Abstract
Background
Vibrio vulnificus is an important pathogen which can cause serious infections in humans. Yet, there is limited knowledge on its virulence factors and the question whether temperate phages might be involved in pathogenicity, as is the case with V. cholerae. Thus far, only two phages (SSP002 and VvAW1) infecting V. vulnificus have been genetically characterized. These phages were isolated from the environment and are not related to Vibrio cholerae phages. The lack of information on temperate V. vulnificus phages prompted us to isolate those phages from lysogenic strains and to compare them with phages of other Vibrio species.
Results
In this study the temperate phage PV94 was isolated from a V. vulnificus biotype 1 strain by mitomycin C induction. PV94 is a myovirus whose genome is a linear double-stranded DNA of 33,828 bp with 5′-protruding ends. Sequence analysis of PV94 revealed a modular organization of the genome. The left half of the genome comprising the immunity region and genes for the integrase, terminase and replication proteins shows similarites to V. cholerae kappa phages whereas the right half containing genes for structural proteins is closely related to a prophage residing in V. furnissii NCTC 11218.
Conclusion
We present the first genomic sequence of a temperate phage isolated from a human V. vulnificus isolate. The sequence analysis of the PV94 genome demonstrates the wide distribution of closely related prophages in various Vibrio species. Moreover, the mosaicism of the PV94 genome indicates a high degree of horizontal genetic exchange within the genus Vibrio, by which V. vulnificus might acquire virulence-associated genes from other species.
Introduction
Vibrio vulnificus is an estuarine bacterium which occurs in coastal waters worldwide [1]. The species is an important pathogen that can cause severe wound infections with lethal outcome [2]. In addition, Vibrio vulnificus is responsible for death cases caused by consumption of contaminated seafood [3]. In the Unites States it is a leading cause of seafood-related deaths [4]. Environmental factors, such as warm water and moderate salinity, are known to favor the multiplication of the pathogen. Hence, the effect of global warming on seawater temperatures has aroused concerns that V. vulnificus infections will increase in numbers [5], [6]. To date only limited information is available about the pathogenicity of this species and on the question what discriminates highly virulent strains from environmental strains with presumably lower virulence. Even though some virulence factors like e.g. capsular polysaccharide [7] and RTX toxins [8] have been identified, the full spectrum of factors has still to be elucidated. In V. cholerae, bacteriophages play a critical role in pathogenicity as cholera toxin is encoded by a temperate phage residing in the chromosome of toxigenic stains. In addition, K139-related phages which are widely distributed throughout different serogroups and biotypes of V. cholerae might be important for pathogenicity. The secreted virulence determinant gene glo (G protein-like ORF) of K139 lysogens has already been characterized [9]. In contrast to V. cholerae, little is known about V. vulnificus phages. Some studies describe the isolation of V. vulnificus phages from estuarine habitats [10], [11]. In oysters up to 105 phages per g tissue have been found [10]. Based on morphological evidence, the phages belonged to the families Myoviridae, Podoviridae and Siphoviridae. They mainly lysed V. vulnificus strains, even though infection of V. fluvialis and V. parahaemolyticus by single phages has also been reported. Only two V. vulnificus phage genomes have been published until now [12], [13]. While the podovirus VvAW1 isolated from surface water in Hawai, USA, has been suggested to be temperate, the siphovirus SSP002 recovered from the coastel area of the Yellow Sea of South Korea may be virulent [12]. The VvAW1 38 kb genome revealed some similarities to non-Vibrio phages (e.g. Thalassomonas phage Ba3, Pseudomonas phage F116) whereas SSP002 is closely related to the V. parahaemolyticus phage vB_VpaS_MAR10 [14].
In this study we characterized the temperate V. vulnificus phage PV94 isolated from a clinical biotype 1 strain. PV94 is a myovirus whose genome shows a modular organization and mosaicism. The phage is closely related to V. cholerae K139-like phages and phages of other Vibrio species.
Materials and Methods
Isolation and purification of PV94 particles
To isolate PV94, the host strain V. vulnificus VN-094 (human isolate, strain collection of the BfR) was grown in Luria Bertani (LB) broth supplemented with 2% sodium chloride (w/v) to an optical density of 0.2–0.3 (A588). Induction of PV94 was performed by adding 0.25 µg/ml mitomycin C to the culture followed by further shaking for 6 to 18 h at 37°C until lysis occurred. High-titer phage lysates were obtained by preparing 500 ml cultures. Upon prophage induction, lysates were centrifuged for 30 min at 12,000×g and passed through 0.45 µm and 0.22 µm filters (Schleicher & Schüll, Dassel, Germany). Between the filtration steps, 10 mM MgCl2, 1 µg ml−1 DNase I, and RNase A (Roche, Mannheim, Germany) were applied to the lysates, which were incubated at 37°C for 2 h. Phage particles were concentrated by ultracentrifugation at 230,000×g for 2 h. Pellets were suspended in SM buffer and purified through discontinuous cesium chloride (CsCl) gradients (1.35 to 1.65 g ml−1), as previously described [15].
Transmission electron microscopy (TEM)
The morphology of the phage was analysed by TEM using the negative staining procedure. Briefly, CsCl-purified phages were allowed to adsorb on pioloform-carbon-coated, 400-mesh copper grids (Plano GmbH, Wetzlar, Germany) for 10 min. Thereafter, the grids were fixed with a 2.5% aqueous glutaraldehyde (Taap Laboratories, Aldermaston, United Kingdom) solution for 1 min, and stained with 2% aqueous uranyl acetate (Merck, Darmstadt, Germany) solution for 1 min. The specimens were examined by TEM using a JEM-1010 (JEOL, Tokyo, Japan) at 80 kV accelerated voltage.
Mass spectrometrical analysis of structural proteins
For the determination of PV94 structural proteins, CsCl-purified phages were disintegrated by boiling in SDS buffer for 10 min. Proteins were subjected to SDS-PAGE as described previously [16]. Bands of interest were excised and prepared for tryptic in gel-digests. The extracted peptides were dried, reconstituted and subjected to tandem matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF-TOF MS/MS) analysis. Mass spectra were analyzed and interpreted using the Mascot software (Matrix Science Ltd., London, United Kingdom) followed by NCBInr database (NCBI) searches.
Sequence determination and analysis of the PV94 genome
First sequence data of the phage were obtained by molecular cloning and sequencing of EcoRI and HindIII restriction fragments. For this purpose phage DNA was isolated from purified particles applying standard protocols [15]. Upon cleavage with EcoRI and HindIII (Thermo Scientific, Hudson, NA), restriction fragments were ligated to the corresponding sites of the vector pIV2 [17] and introduced into the E. coli strain Genehogs (Life Technologies, Darmstadt, Germany) by electroporation. Transformants harboring restriction fragments of the phage were identified by blue/white selection on LB agar containing 100 µg ml−1 neomycin. The inserts of recombinant plasmids were sequenced with vector primers and primers deduced from determined sequences. Gaps between the respective PV94 restriction fragments were closed by primer walking using phage DNA as template.
Sequence analyses and alignments were carried out using the Accelrys DS Gene software package of Accelrys Inc. (USA). ORF Finder of the NCBI database was used to predict putative open reading frames (ORFs) on the genome of PV94. Similarity and identity values nucleotide and amino acid sequences were determined using the algorithms of BLAST search (NCBI) [18]. Putative Rho-independent transcription terminators were identified using TransTerm [19], [20]. The complete nucleotide sequence of PV94 was compared with the genomes of the phages K139 (Accession number: AF125163) [21], kappa (AB374228) and VD1 (JF974301) and with a prophage residing in chromosome 1 of the V. furnissii strain NCTC 11218 (NC_016602, chr 1 positions ∼2,595,000 to ∼2,625,000) [22].
Analysis of the cohesive ends
Cohesive ends of PV94 were identified by digestion of phage DNA with AdeI, BglII and SspI (Thermo Scientific, Hudson, NA) followed by an incubation of the digests at 70°C for 10 min. Thereafter, the obtained restriction patterns were compared to unheated control digests. In a further experiment, PV94 DNA that had been treated before with T4-ligase was digested and analysed. To determine the exact sequence of cos, primers deduced from a region close to the cohesive ends were used for direct sequencing of T4-treated and untreated phage DNA.
Results and Discussion
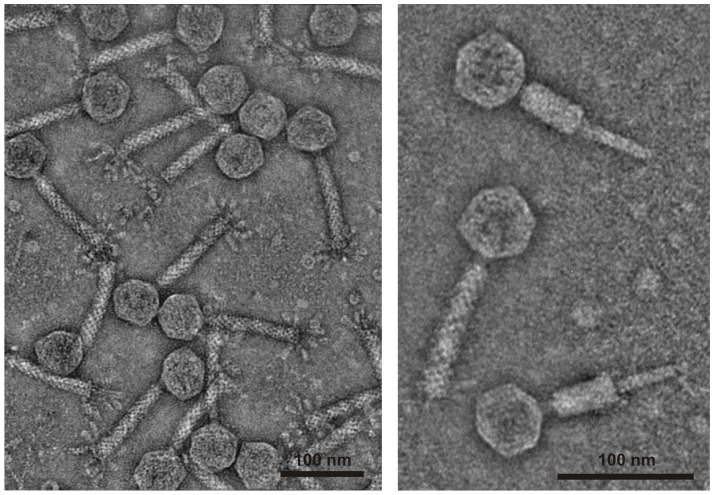
Phage PV94 was isolated from the V. vulnificus biotype 1 strain VN-094 by mitomycin C induction. Similar to other Vibrio strains, a very low concentration of the agent was optimal for phage release. PV94 particles revealed typical characteristics of a myovirus, an isometric head (56 to 58 nm in diameter) and a contractile tail (approximately 114 nm in length and 18 nm in width). At the end of the tail, a bundle of up to eight thick tail fiber-like structures was visible (Fig. 1). Although the virions appeared intact, no indicator strain suitable for phage propagation could be found. For this study 58 V. vulnificus and 25 V. cholerae strains, half of each clinical and environmental isolates, were tested by spot assays. To obtain high numbers of phages sufficient for purification by CsCl step gradients, large volumes of the host strain were induced with mitomycin C (see Materials and Methods). By this procedure a large amount of phage was isolated from which linear, double-stranded DNA was extracted.
Figure 1. Morphology of phage PV94.
Transmission electron micrographs of PV94 particles.
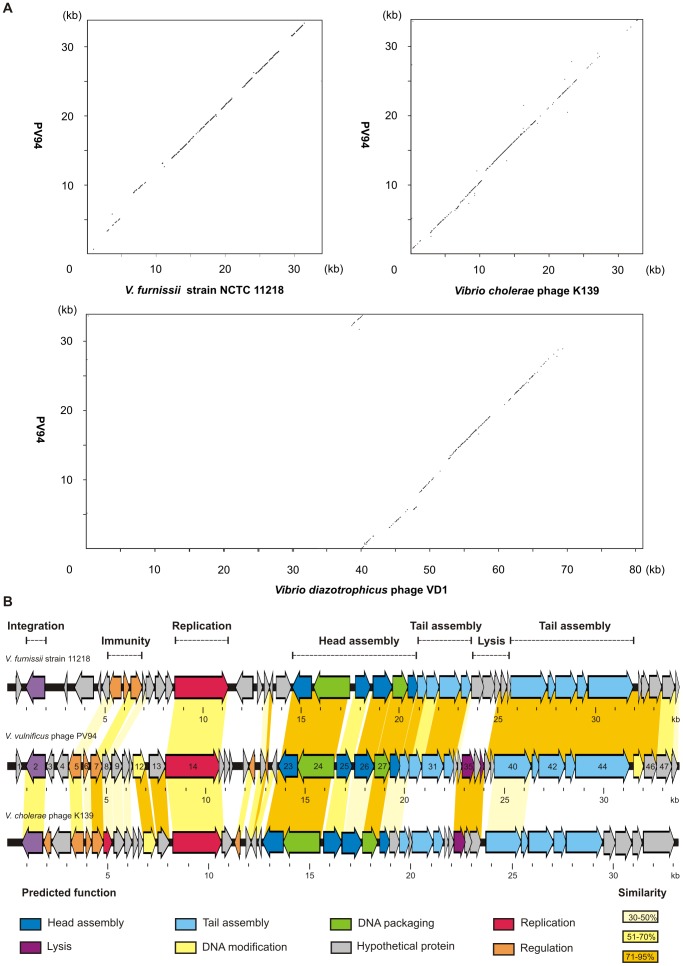
The PV94 genome consists of 33,828 bp with a G+C content of 48.2%, slightly higher than those of its host. We identified 48 possible Open Reading Frames (ORFs) of which 39 are located on one strand and the remaining nine on the second and 21 putative transcription terminators (Table S1 and Table S2). The strongest overall DNA homologies were detected to the V. cholerae phage K139 and related phages belonging to the kappa family, to the V. diazotrophicus phage VD1 and to a prophage residing in the genome of V. furnissii strain NCTC 11218 (Fig. 2A). In addition, PV94 shows some relationship to the Aeromonas media phage ΦO18P [23]. For 29 of the predicted PV94 gene products functional assignments could be made. Twenty-six and 28 products are similar to proteins of K139 and the prophage of V. furnissii NCTC 11218, respectively, with average identity values of 62.5 and 71.4% (Table S1). However, while the left part of the PV94 genome is more similar to K139, the genes of the right side are much more homologous to the V. furnissii prophage (Fig. 2B).
Figure 2. Relationship of PV94 to other Vibrio phages.
(A) Dot plots of the genome of PV94 with genomes of V. cholerae phage K139, V. diazotrophicus phage VD1 and a cryptic prophage of the V. furnissii strain NCTC 11218. The axes of abscissas and ordinates show the coordinates of the respective phage genomes. (B) Genome organization of the phages PV94, K139 and the prophage of V. furnissii strain NCTC 11218. Colours indicate the predicted functions of gene products. Related genes of the phages are connected by coloured shading.
As in these two phages, the PV94 integrase gene is located close to the left genome end. Yet, its product revealed only similarity to the K139 integrase and not to that of the V. furnissii prophage. Next to the integrase gene the PV94 genome harbors an immunity region which contains ORFs whose products are related to the prophage repressor CI, lytic repressor Cox and regulatory protein CII of other Vibrio phages. However, we did not detect a K139 glo-like gene that was suggested to encode a virulence-associated protein [9].
The main replication protein of PV94 is obviously encoded by ORF14. Its product shows significant homology (53 to 66% identity) to replication proteins of other Vibrio phages. Interestingly the protein is also related to protein A of P2 (33% identity) known to replicate by rolling circle replication [24]. Three conserved motifs (A, B and C) including two tyrosine residues within motif C which are part of the active site of the P2 protein A [25], also exist in the PV94 ORF14 product (Fig. 3). Moreover, the possible PV94 origin of replication (ori) which is nearly identical to the P2 ori located in the A gene [26] was found within ORF14. Though, a product similar to P2 protein B that is needed for lagging-strand synthesis during lytic replication [27] was not identified in PV94.
Figure 3. Comparison of the PV94 and P2 replication proteins and origins of replication.
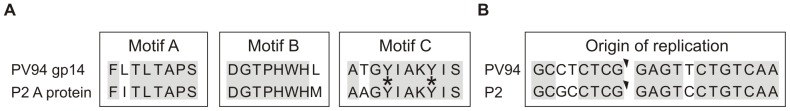
(A) Conserved motifs (A, B and C) of the replication proteins. The two tyrosine residues within motif C which are part of the active site of P2 protein A are marked by an asterisk. (B) Alignment of the P2 origin of replication located within gene A with the corresponding region of PV94 ORF14. Arrows indicate the cleavage site at the replication origin.
As with the replication proteins the probable PV94 large and small terminase subunits encoded by ORF24 and ORF27, respectively, are very similar (85 and 80% identity) to the respective proteins of K139 and they are also closely related to the terminase subunits of the V. furnissii NCTC 11218 prophage. In K139 terminal redundant ends diverging from the cos-ends of P2-like phages have been identified [9] even though the large terminase subunits of these phages fall into the same group [28]. We analysed the ends of the PV94 genome by restriction analyses and direct sequencing of phage DNA. As shown in Fig. 4A restriction fragments appeared in heated PV94 DNA that were absent or hardly visible in the unheated control digests. Upon treatment of the phage DNA with T4-ligase these additional fragments did not emerge (data not shown). Direct sequencing of phage DNA using primers that bound adjacent to the genomic ends confirmed the presence of 5′-overhangs 16 bp in length (Fig. 4B). This clearly showed that despite the close relationship of their terminases, phage PV94 and K139 possess different genome end structures. Thus, while K139 DNA might circularize after infection by recombination of its terminally redundant ends, in PV94 circularization is obviously accomplished by cos.
Figure 4. Analysis of the PV94 cohesive ends.
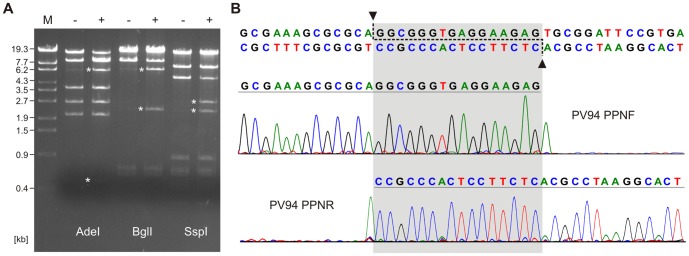
(A) AdeI, Bgll and SspI restriction patterns of PV94 phage DNA. Lane M, DNA size marker (λ DNA Eco130I), lanes (−), unheated phage DNA, lanes (+), phage DNA that had been heated before digestion. (B) Determination of protruding nucleotides. Chromatograms of run-off sequencing reactions using phage DNA as template and primers (PPNF and PPNR) binding close to the genomic ends. The 16 bp overhanging sequences are shaded and marked by arrows.
The PV94 terminase genes are embedded into a cluster of ORFs probably coding for head proteins (Fig. 2B). A portal protein (ORF23), a capsid scaffolding protein (ORF25), a major capsid protein (ORF26) and a head completion protein (ORF28) have been determined (Table S1), of which the major capsid protein has also been identified by MALDI-TOF (data not shown). Yet, it is striking that among the head proteins only the predicted portal protein is very similar (86% identity) to its K139 counterpart, whereas the other head proteins encoded by genes to the right showed only identity values between 30 to 63% (Fig 2B). Furthermore, most PV94 tail proteins whose genes (ORFs 29, 31, 32, 40, 42, 43, 44) are residing downstream from the predicted head assembly genes are only distantly or not related to K139 proteins. While the overall similarity of this part of the PV94 genome to K139 is rather low (exempt from the lysis gene cluster, see below), its similarity to the V. furnissii NCTC 11218 prophage is strong (Table S1). None of the identified head and tail proteins is less than 61%, most of them, however, more than 80% identical to the respective proteins of V. furnissii NCTC 11218. Unfortunately, there is no information available about the activity of the prophage in this strain. As mentioned above we could not find an indicator strain for PV94. Neither the morphology nor the gene content of the phage gave any clue for a possible defect. We found ORFs for structural tail proteins, a tape measure protein, and tail assembly proteins, not homologous but similar in size as their counterparts in K139. The tape measure protein (ORF40) and a baseplate assembly protein (ORF42) have also been identified by mass spectrometry (data not shown). Though, unlike with K139-related phages the predicted PV94 tail fiber protein did not show a mosaic-like structure supposed to determine the host range of V. cholerae phages [21]. Hence, it is likely that PV94 is highly specific with respect to its host or that the phage contains a yet unidentified mutation that prevents its infectiousness. Such a defect would probably not be caused by a mutation or lack of lysis genes as a cluster of ORFs whose products are closely related to the proposed phage lysin, holin and a lytic accessory protein of K139 is embedded in the tail gene region. It is striking that this short stretch of DNA shows significant homology to K139 but not to the V. furnissii NCTC 11218 prophage and further demonstrates the mosaic structure of the PV94 genome.
In conclusion this is the first description of a temperate myovirus of V. vulnificus. Phage PV94 did not reveal relationship to the V. vulnificus phages SSP002 and VvAW1 that have been recently described [12], [13]. Instead it is similar to temperate phages of other Vibrio species. The PV94 genome contains several clusters of genes responsible for specific functions, e.g. head and tail assembly, host cell lysis or the genetic switch for the lytic and lysogenic cycle. The modular organization of the PV94 genome and the varying homologies to other phages suggest recombinations to be responsible for this mosaicism. As most of the PV94 structural proteins are much more distantly related to proteins of K139-like phages than e.g. the terminase subunits or the replication protein, it is not surprising that PV94 did not infect V. cholerae strains. Why it also did not lyse V. vulnificus is still obscure. We checked the tested indicator strains for own prophages which may cause homoimmunity to PV94 but could not isolate any phage from these strains. Notably, we induced some prophages in other V. vulnificus strains which showed similar restriction patterns to PV94 (data not shown). Also for these phages to date suitable indicator bacteria have not be identified. So while prophages are obviously widespread in V. vulnificus, their host specificity and the role which they play for pathogenicity are still unclear. Even though a glo-like gene or other phage-associated virulence genes (e.g. for toxins, proteins that thwart the host defence, tissue invasion factors) have not been identified in PV94, the phage may contain genes which trigger the virulence of its host. Since for many PV94 gene products a functional assignment could not be made as yet, it can not be excluded that prophages may modulate the virulence also in V. vulnificus.
Nucleotide sequence accession number
The genome sequence of PV94 is available under Genbank accession number HG803181.
Supporting Information
ORF analysis of the PV94 genome.
(XLS)
PV94 transcription terminators.
(XLS)
Acknowledgments
We thank Maria Margarida Vargas for excellent technical assistance in the electron microscopical investigations.
Funding Statement
This work was supported by the Federal Ministry of Education and Research (VibrioNet, BMBF grant 01KI1015A). The work of JAH was supported by a grant of the Federal Ministry of Education and Research (SiLeBAT, project no. 13N11202). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Harwood VJ, Gandhi JP, Wright AC (2004) Methods for isolation and confirmation of Vibrio vulnificus from oysters and environmental sources: a review. J Microbiol Methods 59: 301–316. [DOI] [PubMed] [Google Scholar]
- 2.Gulig PA, Bourdage KL, Starks AM (2005) Molecular Pathogenesis of Vibrio vulnificus. J Microbiol 43 Spec No: 118–131. [PubMed]
- 3. Oliver JD (2013) Vibrio vulnificus: Death on the Half Shell. A Personal Journey with the Pathogen and its Ecology. Microb Ecol 65: 793–799. [DOI] [PubMed] [Google Scholar]
- 4. Daniels NA (2011) Vibrio vulnificus oysters: pearls and perils. Clin Infect Dis 52: 788–792. [DOI] [PubMed] [Google Scholar]
- 5. Baker-Austin C, Stockley L, Rangdale R, Martinez-Urtaza J (2010) Environmental occurrence and clinical impact of Vibrio vulnificus and Vibrio parahaemolyticus: A European perspective. Environmental Microbiology Reports 2: 7–18. [DOI] [PubMed] [Google Scholar]
- 6. Baker-Austin C, Trinanes JA, Taylor NGH, Hartnell R, Siitonen A, et al. (2012) Emerging Vibrio risk at high latitudes in response to ocean warming. Nature Clim Change 3: 73–77. [Google Scholar]
- 7. Wright AC, Simpson LM, Oliver JD, Morris JG Jr (1990) Phenotypic evaluation of acapsular transposon mutants of Vibrio vulnificus . Infect Immun 58: 1769–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim YR, Lee SE, Kook H, Yeom JA, Na HS, et al. (2008) Vibrio vulnificus RTX toxin kills host cells only after contact of the bacteria with host cells. Cell Microbiol 10: 848–862. [DOI] [PubMed] [Google Scholar]
- 9. Reidl J, Mekalanos JJ (1995) Characterization of Vibrio cholerae bacteriophage K139 and use of a novel mini-transposon to identify a phage-encoded virulence factor. Mol Microbiol 18: 685–701. [DOI] [PubMed] [Google Scholar]
- 10. DePaola A, Motes ML, Chan AM, Suttle CA (1998) Phages infecting Vibrio vulnificus are abundant and diverse in oysters (Crassostrea virginica) collected from the Gulf of Mexico. Appl Environ Microbiol 64: 346–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pelon W, Siebeling RJ, Simonson J, Luftig RB (1995) Isolation of bacteriophage infectious for Vibrio vulnificus . Curr Microbiol 30: 331–336. [DOI] [PubMed] [Google Scholar]
- 12. Lee HS, Choi S, Choi SH (2012) Complete genome sequence of Vibrio vulnificus bacteriophage SSP002. J Virol 86: 7711 86/14/7711 [pii];10.1128/JVI.00972-12 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nigro OD, Culley AI, Steward GF (2012) Complete genome sequence of bacteriophage VvAW1, which infects Vibrio vulnificus . Stand Genomic Sci 6: 415–426 10.4056/sigs.2846206 [doi];sigs.2846206 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Alanis VA, Kropinski AM, Abbasifar R, Abbasifar A, Griffiths MW (2012) Genome sequence of temperate Vibrio parahaemolyticus bacteriophage vB_VpaS_MAR10. J Virol 86: 13851–13852 86/24/13851 [pii];10.1128/JVI.02666-12 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sambrook J, Russel D (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- 16. Hertwig S, Klein I, Schmidt V, Beck S, Hammerl JA, et al. (2003) Sequence analysis of the genome of the temperate Yersinia enterocolitica phage PY54. J Mol Biol 331: 605–622. [DOI] [PubMed] [Google Scholar]
- 17.Strauch E, Voigt I, Broll H, Appel B (2000) Use of a plasmid of a Yersinia enterocolitica biogroup 1A strain for the construction of cloning vectors. J Biotechnol 79: : 63–72. S0168-1656(00)00216-9 [pii]. [DOI] [PubMed] [Google Scholar]
- 18. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brown CM, Dalphin ME, Stockwell PA, Tate WP (1993) The translational termination signal database. Nucleic Acids Res 21: 3119–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ermolaeva MD, Khalak HG, White O, Smith HO, Salzberg SL (2000) Prediction of transcription terminators in bacterial genomes. J Mol Biol 301: 27–33. [DOI] [PubMed] [Google Scholar]
- 21. Kapfhammer D, Blass J, Evers S, Reidl J (2002) Vibrio cholerae phage K139: complete genome sequence and comparative genomics of related phages. J Bacteriol 184: 6592–6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lux TM, Lee R, Love J (2011) Complete genome sequence of a free-living Vibrio furnissii sp. nov. strain (NCTC 11218). J Bacteriol 193: 1487–1488 JB.01512-10 [pii];10.1128/JB.01512-10 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Beilstein F, Dreiseikelmann B (2008) Temperate bacteriophage PhiO18P from an Aeromonas media isolate: characterization and complete genome sequence. Virology 373: 25–29 S0042-6822(07)00770-2 [pii];10.1016/j.virol.2007.11.016 [doi] [DOI] [PubMed] [Google Scholar]
- 24. Koonin EV, Ilyina TV (1993) Computer-assisted dissection of rolling circle DNA replication. Biosystems 30: 241–268. [DOI] [PubMed] [Google Scholar]
- 25. Odegrip R, Haggard-Ljungquist E (2001) The two active-site tyrosine residues of the a protein play non-equivalent roles during initiation of rolling circle replication of bacteriophage P2. J Mol Biol 308: 147–163 10.1006/jmbi.2001.4607 [doi];S0022-2836(01)94607-8 [pii] [DOI] [PubMed] [Google Scholar]
- 26. Liu Y, Haggard-Ljungquist E (1994) Studies of bacteriophage P2 DNA replication: localization of the cleavage site of the A protein. Nucleic Acids Res 22: 5204–5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Funnell BE, Inman RB (1983) Bacteriophage P2 DNA replication. Characterization of the requirement of the gene B protein in vivo. J Mol Biol 167: 311–334. [DOI] [PubMed] [Google Scholar]
- 28. Casjens SR, Gilcrease EB, Winn-Stapley DA, Schicklmaier P, Schmieger H, et al. (2005) The generalized transducing Salmonella bacteriophage ES18: complete genome sequence and DNA packaging strategy. J Bacteriol 187: 1091–1104 187/3/1091 [pii];10.1128/JB.187.3.1091-1104.2005 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ORF analysis of the PV94 genome.
(XLS)
PV94 transcription terminators.
(XLS)