Fig. 4.
Interaction of SAA1 with HDL and heparin. (A and B) Electrostatic surface representation showing the positively charged patches near the center (A) and apex (B) of the SAA1.1 hexamer. (C) Cartoon of HDL and heparin binding sites on SAA. (D) Concentration-dependent binding of wild-type SAA (black) and the center (red) and apex (green) charge cluster mutants to immobilized heparin as measured by Biacore. RU, response units. (E) Binding of recombinant hexameric (black), trimeric (red), and monomeric (green) SAA1.1, acute-phase serum SAA (cyan), and MBP-SAA189 (blue) to HDL. (F) The binding of wild-type (black, red, pink), center mutant (green, blue), and apex mutant (cyan) to immobilized HDL in the absence (black, green, cyan) or presence (red, blue) of soluble heparin or in the presence of soluble HDL (pink). The error bars are calculated as SDs of three experiments.
HDL Binding Site on SAA1.1.
In the serum, SAA1 normally associates with HDL. The HDL binding site on SAA and its release during the pathogenic formation of AA amyloid remain unclear. Previous work showed that a mutation or truncation in helix 1 resulted in the loss of HDL binding, suggesting that the N terminus is involved in HDL binding (26). To address whether the dissociation of the SAA hexamer into multiple oligomeric forms contributes to the SAA release from HDL, we measured the binding of HDL to the hexameric, trimeric, and monomeric SAA1 by ELISA. The results showed that HDL bound all three oligomeric forms of SAA (Fig. 4E), consistent with an earlier observation of HDL binding by the SAA monomer (27). Earlier work showed that heparan sulfate promoted mouse SAA dissociation from HDL (25), suggesting a potential overlap in GAG and HDL binding sites on SAA. To further investigate the HDL association, we examined HDL binding of the two SAA1 charge cluster mutations, both with reduced GAG binding. Whereas the center-pore triple-charge mutant retained HDL binding, the apex charge mutant exhibited a near-complete loss of HDL binding, indicating the involvement of the apex charge residues (Arg-1, Arg-62, and His-71) in SAA1.1 binding to HDL (Fig. 4F). This HDL binding site includes part of the N-terminal region and is consistent with the previous mutational results (26). Because the trimer apex region is one of the two GAG binding sites, the shared binding site between HDL and HS provides a competitive binding mechanism for GAG-mediated release of SAA from HDL. Indeed, heparin blocked the binding of both the wild-type and center-pore charge mutant SAA1.1 to HDL (Fig. 4F). It is worth noting that the constellation of this three-charge cluster is sensitive to the native conformation of SAA with Arg-1 in helix 1, Arg-62 in helix 3, and His-71 in the loop between helices 3 and 4. Consistently, HDL binding was significantly reduced for the C-terminal truncated maltose binding protein (MBP)-SAA189 mutant, despite having all three binding residues (Fig. 4E). This loss of HDL binding could be the result of a conformational disruption from the removal of the C-terminal tail or steric hindrance from MBP. Interestingly, SAA in acute-phase serum showed significant binding, albeit reduced compared with recombinant SAA, to HDL (Fig. 4E), suggesting either that there is some free (non–HDL-associated) SAA in the serum or that there are additional binding sites on HDL-associated SAA, such as the presence of three trimer apex charge clusters in each SAA hexamer.
Amyloidogenic Core of SAA1.
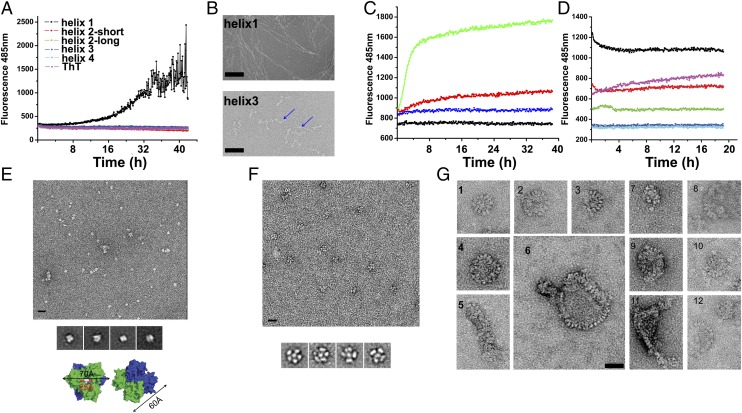
A general character of amyloid formation is the involvement of conformational transitions or structural rearrangements of proteins from native, nonaggregated structures to aggregated amyloid fibrils mediated by short interlocking β-strands, termed steric zippers (10, 28). Although we expected the presence of similar short β-strands in SAA1.1, it was to our surprise that the native SAA1.1 is an all–α-helical structure with no detectable β-strands. Where is the amyloidogenic element of SAA1.1 and how does an all–α-helical structure transit to amyloid fibrils? Previous studies have suggested the N-terminal residues of SAA1.1 to be amyloidogenic (26). To investigate the amyloidogenic region in SAA1.1, we synthesized the peptides that correspond to each of the four helices of SAA1.1 and carried out fibril formation assays using thioflavin T (ThT) staining (29). Among these peptides, the helix 1-corresponding peptide exhibited the most ThT fluorescence enhancement as a function of time at a concentration as low as 50 μg/mL, indicating a spontaneous structural transition and cross-polymerization (Fig. 5A). This result is consistent with previous studies showing that deletion of the first 11 residues of SAA1 dramatically decreased its capability of forming amyloid fibrils (26, 30). Helix 3 peptide showed ThT binding in a dose-dependent manner at higher concentrations (Fig. S4C). These results are consistent with the 3D profile-based prediction algorithms ZipperDB and TANGO (31, 32), which identified the N-terminal segments of helix 1 (residues 2SFFSFLG8) and helix 3 (residues 52VWAAEAIS59) as likely amyloidogenic peptides. The presence of fibrils in these peptides was further examined using electron microscopy (EM). Consistent with the ThT staining, the EM images showed the presence of fibril structures associated with helix 1, but much shorter protofibril-like structures associated with helix 3 (Fig. 5B).
Fig. 5.
Amyloidogenic core of SAA1 and amyloid formation. (A) Amyloid formation of SAA1 peptides using ThT binding assays. (B) EM images of fibrils formed by helix 1 (Upper) and 3 (Lower, and the fibrils are indicated by arrows) peptides. (C) Binding of ThT to hexameric (green, red) and trimeric (blue) SAA1.1 in the presence (green) and absence of cathepsin B. ThT alone is shown in black. (D) Binding of ThT to 70 µg/mL aggregated (black) or monomeric (red) MBP-SAA189, hexameric SAA1.1 (green), MBP (blue), and aggregated IgG1 (pink). (E) EM image of negatively stained his6SAA1 protein (Top) and the class averaging of his6SAA1 particles (Middle). (Bottom) Surface rendering of the his6SAA1 hexamer in two views, with each trimer colored in green and blue. (F) EM image of negatively stained MBP-SAA189 oligomers (Upper) and their class average (Lower). (G) Images of MBP-SAA189 aggregates showing repeating nods in either circular or linear assemblies. (Scale bars, 20 nm.)
The Structural Mechanism of SAA1-Mediated Amyloidogenesis.
Both helices 1 and 3 are involved in oligomerization and are secluded in the SAA1.1 hexamer, suggesting that the native structure is not amyloidogenic and that hexamer dissociation precedes amyloidogenesis (27) (Fig. 2B). Previously, peptides corresponding to residues 1–50, 1–64 or 69, 1–76, and 1–80 or 89 of SAA1.1 were identified from AA amyloid fibrils (12). These peptides match the secondary-structure boundaries between helices 2 and 3, 3 and 4, and at the end of helix 4 (Fig. 1). Consistent with previous reports (12), cathepsin B digestion of SAA resulted in enhanced ThT incorporation (Fig. 5C). To address whether removal of the C-terminal tail is sufficient to destabilize the SAA four-helix bundle structure and lead to amyloidogenic conformational changes, we expressed a four-helix bundle–only form of SAA1.1 (residues 1–89) fused to a maltose binding protein (MBP-SAA189). This truncated SAA is present in various oligomeric forms (Fig. 3D and Fig. S3C). In particular, the aggregate form exhibited an enhanced ThT fluorescence intensity at 485 nm and a characteristic red shift in Congo red absorption from ∼480 nm to ∼530 nm compared with the native SAA1 (Fig. 5D and Fig. S3D). The structure of MBP-SAA189 was then examined by EM imaging. The native SAA1.1 hexamer was visible as mostly single particles, whose class averaging resulted in a two-layered, doughnut-shaped object ∼70 Å in diameter and 60 Å in height with a 25-Å central opening, matching that of the SAA hexamer in the crystal (Fig. 5E). In contrast, distinct particle shapes from the doughnut-shaped native SAA were observed in both the oligomeric and aggregated MBP-SAA189 samples (Fig. 5). In particular, aggregated MBP-SAA189 contained various-sized and -shaped amorphous particles not present in the native SAA sample (Fig. 5G). Some form circular assemblies 22–60 nm in diameter containing 12–50 similar nod-like structures; others show a chain linked of such nods. Regardless of the size and shape of their assembly, the individual nod-like structures are spaced apart evenly with a 4.5- to 5.8-nm pitch, similar to that of amyloid-β protofibrils (33). One structural interpretation of these EM images is that MBP molecules are chain-linked by SAA amyloid-like structures into 600–2,000 kDa particles (Fig. 3D).
Discussion
Secondary amyloidosis, characterized by the deposition of AA amyloid in tissues and organs, is a major complication of many chronic inflammatory diseases (1, 2). However, the structural basis of AA amyloidosis remains largely unknown. Here we present a crystal structure of human SAA1.1, showing a unique cone-shaped up-down-up-down four-helix bundle fold, partially resembling the four-helix bundle of ApoE except that the positions of helices 3 and 4 are switched. Whereas the four-helix bundle fold of SAA is likely preserved throughout the family, residues involved in oligomerization of SAA1.1 vary significantly, suggesting individual members may exhibit different oligomerization states. The topological resemblance of SAA to ApoE is interesting because both associate with HDL, and it raises the question of whether they share a common mechanism in glycosaminoglycan and HDL binding. Despite the structural solution of ApoE over 20 y ago (16, 34), the mechanism of its binding to HDL remains unclear (35). The association of SAA with glycosaminoglycan and HDL is shown primarily to be charge-dependent (23–26). There are two patches of positively charged surfaces on SAA, one located near the central hexamer pore and the other on the apex of each trimer. Alanine mutations of the two charge clusters showed that both contribute to heparin binding. Two of the three charged residues are conserved between human and mouse in both the center (Arg-15, Arg-19, and Arg-47) and apex (Arg-1, Arg-62, and His-71) charge clusters, suggesting additional charged residues involved in mouse SAA binding to GAG. Notably, some of the charged residues implicated in mouse SAA and HS association are located near either the center or the apex charge clusters. Interestingly, only the apex charge cluster appears critical to HDL binding, suggesting that HDL binding also requires structural elements. Consistently, the C-terminal truncated SAA lost its ability to bind HDL despite the presence of wild-type charged residues. The shared binding site between HDL and HS suggests a GAG-mediated competitive release of SAA from HDL at the site of inflammation.
Unlike many amyloid-prone structures, human SAA1.1 contains no β-strands. The seclusion of the N-terminal amyloidogenic peptide obligates hexamer dissociation prior to amyloid formation. It is likely that various oligomeric forms of SAA differ in their function and pathogenicity. For example, the monomeric and trimeric but not hexameric forms are likely amyloidogenic. It is conceivable that the concentration of these potentially pathogenic forms of SAA increase dramatically during inflammation and infection. Unique to the SAA four-helix bundle is the presence of a C-terminal loop, residues 89–104, forming extensive salt bridges and hydrogen bonds with its four-helix bundle. Although it is itself outside of the helix bundle, the presence of extensive interactions suggests that the C-terminal loop is critical to the stability of the helix bundle. Indeed, the four-helix bundle–only truncation SAA189 exhibited an enhanced tendency to form amyloid-like aggregates visible by EM.
In conclusion, the combined structural, mutational, and binding studies suggest a mechanism for human SAA1.1-mediated AA amyloid formation in which the native SAA is dissociated from HDL by binding to tissue-associated heparan sulfate proteoglycans. Further dissociation of the hexamer and proteolytic cleavage would expose the amyloidogenic peptides and initiate conformational changes.
Experimental Procedures
Detailed methods are provided in SI Experimental Procedures. In brief, mature human SAA1.1 peptide (residues 1–104) was cloned into a pET30a vector and expressed with (his6SAA1) or without (SAA1) an N-terminal histidine (his6) tag in E. coli (BL21) and purified by either a HisTrap HP column (GE Healthcare) or a DEAE column and gel-filtration column. All mutations were generated using a QuikChange Mutagenesis Kit (Qiagen). SAA1 and histidine-tagged his6SAA1 were crystallized in P21212 and R32 space groups, respectively. The structures were solved by initial selenomethionine phasing of P21212 crystals and subsequent molecular replacement phasing of the hexamer structure in R32 crystals. The structures were refined using REFMAC (36) and PHENIX (37) software packages. The AUC experiments were performed using a Beckman Coulter XL-A centrifuge, and the data were analyzed using SEDFIT and SEDPHAT software packages (38). Amyloid fibril formation of various SAA peptides and purified proteins was detected by thioflavin T fluorescence and Congo red absorption using a Synergy H1 hybrid reader (BioTek). Binding experiments of SAA to heparin and HDL were carried out either using a Biacore 3000 instrument or by ELISA. Negative-staining electron microscopy images were acquired on a Tecnai T20 microscope (FEI) at 120 kV using a 4K × 4K CCD camera (Gatan).
Supplementary Material
Acknowledgments
We thank Dr. Terry W. Du Clos for providing SAA1-containing human serum and Kiyoshi Egami for performing EM imaging on the helix 1 and 3 peptide samples. We thank the shared Biomedical Engineering and Physical Science Resource at the National Institutes of Health for help with the ultracentrifugation assays. This research was supported in part by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The X-ray crystallographic data and coordinates reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4IP8 and 4IP9).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1322357111/-/DCSupplemental.
References
- 1.Pettersson T, Konttinen YT, Maury CP. Treatment strategies for amyloid A amyloidosis. Expert Opin Pharmacother. 2008;9(12):2117–2128. doi: 10.1517/14656566.9.12.2117. [DOI] [PubMed] [Google Scholar]
- 2.Bhat A, Selmi C, Naguwa SM, Cheema GS, Gershwin ME. Currents concepts on the immunopathology of amyloidosis. Clin Rev Allergy Immunol. 2010;38(2-3):97–106. doi: 10.1007/s12016-009-8163-9. [DOI] [PubMed] [Google Scholar]
- 3.Tuglular S, et al. A retrospective analysis for aetiology and clinical findings of 287 secondary amyloidosis cases in Turkey. Nephrol Dial Transplant. 2002;17(11):2003–2005. doi: 10.1093/ndt/17.11.2003. [DOI] [PubMed] [Google Scholar]
- 4.Koivuniemi R, Paimela L, Suomalainen R, Tornroth T, Leirisalo-Repo M. Amyloidosis is frequently undetected in patients with rheumatoid arthritis. Amyloid. 2008;15(4):262–268. doi: 10.1080/13506120802524676. [DOI] [PubMed] [Google Scholar]
- 5.Cocco E, et al. Serum amyloid A (SAA): A novel biomarker for uterine serous papillary cancer. Br J Cancer. 2009;101(2):335–341. doi: 10.1038/sj.bjc.6605129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chambers RE, MacFarlane DG, Whicher JT, Dieppe PA. Serum amyloid-A protein concentration in rheumatoid arthritis and its role in monitoring disease activity. Ann Rheum Dis. 1983;42(6):665–667. doi: 10.1136/ard.42.6.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zahedi K, et al. Major acute-phase reactant synthesis during chronic inflammation in amyloid-susceptible and -resistant mouse strains. Inflammation. 1991;15(1):1–14. doi: 10.1007/BF00917905. [DOI] [PubMed] [Google Scholar]
- 8.Findeisen P, et al. Serum amyloid A as a prognostic marker in melanoma identified by proteomic profiling. J Clin Oncol. 2009;27(13):2199–2208. doi: 10.1200/JCO.2008.18.0554. [DOI] [PubMed] [Google Scholar]
- 9.Uhlar CM, Whitehead AS. Serum amyloid A, the major vertebrate acute-phase reactant. Eur J Biochem. 1999;265(2):501–523. doi: 10.1046/j.1432-1327.1999.00657.x. [DOI] [PubMed] [Google Scholar]
- 10.Nelson R, Eisenberg D. Recent atomic models of amyloid fibril structure. Curr Opin Struct Biol. 2006;16(2):260–265. doi: 10.1016/j.sbi.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 11.Dember LM, et al. Eprodisate for AA Amyloidosis Trial Group Eprodisate for the treatment of renal disease in AA amyloidosis. N Engl J Med. 2007;356(23):2349–2360. doi: 10.1056/NEJMoa065644. [DOI] [PubMed] [Google Scholar]
- 12.Röcken C, et al. Proteolysis of serum amyloid A and AA amyloid proteins by cysteine proteases: Cathepsin B generates AA amyloid proteins and cathepsin L may prevent their formation. Ann Rheum Dis. 2005;64(6):808–815. doi: 10.1136/ard.2004.030429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamada T, Kluve-Beckerman B, Liepnieks JJ, Benson MD. Fibril formation from recombinant human serum amyloid A. Biochim Biophys Acta. 1994;1226(3):323–329. doi: 10.1016/0925-4439(94)90044-2. [DOI] [PubMed] [Google Scholar]
- 14.Chou KC, Maggiora GM, Némethy G, Scheraga HA. Energetics of the structure of the four-alpha-helix bundle in proteins. Proc Natl Acad Sci USA. 1988;85(12):4295–4299. doi: 10.1073/pnas.85.12.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun PD, Foster CE, Boyington JC. Overview of protein structural and functional folds. Curr Protoc Protein Sci. 2004;35:17.1.1–17.1.189. doi: 10.1002/0471140864.ps1701s35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson C, Wardell MR, Weisgraber KH, Mahley RW, Agard DA. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science. 1991;252(5014):1817–1822. doi: 10.1126/science.2063194. [DOI] [PubMed] [Google Scholar]
- 17.Wang L, Lashuel HA, Walz T, Colon W. Murine apolipoprotein serum amyloid A in solution forms a hexamer containing a central channel. Proc Natl Acad Sci USA. 2002;99(25):15947–15952. doi: 10.1073/pnas.252508399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, et al. Serum amyloid A 2.2 refolds into a octameric oligomer that slowly converts to a more stable hexamer. Biochem Biophys Res Commun. 2011;407(4):725–729. doi: 10.1016/j.bbrc.2011.03.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, Lashuel HA, Colón W. From hexamer to amyloid: Marginal stability of apolipoprotein SAA2.2 leads to in vitro fibril formation at physiological temperature. Amyloid. 2005;12(3):139–148. doi: 10.1080/13506120500223084. [DOI] [PubMed] [Google Scholar]
- 20.Srinivasan S, et al. Pathogenic serum amyloid A 1.1 shows a long oligomer-rich fibrillation lag phase contrary to the highly amyloidogenic non-pathogenic SAA2.2. J Biol Chem. 2013;288(4):2744–2755. doi: 10.1074/jbc.M112.394155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Snow AD, Kisilevsky R, Stephens C, Anastassiades T. Characterization of tissue and plasma glycosaminoglycans during experimental AA amyloidosis and acute inflammation. Qualitative and quantitative analysis. Lab Invest. 1987;56(6):665–675. [PubMed] [Google Scholar]
- 22.Snow AD, Bramson R, Mar H, Wight TN, Kisilevsky R. A temporal and ultrastructural relationship between heparan sulfate proteoglycans and AA amyloid in experimental amyloidosis. J Histochem Cytochem. 1991;39(10):1321–1330. doi: 10.1177/39.10.1940305. [DOI] [PubMed] [Google Scholar]
- 23.Ancsin JB, Kisilevsky R. The heparin/heparan sulfate-binding site on apo-serum amyloid A. Implications for the therapeutic intervention of amyloidosis. J Biol Chem. 1999;274(11):7172–7181. doi: 10.1074/jbc.274.11.7172. [DOI] [PubMed] [Google Scholar]
- 24.Elimova E, Kisilevsky R, Ancsin JB. Heparan sulfate promotes the aggregation of HDL-associated serum amyloid A: Evidence for a proamyloidogenic histidine molecular switch. FASEB J. 2009;23(10):3436–3448. doi: 10.1096/fj.09-134981. [DOI] [PubMed] [Google Scholar]
- 25.Noborn F, Ancsin JB, Ubhayasekera W, Kisilevsky R, Li JP. Heparan sulfate dissociates serum amyloid A (SAA) from acute-phase high-density lipoprotein, promoting SAA aggregation. J Biol Chem. 2012;287(30):25669–25677. doi: 10.1074/jbc.M112.363895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel H, Bramall J, Waters H, De Beer MC, Woo P. Expression of recombinant human serum amyloid A in mammalian cells and demonstration of the region necessary for high-density lipoprotein binding and amyloid fibril formation by site-directed mutagenesis. Biochem J. 1996;318(Pt 3):1041–1049. doi: 10.1042/bj3181041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, Colón W. The interaction between apolipoprotein serum amyloid A and high-density lipoprotein. Biochem Biophys Res Commun. 2004;317(1):157–161. doi: 10.1016/j.bbrc.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 28.Merlini G, Bellotti V. Molecular mechanisms of amyloidosis. N Engl J Med. 2003;349(6):583–596. doi: 10.1056/NEJMra023144. [DOI] [PubMed] [Google Scholar]
- 29.LeVine H., III Quantification of beta-sheet amyloid fibril structures with thioflavin T. Methods Enzymol. 1999;309:274–284. doi: 10.1016/s0076-6879(99)09020-5. [DOI] [PubMed] [Google Scholar]
- 30.Yamada T, Kluve-Beckerman B, Liepnieks JJ, Benson MD. In vitro degradation of serum amyloid A by cathepsin D and other acid proteases: Possible protection against amyloid fibril formation. Scand J Immunol. 1995;41(6):570–574. doi: 10.1111/j.1365-3083.1995.tb03609.x. [DOI] [PubMed] [Google Scholar]
- 31.Fernandez-Escamilla AM, Rousseau F, Schymkowitz J, Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat Biotechnol. 2004;22(10):1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- 32.Thompson MJ, et al. The 3D profile method for identifying fibril-forming segments of proteins. Proc Natl Acad Sci USA. 2006;103(11):4074–4078. doi: 10.1073/pnas.0511295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lashuel HA, et al. Mixtures of wild-type and a pathogenic (E22G) form of Abeta40 in vitro accumulate protofibrils, including amyloid pores. J Mol Biol. 2003;332(4):795–808. doi: 10.1016/s0022-2836(03)00927-6. [DOI] [PubMed] [Google Scholar]
- 34.Dong LM, et al. Novel mechanism for defective receptor binding of apolipoprotein E2 in type III hyperlipoproteinemia. Nat Struct Biol. 1996;3(8):718–722. doi: 10.1038/nsb0896-718. [DOI] [PubMed] [Google Scholar]
- 35.Chen J, Li Q, Wang J. Topology of human apolipoprotein E3 uniquely regulates its diverse biological functions. Proc Natl Acad Sci USA. 2011;108(36):14813–14818. doi: 10.1073/pnas.1106420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 11):1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 38.Brown PH, Schuck P. A new adaptive grid-size algorithm for the simulation of sedimentation velocity profiles in analytical ultracentrifugation. Comput Phys Commun. 2008;178(2):105–120. doi: 10.1016/j.cpc.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.