Significance
Misregulation of cell growth and proliferation leads to the onset of various diseases, including cancer. Two proteins crucial for proper cellular control that were recently shown to affect cellular proliferation are Bcl-2, well-known for its role in programmed cell death, and the newly identified iron-sulfur protein NAF-1, localized near the mitochondrial outer membrane. In this report, we use a strategy utilizing a combination of experimental and computational techniques that provides valuable information to enable us to determine a molecular picture of the NAF-1–Bcl-2 interaction interface that is more complete than that obtained from any one technique alone. This interaction interface provides the basis from which novel drugs can be developed for the treatment of diseases such as cancer.
Keywords: Cisd1, Cisd2, mitoNEET, Miner1, CDGSH
Abstract
Life requires orchestrated control of cell proliferation, cell maintenance, and cell death. Involved in these decisions are protein complexes that assimilate a variety of inputs that report on the status of the cell and lead to an output response. Among the proteins involved in this response are nutrient-deprivation autophagy factor-1 (NAF-1)- and Bcl-2. NAF-1 is a homodimeric member of the novel Fe-S protein NEET family, which binds two 2Fe-2S clusters. NAF-1 is an important partner for Bcl-2 at the endoplasmic reticulum to functionally antagonize Beclin 1-dependent autophagy [Chang NC, Nguyen M, Germain M, Shore GC (2010) EMBO J 29(3):606–618]. We used an integrated approach involving peptide array, deuterium exchange mass spectrometry (DXMS), and functional studies aided by the power of sufficient constraints from direct coupling analysis (DCA) to determine the dominant docked conformation of the NAF-1–Bcl-2 complex. NAF-1 binds to both the pro- and antiapoptotic regions (BH3 and BH4) of Bcl-2, as demonstrated by a nested protein fragment analysis in a peptide array and DXMS analysis. A combination of the solution studies together with a new application of DCA to the eukaryotic proteins NAF-1 and Bcl-2 provided sufficient constraints at amino acid resolution to predict the interaction surfaces and orientation of the protein–protein interactions involved in the docked structure. The specific integrated approach described in this paper provides the first structural information, to our knowledge, for future targeting of the NAF-1–Bcl-2 complex in the regulation of apoptosis/autophagy in cancer biology.
Life requires a controlled balance of energy conversion and utilization. These critical processes are governed by an elaborate set of reactions involving numerous protein–protein interactions. Among them is the ability of organisms to control the recycling of high-energy compounds and to control cell proliferation. These processes are, at least in part, under the control of cell survival and programmed cell death (autophagic and apoptotic) processes. Misregulation of these processes leads to many diseases, including cancer. Among the key proteins involved in these processes are Bcl-2 (1, 2) and the more recently identified iron-sulfur (Fe-S) protein nutrient-deprivation autophagy factor-1 (NAF-1) (also known as Cisd2, Miner1, Eris, and Noxp70) (3–5).
NAF-1 is important for human health and disease. Missplicing of NAF-1 causes Wolfram syndrome 2 (6). NAF-1 is also functionally linked to the regulation of autophagy in cancer, and aging (3–5, 7, 8). This protein is a member of the 2Fe-2S cluster NEET family. NAF-1 has a similar backbone fold and 3Cys-1His coordination of the 2Fe-2S cluster as found in the founding member of the NEET family, mitoNEET (mNT). NAF-1 differs from mNT in the distribution of charged and aromatic surface residues (9, 10). These differences alter the 3D shape and electrostatics of the surfaces of mNT and NAF-1, leading to interactions with distinct binding partners. In fact, recent work identified NAF-1 as a Bcl-2/Bcl-XL binding partner (4) at a branch point between autophagy and apoptosis, life and death, under nutrient-deprived and oxidative stress conditions in vivo (4, 7). This unusual interaction between a 2Fe-2S protein with the apoptotic/autophagic response pathways provides a new target for designing pro- and antiapoptotic therapies. Structural studies of the molecular determinants for these newly discovered protein–protein interactions are important for the specific design of modulators of the apoptotic/autophagic pathways.
Here we report on how a combination of peptide array, deuterium exchange mass spectrometry (DXMS), functional studies, and an unconventional application of direct coupling analysis (DCA) for eukaryotic complexes was used to determine both the dominant docked interface and functional consequences of NAF-1–Bcl-2 interaction. We show that NAF-1 binds to the BH3 and BH4 regions of Bcl-2 and that Bcl-2 binds to the NAF-1 groove formed between the β-cap and iron-sulfur cluster binding domains, with the strongest coupled interactions to the cluster binding domain. Binding of specific Bcl-2 peptides destabilizes the 2Fe-2S clusters of NAF-1 and affects protein functionality. Our unique integrated strategy that combines peptide array, DXMS, and DCA provides a set of data sufficient to obtain a level of structural detail that is not possible from any one individual technique. Furthermore, this robust strategy may be of general use in structural biology, in particular for interactions that may not be easily amenable to crystallography or NMR studies. The results presented provide structural information for future targeting of the novel NAF-1–Bcl-2 protein pair in disease management, such as in the regulation of apoptosis/autophagy in cancer.
Results
Identification of the NAF-1 Binding Interface with Bcl-2 by Peptide Array Screening.
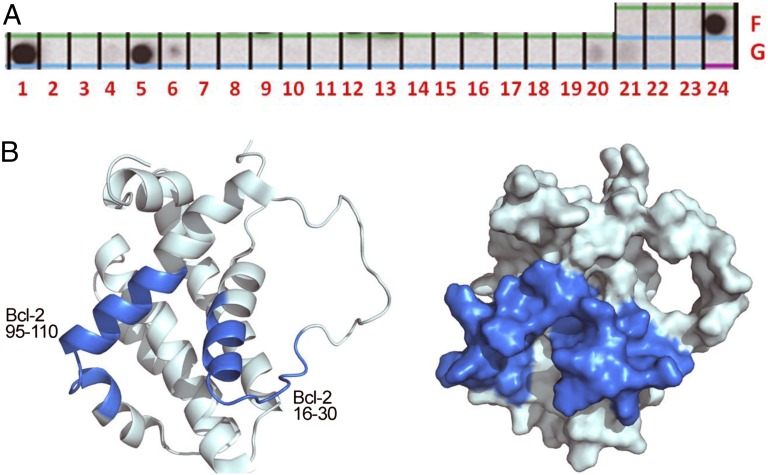
We designed an array composed of 27 partially overlapping peptides derived from Bcl-2 to identify the binding interface between regions in Bcl-2 that mediate the protein–protein interaction with NAF-1. Peptide length varied between 8 and 21 residues (Table 1). Peptides were designed based on the secondary and tertiary structures of the Bcl-2 NMR structure [Protein Data Bank (PDB) ID code 1YSW]. NAF-1 (His-tagged NAF-1 57–135 S92C) was expressed, purified, and screened for binding to the peptide array (Fig. 1A and Table 1). NAF-1 bound Bcl-2 peptides from two distinct regions of the full-length protein (Fig. 1B). The first region was composed of peptides derived from the N-terminal α-helix and from the loop that follows (Bcl-2 peptides 16–30 and 23–44). This site corresponds to the BH4 domain, which is conserved among antiapoptotic Bcl-2 family members, and is essential for apoptosis inhibition. The second region consists of parts of helices 2 and 3, represented by the binding peptide Bcl-2 95–110, part of the BH3 domain. This site overlaps with a known binding site for proapoptotic members of the Bcl-2 family such as Bak and Bad (2). The Bcl-2 interaction surface deduced from the peptide array data is shown in Fig. 1B. The regions are structurally contiguous and overlap both regions associated with known pro- and antiapoptotic binding sites. For studies on the effects of peptide binding on the properties of NAF-1, we initiated our analyses using the strongly interacting Bcl-2 16–30 peptide.
Table 1.
Peptides used in the array screen
| Array no. | Sequence | Bcl-2 residues |
| F21 | HAGRTGYD | 1–8 |
| F22 | HAGRTGYDNREIVMK | 1–15 |
| F23 | YDNREIVMKYIHYKLSQR | 7–24 |
| F24 | YIHYKLSQRGYEWDA | 16–30 |
| G1 | QRGYEWDAGDDVEENRTEAPEG | 23–44 |
| G2 | AGDDVEENRTEAPEG | 30–44 |
| G3 | EAPEGTESEVVHLTL | 40–94* |
| G4 | TESEVVHLTLRQAGDDFSRR | 45–104* |
| G5 | RQAGDDFSRRYRRDF | 95–108 |
| G6 | SRRYRRDFA | 102–110 |
| G7 | RYRRDFAEMSSQL | 104–116 |
| G8 | RYRRDFAEMSSQLHLT | 105–119 |
| G9 | FAEMSSQLHLT | 109–119 |
| G10 | LHLTPFTARGRFATV | 116–130 |
| G11 | PFTARGRFATVVEEL | 120–134 |
| G12 | RFATVVEELFRDGVN | 126–140 |
| G13 | LFRDGVNW | 134–141 |
| G14 | LFRDGVNWGRIVAFF | 134–148 |
| G15 | WGRIVAFFEFGGVMCVESVNR | 141–161 |
| G16 | FEFGGVMCVESVNR | 148–161 |
| G17 | GVMCVESVNREMSPL | 152–166 |
| G18 | EMSPLVDNIALWMTE | 162–176 |
| G19 | LVDNIALWMTEYLNR | 166–180 |
| G20 | EYLNRHLHTWIQDNG | 176–190 |
| G21 | RHLHTWIQDNGG | 180–191 |
| G22 | NGGWDAFVELYG | 189–200 |
| G23 | WDAFVELYGPSMR | 192–204 |
Residues in bold are interacting regions from overlapping peptides.
These peptides are lacking residues 49–88, which is part of a disordered loop of Bcl-2.
Fig. 1.
Mapping the binding sites of the peptides interacting with NAF-1 on the Bcl-2 protein. (A) An array consisting of partly overlapping peptides derived from Bcl-2 was screened for binding to NAF-1. Each dark spot represents binding of NAF-1 to a specific peptide (Table 1). (B) The binding sites of the peptides discovered in the peptide array screening (described in A) are colored on the 3D structure of Bcl-2. The interaction surface involves regions previously shown to be involved in interactions with both pro- and antiapoptotic proteins.
The NAF-1 2Fe-2S Cluster Is Destabilized by Interaction with the Bcl-2 16–30 Peptide.
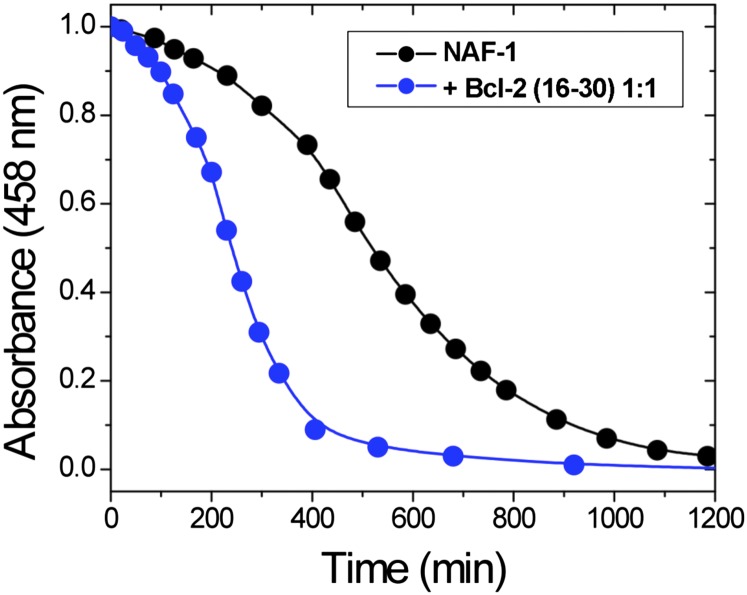
Small-molecule binding to NEET proteins can either stabilize (11) or destabilize (12) the 2Fe-2S clusters. To test whether the binding of the Bcl-2 peptide affects the stability of the NAF-1 2Fe-2S clusters, we incubated NAF-1 with the Bcl-2 16–30 peptide and measured the absorbance at 458 nm [characteristic of the intact 2Fe-2S cluster of NAF-1 (9)] over time and compared the rate of cluster release with the NAF-1 protein in the absence of added peptide. The absorbance at 458 nm decreased faster in the presence of the Bcl-2 peptide (Fig. 2, blue) than without the peptide (black). This indicates that the NAF-1 protein binds the Bcl-2 16–30 peptide and that its cluster is destabilized by the interaction.
Fig. 2.
Bcl-2 16–30 peptide destabilizes the 2Fe-2S cluster of NAF-1. UV-vis spectroscopy was used to observe the 2Fe-2S cluster stability of NAF-1 in the absence and presence of stoichiometric amounts of the Bcl-2 16–30 peptide. The Bcl-2 peptide interaction accelerates the cluster loss by a factor of two. All UV-vis spectra were measured from 250 to 800 nm on a Cary 50 spectrophotometer (Varian). Assay conditions were 100 µM NAF-1 with or without 100 µM Bcl-2 16–30 peptide in 50 mM bis-Tris buffer (pH 6.0) and 100 mM NaCl.
NAF-1’s 2Fe-2S Cluster Transfer Is Accelerated by the Bcl-2 16–30 Peptide.
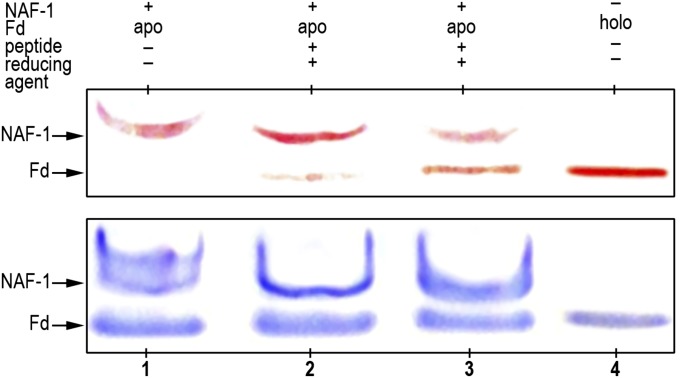
We established that NAF-1 is a cluster donor protein in vitro and in vivo (11). In an effort to determine whether binding of the Bcl-2 16–30 peptide alters these functional properties, we tested the ability of NAF-1 to transfer its cluster to the apo-acceptor protein ferredoxin (Fd) as described previously (11). NAF-1 and apo-Fd with and without the Bcl-2 16–30 peptide were incubated for 20 min at room temperature with 5 mM DTT (to keep the cysteines of apo-Fd reduced). The progress of the reaction was assessed by native gel electrophoresis, as previously described (11) (Fig. 3). As can be seen in the nondyed gel (Upper), the intensity of the red band of NAF-1 is decreased concomitant with an increase in the intensity of the red band of Fd in the presence of Bcl-2 16–30 (compare lane 2 with lane 3), suggesting that cluster transfer is accelerated in the presence of the Bcl-2 16–30 peptide. Coomassie staining of this gel (Lower) shows that the levels of NAF-1 and apo/holo-Fd do not change during the cluster transfer assay. Holo-Fd was run as a reference (holo).
Fig. 3.
NAF-1’s 2Fe-2S cluster transfer is enhanced by the Bcl-2 16–30 peptide. NAF-1 was incubated with apo-Fd for 20 min at room temperature under different conditions, and the products were run on a native gel. Holo-Fd was run as a reference (line 4). Prereduction of the acceptor Cys ligand residues of apo-Fd with 5 mM DTT ensures transfer from NAF-1 to apo-Fd. Cluster transfer was enhanced by the addition of Bcl-2 16–30 peptide (line 3). (Upper) The gel is not stained (the red is from the 2Fe-2S in the cluster). (Lower) Coomassie blue staining of the gel verifying similar protein levels in all lanes. NAF-1 and Fd are indicated.
The Bcl-2 16–30 Peptide Remains Bound to NAF-1 Following Cluster Transfer.
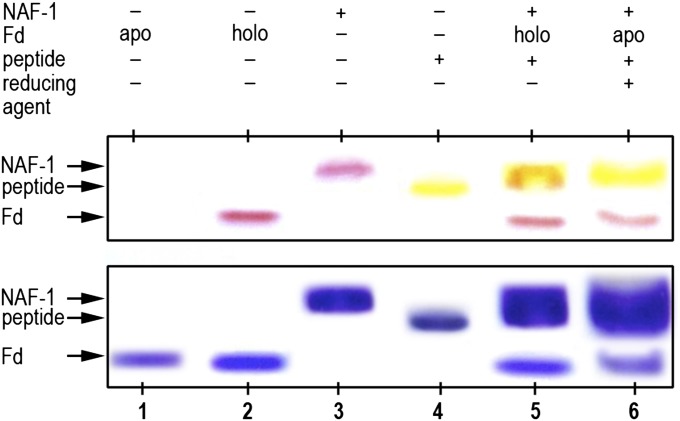
Fluorescein-labeled Bcl-2 16–30 peptide was used to monitor the fate of the peptide following transfer of NAF-1’s 2Fe-2S cluster to the apo-Fd acceptor protein. The results are presented in Fig. 4 and show that the Bcl-2 peptide (yellow band in lane 4) binds to NAF-1 when added to the reaction mix (upper red band in lane 5) as the position of the yellow fluorescent band comigrates with the NAF-1 band. The binding of the Bcl-2 16–30 peptide is specific to NAF-1, as no interactions with apo/holo-Fd are detected. Upon transfer of the 2Fe-2S cluster from NAF-1 to apo-Fd, the yellow Bcl-2 16–30 peptide remains bound to NAF-1 (lane 6); the lower red band demonstrates that the 2Fe-2S cluster is transferred to form holo-red-Fd under these conditions.
Fig. 4.
Bcl-2 16–30 peptide remains bound to NAF-1 following 2Fe-2S cluster transfer. Lane 1, apo-Fd; lane 2, holo-Fd; lane 3, NAF-1; lane 4, fluorescein-labeled Bcl-2 16–30 peptide; lane 5, mixture of NAF-1, holo-Fd, and peptide; lane 6, mixture of NAF-1, apo-Fd, and peptide under conditions to promote cluster transfer (as observed by the presence of the red band at the Fd position). These results show that the Bcl-2 peptide binds to NAF-1 and remains bound following 2Fe-2S cluster transfer to apo-Fd. (Upper) The gel is not stained (the red is from the 2Fe-2S in the cluster, and the yellow is from the fluorescein-labeled peptide). (Lower) Stained with Coomassie.
Direct Coupling Analysis Identifies Putative Interfacial Residues.
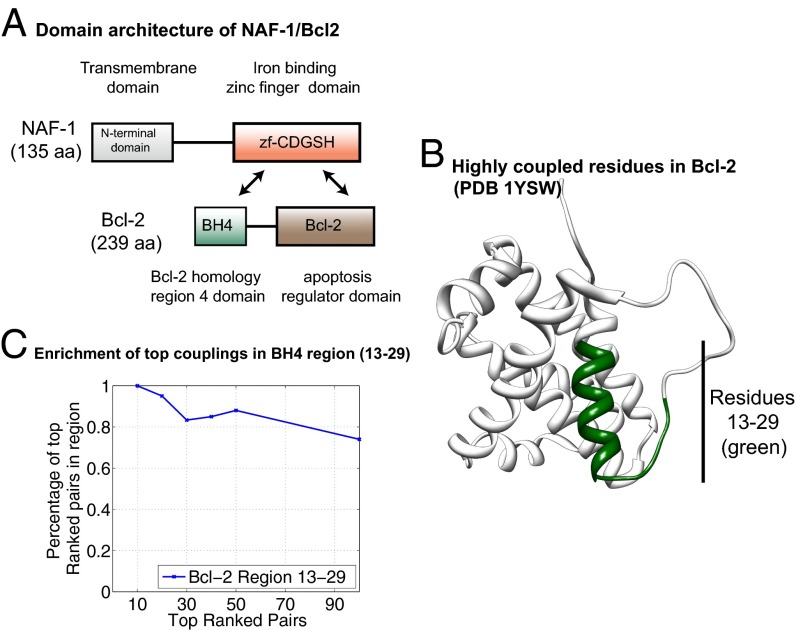
Because the experimental results showed an interaction between NAF-1 and Bcl-2, we used DCA (13) to identify important residues for the interaction based on the coevolution of the amino acid side chains between the two proteins (using sequence data for Pfam domains zf-CDGSH, Bcl-2, and BH4). A distinction with respect to previous applications of DCA to protein interactions (14, 15) is that our criterion for sequence pairing is based on the presence of both proteins (sequences) in the same organism rather than genome adjacency. DCA computes direct information (DI) values for each potential interresidue pair, providing a metric of the most directly coupled residue–residue interactions. DCA found that the top 30 of 1,000 potential interdomain residue–residue interactions involved the 13–29 amino acid peptide region of Bcl-2, consistent with the tight binding observed in the experimental measurements described above (Figs. 1–4). Moreover, the DCA determined that this region of Bcl-2 interacts with the region near the 2Fe-2S clusters of NAF-1; this interaction is shown in Fig. 5. We also verified that the average DI values per residue pair for the interaction region between domains zf-CDGSH and BH4 (amino acids 7–33) are higher than the average DI values for the potential interaction between domains zf-CDGSH and Bcl-2 (amino acids 97–195). This provides additional support that the BH4 region is part of the interaction interface.
Fig. 5.
Direct coupling analysis for protein–protein recognition in the NAF-1–Bcl-2 system. (A) NAF-1 has a domain architecture including an N-terminal (PF10660) transmembrane domain and an iron binding zinc finger domain, zf-CDGSH (PF09360). Bcl-2 has a two-domain architecture as well as other unstructured regions. The domain BH4 (Bcl-2 homology region 4; PF02180) is found in several proteins analogous to Bcl-2 as well as its apoptosis regulator domain Bcl-2 (PF00452). Interaction between the zinc finger domain (zf-CDGSH) and the Bcl-2 domains (BH4 and Bcl-2) was evaluated by pairing and analyzing the genomic sequences of these domain families. (B) Bcl-2 structure (PDB ID code 1YSW). The green section shows the region with the DCA highest couplings for the domain pair zf-CDGSH–BH4. (C) Residues in region 13–29 of Bcl-2 constitute the highest percentage of residues in the top-ranked pairs. This suggests that this region has a higher probability of forming interfacial contacts between the iron binding zinc finger domain of NAF-1 and the BH4 domain of Bcl-2.
Deuterium Exchange Mass Spectrometry Analysis.
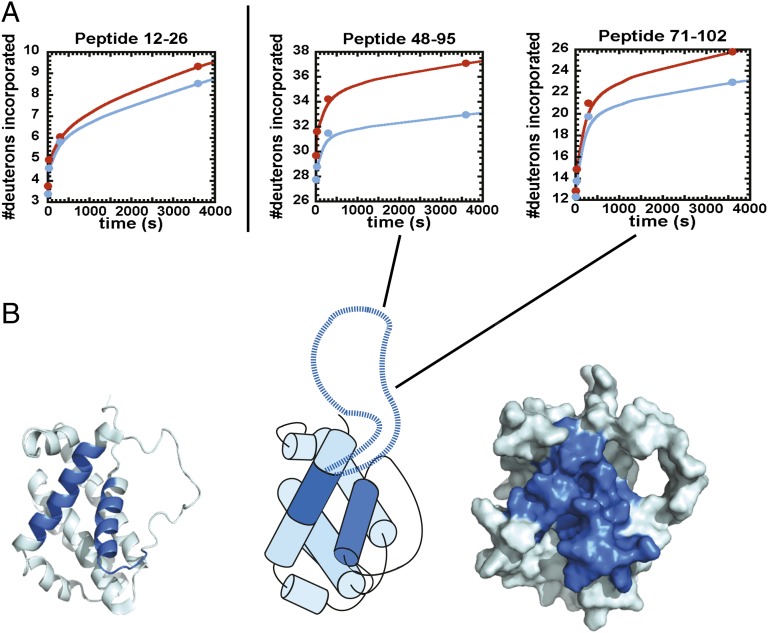
Protein–protein interactions result in changes in the stability of the backbone amide protons to solvent deuterium exchange at the binding interface and, in some cases, distal to the binding interface (16). To assess the changes upon NAF-1–Bcl-2 complex formation, we used a DXMS method. This method is powerful for analyzing protein–protein interactions and also provides an experimental test of the predictions from the DCA presented above. Importantly, DXMS allows an assessment of the role of the signature long loop in helix α2 of Bcl-2 that cannot be evaluated by DCA because it is outside of the evolving core sequence. Also, this loop is not present in the solution structure of the protein (17). In the DXMS experiment a fragmentation map is established for the protein, as described (18). The nondeuterated protein is exposed to deuterated buffer for varying amounts of time, and the mass of each peptide probe is measured as a function of incubation time to determine the number of in-exchanged deuterons incorporated with respect to fully protonated and deuterated probes (Materials and Methods). Formation of a protein–protein complex between NAF-1 and Bcl-2 resulted in a significant reduction in the rate of deuteron incorporation into the backbone amides of specific regions within each protein compared with the same regions of the respective proteins free and in complex (Fig. S1). Data for the protection of specific regions are shown for representative probes from Bcl-2 in Fig. 6A. Deuteron incorporation into these peptide probes is significantly reduced over time in the presence of NAF-1. These time-dependent analyses were performed on fragments that cover ∼90% of the protein’s sequence. Two regions in Bcl-2 that showed the greatest change in protection from solvent exchange upon complex formation are nearly identical to those identified by peptide array, and are mapped onto the structure in Fig. 6B. In addition, the long loop deleted in the structure (residues 49–88) shows significant protection from exchange upon complex formation. Complementary studies on the NAF-1 protein are shown in Fig. 7 and are in good agreement with the areas predicted by DCA.
Fig. 6.
Effects of NAF-1–Bcl-2 protein complex formation on time-dependent solvent deuterium incorporation into Bcl-2 peptide probes. (A) Peptides 12–27, 48–85, and 71–102 in Bcl-2 show increased protection from deuterium incorporation (blue) upon complex formation. Residues 49–88 are missing from the structures available. (B) The peptides that undergo protection from solvent exchange (blue) are mapped onto the structure of Bcl-2 and are in good agreement with those identified in the peptide array and DCA experiments (Figs. 1 and 4). (Center) A representation of the helical Bcl-2 structure with the long loop that is protected from exchange upon complex formation.
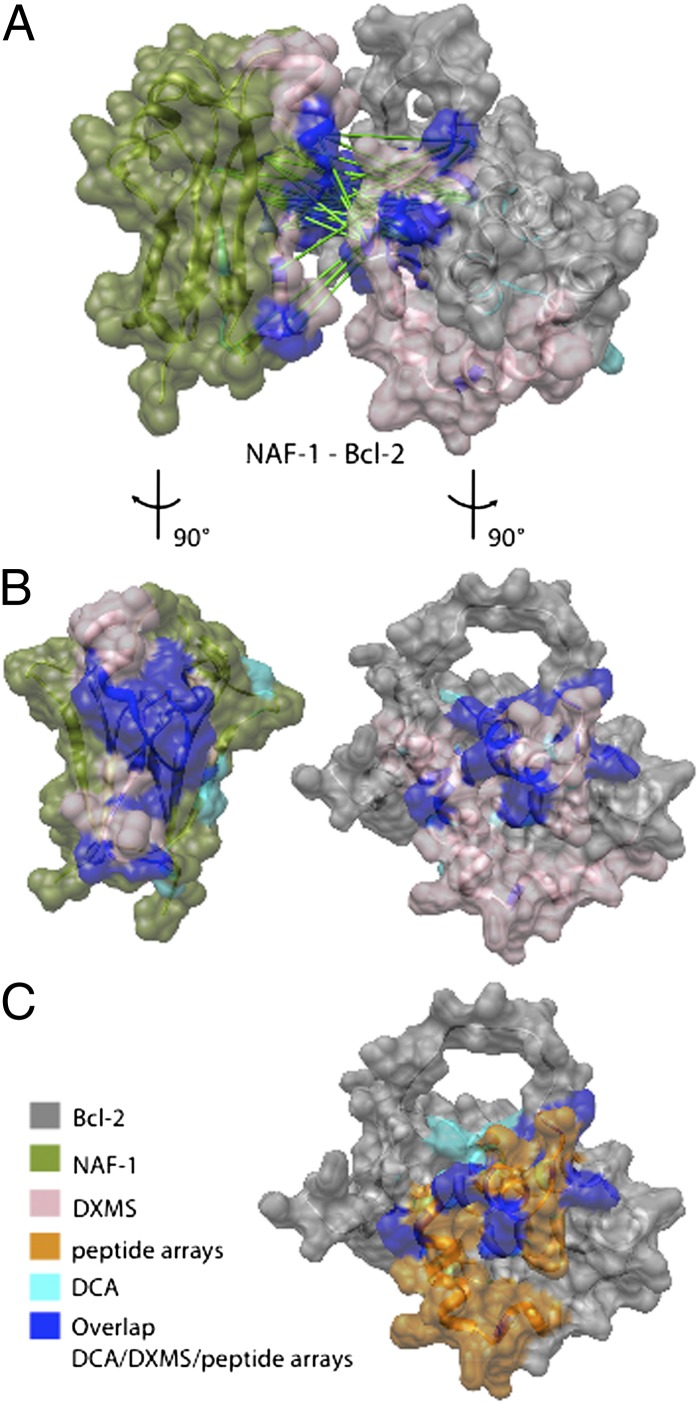
Fig. 7.
Predicted interacting surface of NAF-1 and Bcl-2. (A) A putative interacting surface determined from DCA constraints (green links) is shown in dark blue and cyan. The DXMS results showing all protected residues are highlighted in pink and dark blue. Dark blue is the overlap between DXMS and DCA. The DCA residues are the top 30 DI pairs from each of the family pairings zf-CDGSH–BH4 (Table S1) and zf-CDGSH–Bcl-2 (Table S2). (B) NAF-1 and Bcl-2 are rotated by 90°. (C) Bcl-2 with interacting peptides from the array is shown in orange, and DCA residues are in cyan. Dark blue is the overlap between the peptide array and DCA.
Discussion
Bcl-2 and NAF-1 are recognized players in autophagy and apoptosis. NAF-1, in particular, is an emerging target in cancer and aging-related diseases. The NAF-1–Bcl-2 interaction has been refractory to direct structural determination by NMR or X-ray data collection on the protein complex. In this study, we expand upon the traditional approach of elucidating structural interfaces by using a unique strategy of combining the power of experimental peptide array and DXMS data together with an unconventional use of the theoretical DCA calculation for eukaryotic complexes to help determine the orientation of the dominant docking interface of this important NAF-1–Bcl-2 target. By introducing DCA to eukaryotic protein interactions, we provide key coevolutionary constraints on the possible interaction interface orientations consistent with experimental data obtained for Bcl-2 and NAF-1. The integrated approach presented is synergistic, as the DXMS validates DCA results while DCA orients the multiple interfaces consistent with experimental results. Furthermore, we show that the interaction between the Bcl-2 peptides and NAF-1 causes changes to the characteristics and function of the 2Fe-2S cluster of NAF-1. Because Bcl-2 has been implicated in both the cell-survival autophagic process and the programmed cell-death apoptotic process, interaction between NAF-1 and Bcl-2 places it at the interface between life, death, and disease.
NAF-1–Bcl-2 Interaction Interface.
Peptide array screening has become an established method to determining the binding site between proteins (19, 20). In the present study, we focused on the binding of NAF-1 to an array of Bcl-2 peptides. The binding site of NAF-1 on Bcl-2 was localized to the BH4 domain and part of the BH3 domain of Bcl-2 (Fig. 1). Because peptides F24 and G1 are bound by NAF-1, the binding domain is most likely composed of the QRGYEWDA shared peptide region corresponding to amino acid residues 23–30 of the BH4 region. In addition, binding of NAF-1 was to G5 but not G4 or G6. Thus, the BH3 epitope is likely composed of the sequence DDFSRRYRR (amino acids 98–106) that is complete only in G5. Because G4 and G6 contain only part of the sequence, the binding may have been below detection limits; consistent with this idea is the weaker signal observed for G6 and possibly G4. Although these peptides are in separate regions of the primary sequence, they are in close juxtaposition in the structure of Bcl-2 (Fig. 1). The importance of the NAF-1–Bcl-2 interaction, in particular that with the BH4 binding to NAF-1, is apparent from the observed changes to the known properties of NAF-1 in the absence and presence of the Bcl-2 16–30 peptide. The Bcl-2 16–30 peptide decreases the stability of the 2Fe-2S clusters of NAF-1, indicating that peptide binding affects the properties of the cofactor of NAF-1. Moreover, in the presence of the peptide, the NAF-1 2Fe-2S cluster is transferred to an apo-acceptor protein more rapidly (Figs. 3 and 4).
To gain further insight into the specific amino acid side chains that interact with the BH4 region of Bcl-2, we applied the methodology of DCA to identify the regions of these interacting eukaryotic partners through the coevolution of Bcl-2 and NAF-1 amino acid residues. Amino acid side chains located near the 2Fe-2S cluster of NAF-1 were identified as being involved in many of the top-scoring pairs, as shown in Fig. 7. NAF-1 has a groove that appears to form a favorable binding interface with the helix-loop region of Bcl-2. The lower edge of this groove contains the sequence TFPACDG, with the Cys being one of the ligands for 2Fe-2S coordination. We also probed the protein–protein interaction with DXMS experiments (Figs. 6 and 7). Strikingly, DXMS validates the peptide array and DCA results, but gives additional important information. The long loop (residues 48–88) that is inserted into α2 in wild-type Bcl-2 shows significant protection from exchange upon complex formation. This region is not analyzable by DCA because it is not part of the evolving core sequence. Thus, each technique gives unique information. It is only by using this integrated approach, in which the powers of theoretical and experimental methodologies complement each other, that we could obtain such valuable and validated insights for the interaction interface between NAF-1 and Bcl-2.
Implications for Life and Death Processes.
There are four currently identified proteins that are involved in both autophagy and apoptosis and are linked to cell survival and cell death: Bcl-2, Beclin, Ambra1, and NAF-1 (21). Previous reports have shown the interaction of NAF-1 with Beclin and Bcl-2 (4). Here we establish the detailed molecular interface for the NAF-1–Bcl-2 interaction. Of these proteins, only one (NAF-1) has a known cofactor that can sense (i) ambient redox potential and (ii) Fe and/or Fe-S levels in the cell. Because NAF-1 itself contains a 2Fe-2S center, which can undergo various reactions with ROS and RNS, it becomes a potential sensor of these signaling molecules as well. The idea to “sense” redox/Fe/Fe-S/reactive oxygen and nitrogen species seems critical for the decision of promoting cell survival or deciding upon cell death.
Future Directions.
Up-regulation of NAF-1 occurs in cancer cells (5, 22), indicating that cancer may hijack control over NAF-1 expression. This up-regulation is key to uncontrolled growth of cells without the proper checks and balances. As such, NAF-1 becomes a novel target of therapeutics for the treatment of cancer and other cell division-related diseases. Our results provide a structural platform for future NAF-1–directed drug design as well as a general methodology for approaching structure-based design.
Materials and Methods
Peptide Array Screening.
An array of partially overlapping peptides derived from the Bcl-2 sequence (PDB ID code 1YSW) (23) was synthesized by INTAVIS Bioanalytical Instruments. The peptide array was immersed for 4 h in Tris buffered saline Tween (TBST) [50 mM Tris⋅HCl (pH 7.5), 0.15 M NaCl, 0.05% (vol/vol) Tween 20, nonfat dry milk 2.5% (wt/vol)] and prewashed three times in TBST. His-tagged NAF-1 at a final concentration of 10 μM was diluted with BS and incubated with the array overnight at 4 °C. Washing steps included two times for 5 min in BS and three times for 5 min in TBST. Binding was detected with anti–His-tagged HRP-conjugated mouse monoclonal antibody (Santa Cruz Biotechnology) using the chemiluminescence blotting substrate SuperSignal reagent (Biological Industries) according to the manufacturer’s instructions.
Peptide Synthesis, Labeling, and Purification.
Bcl-2–derived peptides were synthesized using solid-phase peptide synthesis on a Liberty microwave-assisted peptide synthesizer (CEM) using standard Fmoc chemistry as described (19, 20). The peptides were labeled using 5′- and 6′-carboxyfluorescein succinimidyl ester (Molecular Probes) at their N terminus, as described (19, 20). The peptides were purified on a Gilson HPLCy using a reverse-phase C8 semipreparative column (ACE; Advanced Chromatography Technologies) with varying gradients of acetonitrile in water [both containing 0.001% (vol/vol) trifluoroacetic acid]. The peptides were analyzed by mass spectrometry (Voyager DE-PRO; Applied Biosystems). The Bcl-2 peptides G3 and G4 lack residues 49–88, an unstructured loop that was deleted in the NMR structure.
Expression and Purification of NAF-1 and Apo-Fd Proteins.
The soluble part of NAF-1 (amino acids 57–135) was expressed and purified as previously described (9). Apo-NAF-1 was produced by removing the cluster by dialysis with 20 mM Tris⋅HCl (pH 6.0), 100 mM NaCl, and 0.02% (wt/vol) NaN3 at 30 °C for 24–48 h. Apo-Fd was expressed and purified as previously described (24).
Native-PAGE 2Fe-2S Cluster Transfer in Vitro Assay.
NAF-1 (200 μM) was incubated and shaken at room temperature with 400 μM apo-ferredoxin (apo-Fd) with and without 500 µM Bcl-2 16–30 peptide in 20 mM Tris⋅HCl (pH 8.0), 100 mM NaCl, 5 mM DTT, and 0.02% NaN3 for 20 min. DTT was added to keep the disulfide bonds in apo-Fd reduced. Transfer of the 2Fe-2S cluster from NAF-1 to apo-Fd was then analyzed by native gel as previously described (11).
Cluster Stability Analysis.
An amount of 100 μM NAF-1 was incubated either with or without 100 μM Bcl-2 16–30 peptide in 50 mM bis-Tris (pH 6.0) and 100 mM NaCl at 37 °C. The absorbance at 458 nm was measured every 10 min on a Varian Cary 50 UV-visible spectrophotometer with temperature control (Agilent).
Direct Coupling Analysis.
DCA was performed using the mean-field formulation of DCA as previously described (13). When using DCA for protein–protein interactions, there is a sequence-pairing procedure that combines the domains of interest. Similar pairings have been used previously to devise interaction interfaces (14, 15). Here the criterion used to pair proteins was to select domain pairs whose proteins belong to the same organism, the goal being to determine the most-coupled coevolving residue pairs between the two domains and hence the two proteins. The input sequences come from the Pfam (15) zinc finger domain zf-CDGSH (PF09360) in NAF-1, the Bcl-2 homology region 4 BH4 (PF02180) domain, and the Bcl-2 (PF00452) domain in Bcl-2 protein. DI values were computed for all interprotein residue–residue pairs and ranked according to their value. The top-ranked pairs have a higher probability of being in contact during protein–protein binding.
The model in Fig. 7 was created by minimizing the length of the green links representing the top coevolving residue pairs obtained from DCA. Rather than a physics-based docking, geometrical constraints were used to satisfy the DCA contacts but also to include interaction regions obtained experimentally. The docked complex is depicted with a separation amenable for analysis and is not intended to represent an in vivo complex comparable to those obtained via crystallization or NMR. The model was created using UCSF Chimera software (25).
Deuterium Exchange Mass Spectrometry.
Instrument setup and operation were described previously (26). All frozen samples were thawed and run using the conditions determined during fragmentation optimization.
Fragmentation conditions.
The initial fragmentation conditions for the sample composition and instrument parameters were determined before starting the exchange time-course experiments. Stocks (1.4 mg/mL) of wild-type proteins were diluted with storage buffer (50 mM Tris, 150 mM NaCl, 10% glycerol, 2 mM DTT, pH 7.0) at room temperature and quenched with 0.5% formic acid, 16.6% glycerol, and 3.2 M guanidine⋅HCl (quench buffer) at 0 °C, and then immediately frozen on dry ice and stored at −80 °C until analysis as described (27).
Deuterium on-exchange time courses.
The exchange time-course experiments for NAF-1, Bcl-2, and NAF-1 + Bcl-2 were all performed simultaneously at 25 °C with the following procedure. A full time-course experiment was initiated by adding 100 μL of protein in 10 mM sodium phosphate buffer (pH 7.4) with 20 mM NaCl to 300 μL of the equivalent deuterated exchange buffer for a final % D2O of 75%. The exchange was monitored over intervals of 10, 30, 90, 300, 900, 3,600, and 5,600 s. Aliquots were removed and quenched at 0 °C in a final concentration of 0.5 M Gdn⋅DCl, 10% glycerol, and 0.5% formic acid. Quenched time points were flash-frozen and stored at −80 °C until analysis. In- and back-exchange controls were performed as previously described (28, 29).
Sequence identification of peptide fragments.
The identity of the parent peptide ions was determined using the SEQUEST software program (Thermo Finnigan) and MS1 and MS2 data. The quality of each peptide was monitored by individually examining each measured isotopic envelope spectrum for the entire time-course exchange. The deuterium content was calculated for each time point by using specialized software as previously described (26, 30).
Supplementary Material
Acknowledgments
This work is supported by the Israeli Science Foundation (ISF863/09 and ISF865/13; to R.N.); the European Research Council (ERC) under the European Community’s Seventh Framework Programme (FP7/2007-2013)/ERC Grant Agreement 203413 (to A.F.); the National Institutes of Health (Grants GM54038 and GM101467; to P.A.J.); and the Cancer Prevention and Research Institute of Texas and by the Center for Theoretical Biological Physics sponsored by the National Science Foundation (NSF) (Grants PHY-1308264 and MCB-1214457) (J.N.O.). A.F. and R.N. are members of The Minerva Center for Bio-Hybrid Complex Systems and acknowledge funds received from the center. J.N.O. is a Cancer Prevention Research Institute of Texas Scholar.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1403770111/-/DCSupplemental.
References
- 1.Pattingre S, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122(6):927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Youle RJ, Strasser A. The BCL-2 protein family: Opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9(1):47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 3.Chang NC, et al. Bcl-2-associated autophagy regulator Naf-1 required for maintenance of skeletal muscle. Hum Mol Genet. 2012;21(10):2277–2287. doi: 10.1093/hmg/dds048. [DOI] [PubMed] [Google Scholar]
- 4.Chang NC, Nguyen M, Germain M, Shore GC. Antagonism of Beclin 1-dependent autophagy by BCL-2 at the endoplasmic reticulum requires NAF-1. EMBO J. 2010;29(3):606–618. doi: 10.1038/emboj.2009.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sohn YS, et al. NAF-1 and mitoNEET are central to human breast cancer proliferation by maintaining mitochondrial homeostasis and promoting tumor growth. Proc Natl Acad Sci USA. 2013;110(36):14676–14681. doi: 10.1073/pnas.1313198110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amr S, et al. A homozygous mutation in a novel zinc-finger protein, ERIS, is responsible for Wolfram syndrome 2. Am J Hum Genet. 2007;81(4):673–683. doi: 10.1086/520961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen YF, et al. Cisd2 deficiency drives premature aging and causes mitochondria-mediated defects in mice. Genes Dev. 2009;23(10):1183–1194. doi: 10.1101/gad.1779509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiley SE, et al. Wolfram syndrome protein, Miner1, regulates sulphydryl redox status, the unfolded protein response, and Ca2+ homeostasis. EMBO Mol Med. 2013;5(6):904–918. doi: 10.1002/emmm.201201429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conlan AR, et al. Crystal structure of Miner1: The redox-active 2Fe-2S protein causative in Wolfram syndrome 2. J Mol Biol. 2009;392(1):143–153. doi: 10.1016/j.jmb.2009.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nechushtai R, et al. Characterization of Arabidopsis NEET reveals an ancient role for NEET proteins in iron metabolism. Plant Cell. 2012;24(5):2139–2154. doi: 10.1105/tpc.112.097634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamir S, et al. Nutrient-deprivation autophagy factor-1 (NAF-1): Biochemical properties of a novel cellular target for anti-diabetic drugs. PLoS ONE. 2013;8(5):e61202. doi: 10.1371/journal.pone.0061202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuris JA, et al. NADPH inhibits [2Fe-2S] cluster protein transfer from diabetes drug target mitoNEET to an apo-acceptor protein. J Biol Chem. 2012;287(15):11649–11655. doi: 10.1074/jbc.M111.319731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morcos F, et al. Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc Natl Acad Sci USA. 2011;108(49):E1293–E1301. doi: 10.1073/pnas.1111471108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng RR, Morcos F, Levine H, Onuchic JN. Toward rationally redesigning bacterial two-component signaling systems using coevolutionary information. Proc Natl Acad Sci USA. 2014;111(5):E563–E571. doi: 10.1073/pnas.1323734111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schug A, Weigt M, Onuchic JN, Hwa T, Szurmant H. High-resolution protein complexes from integrating genomic information with molecular simulation. Proc Natl Acad Sci USA. 2009;106(52):22124–22129. doi: 10.1073/pnas.0912100106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersen MD, Shaffer J, Jennings PA, Adams JA. Structural characterization of protein kinase A as a function of nucleotide binding. Hydrogen-deuterium exchange studies using matrix-assisted laser desorption ionization-time of flight mass spectrometry detection. J Biol Chem. 2001;276(17):14204–14211. doi: 10.1074/jbc.M011543200. [DOI] [PubMed] [Google Scholar]
- 17.Petros AM, et al. Solution structure of the antiapoptotic protein Bcl-2. Proc Natl Acad Sci USA. 2001;98(6):3012–3017. doi: 10.1073/pnas.041619798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hailey KL, et al. Pro-interleukin (IL)-1beta shares a core region of stability as compared with mature IL-1beta while maintaining a distinctly different configurational landscape: A comparative hydrogen/deuterium exchange mass spectrometry study. J Biol Chem. 2009;284(38):26137–26148. doi: 10.1074/jbc.M109.027375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katz C, et al. Studying protein-protein interactions using peptide arrays. Chem Soc Rev. 2011;40(5):2131–2145. doi: 10.1039/c0cs00029a. [DOI] [PubMed] [Google Scholar]
- 20.Rotem S, et al. The structure and interactions of the proline-rich domain of ASPP2. J Biol Chem. 2008;283(27):18990–18999. doi: 10.1074/jbc.M708717200. [DOI] [PubMed] [Google Scholar]
- 21.Decuypere J-P, Parys JB, Bultynck G. Regulation of the autophagic Bcl-2/Beclin 1 interaction. Cells. 2012;1(4):284–312. doi: 10.3390/cells1030284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salem AF, Whitaker-Menezes D, Howell A, Sotgia F, Lisanti MP. Mitochondrial biogenesis in epithelial cancer cells promotes breast cancer tumor growth and confers autophagy resistance. Cell Cycle. 2012;11(22):4174–4180. doi: 10.4161/cc.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oltersdorf T, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435(7042):677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 24.Fish A, Danieli T, Ohad I, Nechushtai R, Livnah O. Structural basis for the thermostability of ferredoxin from the cyanobacterium Mastigocladus laminosus. J Mol Biol. 2005;350(3):599–608. doi: 10.1016/j.jmb.2005.04.071. [DOI] [PubMed] [Google Scholar]
- 25.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 26.Barkho S, et al. Distal loop flexibility of a regulatory domain modulates dynamics and activity of C-terminal Src kinase (Csk) PLOS Comput Biol. 2013;9(9):e1003188. doi: 10.1371/journal.pcbi.1003188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamuro Y, et al. Dynamics of cAPK type IIbeta activation revealed by enhanced amide H/2H exchange mass spectrometry (DXMS) J Mol Biol. 2003;327(5):1065–1076. doi: 10.1016/s0022-2836(03)00234-1. [DOI] [PubMed] [Google Scholar]
- 28.Wong L, Jennings PA, Adams JA. Communication pathways between the nucleotide pocket and distal regulatory sites in protein kinases. Acc Chem Res. 2004;37(5):304–311. doi: 10.1021/ar020128g. [DOI] [PubMed] [Google Scholar]
- 29.Hamuro Y, et al. Phosphorylation driven motions in the COOH-terminal Src kinase, CSK, revealed through enhanced hydrogen-deuterium exchange and mass spectrometry (DXMS) J Mol Biol. 2002;323(5):871–881. doi: 10.1016/s0022-2836(02)01003-3. [DOI] [PubMed] [Google Scholar]
- 30.Weinreb PH, et al. Dynamic structural changes are observed upon collagen and metal ion binding to the integrin α1 I domain. J Biol Chem. 2012;287(39):32897–32912. doi: 10.1074/jbc.M112.354365. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.