Significance
For the first time we describe a physiological role for HLA-F adjacent transcript 10 (FAT10) in metabolism, obesity, and aging in mammals. We show that FAT10 knockout prevents the development of age-associated obesity in mice while extending lifespan and vigor without the appearance of deleterious developmental effects. In addition, we did not observe an increase in cancer incidence. If the role of FAT10 in humans is similar to mice, then targeting of FAT10 may hold promising therapeutic impact for the treatment of various diseases including obesity and obesity-related diseases and aging associated diseases.
Keywords: longevity, obesity, mammals
Abstract
The HLA-F adjacent transcript 10 (FAT10) is a member of the ubiquitin-like gene family that alters protein function/stability through covalent ligation. Although FAT10 is induced by inflammatory mediators and implicated in immunity, the physiological functions of FAT10 are poorly defined. We report the discovery that FAT10 regulates lifespan through pleiotropic actions on metabolism and inflammation. Median and overall lifespan are increased 20% in FAT10ko mice, coincident with elevated metabolic rate, preferential use of fat as fuel, and dramatically reduced adiposity. This phenotype is associated with metabolic reprogramming of skeletal muscle (i.e., increased AMP kinase activity, β-oxidation and -uncoupling, and decreased triglyceride content). Moreover, knockout mice have reduced circulating glucose and insulin levels and enhanced insulin sensitivity in metabolic tissues, consistent with elevated IL-10 in skeletal muscle and serum. These observations suggest novel roles of FAT10 in immune metabolic regulation that impact aging and chronic disease.
The twin selection pressures of starvation and infection have driven the evolution of proteins that coordinate nutrient homeostasis, fuel use, and immune/inflammatory responses (1–3). Signaling pathways regulated by these proteins are conserved across phyla and include insulin/IGF1, p53, Toll, peroxisome proliferator-activated receptor (PPAR), NF-κB, mitogen-activated protein kinase (MAPK), and AMP kinase (AMPK)/target of rapamycin (TOR)/forkhead box O (FOXO) pathways (1–4). Importantly, chronic dysregulation or imbalance among immune metabolic signaling networks (like in chronic overnutrition) is increasingly appreciated as an underlying cause of aging as well as chronic diseases of humans, including type 2 diabetes, atherosclerosis, inflammatory bowel disease, nonalcoholic steatohepatitis, and cancer (4, 5).
The HLA-F adjacent transcript 10 (FAT10) gene was initially cloned as part of an effort to identify additional genes from the human MHC, and it was named according to its proximity to the HLA-F locus (6). FAT10 is a vertebrate-specific member of the eukaryotic ubiquitin-like (UBL) protein family, containing two UBL domains arranged in tandem with a C-terminal diglycine motif (6). Unlike the posttranslational cleavage required to expose the terminal diglycine motif of ubiquitin, the FAT10 protein is synthesized with an accessible terminal diglycine motif. FAT10 protein becomes coupled to other proteins (FAT10ylation) (7) through the action of UBL modifier activating proteins UBA6 (8, 9) and USE1 (10), which also activate ubiquitin. Both FAT10 and its conjugates are unstable and degraded by the proteasome (11, 12). Currently, no de-FAT10ylating enzymes (analogous to deubiquitinating enzymes) have been identified, suggesting that FAT10ylation is an irreversible process.
Despite intense recent efforts to identify FAT10 substrates (10, 13, 14), physiological role(s) of FAT10 remain obscure. FAT10 mRNA is absent or expressed at very low levels in most normal tissues but constitutively expressed during lymphocyte and dendritic cell maturation in some cell culture lines derived from B cells and several types of neoplasms (15). Notably, FAT10 is ubiquitously induced in response to IFN-γ or TNF-α, with FAT10 mRNA being one of the most highly up-regulated transcripts in some models of inflammation (16). FAT10 may participate in protein quality control based on the preferential binding of FAT10 to polyglutamine-modified huntingtin protein (17) and the observation of FAT10-enhanced formation of hepatocyte Mallory–Denk bodies in a model of drug-induced chronic hepatitis (18, 19).
We initially reported that young FAT10-deficient (KO) mice generated in our laboratory seemed normal in all respects but were more susceptible to LPS-induced death (20). Because sensitivity to sepsis was shown to increase with age in mammals (21–23), we established aging colonies of KO mice and WT littermates. Surprisingly, older KO mice evinced a clear phenotype of delayed aging and extended lifespan. Complementary studies in younger mice revealed global alterations in energy and fuel metabolism, adiposity, glucose–insulin homeostasis, and inflammatory gene expression that were all consistent with delayed aging and lifespan extension. These observations identify FAT10 as a modulator of immune and metabolic homeostasis and implicate these actions in the regulation of vertebrate lifespan by FAT10.
Results
Knockout of the FAT10 Gene Prolongs Lifespan and Reduces Age-Associated Biomarkers.
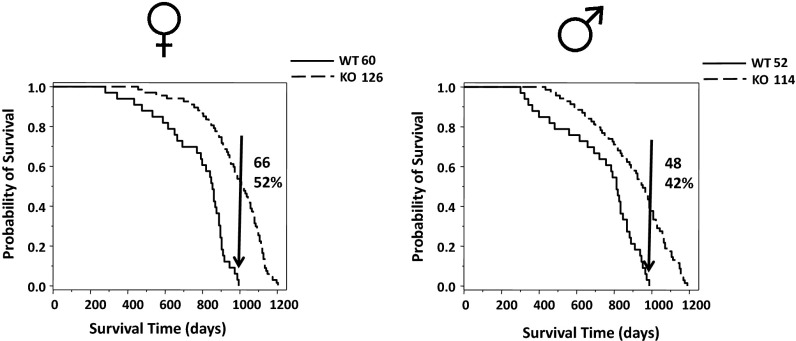
Aged FAT10ko mice appeared younger than their age- and sex-matched controls, with maintenance of muscle mass (delayed sarcopenia), denser, smoother, and darker fur (Fig. S1), and absence of tumors. To assess impact on lifespan, we conducted Kaplan–Meier analysis (24) on mortality patterns of the two sexes compared with WT mice (Fig. 1). The analysis showed that male and female FAT10ko mice had a 20% increase in both median and overall lifespan. Notably, 42% percent of male KOs and 52% of female KOs remained alive after the death of the last WT mouse. Wilcoxon (25), Mantel–Haenszel (26), and Taron–Ware (27) tests (set for a probability of 99%) generated significant P values for females (6.6 × 10−9, 5.99 × 10−11, and 6.42 × 10−10, respectively) and males (4.07 × 10−5, 9.83 × 10−7, and 7.45 × 10−6, respectively) (Dataset S1 A and B). Survival among females in the middle part of the study (600–800 d) was slightly greater than among males (Wilcoxon test, P = 0.036) (Dataset S1C).
Fig. 1.
FAT10 KO mice live longer than littermate controls. (Right) Males and (Left) females are shown separately. The number of animals in each group is shown near the upper right corner of each graph. Arrows in the graphs represent the population of KO mice that exceeded the total lifespan of the WT littermate controls. The actual number and percentile of this population are shown to the right of the arrow inside the graphs’ areas. Kaplan–Meier analysis was performed on these data. The probability of survival is normalized to the final size of each group.
FAT10ko Mice Are Leaner than Their Control Counterparts.
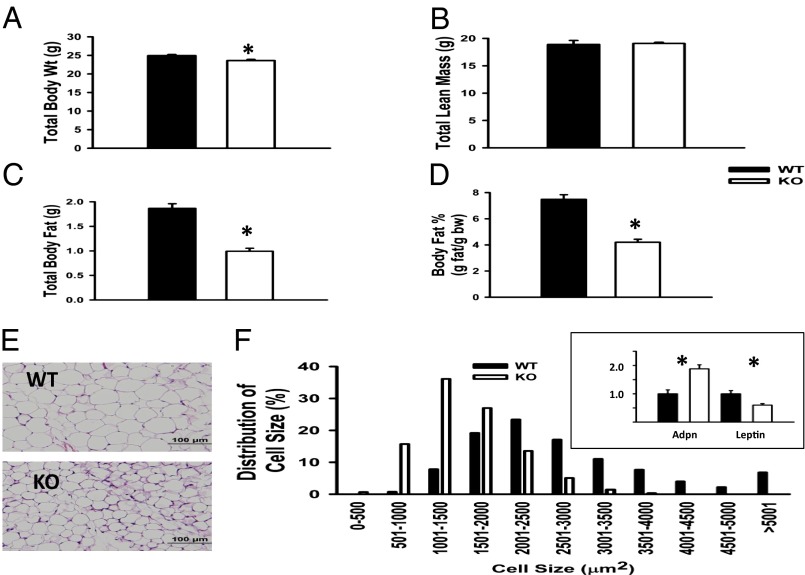
Increased lifespan in KO mice was associated with dramatically reduced adiposity. KO mice, independent of sex, gained significantly less weight than WT mice (Fig. S2A). Differences in weight became more pronounced with age. Whereas WT mice gained weight through their lifespan, the preponderance of weight gain in KO mice occurred by 12 mo of age. By 3 mo of age, KO mice weighed on average 1 g less than WT mice, reflecting an ∼50% reduction in white adipose tissue (WAT) (Fig. 2 A–D). However, there was no difference in total lean mass between genotypes (Fig. 2B). Necropsy revealed that FAT10ko mice have reduced subcutaneous fat depots compared with WT mice (Fig. S2 B and C), with significantly less visceral periovarian adipose tissue (epididymal in males) in the KO mice compared with the WT controls. Histological examination of epididymal WAT (eWAT) revealed an ∼50% reduction in average adipocyte size (Fig. 2E), reflecting a greater proportion of smaller adipocytes (<3,000 μm2) and a coincident decrease in larger adipocytes (3,000–5,000 μm2) (Fig. 2F). There was no significant difference in epididymal adipocyte number between genotypes (KO: 3.46 ± 0.19 × 106 adipocytes per depot; WT: 3.75 ± 0.22 × 106 adipocytes per depot; n = 6, P > 0.05). Therefore, reduced adiposity in KO mice is caused by decreased adipocyte size (i.e., decreased triglyceride per cell). Consistent with smaller adipocyte size, adiponectin gene expression was greater and leptin gene expression was reduced compared with adipocytes of WT mice (Fig. 2F, Inset). However, circulating adiponectin levels were lower than in WT (Table S1), reflecting overall reduction of adipose mass in the KO.
Fig. 2.
Adipose mass and adipocyte size are reduced in FAT10ko mice. (A) Body weight, (B) lean mass, (C) body fat, and (D) percent body fat of 12-wk-old WT and KO mice. (E) Representative H&E-stained sections of eWAT. (F) Adipocyte size distributions determined from eWAT histomorphometry and image analysis; (Inset) adiponectin (Adpn) and leptin gene expression in eWAT. *P < 0.05 (n = 6 per group).
Altered Energy Expenditure and Fuel Use in FAT10ko Mice.
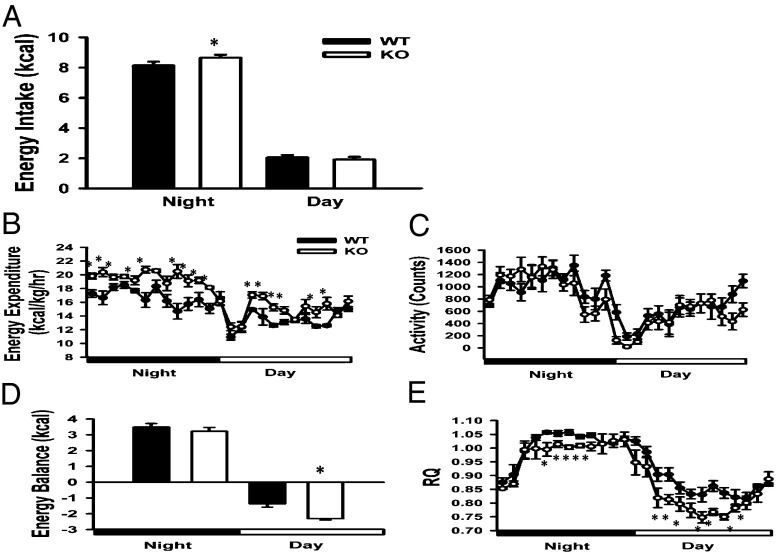
KO mice consume more energy (per body weight) than WT mice during the night (period of active feeding) and ingest comparable levels during the day (period of sleep/intermittent feeding) (Fig. 3A). However, FAT10ko mice expend more energy throughout 24 h than WT mice (Fig. 3B), despite comparable locomotor activity (Fig. 3C). Importantly, increased energy expenditure during the day in the absence of increased energy intake result in a greater daytime negative energy balance in FAT10ko mice compared with WT mice (Fig. 3D). Reduced adipose stores in FAT10ko mice are consistent with this greater daytime energy deficit.
Fig. 3.
The repatterning of energy metabolism in KO mice is shown by nighttime and daytime values for (A) energy intake, (B) energy expenditure, (C) locomotor activity, (D) energy balance, and (E) respiratory quotient in WT and KO mice. Data are presented as means ± SEMs. *P < 0.05 (n = 8 per group).
Relative to WT mice, FAT10ko mice preferentially metabolize fat (as opposed to carbohydrate) for energy, resulting in a reduced respiratory quotient (RQ; RQ = CO2/O2), even during active night feeding. This lower RQ becomes more pronounced in KO mice during the light phase, when decreased food intake results in mobilization of fat stores in WT and KO animals (Fig. 3E). Because fat is the exclusive fuel used for adaptive (nonshivering) thermogenesis in mice, we housed KO and WT mice at thermoneutrality (30 °C) for 5 wk and measured increases in body weight and adiposity (by MRI) in response to this reduced thermogenic demand. KO mice did not gain proportionally more weight or fat than WT mice (Fig. S3), indicating that greater energy expenditure in KO mice is not caused by greater demands of adaptive thermogenesis.
Genes Promoting β-Oxidation Are Up-Regulated in Skeletal Muscle of FAT10ko Mice.
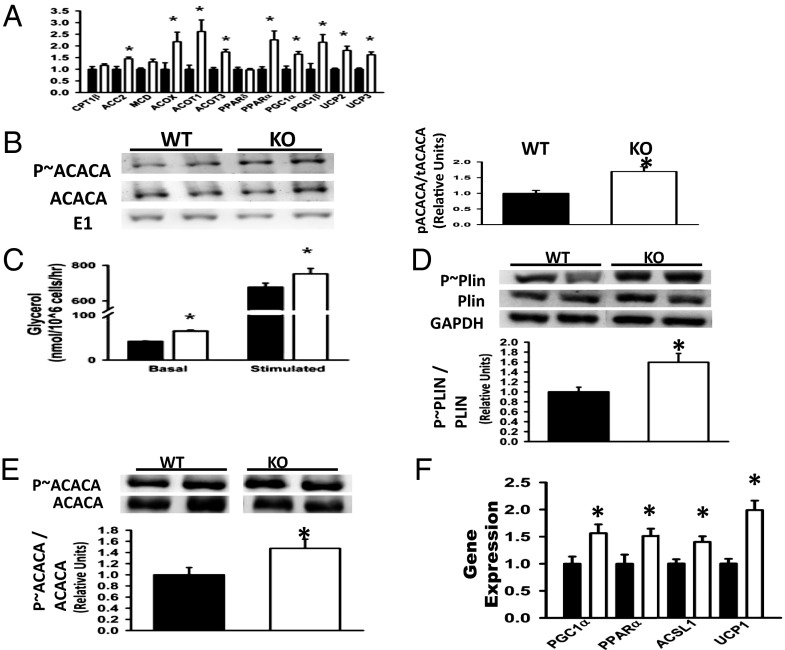
To elucidate molecular mechanisms underlying enhanced use of lipid as a metabolic fuel in KO mice, we compared the expression profiles of metabolic genes in skeletal muscles from WT and KO mice by quantitative PCR (QPCR). Skeletal muscle of FAT10ko mice up-regulated transcription factors [peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) and PPARα] and target enzymes promoting β-oxidation [acyl-CoA synthetase long-chain family member 1 (ACSL1), acyl-CoA oxidase (ACOX), and acyl-CoA thioesterase 1 & 3 (ACOT1 & ACOT3)] and uncoupling proteins (UCP2 and UCP3) were examined (Fig. 4A). Evidence for elevated β-oxidation in KO mice is provided at the protein level by the proportionally greater level of phosphorylated (inactive) acetyl-CoA carboxylase (ACACA) (Fig. 4B) and decreased triglyceride content (Table S1). Liver triglyceride levels were comparable in WT and KO mice (Table S1). Overall, these data indicate that global deletion of FAT10 induces a skeletal muscle transcriptional program promoting fat oxidation.
Fig. 4.
Molecular signatures promoting fatty acid oxidation are up-regulated in skeletal muscle of FAT10ko mice. (A) Expression of genes promoting fatty acid oxidation in quadriceps of WT and KO mice. (B) Total ACACA and phosphorylated ACACA (p∼ACACA) in quadriceps of WT and KO mice. Ubiquitin-E1 (E1) levels were used as a nonaffected reference. Representative blots are shown in Left. Densitometry data from multiple blots are summarized in Right. (C) Triglyceride turnover increase in adipocytes of KO mice is shown by increased in vitro basal and PKA-stimulated lipolysis (glycerol release) in KO adipocytes. (D) Western blots of eWAT lysates showing elevated levels of phosphorylated perilipin (p∼Plin) protein in KO mice with densitometry. (E) Western blots of eWAT lysates showing enhanced levels of p∼ACACA vs. total ACACA in KO mice with densitometry. (F) Up-regulation of genes promoting β-oxidation in epididymal AT of KO mice. Data are presented as means ± SEM. *P < 0.05 (n = 6 per group).
Increased Triglyceride Breakdown (Lipolysis) in Adipocytes of FAT10ko Mice.
Consistent with the preferential use of fatty acids as metabolic fuel, data indicate that triglyceride hydrolysis (lipolysis) was increased in adipocytes of KO mice. First, levels of circulating nonesterified fatty acids (NEFAs) were greater in KO compared with WT mice (Table S1). Second, basal and stimulated ex vivo lipolysis, indicated by glycerol release, were significantly increased in isolated KO adipocytes (Fig. 4C). In addition, hallmarks of lipolysis were elevated in adipose tissue, including phosphorylation of the lipid droplet-associated protein perilipin [the rate-controlling event in catecholamine (fasting) -induced lipolysis] (28, 29) (Fig. 4D), increased β-oxidation manifest as inhibitory phosphorylation of ACACA-1 (Fig. 4E), and up-regulation of PGC1α, PPARα, ACSL1α, and UCP1 gene expression (Fig. 4F) (30, 31). Thus, increased fatty acid release and triglyceride turnover provide a proximate physiological mechanism to explain the reduced adipocyte size and adipose mass of KO mice.
Enhanced Glucose Insulin Homeostasis in FAT10ko Mice.
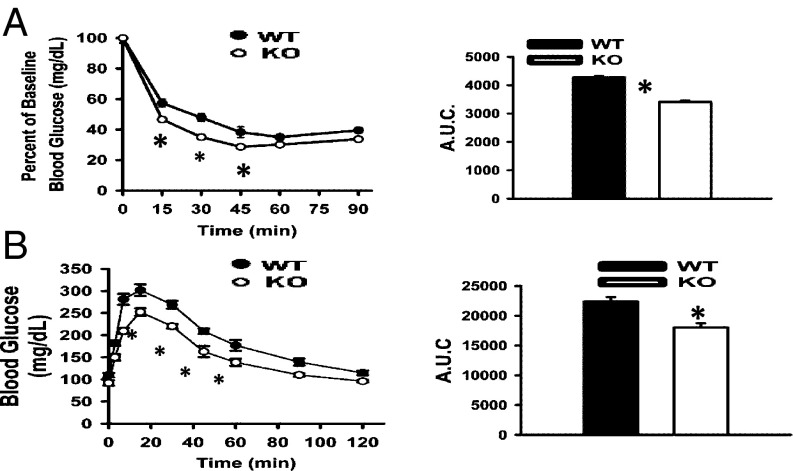
Leanness is associated with enhanced glucose–insulin homeostasis, which can independently promote longevity in mice (32). KO mice maintained lower circulating glucose and insulin concentrations than WT mice (Table S1), resulting in significantly lower homeostatic assessment of insulin resistance values (KO: 1.93 ± 0.37; WT: 5.56 ± 0.38; P < 0.001). Improved insulin action in KO mice was confirmed at the whole-body level by more rapid glucose disposal during the i.p. insulin tolerance test (ITT). Blood glucose stabilization 1 h after insulin injection was similar in WT and KO mice (Fig. 5A), suggesting comparable hepatic insulin clearance. KO mice were also more glucose tolerant than WT mice (Fig. 5B), showing more efficient clearance of glucose despite lower insulin levels. During the glucose tolerance test (GTT), relative increases over baseline in first- and second-phase glucose-stimulated insulin responses were comparable between KO and WT mice, indicating that reduced insulin levels in KO mice do not reflect a defect in insulin secretion. In vivo insulin signaling assays suggested that greater insulin action in KO mice reflected enhanced capacity for insulin signaling (i.e., insulin-stimulated AKT phosphorylation) in adipose, liver, and skeletal muscle.
Fig. 5.
Increased efficiency of glucose–insulin regulation in FAT10ko mice. (A) I.p. ITTs (Left) and areas under curves (AUCs; Right; n = 8 per group). (B) I.p. GTTs (Left) and AUCs (Right) for WT and KO mice (n = 8 per group).
Inflammation in Metabolic Tissues of FAT10ko Mice.
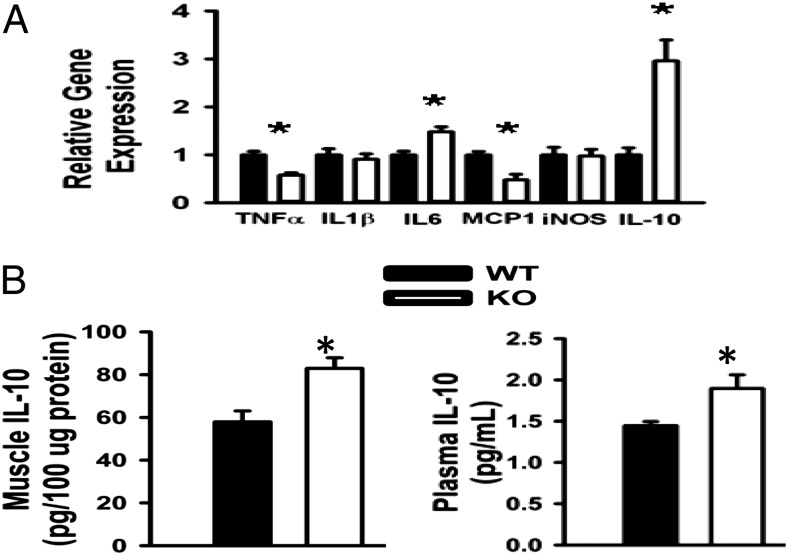
FAT10 has been implicated in NF-κB–dependent inflammatory gene expression (16), which can impair insulin signaling and promote insulin resistance in metabolic tissues (33, 34). Consistent with enhanced insulin action and sensitivity (Figs. 4 and 5), we observed a tissue-specific pattern of altered inflammatory gene expression in metabolic tissues of FAT10ko mice. In quadriceps, transcript levels of TNF-α and monocyte chemotactic protein-1 (MCP-1) were decreased, whereas IL-6 and IL-10 were increased (Fig. 6A). Proinflammatory genes were generally down-regulated in eWAT of FAT10ko mice, with the exception of the induced nitric oxide synthase (iNOS) gene (Fig. S4). Thus, although proinflammatory gene expression was generally reduced in metabolic tissues of FAT10ko mice, effects on specific NF-κB–regulated genes (e.g., IL-1β, IL-6, IL-10, iNOS, and MCP-1) varied. The most highly up-regulated transcript in our inflammatory panel was the antiinflammatory cytokine, IL-10. Moreover, IL-10 protein levels were significantly increased in both skeletal muscle and serum of KO mice (Fig. 6B). These results suggest that deletion of FAT10 promotes an inflammation-suppressive milieu in skeletal muscle and perhaps, systemically as well.
Fig. 6.
Altered profiles of inflammatory gene expression in FAT10ko mice are consistent with enhanced glucose–insulin homeostasis and delayed aging. (A) Attenuated proinflammatory gene expression in muscle of KO mice. (B) IL-10 protein in quadriceps (Left) and plasma (Right). Data are presented as means ± SEMs. *P < 0.05 (n = 6 per group).
New Aging Biomarker—FAT10 Expression Increases with Age.
Aging is associated with increased expression of inflammatory cytokines in adipose tissue, concordant with impaired metabolic homeostasis. We assessed FAT10 expression in WAT obtained from young and old WT mice. FAT10 expression was evaluated compared with TNF-α expression, an established physiological biomarker of chronic inflammation in aging (35–38). QPCR analysis indicated that FAT10 mRNA expression increases with age in WAT of WT mice, comparable with the increase in TNF-α expression (Fig. 7). These data suggest that FAT10-mediated processes are up-regulated in adipose tissue with aging and age-related increases in inflammation.
Fig. 7.
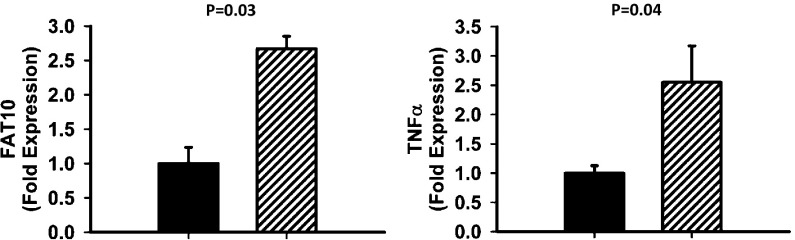
FAT10 and TNF-α gene expressions are up-regulated in perigonadal adipose tissue of old WT mice, which is shown by RNA extracted from perigonadal adipose tissue of 2- (black bars) and 22-mo-old (striped bars) C57BL/6N males. RNA samples were assayed by QPCR to evaluate the expression levels of TNF-α and FAT10 using β-actin as a reference gene. t Test P values shown in graphs were obtained by EXCEL.
Discussion
FAT10 is a pleiotropic UBL protein that has evolved with the vertebrate MHC, and it participates in protein quality control, signal transduction, and gene transcription, typically in the setting of inflammation or other cell/tissue insult (39, 40). Apart from ubiquitin, FAT10 is the only member of the UBL family known to promote proteosomal degradation, which is required for peptide presentation by MHC-I (11). However, FAT10 modification is distinct from ubiquitination and an alternative mechanism for antigen presentation by MHC-I (41). FAT10 modification was found to be more efficient than ubiquitination in transferring proteins to the proteosome; whereas polyubiquitination of proteins is needed to enable proteosomal activity, the binding of one FAT10 molecule to its target is sufficient (42). The induction of FAT10 expression by IFN-γ and TNF-α combined with its highly efficient role in proteosomal antigen processing may suggest that the genomic location of FAT10 within the MHC locus is the result of a coevolution process based on shared function.
Coordinate integration of nutrient sensing, energy storage, and immune/inflammatory responses has evolved under the twin selection pressures of nutrient deprivation and infection. A current paradigm of human disease pathogenesis posits that persistent dysregulation of this immunometabolic integration promotes normal aging and the development of chronic age-related disabilities, including type 2 diabetes, atherosclerosis, and cancer (2–5). Here, we identify FAT10 as a novel regulator of immune metabolic homeostasis and lifespan in healthy mice. Specifically, targeted gene deletion of FAT10 delayed the appearance of aging biomarkers and extended both median and maximum lifespan in mice. Importantly, these salutary effects on aging were coincident with global reprogramming of metabolic and inflammatory gene expression in key metabolic tissues and alterations in body composition, whole-body nutrient and energy metabolism, and glucose–insulin homeostasis that are hallmarks of delayed aging and lifespan extension.
Longevity is inversely correlated with adiposity in humans and animal models (43, 44). FAT10ko mice had 50% less white adipose mass than WT counterparts, despite comparable energy intake. This reduction in adiposity reflected elevated rates of triglyceride hydrolysis (lipolysis) in adipocytes that were manifest as increased levels of circulating NEFAs in the fasted state and up-regulation of a counterregulatory program of fat oxidation in adipose tissues (31). A proximate mechanism to explain increased adipocyte lipolysis in KO mice is enhanced phosphorylation of the lipid droplet protein perilipin (28, 29). This rate-determining step in lipolysis is cAMP/protein kinase A (PKA)-dependent and tightly regulated by stimulatory actions of adrenergic signaling and glucagon as well as reciprocal inhibitory actions of insulin and adenosine on cAMP levels (45). Future studies will be required to determine whether FAT10 regulates perilipin phosphorylation by direct interaction with components of one or more of these signaling pathways in adipocytes or indirectly by modulating adrenergic tone and/or adenosine or insulin levels.
Reduced adipose depot mass in KO mice was not associated with ectopic lipid deposition in skeletal muscle or liver, suggesting that NEFAs were being mobilized as fuel in skeletal muscle. This notion was confirmed by indirect calorimetry (i.e., reduced RQ) by a skeletal muscle gene expression signature of enhanced mitochondrial respiration, β-oxidation and uncoupling, and increased cAMP protein kinase (AMPK)-dependent phosphorylation of skeletal muscle ACACA-2 (reviewed in ref. 46). These alterations in skeletal muscle gene expression and metabolic function are also observed in response to exercise training muscle (47). Notably, these coordinate increases in skeletal muscle fat oxidation and uncoupling combined with AMPK activation are themselves proposed as a critical mechanism regulating the rate of aging and lifespan (uncoupling to survive) (48). Thus, the effects of FAT10 KO on aging and lifespan are likely to reflect the combined benefits of reduced adipose mass and enhanced metabolism in skeletal muscle.
Enhanced glucose–insulin homeostasis is typically associated with reduced adiposity and itself a hallmark of lifespan extension (32). FAT10ko mice maintained normoglycemia and showed enhanced insulin action (clearance of a glucose bolus in the GTT), despite reduced circulating insulin levels.
Enhanced insulin action in the presence of relatively low levels of insulin reflects enhanced insulin signaling [i.e., insulin-stimulated AK thymoma (AKT)/protein kinase B phosphorylation] in metabolic tissues of KO mice. How the absence of FAT10 results in improved insulin signaling is currently unclear. Our data implicate altered inflammatory gene expression—in particular, elevated IL-10 production by skeletal muscle—in the enhanced insulin sensitivity in FAT10ko mice. IL-10 suppresses inflammation and enhances tissue and whole-body insulin sensitivity by inhibiting the expression of proinflammatory cytokines and antagonizing IKK/NF-κB signaling and ER stress (28, 49). Muscle-specific transgenic overexpression of IL-10 at levels comparable with those levels measured in KO mice (present study) was shown to enhance whole-body insulin sensitivity in both lean and obese mice (50). Up-regulation of IL-10 in skeletal muscle of KO mice may, in part, reflect the coincident modest increase in IL-6, a robust inducer of IL-10 gene expression in skeletal muscle (51). As noted above for enhanced mitochondrial oxidative function, elevated expression of IL-10 and IL-6 are hallmarks of skeletal muscle adaptation to exercise (52). These observations further suggest that FAT10 abrogation and its benefits on metabolic health and lifespan, in part, mimic the exercise-trained state.
Evidence presented here for important roles of FAT10 in metabolic programming and lifespan determination extends and informs recent identification of numerous FAT10 target proteins and interacting partners with acknowledged roles in energy sensing, nutrient and bile acid metabolism, and insulin-, PI3K/Akt/mTOR-, and cAMP- dependent signaling as well as NF-κB–dependent gene expression (10, 13, 14). Previous studies identified p53 and p62/sequestosome1 as FAT10 targets (7, 53), and both target proteins are known to modulate energy metabolism, mitochondrial activity, adiposity, glucose–insulin homeostasis, cell stress, and aging (54, 55). Together, these myriad and diverse functions of FAT10 substrates in metabolism and immune function support the recognition of FAT10 as a new and important node in protein networks regulating immunometabolic homeostasis. Prior in vitro studies have also suggested roles for FAT10 in mitosis (14) and chromosome abnormalities that were seen when FAT10 was overexpressed (56). However, the normal development, relative absence of malignancies, and extended lifespan of FAT10ko mice (13) question the roles for FAT10 in these processes in vivo.
In summary, our results suggest that constitutive actions of FAT10 promote adiposity, insulin resistance, and inflammation, while attenuating fat oxidation, uncoupling, and AMPK activation in skeletal muscle. Whereas these actions may enhance survival in response to starvation or pathogen challenge, they could conceivably promote obesity and its inflammatory complications in our current environment of chronic overnutrition and sedentary lifestyle. Moreover, increases in FAT10 expression are hallmarks of chronic pathologic states, including various cancers (11, 40), liver cirrhosis (9, 10), and HIV-associated nephropathy (57). These observations, in concert with the beneficial global effects of FAT10 abrogation on metabolic function, adiposity, inflammation, and tumorogenesis, suggest that targeting the FAT10 pathway may be an effective therapeutic approach to chronic diseases.
A number of animal models have been created that increase the lifespan of mice (58). In one group of models, such as the FIRKO mouse (59, 60), selective removal of a gene from a certain tissue results in increased lifespan. In a second group, animals with systemic alterations in genes, such as defects in growth hormone production or abrogation of S6 kinase (58), the animal is abnormal in either size or some other way. FAT10ko belongs to the considerably smaller group of model systems, in which complete removal of a gene prolongs lifespan without developmental deleterious effects during growth and with prolonged maintenance of vigor.
Methods
The generation of FAT10ko mice has been previously described (20). Indirect calorimetric evaluation was performed as previously described (61). ITTs and GTTs were performed on nonanesthetized animals, and body composition was determined by MRI as described (62). Antibodies were purchased from Cell Signaling Technologies. Detailed information is in SI Methods.
Supplementary Material
Acknowledgments
For reading the manuscript and providing very useful remarks, we would like to thank Prof. Diane Krause (Department of Laboratory Medicine, Cell Biology and Pathology), Dr. Stacey N. Brown (Department of Internal Medicine, Section of Endocrinology), and Dr. Victoria E. Clark (Department of Neurosurgery) from Yale School of Medicine. We thank the late Prof. William Prusoff (Department of Pharmacology, Yale School of Medicine) and the William Prusoff Foundation for supporting the studies in this manuscript. Additional funding came from National Institutes of Health Grants T32DK062032 (to J.D.) and R01DK074979 (to M.S.O.) and US Department of Agriculture Agricultural Research Service Contract 58-1950-7-70.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1323426111/-/DCSupplemental.
References
- 1.Rynes J, et al. Activating transcription factor 3 regulates immune and metabolic homeostasis. Mol Cell Biol. 2012;32(19):3949–3962. doi: 10.1128/MCB.00429-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerriets VA, Rathmell JC. Metabolic pathways in T cell fate and function. Trends Immunol. 2012;33(4):168–173. doi: 10.1016/j.it.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson AR, Milner JJ, Makowski L. The inflammation highway: Metabolism accelerates inflammatory traffic in obesity. Immunol Rev. 2012;249(1):218–238. doi: 10.1111/j.1600-065X.2012.01151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salminen A, Hyttinen JM, Kaarniranta K. AMP-activated protein kinase inhibits NF-κB signaling and inflammation: Impact on healthspan and lifespan. J Mol Med (Berl) 2011;89(7):667–676. doi: 10.1007/s00109-011-0748-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mathis D, Shoelson SE. Immunometabolism: An emerging frontier. Nat Rev Immunol. 2011;11(2):81. doi: 10.1038/nri2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bates EE, et al. Identification and analysis of a novel member of the ubiquitin family expressed in dendritic cells and mature B cells. Eur J Immunol. 1997;27(10):2471–2477. doi: 10.1002/eji.1830271002. [DOI] [PubMed] [Google Scholar]
- 7.Li T, et al. FAT10 modifies p53 and upregulates its transcriptional activity. Arch Biochem Biophys. 2011;509(2):164–169. doi: 10.1016/j.abb.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Groettrup M, Pelzer C, Schmidtke G, Hofmann K. Activating the ubiquitin family: UBA6 challenges the field. Trends Biochem Sci. 2008;33(5):230–237. doi: 10.1016/j.tibs.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 9.Pelzer C, Groettrup M. FAT10: Activated by UBA6 and functioning in protein degradation. Subcell Biochem. 2010;54:238–246. doi: 10.1007/978-1-4419-6676-6_19. [DOI] [PubMed] [Google Scholar]
- 10.Aichem A, et al. USE1 is a bispecific conjugating enzyme for ubiquitin and FAT10, which FAT10ylates itself in cis. Nat Commun. 2010;1:13. doi: 10.1038/ncomms1012. [DOI] [PubMed] [Google Scholar]
- 11.Schmidtke G, Kalveram B, Groettrup M. Degradation of FAT10 by the 26S proteasome is independent of ubiquitylation but relies on NUB1L. FEBS Lett. 2009;583(3):591–594. doi: 10.1016/j.febslet.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 12.Mehta R, et al. Proteasomal regulation of the hypoxic response modulates aging in C. elegans. Science. 2009;324(5931):1196–1198. doi: 10.1126/science.1173507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leng L, et al. A proteomics strategy for the identification of FAT10-modified sites by mass spectrometry. J Proteome Res. 2014;13(1):268–276. doi: 10.1021/pr400395k. [DOI] [PubMed] [Google Scholar]
- 14.Merbl Y, Refour P, Patel H, Springer M, Kirschner MW. Profiling of ubiquitin-like modifications reveals features of mitotic control. Cell. 2013;152(5):1160–1172. doi: 10.1016/j.cell.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee CG, et al. Expression of the FAT10 gene is highly upregulated in hepatocellular carcinoma and other gastrointestinal and gynecological cancers. Oncogene. 2003;22(17):2592–2603. doi: 10.1038/sj.onc.1206337. [DOI] [PubMed] [Google Scholar]
- 16.Gong P, et al. The ubiquitin-like protein FAT10 mediates NF-kappaB activation. J Am Soc Nephrol. 2010;21(2):316–326. doi: 10.1681/ASN.2009050479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagashima Y, Kowa H, Tsuji S, Iwata A. FAT10 protein binds to polyglutamine proteins and modulates their solubility. J Biol Chem. 2011;286(34):29594–29600. doi: 10.1074/jbc.M111.261032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.French BA, Oliva J, Bardag-Gorce F, French SW. The immunoproteasome in steatohepatitis: Its role in Mallory-Denk body formation. Exp Mol Pathol. 2011;90(3):252–256. doi: 10.1016/j.yexmp.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.French SW, Bardag-Gorce F, French BA, Li J, Oliva J. The role of innate immunity in the pathogenesis of preneoplasia in drug-induced chronic hepatitis based on a mouse model. Exp Mol Pathol. 2011;91(3):653–659. doi: 10.1016/j.yexmp.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Canaan A, et al. FAT10/diubiquitin-like protein-deficient mice exhibit minimal phenotypic differences. Mol Cell Biol. 2006;26(13):5180–5189. doi: 10.1128/MCB.00966-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruunsgaard H, Pedersen AN, Schroll M, Skinhoj P, Pedersen BK. Impaired production of proinflammatory cytokines in response to lipopolysaccharide (LPS) stimulation in elderly humans. Clin Exp Immunol. 1999;118(2):235–241. doi: 10.1046/j.1365-2249.1999.01045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mouton PR, et al. The effects of age and lipopolysaccharide (LPS)-mediated peripheral inflammation on numbers of central catecholaminergic neurons. Neurobiol Aging. 2012;33(2):e27–e36. doi: 10.1016/j.neurobiolaging.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saito H, Sherwood ER, Varma TK, Evers BM. Effects of aging on mortality, hypothermia, and cytokine induction in mice with endotoxemia or sepsis. Mech Ageing Dev. 2003;124(10-12):1047–1058. doi: 10.1016/j.mad.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 24.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 25.Wilcoxon F. Individual comparisons by ranking methods. Biom Bull. 1945;1(6):80–83. [Google Scholar]
- 26.Wallenstein S, Wittes J. The power of the Mantel-Haenszel test for grouped failure time data. Biometrics. 1993;49(4):1077–1087. [PubMed] [Google Scholar]
- 27.Tarone RE. On heterogeneity tests based on efficient scores. Biometrika. 1985;72:91–95. [Google Scholar]
- 28.Brasaemle DL. Thematic review series: Adipocyte biology. The perilipin family of structural lipid droplet proteins: Stabilization of lipid droplets and control of lipolysis. J Lipid Res. 2007;48(12):2547–2559. doi: 10.1194/jlr.R700014-JLR200. [DOI] [PubMed] [Google Scholar]
- 29.Greenberg AS, et al. Perilipin, a major hormonally regulated adipocyte-specific phosphoprotein associated with the periphery of lipid storage droplets. J Biol Chem. 1991;266(17):11341–11346. [PubMed] [Google Scholar]
- 30.Ahmadian M, et al. Adipose overexpression of desnutrin promotes fatty acid use and attenuates diet-induced obesity. Diabetes. 2009;58(4):855–866. doi: 10.2337/db08-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gauthier MS, et al. AMP-activated protein kinase is activated as a consequence of lipolysis in the adipocyte: Potential mechanism and physiological relevance. J Biol Chem. 2008;283(24):16514–16524. doi: 10.1074/jbc.M708177200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Avogaro A, de Kreutzenberg SV, Fadini GP. Insulin signaling and life span. Pflugers Arch. 2010;459(2):301–314. doi: 10.1007/s00424-009-0721-8. [DOI] [PubMed] [Google Scholar]
- 33.Olefsky JM, Glass CK. Macrophages, inflammation, and insulin resistance. Annu Rev Physiol. 2010;72:219–246. doi: 10.1146/annurev-physiol-021909-135846. [DOI] [PubMed] [Google Scholar]
- 34.Saltiel AR. Derepressing nuclear receptors for metabolic adaptation. Cell. 2011;147(4):717–718. doi: 10.1016/j.cell.2011.10.027. [DOI] [PubMed] [Google Scholar]
- 35.Bruunsgaard H, Skinhøj P, Pedersen AN, Schroll M, Pedersen BK. Ageing, tumour necrosis factor-alpha (TNF-alpha) and atherosclerosis. Clin Exp Immunol. 2000;121(2):255–260. doi: 10.1046/j.1365-2249.2000.01281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fielding RA, et al. Sarcopenia: An undiagnosed condition in older adults. Current consensus definition: Prevalence, etiology, and consequences. International working group on sarcopenia. J Am Med Dir Assoc. 2011;12(4):249–256. doi: 10.1016/j.jamda.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franceschi C, et al. Inflammaging and anti-inflammaging: A systemic perspective on aging and longevity emerged from studies in humans. Mech Ageing Dev. 2007;128(1):92–105. doi: 10.1016/j.mad.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 38.Peake J, Della Gatta P, Cameron-Smith D. Aging and its effects on inflammation in skeletal muscle at rest and following exercise-induced muscle injury. Am J Physiol Regul Integr Comp Physiol. 2010;298(6):R1485–R1495. doi: 10.1152/ajpregu.00467.2009. [DOI] [PubMed] [Google Scholar]
- 39.Ebstein F, et al. Maturation of human dendritic cells is accompanied by functional remodelling of the ubiquitin-proteasome system. Int J Biochem Cell Biol. 2009;41(5):1205–1215. doi: 10.1016/j.biocel.2008.10.023. [DOI] [PubMed] [Google Scholar]
- 40.Lukasiak S, et al. Proinflammatory cytokines cause FAT10 upregulation in cancers of liver and colon. Oncogene. 2008;27(46):6068–6074. doi: 10.1038/onc.2008.201. [DOI] [PubMed] [Google Scholar]
- 41.Ebstein F, Lehmann A, Kloetzel PM. The FAT10- and ubiquitin-dependent degradation machineries exhibit common and distinct requirements for MHC class I antigen presentation. Cell Mol Life Sci. 2012;69(14):2443–2454. doi: 10.1007/s00018-012-0933-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hipp MS, Kalveram B, Raasi S, Groettrup M, Schmidtke G. FAT10, a ubiquitin-independent signal for proteasomal degradation. Mol Cell Biol. 2005;25(9):3483–3491. doi: 10.1128/MCB.25.9.3483-3491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fontana L. Excessive adiposity, calorie restriction, and aging. JAMA. 2006;295(13):1577–1578. doi: 10.1001/jama.295.13.1577. [DOI] [PubMed] [Google Scholar]
- 44.Fontana L, Klein S. Aging, adiposity, and calorie restriction. JAMA. 2007;297(9):986–994. doi: 10.1001/jama.297.9.986. [DOI] [PubMed] [Google Scholar]
- 45.Duncan RE, Ahmadian M, Jaworski K, Sarkadi-Nagy E, Sul HS. Regulation of lipolysis in adipocytes. Annu Rev Nutr. 2007;27:79–101. doi: 10.1146/annurev.nutr.27.061406.093734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hardie DG, Pan DA. Regulation of fatty acid synthesis and oxidation by the AMP-activated protein kinase. Biochem Soc Trans. 2002;30(Pt 6):1064–1070. doi: 10.1042/bst0301064. [DOI] [PubMed] [Google Scholar]
- 47.McGee SL, Hargreaves M. AMPK-mediated regulation of transcription in skeletal muscle. Clin Sci (Lond) 2010;118(8):507–518. doi: 10.1042/CS20090533. [DOI] [PubMed] [Google Scholar]
- 48.Brand MD. Uncoupling to survive? The role of mitochondrial inefficiency in ageing. Exp Gerontol. 2000;35(6-7):811–820. doi: 10.1016/s0531-5565(00)00135-2. [DOI] [PubMed] [Google Scholar]
- 49.Harden LM, et al. Critical role for peripherally-derived interleukin-10 in mediating the thermoregulatory manifestations of fever and hypothermia in severe forms of lipopolysaccharide-induced inflammation. Pflugers Arch. 2013 doi: 10.1007/s00424-013-1371-4. 10.1007/s00424-013-1371-4. [DOI] [PubMed] [Google Scholar]
- 50.Hong EG, et al. Interleukin-10 prevents diet-induced insulin resistance by attenuating macrophage and cytokine response in skeletal muscle. Diabetes. 2009;58(11):2525–2535. doi: 10.2337/db08-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pedersen BK, Fischer CP. Physiological roles of muscle-derived interleukin-6 in response to exercise. Curr Opin Clin Nutr Metab Care. 2007;10(3):265–271. doi: 10.1097/MCO.0b013e3280ebb5b3. [DOI] [PubMed] [Google Scholar]
- 52.Pedersen BK. Exercise-induced myokines and their role in chronic diseases. Brain Behav Immun. 2011;25(5):811–816. doi: 10.1016/j.bbi.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 53.Aichem A, et al. The proteomic analysis of endogenous FAT10 substrates identifies p62/SQSTM1 as a substrate of FAT10ylation. J Cell Sci. 2012;125(Pt 19):4576–4585. doi: 10.1242/jcs.107789. [DOI] [PubMed] [Google Scholar]
- 54.Feng Z, Levine AJ. The regulation of energy metabolism and the IGF-1/mTOR pathways by the p53 protein. Trends Cell Biol. 2010;20(7):427–434. doi: 10.1016/j.tcb.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kwon J, et al. Assurance of mitochondrial integrity and mammalian longevity by the p62-Keap1-Nrf2-Nqo1 cascade. EMBO Rep. 2012;13(2):150–156. doi: 10.1038/embor.2011.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ren J, et al. FAT10 plays a role in the regulation of chromosomal stability. J Biol Chem. 2006;281(16):11413–11421. doi: 10.1074/jbc.M507218200. [DOI] [PubMed] [Google Scholar]
- 57.Snyder A, et al. FAT10: A novel mediator of Vpr-induced apoptosis in human immunodeficiency virus-associated nephropathy. J Virol. 2009;83(22):11983–11988. doi: 10.1128/JVI.00034-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Selman C, et al. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science. 2009;326(5949):140–144. doi: 10.1126/science.1177221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blüher M, Kahn BB, Kahn CR. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science. 2003;299(5606):572–574. doi: 10.1126/science.1078223. [DOI] [PubMed] [Google Scholar]
- 60.Blüher M, et al. Adipose tissue selective insulin receptor knockout protects against obesity and obesity-related glucose intolerance. Dev Cell. 2002;3(1):25–38. doi: 10.1016/s1534-5807(02)00199-5. [DOI] [PubMed] [Google Scholar]
- 61.Danilova OV, et al. Neurogenin 3-specific dipeptidyl peptidase-2 deficiency causes impaired glucose tolerance, insulin resistance, and visceral obesity. Endocrinology. 2009;150(12):5240–5248. doi: 10.1210/en.2009-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vieira Potter VJ, et al. Adipose tissue inflammation and reduced insulin sensitivity in ovariectomized mice occurs in the absence of increased adiposity. Endocrinology. 2012;153(9):4266–4277. doi: 10.1210/en.2011-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.