Significance
Superresolution microscopy has made much progress in improving resolution and imaging speed over the past several years, but the ability to image below the diffraction limit in thick scattering specimens has not kept pace. In many interesting samples, such as Caenorhabditis elegans, Drosophila melanogaster, mouse, or human tissues, resolution is limited primarily by scattering rather than diffraction. In this paper, we show that the combination of multiphoton excitation with multifocal structured illumination microscopy gives high quality resolution-doubled images even in thick opaque samples, which until now have resisted superresolution techniques. Since the majority of model organisms and human tissues are opaque to some degree, this advance brings superresolution imaging to a substantial fraction of biological problems.
Keywords: multiphoton, superresolution
Abstract
Multifocal structured illumination microscopy (MSIM) provides a twofold resolution enhancement beyond the diffraction limit at sample depths up to 50 µm, but scattered and out-of-focus light in thick samples degrades MSIM performance. Here we implement MSIM with a microlens array to enable efficient two-photon excitation. Two-photon MSIM gives resolution-doubled images with better sectioning and contrast in thick scattering samples such as Caenorhabditis elegans embryos, Drosophila melanogaster larval salivary glands, and mouse liver tissue.
Fluorescence microscopy is an invaluable tool for biologists. Protein distributions in cells have an interesting structure down to the nanometer scale, but features smaller than 200–300 nm are blurred by diffraction in widefield and confocal fluorescence microscopes. Superresolution techniques like photoactivated localization microscopy (1), stochastic optical reconstruction microscopy (2), or stimulated emission depletion (STED) (3) microscopy allow the imaging of details beyond the limit imposed by diffraction, but usually trade acquisition speed or straightforward sample preparation. And although STED can provide resolution down to 40 nm, STED-specific fluorophores are recommended and it often requires light intensities that are orders of magnitude above widefield and confocal microscopy. On the other hand, structured illumination microscopy (SIM) (4) gives twice the resolution of a conventional fluorescence microscope with light intensities on the order of widefield microscopes and can be used with most common fluorophores. SIM uses contributions from both the excitation and emission point spread functions (PSFs) to substantially improve the transverse resolution and is generally performed by illuminating the sample with a set of sharp light patterns and collecting fluorescence on a multipixel detector, followed by image processing to recover superresolution detail from the interaction of the light pattern with the sample. A related technique, image scanning microscopy (ISM), uses a scanned diffraction-limited spot as the light pattern (5, 6). Multifocal SIM (MSIM) parallelizes ISM by using many excitation spots (7), and has been shown to produce optically sectioned images with ∼145-nm lateral and ∼400-nm axial resolution at depths up to ∼50 µm and at ∼1 Hz imaging frequency. In MSIM, images are excited with a multifocal excitation pattern, and the resulting fluorescence in the multiple foci are pinholed, locally scaled, and summed to generate an image [multifocal-excited, pinholed, scaled, and summed (MPSS)] with root 2-improved resolution relative to widefield microscopy, and improved sectioning compared with SIM due to confocal-like pinholing. Deconvolution is applied to recover the final MSIM image which has a full factor of 2 resolution improvement over the diffraction limit.
MSIM works well in highly transparent samples (such as zebrafish embryos), but performance degrades in light scattering samples (such as the Caenorhabditis elegans embryo). Imaging in scattering samples can be improved by two-photon microscopy (8) and although the longer excitation wavelength reduces the resolution in nondescanned detection configurations, this can be partially offset by descanned detection and the addition of a confocal pinhole into the emission path. Whereas the nondescanned mode collects the most signal, the addition of a pinhole in the emission path of a point-scanning system can improve resolution when the pinhole is closed (9). In practice this is seldom done for biological specimens because signal-to-noise decays as the pinhole diameter decreases (9–11).
SIM is an obvious choice in improving resolution without a dramatic loss in signal-to-noise, but the high photon density needed for efficient two-photon excitation is likely difficult to achieve in the typical widefield SIM configuration. This has led to other methods, such as line scanning (12) to achieve better depth penetration than confocal microscopy and up to twofold improvements in axial resolution (but with only ∼20% gain in lateral resolution). Multiphoton Bessel plane illumination (13) achieved an anisotropic lateral resolution of 180 nm (only in one direction) but requires an instrument design with two objectives in an orthogonal configuration. Cells and embryos can be readily imaged, but the multiaxis design may hinder the intravital imaging of larger specimens. Here, a combination of multiphoton excitation with MSIM is shown to improve both lateral and axial resolutions twofold compared with conventional multiphoton imaging while improving the sectioning and contrast of MSIM in thick, scattering samples.
Results
MSIM excitation was originally implemented with a digital micromirror device (DMD) which is very inefficient in light transmission as less than 1% of the input illumination is directed toward the sample. To accommodate the need for better throughput, the DMD was replaced with an efficient microlens array in the two-photon MSIM (2P-MSIM) implementation used here. A 2D galvanometric mirror system was used to step the illumination pattern across the sample (SI Text and Fig. S1), capturing a series of raw images for each 2D slice. A resolution-doubled image is obtained by postprocessing the raw image series with a modified version of the MSIM analysis code, followed by deconvolution. The modified code allows arbitrary scan patterns and corrects for nonuniform illumination of the sample (7) (the code is available at http://code.google.com/p/msim/).
This instrument bears a resemblance to a number of multifocal multiphoton configurations, whose major advantage is that they increase the image acquisition rate over that of point-scanning systems (12, 14–25). Most use widefield detection for parallelized detection of the multiple focal spots and this is important for 2P-MSIM for two critical reasons. First, it maintains the MSIM increase in image acquisition speed compared with ISM (7). Second, widefield detection with multiphoton excitation inherently improves lateral resolution as it depends on the lower wavelength emission PSF instead of the longer wavelength excitation PSF (26). With our 2P-MSIM setup, we were able to achieve 150 ± 20-nm lateral and 440 ± 50-nm axial resolution when imaging 0.1-µm fluorescent beads in water (Fig. S2 and Table S1). The resolutions obtained in the widefield images here compare well with those reported for previous multifocal multiphoton instruments when using a similar numerical aperture (N.A.) objective lens (15, 17). The striped-illumination multibeam two-photon microscope is capable of doubling the axial resolution in deep tissue, but achieves only a 23% improvement in the lateral direction (12). By comparison, 2P-MSIM doubles the resolution in all directions (Figs. S2 and S3, Table S1, and Dataset S1).
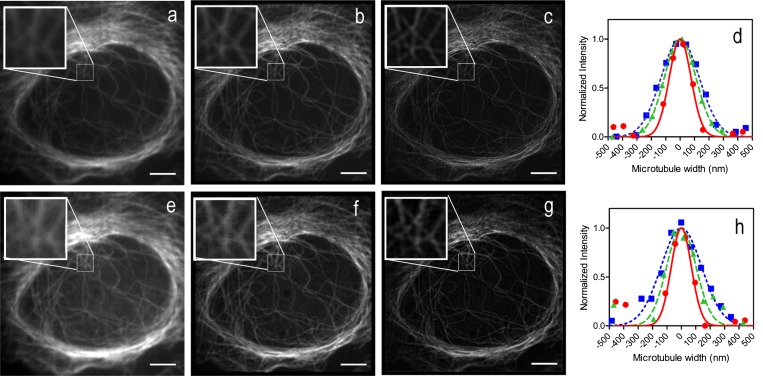
To further test 2P-MSIM resolution, labeled microtubule samples were chosen because the well-defined microtubule diameter of 25 nm makes them convenient tests for superresolution microscopy. Images of immunostained microtubules in fixed U2OS cells (American Type Culture Collection) were collected at different axial positions using 488-nm excitation (Fig. 1 A–D) or 850-nm multiphoton excitation (Fig. 1 E–H). The MPSS images (Fig. 1 B and F) show a visible enhancement of lateral resolution over the widefield images (Fig. 1 A and E), and the MPSS images are further improved by deconvolution in the final one-photon (1P)- and 2P-MSIM images (Fig. 1 C and G, respectively). Full width at half maximum (FWHM) values, obtained from Gaussian fits to the intensity profiles of in-focus microtubules (Fig. 1 D and H), demonstrate the enhancement of lateral resolution. As previously observed (7), measurements of the microtubules in the axial direction indicate that the resolution is improved in 1P images by about twofold over that of the widefield images. We also found the deconvolved 2P-MSIM axial FWHM (500 ± 100 nm) to be improved by about twofold relative to the widefield case (900 ± 100 nm) (Table S2). These data highlight two important points. First, measurements on two-photon widefield images show approximately the same lateral resolution as that of the one-photon images. These show no improvement over the theoretical axial resolution of a two-photon point-scanning microscope but this is consistent with the theoretical treatment of multiphoton widefield detection (26). Second, the twofold improvement of resolution requires the combination of MPSS and deconvolution to yield the twofold improvement over widefield resolution.
Fig. 1.
Both 1P- and 2P-MSIM double resolution. U2OS cells were labeled with an anti-tubulin antibody and imaged by (A–D) one- or (E–H) two-photon excitation. Widefield images of the same cell were produced by summing the fluorescence from the unprocessed images collected under (A) one- or (E) two-photon imaging. The processed MPSS images show the results of pinholing, scaling, and summing the data collected under (B) one- or (F) two-photon excitation. The images in B and F were deconvolved to produce the final (C) 1P- or (G) 2P-MSIM images. The Insets display magnified regions. The widths of microtubule filaments were measured for each of the three image types and example plots with their corresponding Gaussian fits are shown for (D) 1P- and (H) 2P-MSIM imaging. The widefield measurements (blue squares) and 1D Gaussian fits (dotted blue lines) show average FWHM ± SD of 350 ± 40 nm for 1P-MSIM imaging and 360 ± 30 nm for 2P-MSIM. Measurements on the intermediate MPSS images for 1P-MSIM (D, green triangles) and 2P-MSIM (H, green triangles) show an average of 240 ± 20 nm or 260 ± 30 nm FWHM determined from their respective fits (D and H, dashed green lines). Measurements on 1P-MSIM (D, red circles) and 2P-MSIM (H, red circles) images show average FWHM of 150 ± 20 nm and 160 ± 20 nm from their respective 1D Gaussian fits (D and H, solid red lines). For all measurements, the sample size is ≥30. (Scale bars: 5 µm.)
Because 2P-MSIM performed similarly to 1P-MSIM on thin specimens, we performed tests to check the resolution enhancement in highly scattering samples and thick specimens where one-photon excitation performance degrades. We compared the performance of 2P-MSIM with 1P-MSIM on an artificial scattering sample. A suspension of yellow-green fluorescent microspheres (∼0.1-µm diameter) in 3% (wt/vol) agarose gel was made to distribute subdiffractive fluorescent objects sparsely throughout a thick specimen. Scattering was introduced by adding increasing concentrations of 1-µm nonfluorescent polystyrene beads. One- and two-photon excitation powers were set to equalize the photobleaching rates (per acquisition volume) observed with a thick test specimen, 50 µm volumes of Drosophila melanogaster salivary glands immunostained for lamin-C (Fig. S4), and these powers were used for the majority of this work.
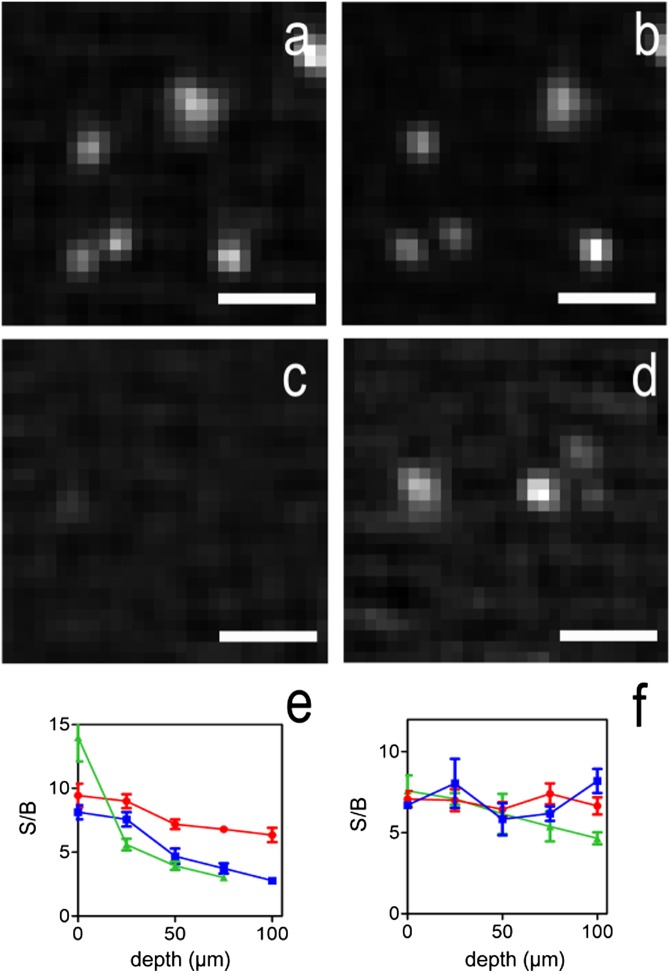
Images from the bead samples showed the scattering effects on the 1P- and 2P-MSIM images. When the sample is minimally scattering (0% or 0.13% nonfluorescent polystyrene beads), both 1P- and 2P-MSIM images revealed beads even ∼100 µm deep into the sample (Fig. 2 A and B). As the depth increases in the most highly scattering sample (0.26% nonfluorescent polystyrene beads), the background in the 1P-MSIM images overwhelmed the fluorescence from the beads, drastically reducing contrast, whereas the 2P-MSIM images retained high contrast (Fig. 2 C and D). The signal-to-background ratio (S/B) (defined as the ratio of the amplitude and offset derived from Gaussian fits of the bead intensity profiles) of 1P-MSIM was quantitatively worse than that of 2P-MSIM for scattering samples at greater depths (Fig. 2 E and F), showing fewer beads than the 2P-MSIM images. No beads were observed at 100-µm depth for the most highly scattering sample when using 1P-MSIM, whereas 2P-MSIM detected beads at all measured depths. The resolution obtained from visible beads in the different excitation modes was independent of depth, remaining almost constant for both one- and multiphoton excitation (Fig. S5).
Fig. 2.
One- and two-photon MSIM imaging at different depths in scattering artificial samples. Fluorescent beads (yellow-green 0.1 µm) suspended in 3% agarose gel were imaged as a function of depth and scattering. The imaging laser powers were chosen to give similar fluorescence between the two imaging modes in the absence of scattering beads. Images were collected in 5-µm volumes on the same fields of beads for each imaging mode at depths of 0, 25, 50, 75, and 100 µm with the addition of 0%, 0.13%, or 0.26% polystyrene nonfluorescent scattering beads. In the absence of scattering beads at 50-µm depth, beads are visible in both (A) 1P- (B) and 2P-MSIM modes. In contrast, in the presence of 0.26% scattering beads, fewer beads are visible when imaging with 1P-MSIM (C), whereas beads are readily observable with 2P-MSIM (D). The mean and SEs of the S/B ratios, as defined by the ratio of the amplitude to the offset of a Gaussian fit to the 1D intensity profiles of beads in samples containing 0% (red circles), 0.13% (blue squares), or 0.26% (green triangles) nonfluorescent beads are plotted as a function of imaging depth for (E) 1P- and (F) 2P-MSIM. The S/B ratio as a function of depth of 1P-MSIM decreases faster than 2P-MSIM in scattering samples. (Scale bars: 1 µm.)
The scattering tests highlight what are often considered the disadvantages of widefield detection compared with nondescanned detection for multiphoton excitation. One disadvantage is the increasing difficulty of detecting and accurately assigning the scattered light to its correct location. In the multifocal multiphoton microscope configuration used here, the scattered photons collected are not properly assigned to their original position and hence lead to decreased signal compared with nondescanned microscopes. A second disadvantage is that the scattered light can exacerbate the decreased S/B. If the light from one focal spot is scattered, it can be collected by the objective and detected elsewhere on the camera chip, which will lead to increased background. These scattered photons are collected and can increase background in the 2P-MSIM experiments here, but each image shows the signal from the array of focal spots making up a small percentage (30%) of the total pixel area. Therefore, scattered photons have only a ∼30% chance of falling on an area of the camera chip detecting a focal spot. Importantly, MSIM image analysis relies on masking the background signal between the focal points (i.e., pinholing) before scaling and summing the images. As a consequence, the background is reduced and 2P-MSIM images suffer S/B decreases much less than most multiphoton images collected under widefield detection. This is evident even for an image collected at a shallow depth where processing by pinholing and scaling improves the S/B by ∼10-fold over a widefield image and subsequent deconvolution into an MSIM image further improves this parameter (Fig. S6).
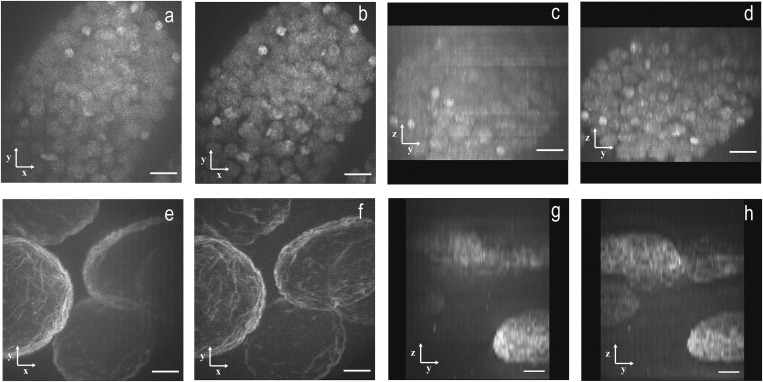
Avoiding a high background is important when imaging thicker biological specimens. To determine if improvements observed with the 1P- and 2P-MSIM comparisons on bead phantoms would translate into improved tissue imaging, we imaged live C. elegans embryos expressing H2B-EGFP, a histone marker. Both 1P- and 2P-MSIM (Fig. 3 A and B, respectively) exhibit substantially higher resolution than widefield images (Fig. S7). Comparing yz projections (Fig. 3 C and D) reveals that 2P-MSIM offers substantially better detail throughout the imaging volume compared with 1P-MSIM. Even at ∼10-µm depths, individual nuclei have reduced contrast in the 1P-MSIM image (Fig. 3C) and at a 20- to 25-µm depth nuclei that are visible in 2P-MSIM are barely visible in 1P-MSIM (Fig. 3D). Although the signal in 1P-MSIM might be improved by increasing the 488-nm laser power, this would also introduce more photobleaching than 2P-MSIM, and would not change the observed underlying one photon-induced depth-dependent signal degradation.
Fig. 3.
One- and two-photon MSIM imaging of C. elegans embryo and D. melanogaster salivary glands. The 1P- and 2P-MSIM imaging was performed on a C. elegans embryo expressing H2B-EGFP (A–D) using (A and C) 1P- and (B and D) 2P-MSIM. The images in a 25-µm volume are shown here as maximum-intensity (A and B) xy and as yz projections. Imaging was also performed on an Alexa Fluor 488 lamin-C–labeled D. melanogaster salivary gland using (E and G) 1P- and (F and H) 2P-MSIM, shown here as maximum-intensity xy and yz projections from a 50-μm volume. The optical axis is indicated by the z axis in YZ projections with increasing depth into the specimen displayed from bottom to top. (Scale bars: 5 µm.)
Salivary glands isolated from larvae of D. melanogaster and labeled with lamin-C presented an even more challenging sample because they are thicker and more scattering than C. elegans (Fig. 3 E–H and Fig. S8). Lamin-C is a nuclear lamin that labels the nuclear envelope of the nuclei. In the salivary glands, the multiphoton-excited lamin-C fluorescence signal outperforms the 488-nm excited signal as the imaging plane moves deeper into the specimen (Fig. 3 G and H and Figs. S8 and S9). Neither one- nor two-photon widefield images provide detail comparable to 1P- and 2P-MSIM (Fig. 3 E and F, respectively) images (Fig. S9). As in the nematode embryos, yz projection comparisons of 1P- and 2P-MSIM (Fig. 3 G and H, respectively) indicate that 2P-MSIM provides better detail at depth (up to 50 µm).
We also inspected liver samples from myosin IIA-EGFP–expressing mice, an even more challenging, nearly opaque specimen (Fig. S10). None of the imaging modes we tested were able to reliably discern structure more than ∼25 µm into this sample. Nevertheless, 2P-MSIM performed better than 1P-MSIM at all depths. At shallower depths, 2P-MSIM compared well and provided better resolution and S/B than conventional point-scanning microscopy. The point-scanning system images were always noisier than 2P-MSIM but had better contrast at greater depth.
Discussion
SIM techniques have undergone rapid development over the past few years, but SIM progress in scattering samples has lagged behind. The work presented here demonstrates that 2P-MSIM produces similar resolution to 1P-MSIM and better S/B in images collected deep within scattering samples. The improvement over 1P-MSIM is likely due to two major multiphoton advantages: less scattering due to the longer wavelength excitation light and lower background fluorescence due to the inherent optical sectioning provided by the two-photon effect. Thus, compared with 1P-MSIM, multiphoton excitation and its advantageous characteristics are largely maintained in 2P-MSIM.
Although faster than point-scanning multiphoton microscopes, multifocal multiphoton microscopes using parallelized excitation and detection, such as the one used here, possess some disadvantages. These include the loss of signal as well as increased background associated with the inability to properly assign scattered light (17). Although 2P-MSIM does sacrifice proper assignment of the scattered light signal, it does not suffer the same level of increased background normally associated with multifocal multiphoton microscopy. This is due to the digital pinholing and deconvolution during image analysis (7) which masks and thus removes the scattered light from the background between the excitation spots (Fig. S6). And although 2P-MSIM is slower than many other multifocal multiphoton techniques (12, 14–25), the resolution improvement is similar to that of 1P-MSIM despite using a much longer wavelength of light.
Finally, 2P-MSIM improves resolution compared with the reported values for point-scanning multiphoton techniques. In addition to the MSIM image processing, widefield detection with an EMCCD may play a role in this improvement. This should inherently improve multiphoton lateral resolving power over the nondescanned method because the PSF is dependent on the emission wavelength and not a convolution of emission and excitation (26). Unfortunately, the improvement via widefield detection does not apply to the axial direction, but MSIM provides a doubling of the resolution in this direction also (Fig. S2 and Table S1). It is noteworthy that the addition of a pinhole in the emission path of a point-scanning system can improve resolution by stopping down the pinhole. So confocal multiphoton microscopy can in principle provide equivalent lateral resolution and better axial resolution than 2P-MSIM (27). However, stopping down the pinhole is seldom practical for biological specimen with limited fluorescence signals because the resulting degradation in signal to noise results in a useless image. In practice, 2P-MSIM provides improved resolution relative to point scanning and other modes of multiphoton imaging, while retaining many of the advantages of these methods.
Methods
Instrumentation.
Multifocal, multiphoton illumination was achieved by modifying an Olympus IX-71 widefield microscope. In the case of multiphoton excitation, the illumination was provided by a 3-W tunable multiphoton Ti:sapphire laser (Coherent Inc.; Chameleon Vision II). The power output of this laser was controlled using a variable attenuator consisting of an achromatic zero order half-wave plate designed for use at 700–1,000 nm wavelengths (Newport; 10RP52-2) and a Glan-Laser Calcite Polarizer (Newport; 10GL08AR.16). The half-wave plate was mounted on a motorized rotation stage (Thorlabs Inc.; PRM1Z8E) controlled with software provided by the manufacturer to rotate the polarization of the beam while the polarizer separates the P and S components. Beam blanking was performed with a mechanical shutter (Vincent Associates; LS 6Z2) under Micro-Manager control (28). The beam was then passed through a 2× beam expander composed of a pair of lenses with focal length (f) = 100 mm and f = 200 mm (Thorlabs Inc.; AC254-B). A 50-mW 488-nm laser served as the illumination source for the one-photon experiments (Oxxius; LBX-488–50-CIR-PP). The laser light was filtered with a 488-nm MaxLine laser clean-up filter (Semrock Inc.; LL01-488-25) before collimation with a 1× excitation telescope (Thorlabs Inc.; AC254-100-A). A mechanical shutter and acoustooptic tunable filter (AA Opto-Electronic Inc.; AOTFnC-400.650) under Micro-Manager (28) control allowed for laser power tuning and shuttering. The 488-nm beam was expanded 5× using an f = 40 mm and an f = 200 mm lens pair (Thorlabs Inc.; AC254-A). The one-photon and multiphoton paths were combined at this point using a 705-nm edge multiphoton dichroic mirror (Semrock Inc.; FF705-Di01-25x36). Multifocal illumination was produced by passing the excitation through a chrome-masked microlens array with a focal length of 5.2 mm and a 150-µm pitch (Thorlabs Inc.; MLA150-5C). The microlens array was mounted on a motorized stage (Zaber Technologies Inc.; T-LSM100B-KT04) to allow for correction of focal shift between wavelengths. A 2D scanning galvo system (Thorlabs Inc.; GVSM002), positioned between a pair of 60-mm lenses (Thorlabs Inc.; AC254-B), displaced the beam on the sample without changing the incoming angle. The galvanometric mirrors were stepped using a program written in Python (www.python.org) or a BeanShell (www.beanshell.org/) script written for use in Micro-Manager. A 250-mm tube lens (Thorlabs Inc.; AC254-B) was placed before the entrance to the left-side port of an Olympus widefield IX-71 base containing a custom-designed 3-mm dichroic mirror (Chroma Technology Corp.; zt405/488/561/647/NIR-rpc). The objective used in this study was a UPlanSApo 60×/1.20-W PSF-grade objective (Olympus America Inc.). A C-focus device (Mad City Labs Inc.) compensated for focal drift. z stacks were acquired using a NanoZ-100 (Mad City Labs Inc.), controlled with either a BeanShell script in Micro-Manager (28) or a program written in Python. Imaging was performed using an Andor iXon Ultra EMCCD camera controlled with Andor Solis software provided by the manufacturer or with Micro-Manager (28). Fluorescence emission was magnified 2× using an f = 75 mm and f = 150 = mm lens pair to provide a ∼130-nm pixel size, confirmed using a stage micrometer (Ted Pella, Inc.; 2280–15).
Conventional point-scanning multiphoton imaging was performed on an IX81 inverted confocal microscope (Olympus America Inc.) modified to perform multiphoton microscopy. A tunable Ti:sapphire femtosecond laser, Chameleon Ultra II (Coherent Inc.), was used as a laser source, and the excitation power was modulated using a combination of neutral density filters (Chroma Technology Corp). The size of the beam was expanded (LSM Technology Inc.) and directed into a Fluoview 1000 scanning head (Olympus America Inc.). The emitted signal was directed into a custom-made array of three cooled nondescanned photomultiplier tubes (PMTs) that were installed on the right port of the microscope (LSM Technology Inc.). A 680-nm barrier filter (Chroma Technology Corp.) was used to prevent the scattered IR light from reaching the detectors. The three cooled PMTs were purchased from Hamamatsu (R6060-12), and two associated dichroic mirrors and barrier filters were purchased from Chroma Technology Corp. The excised liver was imaged in the inverted setting and the image was acquired using a UPlanSApo 60× N.A. 1.2 water immersion objective. The GFP fluorescent signal was excited at 930 nm and detected on the second PMT (570-nm dichroic mirror, 505 to 560-nm barrier filter).
One- and Two-Photon MSIM Data Collection.
The square grid of foci was positioned at a 45° tilt relative to the horizontal of the camera chip, so that the nearest neighbor to each focus was along a diagonal line. For each slice, 340 frames were acquired at a rate of 52 Hz for a 256 × 256 pixel-wide field of view. The EMCCD spooled to disk in 16-bit frame transfer mode, with a vertical shift speed of 0.3 µs, a vertical clock voltage amplitude of 4, and a preamplifier gain of 3. A 10-ms exposure was collected after stepping the galvo by a voltage corresponding to a shift of approximately one image pixel. The galvo was stepped repeatedly 22 times to the right, once down, then 22 times to the left, and down again. Timing was controlled by voltages sent to the camera, galvo, and z stage using a 16-bit data acquisition board (Measurement Computing Inc.; USB-3101FS) controlled by a Python script or an ESio AOTF controller (ESImaging) and Micro-Manager BeanShell script (28). Power levels were normalized based on the bleaching rate of a 50-µm deep stack sampled at 1-µm steps of an Alexa 488-labeled lamin-C D. melanogaster salivary gland sample. Under these conditions, the power levels measured at the right-side port of the microscope were 6 µW for 488-nm and 1.1 W for 850-nm light.
One- and Two-Photon MSIM Data Analysis.
Superresolution was achieved by postprocessing the images computationally by pinholing (with a pinhole size of three pixels) the detected spots, scaling the spots by 0.5, and integrating the scaled images over all positions. Postprocessing was based on freely available software (http://code.google.com/p/msim/). Two main modifications were introduced into the original MSIM software. First, the offset vectors were calculated for each individual frame. This modification was necessary because the scan pattern is not a perfect Cartesian grid like the original MSIM implementation. Second, to avoid striping artifacts arising from illuminating areas more than once, the contribution of each illumination spot to the final image is weighted by the local density of scan spots. This artifact also occurs because the scan pattern is not a perfect Cartesian grid and the result is that some regions of the image are sampled more densely or sparsely by the illumination spots. Without correction, the densely sampled regions appear brighter and the sparsely sampled regions appear dimmer, which produces a striped appearance in the image. To correct for this artifact, the set of all scan points is constructed, the area surrounding each scan point is determined by Delaunay triangulation, and each scan point is assigned a weight based on the area occupied by all triangles it touches. A scan point with distant neighbors will touch Delaunay triangles which cover a large area and receive a large weight, and a scan point with close-neighboring scan points will touch a small area and receive a small weight. The modified version of the software used in this study is also freely available at the same Web address above.
Deconvolution.
Deconvolution was performed on the pinholed, scaled, and integrated stacks using a program written in Python (freely available at http://code.google.com/p/msim/) implementing Richardson–Lucy deconvolution (29, 30). Given a measurement M which has been blurred by convolution (indicated by *) with a PSF P and corrupted with Poisson noise, Richardson–Lucy deconvolution iteratively improves an estimate E of the sample density:
This iteration converges to an E which maximizes the likelihood that M was measured given the assumption of Poisson noise. In our experiments, Poisson noise is not the only noise source, but because we use a sensitive EMCCD, Poisson noise is probably the primary noise source.
For P, we use a theoretical PSF approximated by a Gaussian function. The approximate dimensions of the theoretical PSF were obtained from the FWHM of Gaussian fits to the intensity profiles of 100-nm fluorescent beads in x, y, and z directions. We performed between 30–100 iterations using a PSF ∼240 nm in xy and 600 nm in the z direction. For 8-bit display, each 16-bit image is linearly transformed so its maximum and minimum values span the 0–255 pixel value range.
Preparation and Imaging of Beads in Scattering Samples.
Measurement of the FWHM as a function of depth and scattering was performed using yellow-green 0.1-µm fluorescent microspheres (Molecular Probes; F88-03) suspended in 3% agarose gels. The scattering properties of the sample was varied by adding 0.13% or 0.26% 1-µm nonfluorescent polystyrene beads (Polysciences Inc.; 07310) with a refractive index of 1.606 at 486 nm and 1.577 at 833 nm. Imaging was performed by acquiring stacks that covered a range of 5 µm in 0.2-µm steps. A total of five stacks were spaced along the z axis up to a depth of 100 μm. Power levels were normalized in two different ways for these experiments: one normalized based on the photobleaching rate of the D. melanogaster sample (Fig. S4) and the second normalized the power based on the signal intensity of the beads in the nonscattering sample. The photobleaching rate normalization required a 488-nm laser power that was twice that of the intensity normalized level. The intensity normalization experiments were used to allow comparison of the imaging modes based on slices instead of volumes.
Immunostaining Procedure.
U2OS cells adhered to no. 1.5 25-mm-diameter coverslips (Warner Instruments; 64-0715) were fixed in a mixture of 4% formaldehyde and 0.3% Triton X-100 in cytoskeletal buffer (10 mM 3-(N-morpholino) propanesulfonic acid, 138 mM KCl, 3 mM MgCl2, 2 mM EGTA, 0.01% NaN3, and 160 mM sucrose, pH 6.1) for 15 min. Labeling of microtubules with Alexa Fluor 488-conjugated antibody was carried out as described previously (7). Samples were mounted in ProLong (Invitrogen; P36930). Data were acquired using an Olympus PSF-corrected UPlanSApo 60×/1.20-W objective. (“PSF-corrected” is the Olympus nomenclature for objectives which have more symmetrical point spread functions than standard objectives.) A set of 25 images that were 0.2-µm apart was collected for each experiment producing a stack of 5-µm axial depth. FWHM values of individual microtubules were obtained from Gaussian fits of the intensity distribution along a line on the XY plane perpendicular to an in-focus microtubule for the lateral dimension or along the z axis of the stack for the axial dimension.
C. elegans Embryo Preparation.
Strain BV24 (ltIs44 [pie-1p-mCherry::PH(PLC1delta1) + unc-119(+)]; zuIs178 [(his-72 1kb::HIS-72::GFP); unc-119(+)] was cultured as described previously (7). Embryos were imaged at room temperature in PBS (KD Medical Inc.; RGF 3210). A 25-µm-thick stack spaced in 0.5-µm intervals for a total of 50 slices per volume was acquired in a field of view 256 × 256 pixels wide.
D. melanogaster Larval Salivary Glands.
Salivary glands from third instar larvae were dissected and fixed in 4% paraformaldehyde/PBS on ice for 1 h. The glands were washed several times with 0.1% Triton X-100/PBS at room temperature and incubated in Drosophila lamin-C antibody (Developmental Studies Hybridoma Bank-developed under the auspices of the National Institute of Child Health and Human Development and maintained by the Department of Biology, The University of Iowa, Iowa City, IA) overnight in 4 °C. The salivary glands were washed in 0.1% Triton X-100/PBS and incubated in Alexa Fluor 488 Goat Anti-Mouse IgG (H+L) secondary antibody (Invitrogen) for 2 h at room temperature. After several washes in 0.1% Triton X-100/PBS, the glands were mounted in ProLong (Invitrogen). A 50-µm-thick stack spaced in 1-µm intervals for a total of 50 slices per volume was acquired in a field of view 256 × 256 pixels wide.
Liver Tissue Samples.
Mice expressing nonmuscle myosin IIA-GFP (lining the borders of the hepatocytes and outlining the bile canalicular network) were a gift from Robert Adelstein (National Heart, Lung, and Blood Institute, Bethesda). To preserve the morphology and subcellular architecture of the liver, it was rapidly fixed by performing cardiac puncture and perfusing the animals with a normal saline immediately followed by 4% formaldehyde, 0.05% glutaraldehyde, and 0.2 M Hepes buffer (pH 7.3). The liver was then excised and securely placed into a chamber with 1× PBS for imaging. All of the experiments were approved by the National Institute of Dental and Craniofacial Research (National Institutes of Health, Bethesda) Animal Care and Use Committee.
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Programs of the National Institutes of Health including the National Institute of Biomedical Imaging and Bioengineering; the National Institute of Dental and Craniofacial Research; and the National Heart, Lung, and Blood Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.D. is a guest editor invited by the Editorial Board.
Data deposition: The code reported in this paper has been deposited at http://code.google.com/p/msim/.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1314447111/-/DCSupplemental.
References
- 1.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313(5793):1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 2.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3(10):793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hell SW, Wichmann J. Breaking the diffraction resolution limit by stimulated emission: Stimulated-emission-depletion fluorescence microscopy. Opt Lett. 1994;19(11):780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 4.Heintzmann R, Ficz G. Breaking the resolution limit in light microscopy. Brief Funct Genomics Proteomics. 2006;5(4):289–301. doi: 10.1093/bfgp/ell036. [DOI] [PubMed] [Google Scholar]
- 5.Müller CB, Enderlein J. Image scanning microscopy. Phys Rev Lett. 2010;104(19):198101. doi: 10.1103/PhysRevLett.104.198101. [DOI] [PubMed] [Google Scholar]
- 6.Sheppard CJR. Super-resolution in confocal imaging. Optik (Stuttg) 1988;80(2):5354. [Google Scholar]
- 7.York AG, et al. Resolution doubling in live, multicellular organisms via multifocal structured illumination microscopy. Nat Methods. 2012;9(7):749–754. doi: 10.1038/nmeth.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248(4951):73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 9.Gauderon R, Sheppard CJR. Effect of a finite-size pinhole on noise performance in single-, two-, and three-photon confocal fluorescence microscopy. Appl Opt. 1999;38(16):3562–3565. doi: 10.1364/ao.38.003562. [DOI] [PubMed] [Google Scholar]
- 10.Sandison DR, Piston DW, Williams RM, Webb WW. Quantitative comparison of background rejection, signal-to-noise ratio, and resolution in confocal and full-field laser scanning microscopes. Appl Opt. 1995;34(19):3576–3588. doi: 10.1364/AO.34.003576. [DOI] [PubMed] [Google Scholar]
- 11.Gauderon R, Lukins PB, Sheppard CJR. Effect of a confocal pinhole in two-photon microscopy. Microsc Res Tech. 1999;47(3):210–214. doi: 10.1002/(SICI)1097-0029(19991101)47:3<210::AID-JEMT7>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 12.Andresen V, et al. High-resolution intravital microscopy. PLoS ONE. 2012;7(12):e50915. doi: 10.1371/journal.pone.0050915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao L, et al. Noninvasive imaging beyond the diffraction limit of 3D dynamics in thickly fluorescent specimens. Cell. 2012;151(6):1370–1385. doi: 10.1016/j.cell.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nielsen T, Fricke M, Hellweg D, Andresen P. High efficiency beam splitter for multifocal multiphoton microscopy. J Microsc. 2001;201(Pt 3):368–376. doi: 10.1046/j.1365-2818.2001.00852.x. [DOI] [PubMed] [Google Scholar]
- 15.Niesner R, Andresen V, Neumann J, Spiecker H, Gunzer M. The power of single and multibeam two-photon microscopy for high-resolution and high-speed deep tissue and intravital imaging. Biophys J. 2007;93(7):2519–2529. doi: 10.1529/biophysj.106.102459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fittinghoff DN, Squier JA. Time-decorrelated multifocal array for multiphoton microscopy and micromachining. Opt Lett. 2000;25(16):1213–1215. doi: 10.1364/ol.25.001213. [DOI] [PubMed] [Google Scholar]
- 17.Bewersdorf J, Pick R, Hell SW. Multifocal multiphoton microscopy. Opt Lett. 1998;23(9):655–657. doi: 10.1364/ol.23.000655. [DOI] [PubMed] [Google Scholar]
- 18.Fujita K, et al. Real-time imaging of two-photon-induced fluorescence with a microlens-array scanner and a regenerative amplifier. J Microsc. 1999;194(Pt 2-3):528–531. doi: 10.1046/j.1365-2818.1999.00493.x. [DOI] [PubMed] [Google Scholar]
- 19.Straub M, Lodemann P, Holroyd P, Jahn R, Hell SW. Live cell imaging by multifocal multiphoton microscopy. Eur J Cell Biol. 2000;79(10):726–734. doi: 10.1078/0171-9335-00105. [DOI] [PubMed] [Google Scholar]
- 20.Egner A, Hell SW. Time multiplexing and parallelization in multifocal multiphoton microscopy. J Opt Soc Am A Opt Image Sci Vis. 2000;17(7):1192–1201. doi: 10.1364/josaa.17.001192. [DOI] [PubMed] [Google Scholar]
- 21.Grant DM, et al. Optically sectioned fluorescence lifetime imaging using a Nipkow disk microscope and a tunable ultrafast continuum excitation source. Opt Lett. 2005;30(24):3353–3355. doi: 10.1364/ol.30.003353. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi M, et al. Second-harmonic-generation microscope with a microlens array scanner. Opt Lett. 2002;27(15):1324–1326. doi: 10.1364/ol.27.001324. [DOI] [PubMed] [Google Scholar]
- 23.Shimozawa T, et al. Improving spinning disk confocal microscopy by preventing pinhole cross-talk for intravital imaging. Proc Natl Acad Sci USA. 2013;110(9):3399–3404. doi: 10.1073/pnas.1216696110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buist AH, Müller M, Squier J, Brakenhoff GJ. Real time two-photon absorption microscopy using multi point excitation. J Microsc. 1998;192(2):217–226. [Google Scholar]
- 25.Egner A, Jakobs S, Hell SW. Fast 100-nm resolution three-dimensional microscope reveals structural plasticity of mitochondria in live yeast. Proc Natl Acad Sci USA. 2002;99(6):3370–3375. doi: 10.1073/pnas.052545099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bewersdorf J, Schmidt R, Hell SW. Comparison of I5M and 4Pi-microscopy. J Microsc. 2006;222(Pt 2):105–117. doi: 10.1111/j.1365-2818.2006.01578.x. [DOI] [PubMed] [Google Scholar]
- 27.Sheppard CJR, Gu M. Image formation in two-photon fluorescence microscopy. Optik (Stuttg) 1990;86(3):104–106. [Google Scholar]
- 28.Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using µManager. Curr Protoc Mol Biol. 2010;92:14.20.1–14.20.7. doi: 10.1002/0471142727.mb1420s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richardson WH. Bayesian-based iterative method of image restoration. JOSA. 1972;62(1):55–59. [Google Scholar]
- 30.Lucy L. An iterative technique for the rectification of observed distributions. Astron J. 1974;79(6):745–754. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.