Abstract
Myxococcus xanthus undergoes phase variation during growth to produce predominantly two colony phenotypes. The majority are yellow colonies containing swarm-proficient cells and a minority are tan colonies containing swarm-deficient cells. Comparison of the transcriptomes of a yellow variant, a tan variant, and three tan mutants led to the identification of differentially-regulated genes that define key segments of the phase variation pathway. For example, expression of genes for the yellow pigment DKxanthene and the antibiotic myxovirescin was increased significantly in yellow variants. In contrast, expression of the siderophore myxochelin, hemin binding proteins, and iron transport proteins was increased specifically in tan strains. Thus, a consequence of phase variation is that yellow cells shift from producing antibiotic and pigment to producing components involved in acquisition of iron, which may increase fitness during periods of iron limitation. Multiple protein kinases and HTH-Xre DNA-binding proteins identified in this study may be involved in the regulatory hierarchy that governs phase variation.
Introduction
Phase variation (PV) is a form of phenotypic plasticity that allows a single bacterial species to persist in alternate forms by expressing different sets of genes. Organisms that undergo phase variation alter the expression of various cellular components via a genetic [1] or epigenetic [2], [3] switch. Typically, the alternative programs result in production of different combinations of proteins, lipids, or carbohydrates on the cell surface that contribute to phase-specific changes in colony texture, color, or morphology [4].
Pathogenic strains that undergo PV produce cell types that are less likely to be recognized by the immune system, thus affording these pathogens enhanced opportunities to survive in the host. While human pathogenic strains have been the subject of intense research, non-human pathogens similarly benefit from phase switching. Photorhabdus luminescens produces M and P- morphotypes during phase variation. The P-form cells, produce larger colonies, more secondary metabolites, and generate more bioluminescence than the smaller M-form cells. The M form is critical for the life cycle of P. luminescens because it is the form that can persist in the nematode host Heterorhabditis bacteriophora [5].
M. xanthus is a nonpathogenic bacterium with a biphasic life cycle comprised of a vegetative phase and a complex developmental phase that together ensure survival in harsh environments. Multiple studies have documented PV in M. xanthus and its effects on vegetative growth, swarming, and development [6]–[10]. The predominant morphotype, or variant, in M. xanthus produces a rough, matte colony that is yellow due to production of the polyketide pigment DKxanthene (DKX). Yellow variants are proficient at swarming on agar surfaces. Colonies of the other morphotype are tan and generally have a smooth, shiny surface. Tan variants exhibit reduced swarming. The P. luminescens M and P-forms share features in common with the yellow and tan variants of M. xanthus including a property where the one form - the tan variant- can be ingested by bacteriophagous nematodes, while the other form – the yellow variant - is resistant to the nematode [10]. The ability to produce a variant that is resistant to a predator may enhance survival of M. xanthus in its soil habitat.
Yellow and tan colonies do not represent pure populations of either variant type, but rather are dynamic mixtures. Yellow colonies contain cells capable of giving rise to ≈95% yellow (range 75–99%) colonies and 5% tan colonies (range 1–25%); these ratios are reversed in the tan variants. The rate of switching from yellow to tan has been reported to be as high as 10−2 to 10−3 per cell per generation; switching from tan to yellow is higher, so colonies of the yellow variant outnumber those of the tan variant [11].
M. xanthus cells aggregate together to form fruiting bodies filled with heat and desiccation-resistant spores in response to starvation [12]. Cultures that are predominantly yellow form dark fruiting bodies within which roughly 10% of the cells differentiate into spores. Tan variants typically form immature mounds that are light in color because fewer than 0.1% of the initial cell population differentiates into spores. Although the tan variants produce few spores when developed on their own, they contribute disproportionately to the population of heat-resistant spores when allowed to develop in the presence of yellow variants [6], [11]. This suggests that yellow cells provide some factor that the tan cells need to produce viable spores. One factor needed by tan variants is DKX, the secondary metabolite that confers the characteristic color of the yellow variants [13]. Strains with mutations in the dkx biosynthetic operon are tan in color and are deficient in their ability to produce heat-resistant spores. Addition of purified DKX partially rescued the sporulation defect of the dkx mutants [13].
Here we compare the transcriptome of the wild-type tan variant (WT-T) with that of three tan mutants. We identified >200 genes whose expression was increased or decreased at least 7.3 fold in two or more of the strains and found multiple components that can serve as biomarkers for PV. Despite their distinct genetic backgrounds, all four tan strains showed significant increases in expression of genes whose products are needed for acquisition of iron and most showed significant decreases in expression of genes for synthesis of DKX and an antibiotic named myxovirescin (also known as TA). Bioassays confirmed that changes in transcription correlated with changes in the amount of DKX, myxovirescin, and the siderophore myxochelin. Sets of genes encoding serine-threonine protein kinases and helix-turn-helix xenobiotic response elements (HTH-Xre) were up-regulated in tan variants. Results suggest that some of these genes act downstream of the HTH-Xre228 protein, which previously was found to regulate expression of dkx genes [9].
Results
Phase variation affects multiple phenotypes
Phase variation in M. xanthus is pleiotropic: phenotypes including pigment and antibiotic production, swarming, and development are affected. To identify genes and regulatory processes that contribute to these phenotypes, the transcriptomes and behaviors of four tan strains were compared with the yellow WT parent (WT-Y). The tan strains included the wild-type tan variant (WT-T) and three mutants. Two of the tan mutants, xre228 (MXAN_0228) and asgB (MXAN_2913), were chosen because they carry mutations in putative or known regulatory genes. The xre0228 gene encodes a putative member of the HTH-XRE superfamily that is differentially regulated in yellow and tan variants [9]. The asgB gene encodes a 163 amino acid DNA binding protein whose C-terminal region shares similarity with region 4 of the sigma 70 protein family and the HTH-XRE superfamily [14]. The mutant has a profound developmental defect [15]. The asgB mutant constructed for this study was initially yellow but within a few weeks, it was tan and did not revert to yellow. The third tan mutant, dkxG (MXAN_4299::Himar), was chosen as it carries a transposon mutation in a gene that encodes a polyketide synthase required for DKX biosynthesis [9]. The dkxG mutant is tan due to loss of pigment.
Cell cohesion (aka agglutination, aggregation) has been used to differentiate strains on the basis of surface components, particularly fibrils [16], [17]. Loss of fibril material occurs in a subset of mutants, such as dsp/dif, that are defective in social gliding [18]. We compared the cohesive properties of WT-Y against the four tan strains. The WT-Y strain was proficient for cohesion and precipitated from solution (Fig S1). Two of the tan mutants, dkx and xre228 aggregated as well as or better than the WT-Y, respectively. In contrast, the WT-T and asgB mutant strains remained suspended with little change in optical density associated with settling of aggregated cells (Fig S1).
A hallmark of M. xanthus is its ability to form spore-filled multicellular fruiting bodies in nature. In the laboratory, this complex development cycle can be induced and synchronized by removing nutrients from a growing population of cells and simultaneously concentrating the cells to achieve a high cell density [19]. Development can ensue when concentrated, starved cells are placed on a solid surface devoid of nutrients. The WT-Y strain formed dense mounds after 48 hr whereas WT-T formed only soft mounds or ridges. With additional time some of the mounds produced by WT-T cultures darkened slightly but never achieved the dark, dense fruits of the WT-Y. The dkx and asgB tan mutants showed significant delays in fruiting or failed to develop (data not shown, [13], [14]. The xre228 mutant produced near normal fruiting bodies, but the number of viable heat-resistant spores was reduced [9].
Expression of myxochelin and iron acquisition systems is increased in tan variants and mutants
The expression profiles of the WT-T and tan mutants were compared to the WT-Y reference strain variant using a custom DNA tiling array and RT-PCR. These data are summarized in Table 1; data for individual genes are listed in Table S1. Expression of genes involved in acquisition and transport of iron was increased significantly in tan strains. Fig 1 shows the likely relationship between the products of these genes. Expression of all six genes of the regulon encoding hemin transport proteins (MXAN_1316-1321) was increased >5-fold in all four strains. The nine genes of the myxochelin siderophore biosynthesis pathway (mxc, MXAN_3639-3647), which share homology with the genes characterized for myxochelin synthesis and transport from Sorangium [20] increased 5 to 55-fold.
Table 1. Expression of genes for iron acquisition is increased in tan strains.
| Fold change* in | |||||
| MXAN ID | Description | WT-T | asgB | dkxG | xre228 |
| 1316 to 1321 | Genes for hemin transport system | +6 to +13 | +8 to +43 | +6 to +16 | +3 to +6 |
| 1688 to 1690 | TonB family protein, CHP, esterase | +6 | +20 | +16 | +5 |
| 3639 to 3647 | Genes for myxochelin biosynthesis (mxc) | +8 to +55 | +15 to +46 | +18 to +48 | +8 to +17 |
| 4198 to 4201 | Genes for putative OM macrolide efflux, ABC protein family | +75 to +119 | +15 to +35 | +216 to +412 | +60 to +137 |
| 6000 | Fe•ABC periplasmic iron compound-binding transporter | +10 | +17 | +10 | +7 |
| 6911 | TonB-dependent receptor | +61 | +278 | +39 | +6 |
Legend: * for multigene clusters, the range of expression change values is given.
Partial list of genes showing significant expression changes in the M. xanthus tan variant and three tan mutants (12 datasets: four biological replicates with triplicate technical replicates) in DNA tiling array studies relative to the WT-Y variant (data not shown). Expression patterns consistent in four different tan M. xanthus strains (data for individual genes are provided in Table S1). MXAN ID and Description refers to annotation; both are derived from the M. xanthus genome [43]; fold change represents the increase (A) or decrease (B) in expression and is derived from coefficient value.
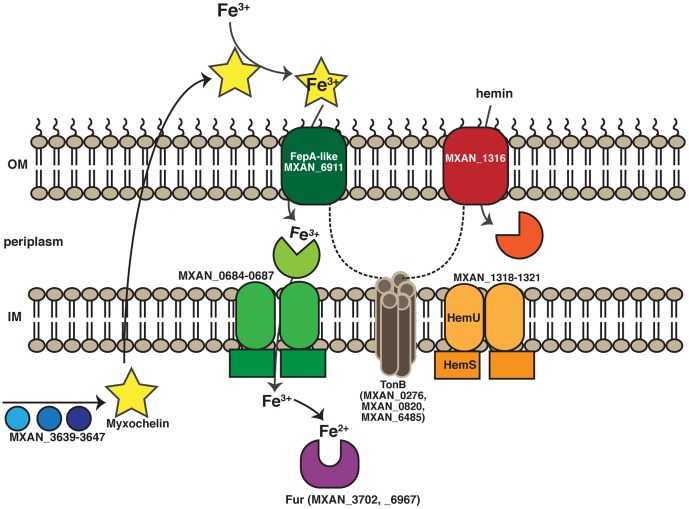
Figure 1. Tan strains activate pathways dedicated to acquisition of iron.
Many of the genes whose expression is increased significantly in the tan strains comprise the siderophore production and transport, hemin transport, and proteins that provide energy for transport of iron-binding compounds. A partial list of components produced in tan cells is shown in this cartoon. The blue/purple circles represent the siderophore myxochelin pathway (MXAN_3639-3647) [39], [40]. Myxochelin (mxc) is shown as a yellow star that binds Fe3+ when exported. Mxc•Fe3+ may be imported by a FepA-like protein (candidate is MXAN_6911; dark green shape), located in the outer membrane [41]. Light green shapes represent the ferric siderophore ABC transporter genes (permease and MXAN_0684-0687 in membrane); brown cylinders represent periplasmic energy transduction TonB proteins (MXAN_0276, MXAN0820, and 6485) that may energize the outer membrane proteins (dashed lines). Red/orange icons represent hemin transporters (MXAN_1318-1321). Not shown are RhtX/FptX siderophore transporter (MXAN_5357), iron ABC transporters (MXAN_0770-0772), iron compound ABC transporter, and a periplasmic iron compound binding protein, Fe•ABC (MXAN_6000). Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) software [42] predicts MXAN_3639 (putative iron-chelator utilization protein in the myxochelin operon) interacts with the ferric siderophore ABC transporter permease (MXAN_0685, 0686).
Many Gram-negative bacteria produce a protein named Fur (ferric uptake regulator) that uses iron (Fe2+) as a cofactor to regulate expression of genes involved in production of siderophores and even toxins [21]. Expression of two Fur-like homologs, MXAN_3702 and MXAN_6967 that share 36% (47/129) and 30% (37/125) identity with the E. coli Fur protein (EHU14036) respectively was increased in tan variants. Membrane components involved in transport of siderophores, including a putative Fe-ABC periplasmic iron compound binding protein transporter, a FepA homolog (TonB-dependent outer membrane receptor), and a TonB-family protein that likely energizes an outer membrane receptor were increased in tan strains.
Siderophore production is increased in tan strains
The results above showed that expression of genes whose products are needed for iron acquisition is increased in tan strains. To determine if increases in transcription correlated with actual changes in production of iron-binding components, levels of siderophore in the WT-Y, WT-T, and mutant strains were assayed. M. xanthus produces a catecholate type of siderophore named myxochelin [20] which was assayed using chrome azurol S (CAS) [22]. When aliquots of 107 cells were spotted on CAS-Fe agar medium, tan M. xanthus strains produced an orange halo that was 2-fold greater (range 1.8 to 2.4-fold increase, n = 8) than that of a yellow strain (Fig S2). The orange halo is produced because the high affinity siderophore secreted by cells removes iron from the CAS-Fe complex, resulting in conversion of the blue CAS-Fe color to the orange CAS color. This plate assay uses a CAS-Fe complex containing the detergent hexadecyltrimethylammonium bromide (HDTMA) to observe production of a halo around individual colonies as they grow.
To ensure that the difference in the siderophore zone was not an artifact caused by increased sensitivity of tan strains to the detergent, we compared the level of siderophore in the spent medium (supernatant) and cell extract of broth-grown cultures. The siderophore units were calculated from the absorbance at 630 nm of cell-free supernatants of WT-Y, WT-T and mutant cultures incubated with CAS-Fe solution [22]. To facilitate direct comparison, samples were harvested when the cultures were at the same cell density (≈5×108 cells ml−1; mid-log phase of growth).
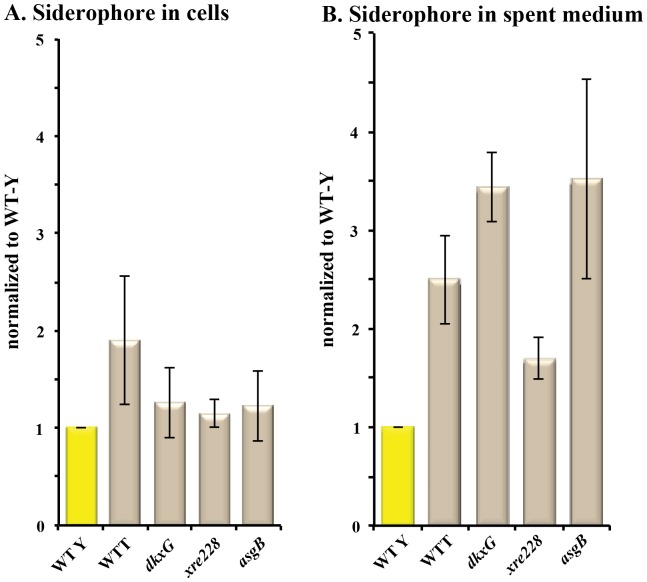
All of the tan strains secreted more siderophore than the WT-Y strain. As shown in Fig 2B, the amount of siderophore detected in the spent medium of the tan strains increased 175% to 350% compared with the WT-Y strain. The abundance of siderophore in the supernatant was not due to leakage of tan strains because the amount of siderophore in the cell pellet, lysed in 1% NP-40 detergent to release bound siderophore, was similar in the tan strains and the WT-Y (Fig 2A). The increased amount of siderophore released by tan strains is consistent with the increased expression of myxochelin genes.
Figure 2. Tan mutants produce and secrete more siderophore.
Chrome azurol S assays were performed as described in Methods. Yellow bars signify the yellow strain; tan bars signify tan strains. (A) The amount of siderophore in 2×108 whole cells after lysis is reported as siderophore units relative to the WT-Y strain (100%). (B) Culture supernate (spent medium) was taken when the density was at ≈5×108 cells ml−1 and was used to quantify the amount of released siderophore. Samples represent the average of three independent assays.
Iron plays a role in regulation of DKX and siderophore
The increase in iron acquisition components in tan cells suggested that phase variation may be a response to iron limitation. To explore the role of iron in phase variation, the effects of iron availability on levels of the PV biomarkers DKX and siderophore were compared. WT-Y and WT-T cells were inoculated into separate batches of rich medium (casitone Tris, phosphate, Mg2+; CTPM) and split into four 12 ml batches, which were supplemented with H2O, FeCl3, dipyridyl, and CuCl2. Aliquots (5 ml) from each batch were transferred to Klett flasks and incubated at 32°C with shaking. Cells and supernates were harvested when the density reached 6×108 cells ml−1.
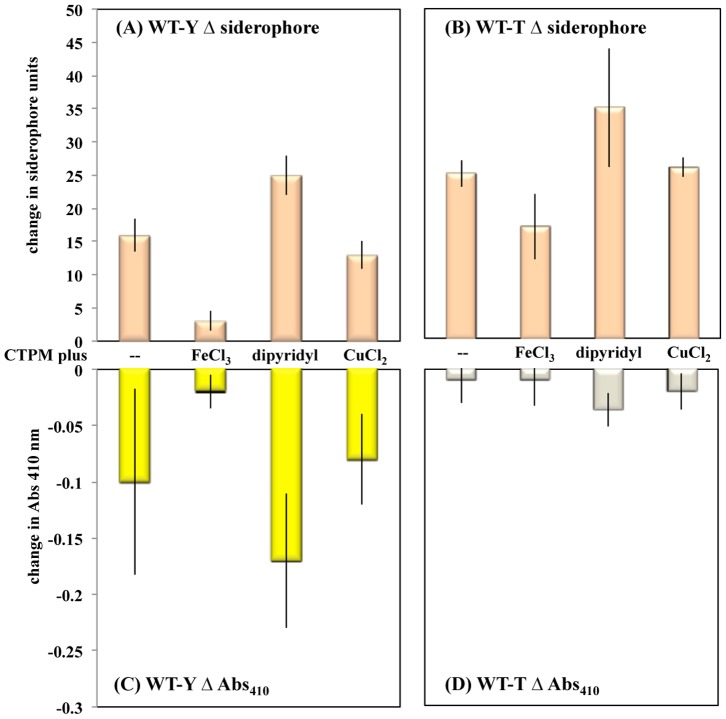
Fig 3 shows that siderophore levels increased and relative DKX levels (estimated from Abs410) decreased in all samples compared with those from the inoculum (4×108 cell ml−1). The changes detected in WT-Y cells growing in CTPM are consistent with a decrease in the ratio of yellow to tan cells in a WT population as it ages. Siderophore (Fig 3A) and DKX (Fig 3C) levels were stable in the presence of FeCl3 compared with the CTPM control. Accumulation of fewer tan variant cells in the population could account for the smaller changes in siderophore and DKX. In contrast, dipyridyl stimulated siderophore production 8X and decreased the relative DKX value 8.5X compared with FeCl3. CuCl2 did not have the same effect as FeCl3, which shows that the response is specific to iron.
Figure 3. Effect of iron on production of PV molecules.
Parallel cultures of WT-Y and WT-T strains were grown with and without supplements as described in Methods; siderophore and Abs410 were assayed when cells reached 6×108 cells ml−1. Data represent the average of duplicate readings obtained after performing three independent tests comparing the endpoint against the cells used as inoculate. (A) change in siderophore WT-Y, (B) change in siderophore WT-T, (C) change in Abs 410 WT-Y, and (D) change in Abs 410 WT-T.
The response of WT-T cells to different concentrations of iron in growth medium followed the same trend as WT-Y, but was less dramatic. Levels of siderophore are elevated in tan variants compared with yellow variants and added iron resulted in only a small decrease compared with the CTPM control (Fig 3B). As expected, the change in DKX (Fig 3D) was minimal. Similar results were seen on solid medium. Colonies of WT-Y on plates containing 0.5 mM FeCl3 remained yellow for more than a week, while colonies on standard medium began to turn brown after the same time period (data not shown). Brown coloration is thought to be due to secretion of the myxochelin siderophore and other compounds.
The results suggest that (1) the switch from yellow to tan variant is delayed significantly by high levels of iron and accelerated by removal of iron and (2) iron does not promote a switch from tan to yellow. With regard to the latter, WT-T cells may have a reduced ability to sense iron.
Addition of iron suppresses the yellow to tan conversion
To test the hypothesis that high levels of iron impede the yellow to tan switch, we assayed the number of yellow and tan colonies recovered on plates after first growing WT-Y cells in medium with and without added iron. WT-Y cells, grown to the same density (5×108 ml−1) in CTPM and CTPM+1 mM FeCl3 yielded 94% (n = 341) and 98% (n = 449) yellow colonies respectively when plated on standard CTPM medium. When the iron-grown cells were plated on CTPM plus FeCl3, 100% of the colonies that arose were yellow (n = 312). In contrast, when cells were plated on CTPM with 25 µM dipyridyl, 59% of the colonies were yellow (n = 380).
To rule out the possibility that dipyridyl-induced lethality of yellow cells was skewing results, a parallel experiment was performed using mixtures of marked and unmarked cells. Equal numbers of KanR tan and KanS yellow variants were mixed and plated on CTPM medium with 25 µM dipyridyl. After 6 days, tan colonies (n = 69) were replica-plated to CTPM and CTPM+Kan media. 100% of colonies (n = 69) grew on CTPM medium; 31 of these also grew on CTPM+Kan. Hence more than half (38/69) of the colonies originated from the yellow WT parent. Indeed, after transfer from CTPM+dipyridyl back to CTPM, 92% of the recovered KanS colonies (35/38) were yellow. This shows that dipyridyl does not selectively kill yellow variants.
To determine if other elements could elicit the same response as iron in preventing the yellow to tan switch, an add-back experiment was performed. WT-Y cells were plated in parallel on CTPM medium with 25 µM dipyridyl plus (a) no addition, (b) 200 µM FeCl3, (c) 200 µM CuCl2, and (d) 200 µM ZnCl2. We found (a) 41% yellow (88/210), (b) 78% yellow (147/188), (c) 56% yellow (149/267) and (d) 49% yellow (114/233). Hence, addition of FeCl3 largely rescued or prevented the yellow to tan switch in the population; copper and zinc provided some protection.
Iron represses expression of genes encoding myxochelin biosynthetic proteins and a FepA-like protein
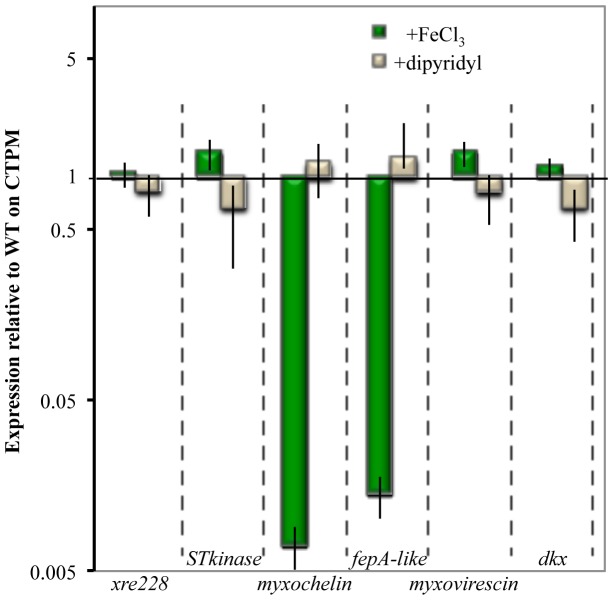
The results shown in Fig 3 showed that different concentrations of iron can affect DKX and siderophore production. To determine if similar changes would affect gene expression, we used RT-PCR to compare the expression of a subset of genes from WT-Y cells grown on CTPM, CTPM+FeCl3 and CTPM+dipyridyl. Fig 4 shows the relative changes in expression of genes in cells grown in CTPM+iron and CTPM+dipyridyl (reduced iron) relative to the CTPM control. Expression of DKX, myxovirescin (Mxv), and ST-kinase (MXAN_7370) was increased in the presence of iron and decreased when iron was removed. The opposite was true for MXAN_3641 (myxochelin) and MXAN_6911 (FepA homolog), both of which are involved in iron acquisition.
Figure 4. Regulation of gene expression by iron.
RNA harvested from WT-Y cells was used to prepare cDNA for qRT-PCR analysis of representative genes whose expression is affected during phase variation; MXAN_0228 (Xre228), MXAN_7370 (ST_kinase), MXAN_3641 (myxochelin), MXAN_6911 (FepA homolog), MXAN_3639 (myxovirescin) and MXAN_4305 (DKX). The results represent three biological and technical replicates; data are from cells grown in CTPM+FeCl3 and CTPM+dipyridyl medium relative to levels from cells grown in CTPM alone (value = 1) are presented.
Data from independent experiments presented thus far show that iron influences gene expression and production of compounds that are linked to PV in M. xanthus. With regard to gene expression and phenotype, depletion of iron is sufficient to phenocopy WT-T in an otherwise WT-Y strain.
Production of DKxanthene and myxovirescin (antibiotic TA) is decreased in tan mutants
The species name for M. xanthus is from the Latin word for yellow, the color of the compound DKxanthene. Loss of pigment in the tan strains was due to a significant decrease in expression of dkx genes (MXAN_4289-4305) in WT-T [9], and the asgB and xre228 mutants (Table 2 and Table S1). Expression of the operon containing dkxG is decreased in the tan dkxG mutant, but expression of genes in other dkx operons was not affected.
Table 2. Expression of DKxanthene and myxovirescin (antibiotic TA) is decreased in tan strains.
| Fold change* in | |||||
| MXAN ID | Description | WT-T | asgB | dkxG | xre228 |
| 3930 to 3950 | Genes for Mxv synthesis | −5 to −208 | −6 to −195 | −1 to −6 | −5 to −176 |
| 4289 to 4305 | Genes for DKX synthesis | −19 to −316 | −21 to −584 | −1 to −59 | −28 to −295 |
*For multigene clusters, the range of expression change values is given.
Legend: See Table 1 legend.
Yellow variants secrete a secondary metabolite called myxovirescin, an inhibitor of type II signal peptidase, to prey on other organisms [23], [24]. Expression of genes involved in synthesis of myxovirescin (mxv; MXAN_3930-3950)(Table 2) was decreased significantly in the WT-T, xre228, and asgB strains but decreased only slightly in the dkxG mutant. Mxv can be detected using a bioassay that measures inhibition of E. coli growth [24]. A zone of inhibition, due to secretion of Mxv, was detected for WT-Y (Fig 5A), but was absent in the WT-T variant (Fig 5B), consistent with the transcriptome data. Similarly, as shown in Fig. 5C, the bioassay confirmed the loss of Mxv in the asgB mutant and presence of Mxv in the dkxG mutant. Surprisingly, although we did not detect mxv transcript in the xre228 mutant, the mutant produced a zone of inhibition (Fig 5C). The zone of clearing was visible around the WT-Y strain 18 hr after overlaying with E. coli while the zone of clearing around the xre228 mutant was visible only after 30 hrs. This suggests that expression of mxv may be delayed in the xre228 mutant or is induced by contact with a solid surface, high cell density, or by exposure to E. coli. Delayed expression would explain why mxv transcript was not detected in the xre228 mutant. Alternatively, an unidentified E. coli inhibitor may be expressed in the xre228 mutant.
Figure 5. WT-T strains do not produce myxovirescin.
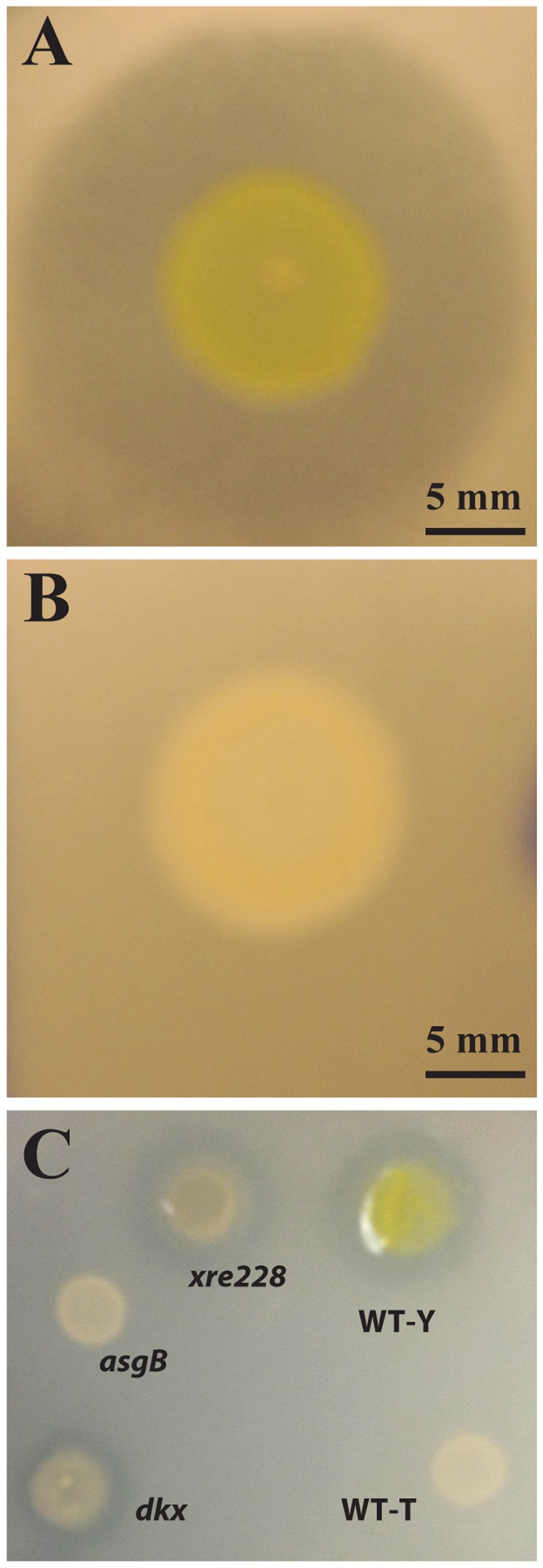
Production of myxovirescin, also known as antibiotic TA, was monitored using a zone of inhibition assay with E. coli DH5α. Myxovirescin was produced by WT-Y variants (panel A), but not WT-T variant (B). (C) Like WT-T, myxovirescin was absent in asgB, but present in dkxG and xre228 mutants. Production was delayed in xre228.
Differences between WT-T and tan mutants: protein kinases and Xre-type DNA-binding proteins
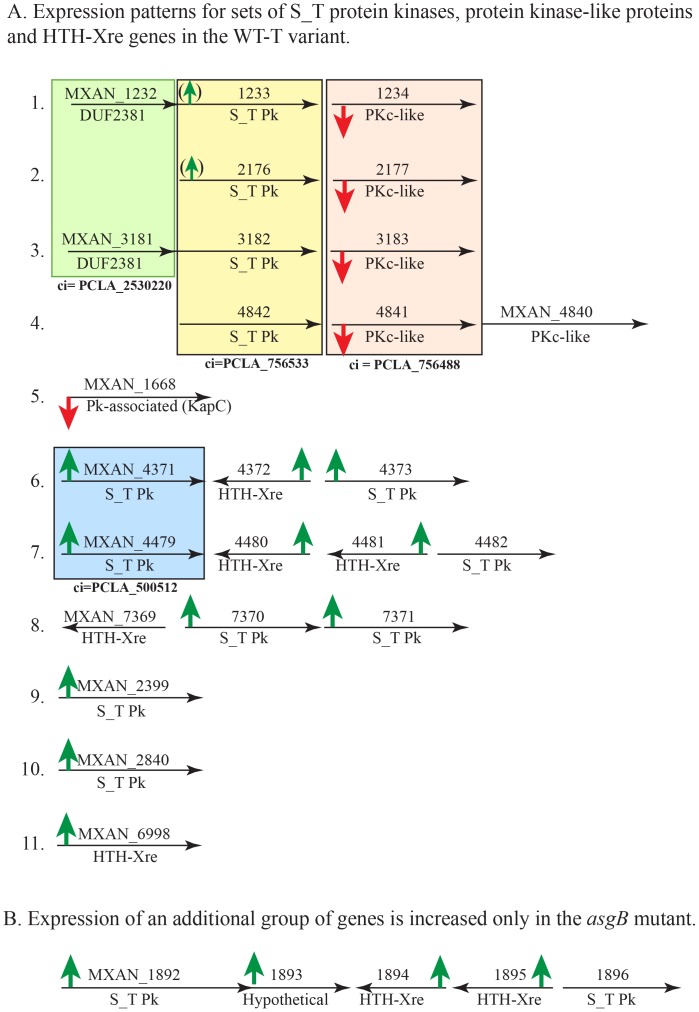
While the expression profiles of the asgB, xre228 and dkxG mutants resemble that of the WT-T variant with regard to genes for iron acquisition, pigment production and antibiotic, other changes in gene expression reveal patterns that highlight differences between these strains. The genome of M. xanthus DK1622 contains over 50 genes annotated as serine-threonine protein kinases (S_TPk) or ST_kinase-like proteins (PKc-like) and as many as 24 genes encoding HTH-Xre DNA binding proteins. When we compared the transcriptomes of tan strains, a pattern emerged for a significant subset of the kinase and HTH-Xre-encoding genes. The data are presented in Table 3; the genomic context for these genes is shown in Fig 6.
Table 3. Clusters of ST_ kinases, PKc-like proteins and HTH-Xre players show altered expression during phase variation.
| A. Genes differentially regulated in two or more tan strains | |||||
| Fold change | |||||
| MXAN ID | Description | WT-T | asgB | dkxG | xre228 |
| 1233 | serine-threonine kinase | +2 | +3 | −3 | +1 |
| 1234 | protein kinase-like (PKc-like) | −41 | −55 | −1 | −12 |
| 1668 | serine-threonine kinase associated protein KapC | −8 | −16 | +2 | +2 |
| 2177 | protein kinase-like (PKc-like) | −35 | −62 | +1 | −24 |
| 2399 | serine-threonine kinase | +11 | +4 | −1 | +5 |
| 2840 | serine-threonine kinase | +57 | +32 | −1 | +1 |
| 3183 | protein kinase like (PKc-like) | −37 | −60 | +1 | −20 |
| 4371 | serine-threonine kinase | +132 | +293 | −3 | −15 |
| 4372 | HTH-Xre DNA-binding protein | +96 | +337 | −1 | −3 |
| 4373 | serine-threonine kinase | +9 | +9 | 1 | −1 |
| 4479 | serine-threonine kinase like | +43 | +42 | −5 | −6 |
| 4480 | HTH- Xre DNA-binding protein | +26 | +51 | −1 | −1 |
| 4481 | HTH- Xre DNA-binding protein | +31 | +50 | +1 | −2 |
| 4841 | protein kinase like (PKc-like) | −46 | −72 | +1 | −17 |
| 6998 | HTH- Xre DNA-binding protein | +109 | +125 | +2 | +1 |
| 7370 | serine-threonine kinase | +187 | +293 | +2 | +2 |
| 7371 | serine-threonine kinase | +99 | +116 | +10 | +20 |
Legend: A description of the microarray is provided in Table 1 legend. Column headings: ID = MXAN # and description refers to annotation; both are derived from the M. xanthus genome [43]; fold change represents the increase or decrease in expression and is derived from coefficient value. + = increased expression; - = decreased expression.
Figure 6. Expression of regulators and signal transduction proteins show phase-specific behavior.
(A1-40 Four genes encoding nearly identical protein kinase-like proteins are down-regulated in WT-T variants. In each case, the PKc-like gene is downstream of a gene annotated as a serine-threonine protein kinase (S_T Pk). The PKc-like genes that share significant similarity with serine-threonine protein kinases. (A5) A putative kapC gene (KapC = protein kinase associated protein) is down-regulated in WT-T. (A6–10) Expression of multiple S_T Pk-encoding genes and HTH-Xre-encoding genes (helix-turn-helix xenobiotic response element family of DNA-binding proteins) is increased in WT-T relative to WT-Y strains. Four colored boxes indicate genes whose products share sufficient identity to be considered a protein cluster (ci = cluster index; NCBI protein cluster database PCLA designation). (B) The asgB mutant behaves similar to the WT-T with regard to genes in A, but differs from WT-T in that it also shows significant increases in four additional genes predicted to encode a S_T Pk, a hypothetical and two HTH-Xre proteins.
Expression of seven S_TPk, five xre genes, four PKc-like, and one kap (kinase associated protein) gene showed significant increases or decreases in the WT-T strain and some tan mutants. Expression of four protein kinase C-like proteins (PKc-like), MXAN_1234, 2177, 3183 and 4841, was decreased 41-, 35-, 37- and 46-fold, respectively in the WT-T variant compared with the WT-Y. Each of these genes encodes a member of the protein kinase-like superfamily (cl09925) and contains the catalytic domain of serine-threonine kinases. These four PKc-like proteins are in the PLCA_756488 identity cluster (Fig 6 pink box); they share >80% identity over the entire length (596–597 residues). Expression of all four PKc-like genes was decreased in the asgB and xre228 mutants, but not in the tan dkxG mutant.
It is significant to note that the genomic context of these genes is very similar, which suggest that gene duplication occurred at some point. Each PKc-like gene is immediately downstream of a gene encoding a serine-threonine protein kinase whose expression was virtually unchanged in the tan strains. The four S-TPk proteins share significant identity (protein cluster 756533; Fig 6 yellow box). The sets of genes diagramed in Fig 6 (1–4), follow the pattern serine-threonine kinase (S_T Pk) and protein kinase-like (Pkc), respectively. In two cases, there is an additional, related gene upstream (Fig 6, green box). Expression of MXAN_1668, encoding a serine-threonine kinase associate protein (KapC; Fig 6 line 5), was decreased in WT-T and the asgB mutant, but was increased slightly in the dkxG and xre228 mutants.
In contrast, expression was increased significantly in WT-T and the asgB mutant for seven serine-threonine protein kinase (S_T Pk)-encoding genes and five HTH-Xre encoding genes (Fig 6, 6–11). In some cases, the S_T Pk genes are adjacent to the HTH-Xre genes. As a rule, expression levels were higher in the asgB mutant than in the WT-T strain (Table 3). Except for MXAN_7371, expression of the S_T Pk and HTH-Xre genes in dkxG and xre228 strains was unchanged (value = 1), increased slightly (MXAN_6998) or even decreased (MXAN_4371 and 4479) which highlights some major distinctions between the tan strains.
Expression of four additional genes, related to the group described above, showed an increase unique to the asgB mutant. As shown in Fig 6B and Table 3B, expression of MXAN_1892 (S_T Pk), 1893 (hypothetical), 1894 (HTH-Xre) and 1895 (HTH-Xre) was up-regulated 40 to 350-fold in the asgB mutant whereas the other three tan strains showed no change or a slight decrease in expression.
The expression profile of genes other than those in Tables 1, 2, and 3 is listed in Table 4. These genes fell into four groups: (A) those affected in WT-T asgB and xre228, (B) those affected in WT-T and asgB, (C) those affected in asgB, dkxG, and/or xre228, but not in WT-T, (D) those affected in asgB only and (E) those showing the opposite response in one or more tan strain. Group A includes several hypotheticals and acyl carrier protein. Group B (WT-T and asgB) includes hypotheticals, bacterial leucyl aminopeptidase and a cytochrome P450 family protein. Most of the genes in Group C, including a lipoprotein and phosphatase, were up-regulated significantly in asgB and to a lesser extent, in dkxG.
Table 4. Outliers: a subset of genes show differential expression profiles that may help distinguish pathways in tan strains.
| MXAN ID | Description | WT-T | asgB | dkx | xre228 |
| Group A: changes in WT-T, asgB and xre228 | |||||
| 3949 | acyl carrier protein | −235 | −172 | −2 | −112 |
| 1697 | conserved hypothetical protein | −51 | −130 | −3 | −65 |
| 3434 | hypothetical protein | −23 | −110 | −2 | −40 |
| 5266 | hypothetical protein | −23 | −64 | −3 | −27 |
| Group B: changes in WT-T and asgB | |||||
| 0519 | conserved hypothetical protein | +10 | +19 | +1 | +1 |
| 4985 | hypothetical protein | +5 | +55 | +1 | +1 |
| 5166 | bacterial leucyl aminopeptidase | −23 | −67 | −1 | −1 |
| 6991 | hypothetical protein | −12 | −49 | −1 | −2 |
| 7298 | cytochrome P450 family protein | −8 | −50 | −1 | −2 |
| Group C: changes in asgB, dkx , and/or xre228 but not in WT-T | |||||
| 0502 | transcriptional regulator, AsnC family | +1 | +1 | +7 | +12 |
| 1238 | conserved hypothetical protein | +1 | +97 | +6 | +1 |
| 1485 | putative cytochrome c | +1 | +70 | +7 | +1 |
| 1486 | HNH endonuclease domain protein | +1 | +114 | +8 | +1 |
| 1499 | ThiF domain protein | +1 | +21 | +4 | +1 |
| 1508 | hypothetical protein | +1 | +57 | +9 | +1 |
| 1509 | Ser-Thr protein phosphatase family protein | +1 | +92 | +6 | +1 |
| 3166 | hypothetical protein | +1 | +73 | +5 | +2 |
| 5986 | putative lipoprotein | +1 | +77 | +21 | +11 |
| 6350 | hypothetical protein | +1 | +35 | +4 | +1 |
| Group D: changes in asgB only | |||||
| 0383 | hypothetical protein; C-terminal catalytic domain BreC (break-rejoining enzymes) | +1 | +124 | +3 | +1 |
| 0384 | quaternary ammonium compound-resistance protein SugE (sugE) | +2 | +93 | +3 | +3 |
| 0455 | conserved domain protein | +1 | +35 | +1 | +1 |
| 1894 | DNA-binding protein | +1 | +46 | +1 | +1 |
| 2912 | hypothetical protein | +2 | +48 | +2 | +1 |
| 5350 | DNA repair protein RecN (recN) | +2 | +23 | +1 | +1 |
| 6264 | conserved hypothetical protein | +2 | +27 | +1 | +1 |
| 7217 | hypothetical protein (BIg domain) | +5 | +2383 | +2 | +5 |
| 7219 | hypothetical protein (BIg domain) | +2 | +1146 | +7 | +5 |
| 7222 | kelch domain protein | +2 | +12 | +2 | +1 |
| Group E: Opposite response in different tan strains | |||||
| 0046 | hypothetical protein | +10 | +11 | −3 | +1 |
| 0047 | hypothetical protein | +9 | +15 | −3 | +1 |
| 0048 | hypothetical protein | +17 | +33 | −2 | +1 |
| 0049 | hypothetical protein | +11 | +22 | −1 | +1 |
| 0050 | hypothetical protein | +11 | +12 | −1 | +2 |
| 0201 | hydrolase, alpha-beta fold family | +71 | +50 | −2 | +10 |
| 0503 | DnaJ domain protein, authentic frameshift | −7 | −133 | +2 | +8 |
| 0504 | hypothetical protein | −12 | −171 | +2 | +7 |
| 0587 | trypsin domain protein | −8 | −144 | −1 | +1 |
| 0947 | RNA polymerase sigma-70 factor, ECF subfamily | +1 | +11 | −2 | −3 |
| 1164 | GAF domain protein | −10 | −176 | +1 | −3 |
| 1650 | peptidase, S1A (chymotrypsin) subfamily | −9 | −99 | +2 | −2 |
| 1725 | hypothetical protein | −11 | −76 | +0 | +2 |
| 1893 | hypothetical protein | +1 | +356 | −1 | −3 |
| 2297 | conserved hypothetical protein | +3 | +142 | −1 | −1 |
| 3676 | hypothetical protein | −11 | −63 | +1 | −2 |
| 3985 | hypothetical protein | +4 | +35 | +5 | −1 |
| 4305 | amino acid adenyltransferase | −52 | −57 | +1 | −69 |
| 4368 | hypothetical protein | +3 | +24 | +7 | −3 |
| 4837 | endoglucanase (celA) | −11 | −105 | −1 | +1 |
| 5970 | peptidase, S8 (subtilisin) family | −7 | −111 | +1 | −2 |
| 7375 | conserved hypothetical protein | +2 | +29 | −1 | −1 |
Table legend: Column headings: fold increase represents the change in expression and is derived from coefficient value; ID = MXAN # and description refers to annotation; both are derived from the M. xanthus genome data at NCBI [43].
Group D (increased mainly in the asgB mutant) contained genes encoding proteins that contain Big (bacterial immunoglobulin-like; pfam13205) domains; some Big proteins have been shown to contain novel Ca2+ binding modules [25]. As shown in Table 4, expression of big genes MXAN_7217 and 7219 was increased over 1000-fold in asgB while almost unchanged in the other tan strains. There were eight more genes that showed a significant (>10 fold) increase relative to the other three tan strains. These include MXAN_0383 (hypothetical, DNA-BreC (breaking rejoining catalytic domain)) and its neighbor MXAN _0384 (quaternary ammonium compound-resistance protein SugE). Finally, Group E genes were generally increased or decreased in WT-T and asgB but showed the opposite response in dkxG and/or xre228. For example, MXAN_ 0503 and 0504 were decreased in WT-T and asgB but increased in dkxG and xre228.
Discussion
Iron is an essential element for microbial growth. Among other things, it is necessary for enzymatic function of ribonucleotide reductase, iron-sulfur proteins, succinate dehydrogenase, and heme-containing enzymes such as cytochromes and peroxidases. The redox versatility of this element accounts for its biological significance but also contributes to problems with acquisition of iron. This is particularly true for microbes such as M. xanthus that thrive in aerobic environments because Fe is very insoluble at neutral pH and in oxic environments (available Fe ≈10−17 M). Most microbes produce extracellular ligands such as siderophores to sequester needed iron from the environment and have dedicated transport systems to bring Fe (III) siderophore complexes into the cell.
The results of this study demonstrate that iron availability plays a role in phase variation of Myxococcus xanthus. First, comparison of otherwise isogenic yellow and tan variants revealed that tan variants up-regulate a core set of genes encoding proteins critical for acquisition of iron, including enzymes for synthesis of the siderophore myxochelin (mxc) and import of Fe•myxochelin and hemin. Biochemical assays confirmed the increase in siderophore in tan variants. Second, addition of iron dramatically inhibited expression of mxc genes and slowed the yellow-to-tan switch significantly, effectively “prolonging” cells in the yellow phase. When the concentration of available iron was reduced, the number of tan variants arising from a wild-type yellow culture increased.
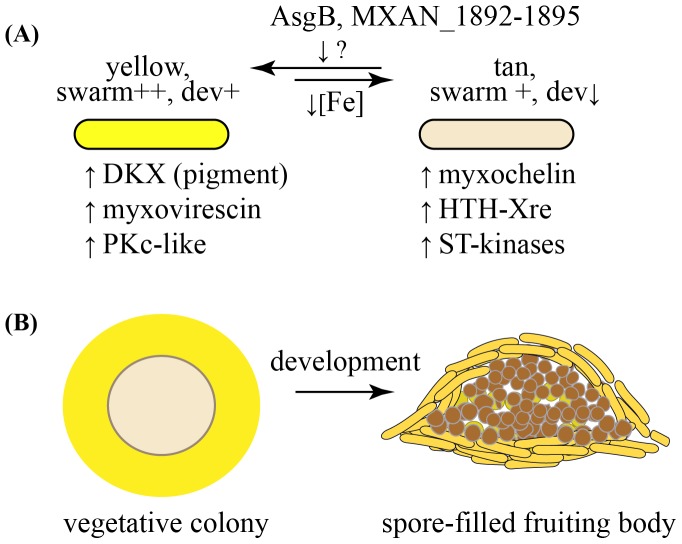
Iron acquisition genes were not the only genes responsive to phase variation. As shown in Fig 7, expression of PV markers, including DKX, antibiotic myxovirescin (Mxv), and kinase-like proteins (PKc-like) decreased while expression of potential regulators, serine threonine kinases and HTH-Xre proteins, increased in WT-T (blue bars). Changes in Mxv and DKX may reflect a metabolic trade-off as cells cease to invest in secondary metabolites when there is shortage of iron. Surprisingly, although swarming is also diminished in tan variants compared with yellow variants, we did not find differences in expression of known gliding genes. General characteristics of yellow and tan variants are summarized in Fig 8A.
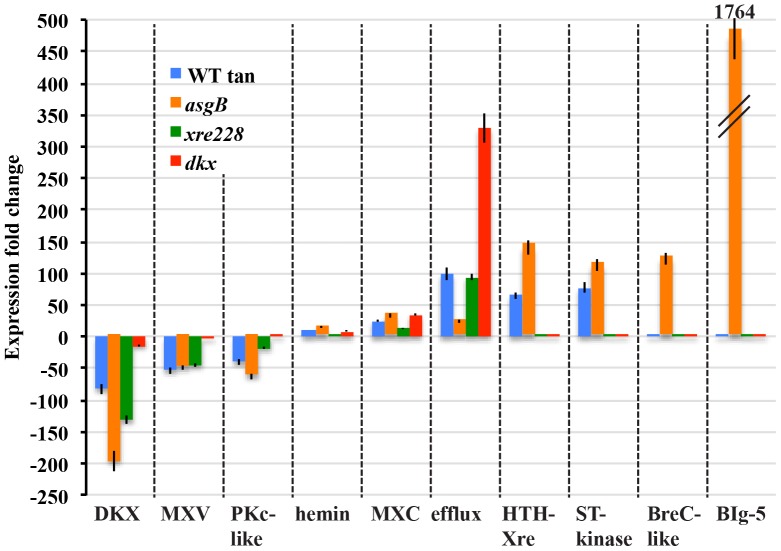
Figure 7. Potential biomarkers of phase variation.
A side-by side comparison illustrates the magnitude of changes between the tan variant and the mutants and highlights some of the outliers. Eight of the ten markers (genes or operons) show significant changes in the WT-T, while the remaining two genes represent changes that are unique to the asgB mutant. The fold change for gene operons represents the average of all genes in the pathway. DKX represents the average of the 14 DKxanthene genes (MXAN_4289-4305), MXV represents the average of 21 myxovirescin genes (MXAN_3930-3950), pKc represents the average of four protein kinase-like genes (MXAN_1234, 2177, 3183, 4841), hemin represents the average of nine myxochelin genes (MXAN_3639-3647), efflux represents the average of four macrolide efflux genes (MXAN_4198-4201), HTH-Xre represents the average of four xenobiotic response element genes (MXAN_4372, 4480, 4481, 6998), and ST_kinase represents the average of seven protein kinases (MXAN_2399 2840, 4371, 4373, 4479, 7370, 7371). BreC is MXAN_0383 and BIg-5 is the average of MXAN_7217 and 7219, which at fold increase of 1764, was off the scale of this graph.
Figure 8. Phase variation yields two cell types.
(A) The data show that the concentration of iron influences the yellow to tan phase switch; in this model, we propose that AsgB or AsgB-dependent products play a key role in the reverse direction switch. The yellow variants produce myxovirescin, the antibiotic needed for predation [23]. Tan variants are mediocre swarmers but are more adept at sequestering iron. (B) The division of labor between yellow and tan variants may be critical for survival of M xanthus in nature. The robust yellow swarmers are enriched at the periphery of a WT colony whereas slow swarming tan variants would be more enriched at the colony center. The ramifications for development are clear: tan cells, which are predisposed to become spores are well positioned to end up in the interior of the fruiting body and are surrounded by cells that produce DKX, the pigment needed to for production of viable spores [13].
To begin to understand the context for these PV markers, three strains that are tan due to mutation were analyzed alongside the WT-T. The dkx mutant (Fig 7 red bars), which is tan due to disruption of the pigment biosynthetic pathway, resembled the WT-T expression profile for mxc, hemin, and macrolide efflux genes. Hence, loss of pigment or disruption of the pigment biosynthetic pathway alone is sufficient to activate the iron acquisition scheme in M. xanthus. The DKX pathway does not autoregulate because disruption of the dkxG gene did not affect expression of genes in other dkx operons. Loss of DKX did not affect production of Mxv or PKc-like proteins, however.
The overall profile of the xre228 mutant (Fig 7 green bars) positions the function of Xre228 ahead of dkx and suggests that Xre228 regulates dkx, mxv, and genes encoding the four PKc-like proteins. Several key differences between xre228 and WT-T helped pinpoint the position of Xre228 in the PV pathway. Specifically, clusters of genes including MXAN_4371-4373, 4479-4481, and 7370 were unchanged in the xre228 mutant, which suggests that the ST-kinases and HTH-Xre act before Xre228 in the PV pathway or are independent of Xre228.
The pattern of the asgB mutant (Fig 7 orange bars) paralleled the WT-T strain for the eight PV biomarkers but showed substantial increases in genes such as breC (putative break-rejoining enzyme) and big (Fig 7) and potential regulators (Fig 6B). AsgB is a DNA-binding protein involved in cell-density dependent signaling [26]. It contains a small (≈30 amino acids), C-terminal HTH-Xre superfamily domain, similar to Xre228 and conserved in region 4 of the sigma 70 proteins (pfam04545) [15]. Although asgB was predicted to be essential [15] we were able engineer stable disruptions of the gene. Additionally, while the asgB mutant has historically been described as a tan mutant, our KanR asgB mutant was initially yellow, but it converted to tan over the period of a few weeks after which time it was a stable tan mutant. This result, combined with similarities in the transcriptome and phenotype data between the WT-T and the asgB mutant lead us to speculate that asgB mutant is blocked in the ability to switch from tan to yellow, as shown in Fig 8A. If this model is correct, AsgB or the genes it controls, including MXAN_1892-1895 (S_TPk and HTH-Xre proteins) may govern one of the main PV switch pathways. The severity of the asgB developmental phenotype may relate to the fact that it is locked in the tan phase. The BreC and BIg-domain proteins whose expression AsgB regulates may play direct roles in PV or may affect developmental processes that are known to be part of a signaling pathway regulated by AsgB [27].
Bacterial serine-threonine protein kinases were first discovered in M. xanthus [28] and have been shown to play critical roles in the complex life cycle of this organism [29], [30]. Two types of kinases, PKc-like and S_TPk, exhibited divergent regulation during PV. Genes for four nearly identical PKc-like proteins were down-regulated in WT-T, asgB and xre228. Gene expression was repressed to a similar extent for each gene, which suggests that their regulation may be coordinated and that they may encode redundant functions. The similarity between these proteins and the proximity of each to a S_TPk suggests that these clusters are the result of gene duplication. Such regions of the chromosome may be targets for gene conversion, a known mechanism for regulating phase variation. In contrast with the PKc-like genes, expression of S_TPk genes was increased in WT-T and asgB. This group of S_TPk may function in pathways involving motility or development, which are impacted in WT-T and asgB.
The ability of M. xanthus to generate two distinct cell types depending on the availability of iron makes sense given the social nature and habitats of this organism. M. xanthus has been compared to higher order social organisms, including bees and ants, that live in large groups and cooperate through a division of labor to accomplish complex tasks. As M. xanthus colonies normally contain a mixture of yellow (robust swarmers) and tan (mediocre swarmers) cell types, phase variation takes on a mutualistic quality. During growth, yellow variants accumulate at the colony edge and surround the slower swarming tan variants (Fig 8B). Elevated levels of DKX and Mxv, which are needed for sporulation and predation, respectively, likely benefit tan variants. In return, elevated levels of Fe•siderophore produced by tan variants may benefit yellow variants. When nutrient depletion induces the development cycle, tan variants enriched near the colony center and bathed in chemicals, such as DKX, from yellow variants are well positioned to differentiate into spores. Maintenance of both yellow and tan cell types with minimal ‘cheating’ may be safeguarded by the fact that the potentially iron-rich tan cells need DKX for efficient spore production. As only yellow cells produce the DKX pigment, their survival is guaranteed.
Materials and Methods
Bacterial strains and growth conditions
A list of strains, plasmids, and oligonucleotides is provided in Table S2. The M. xanthus strains included wild-type strain DK1622 [31]. DK1622 shows characteristic normal gliding motility, swarming, fruiting body morphogenesis, sporulation, and phase variation and was used as the source for yellow and tan phase cells. The asgB gene was disrupted using the Mip (myxo integrative plasmid) technique.
M. xanthus was grown routinely at 32°C at >200 rpm in CTPM liquid medium (1% Casitone, 10 mM Tris, 1 mM potassium phosphate, 5 mM MgSO4, final pH 7.6); according to the manufacturer, a 1% solution of Casitone is 15–45 µM Fe. TPM buffer is identical except that it lacks Casitone. CTPM was supplemented with kanamycin (Kan; 40 µg/ml) when applicable. Dipyridyl (2, 2-dipyridyl) is a bidentate chelating ligand that was added to CTPM medium to deplete iron. Where applicable, CTPM medium was supplemented with ferric citrate, ferric chloride, copper chloride, or dipyridyl. LB was used for growth of E. coli.
Analysis of wild-type phase variants and mutants
To determine viable cell numbers, aliquots were removed from broth cultures at regular intervals and plated on rich medium. Colony forming units (CFU) were determined by removing aliquots of cells and diluting serially 1∶10 in CTPM broth. Viability was checked by plating 4 µl spot aliquots of each dilution or by spread plating 50 µl aliquots of the 10−4 to 10−5 samples. After 4 days growth at 32°C, plates were scored for growth on spot plates and total number of yellow and tan colonies on spread plates. The swarming rate of each mutant was compared with the WT strain on CTPM medium with 0.3% and 1.5% high grade agar as described elsewhere [32]. Cell aggregation, Congo red and trypan blue dye binding were assayed using published protocols [9].
Siderophore: The amount of siderophore in supernate and cell pellet was determined by the liquid CAS assay [22]. Cells were grown to a density of 5×108 cells ml−1 (Klett = 100) in CTPM (+ supplements where appropriate). A 1 ml aliquot was removed and centrifuged in a microfuge tube for 2 min. The supernate was transferred to a fresh tube. The cell pellet was suspended in 1 ml of 1% NP-40 and resuspended by pipetting up and down; samples were incubated for 5 min to completely lyse the cells. Aliquots (0.4 ml each) of supernate and lysed cell suspension were transferred to glass tubes and 0.4 ml of CAS-Fe-HDTMA piperazine assay solution (below) was added to each tube and vortexed immediately. The CAS controls contained 0.4 ml CTPM broth (+ supplements where appropriate) or 1% NP-40. Tubes were incubated at room temperature for 30 min. After reaching equilibrium the absorbance was measured at 630 nm and compared against the cell free controls.
CAS-Fe-HDTMA piperazine solution: the liquid CAS solution was prepared as follows: 7.5 ml of 2 mM CAS was mixed with 1.5 ml of 1 mM FeCl3•6 H2O, and 50 ml HDTMA was added with stirring. The CAS-Fe-HDTMA was brought to 100 ml total volume by addition of piperazine buffer (≈36–40 ml, depending on the about of HCl used to bring pH to 5.6) and H2O to balance. Piperazine buffer was prepared by dissolving 9.7 gm piperazine hexahydrate (or 4.3 gm piperazine) in 30 ml H2O and adjusting the pH to 5.6 with conc HCl; the final volume was 37–40 ml.
Chrome azurol S (CAS) was added to CTPM medium to quantify the production of siderophore on agar medium. Sterile CAS•Fe• hexadecyltrimethylammonium bromide (HDTMA) [33] was added to CTPM agar. Aliquots of M. xanthus strains were spotted on CAS-Fe-HDMA medium and incubated for >18 hrs at 32°C and the zone of color was measured (growth did not occur as cells are sensitive to the HDTMA detergent).
DKxanthene: The amount of DKX was estimated by spectral analysis of log-phase cells (density noted for each experiment) harvested by centrifugation and suspended in 1 ml of TPM. DKX was measured at 410 nm (λmax [13]) using a Spectronic 21D spectrophotometer (Milton Roy).
Changes in siderophore and Abs 410 nm (λmax for DKX) in response to different concentration/types of metals were measured from cells grown in CTPM supplemented with different components. First, CTPM broth-grown WT-Y and WT-T cells (density 4×108 cells ml−1) were diluted 1∶50 in CTPM and immediately divided into four aliquots. Aliquot #1 received H2O, #2 received FeCl3 (100 µM fc), #3 received dipyridyl (25 µM fc) and #4 received CuCl2 (100 µM fc). Each of the four aliquots was split into two culture flasks and incubated at 32°C with shaking until reaching a density of 6×108 cells ml−1. Aliquots were removed and assayed for siderophore and absorbance at 410 nm as described above. Siderophore and Abs410 values for the test samples were compared against the original inoculum (density 4×108 cells ml−1) used for each series. Add-back experiments were performed with a basal medium of CTPM+25 µM dipyridyl to which H2O, CuCl2, FeCl3 or ZnCl2 was added at amounts listed in the text.
Mxv: The production of myxovirescin was determined using the myxovirescin assay described by [24] with the following modifications. Cultures of M. xanthus were harvested by centrifugation and suspended in TPM buffer to 5×108 cells ml−1 (Klett 100). 10 µl aliquots were spotted on TPM agar medium with 0.5% casitone and incubated at 32°C. After 18 hr, plates were overlaid with 3 ml of 0.5% CTPM soft agar containing 100 µl of an overnight culture of E. coli DH5α. The zone of inhibition (ZOI) was measured after 24 hr incubation at 32°C and is reported as the diameter of the clearing zone.
Microarray analysis
A custom Roche NimbleGen 12-plex microarray was designed for M. xanthus based on a consensus sequence from mapping paired end 91 bp Illumina sequence data from the National Genome Research Center for two tan and two yellow clones to the reference genome (dk_1622). Use of this custom array made it possible to generate more refined transcriptome data than previously [9]. The microarray contained 137,799 60-mer probes tiled across the genome, with an average spacing of 65 bp, and representing a possible 6,935 expressed transcripts (probe sets). Probe sets were defined as all probes that mapped to a single gene. Double-stranded cDNAs were synthesized using Invitrogen's SuperScript cDNA Synthesis Kit with the standard protocol using oligo dT primers (Invitrogen, Carlsbad, CA). RNA was harvested from yellow and tan derivatives of the WT (DK1622) and mutants as described previously [9]. cDNAs were fluorescently labeled with Cy3 using the standard NimbleGen Gene Expression Analysis protocol. All samples were hybridized to a single 12-plex chip to reduce technical noise. An 18-hour hybridization was conducted at 42 degrees in a NimbleGen Hybridization System 4 chamber (NimbleGen, Madison, WI). The chip was washed and then scanned on a Roche MS200 scanner. Finally, probe signal intensities were extracted using Roche Nimblegen's NimbleScan Software v2.6. Samples with adjusted p values of <0.05 from the independent array runs (triplicate samples of three biological replicates) were considered statistically significant. All analyses were done in the R programming language [34], raw intensity data were read in and checked for quality using the Limma package [35]. The data were then background corrected using background subtraction and the arrays were normalized with quantile normalization [36]. A simple linear model was fit to the data and variances were corrected using an empirical Bayes approach [37] from the bioconductor limma package. Results were corrected for multiple testing using the false discovery rate procedure of Benjamini and Hochberg (BH) and considered significant if their BH adjusted p-values were <0.05 [38].
Real-time PCR
WT–Y, WT-T variants and mutants were grown to a density of 5×108 cells ml−1 in CTPM medium at 32°C. A 1 ml aliquot was harvested by centrifugation at 13,700xg and the cell pellet was suspended in 0.5 ml PBS buffer. Total mRNA was extracted using RNAprotect Bacterial reagent (with DNase) and RNeasy minikit according to manufacturers instruction (Qiagen, Valencia CA). 500 ng total RNA was used to produce cDNA using a Hexonucleotide Mix (Roche, Indianapolis, IN) as primer for the reaction using the Superscriptase II kit (Invitrogen). The resulting cDNA was diluted 1∶25 or 1∶1250 for probing target genes and 16s rRNA templates respectively. 5 µl of diluted cDNA sample was added to 20 µl of a PCR mixture prepared from Power SYBR Green PCR Master Mix (Applied Biosystems, Carlsbad, CA), which contained each primer at a concentration of 160 nM. Primers, listed in Table S1, were chosen using Primer Express 3.0 software (Applied Biosystems) and were designed to amplify a region of about 150–200 bp within each transcript.
Supporting Information
Cell cohesion (aka agglutination) is a phenotype that can be used to distinguish yellow and tan variants. The cohesion assay was performed as described in Methods. Tan strains (WT-T, asgB, xre228, and dkxG) were compared with the WT-Y variant; data are presented for three independent assays.
(TIF)
The WT-T (right) strain produces a larger siderophore halo than the WT-Y (left) strain. Cultures were grown to 5×108 cells ml−1 (Klett 100) and concentrated 10-fold in TPM buffer. 5 µl aliquots were spotted on CTPM blue agar (containing CAS-Fe) and incubated at 32°C for 4 hr.
(TIF)
DNA microarray data supplement to Tables 1 and 2. Additional data are listed in Tables 3 and 4. Supporting array table legend: Genes showing significant expression changes in at least one of the M. xanthus tan strains (12 datasets: four biological replicates with triplicate technical replicates) in DNA tiling array studies relative to the WT-Y variant. Column headings: fold increase represents the change in expression and is derived from coefficient value; ID = MXAN # and description refers to annotation; both are derived from the M. xanthus genome data at NCBI http://www.ncbi.nlm.nih.gov/genome/1120.
(DOCX)
Strains and molecular reagents.
(DOCX)
Acknowledgments
We would like to dedicate this to Dr. Dziewanowska who died shortly before the submission.
Funding Statement
This work was supported by the following granting agencies: the National Science Foundation (MCB1052525 to PLH) and NM-INBRE/NCGR pilot award (National Center for Research Resources award P20RR016480 and National Institute of General Medical Sciences award P20 GM103451). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. van der Woude MW (2006) Re-examining the role and random nature of phase variation. FEMS Microbiol Lett 254: 190–197. [DOI] [PubMed] [Google Scholar]
- 2. Piggot P (2010) Epigenetic switching: bacteria hedge bets about staying or moving. Curr Biol 20: R480–R482. [DOI] [PubMed] [Google Scholar]
- 3. Chai Y, Norman T, Kolter R, Losick R (2010) An epigenetic switch governing daughter cell separation in Bacillus subtilis . Genes Dev 24: 754–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. van der Woude MW, Baumler AJ (2004) Phase and antigenic variation in bacteria. Clin Microbiol Rev 17: 581–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Somvanshi VS, Sloup RE, Crawford JM, Martin AR, Heidt AJ, et al. (2012) A single promoter inversion switches Photorhabdus between pathogenic and mutualistic states. Science 337: 88–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Laue BE, Gill RE (1995) Using a phase-locked mutant of Myxococcus xanthus to study the role of phase variation in development. J Bacteriol 177: 4089–4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burchard RP, Dworkin M (1966) Light-induced lysis and carotinogenesis in Myxococcus xanthus . J Bacteriol 91: 535–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Burchard RP, Burchard AC, Parish JH (1977) Pigmentation phenotype instability in Myxococcus xanthus . Can J Microbiol 23: 1657–1662. [DOI] [PubMed] [Google Scholar]
- 9. Furusawa G, Dziewanowska K, Stone H, Settles M, Hartzell P (2011) Global analysis of phase variation in Myxococcus xanthus . Mol Microbiol 81: 784–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dahl JL, Ulrich CH, Kroft TL (2011) Role of phase variation in the resistance of Myxococcus xanthus fruiting bodies to Caenorhabditis elegans predation. J Bacteriol 193: 5081–5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Laue BE, Gill RE (1994) Use of a phase variation-specific promoter of Myxococcus xanthus in a strategy for isolation a phase-locked mutant. J Bacteriol 176: 5341–5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kaiser D (2003) Coupling cell movement to multicellular development in myxobacteria. Nat Rev Microbiol 1: 45–54. [DOI] [PubMed] [Google Scholar]
- 13. Meiser P, Bode HB, Muller R (2006) The unique DKxanthene secondary metabolite family from the myxobacterium Myxococcus xanthus is required for developmental sporulation. Proc. Natl Acad Sci USA 103: 19128–19133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mayo KA, Kaiser D (1989) asgB, a gene required early for developmental signalling, aggregation, and sporulation of Myxococcus xanthus . Mol & Gen Gene 218: 409–418. [DOI] [PubMed] [Google Scholar]
- 15. Plamann L, Davis JM, Cantwell B, Mayor J (1994) Evidence that asgB encodes a DNA-binding protein essential for growth and development of Myxococcus xanthus . J Bacteriol 176: 2013–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dana JR, Shimkets LJ (1993) Regulation of Cohesion-Dependent Cell Interactions in Myxococcus xanthus . J Bacteriol 175: 3636–3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shimkets LJ (1986) Correlation of energy-dependent cell cohesion with social motility in Myxococcus xanthus . J Bacteriol 166: 837–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Black WP, Xu Q, Yang Z (2006) Type IV pili function upstream of the Dif chemotaxis pathway in Myxococcus xanthus EPS regulation. Mol Microbiol 61: 447–456. [DOI] [PubMed] [Google Scholar]
- 19. Kroos L, Kuspa A, Kaiser D (1986) A global analysis of developmentally regulated genes in Myxococcus xanthus . Dev Biol 117: 252–266. [DOI] [PubMed] [Google Scholar]
- 20. Silakowski B, Kunze B, Nordsiek G, Blocker H, Hofle G, et al. (2000) The myxochelin iron transport regulon of the myxobacterium Stigmatella aurantiaca Sg a15. Eur J Biochem (FEBS) 267: 6476–6485. [DOI] [PubMed] [Google Scholar]
- 21. Ducey TF, Jackson L, Orvis J, Dyer DW (2009) Transcript analysis of nrrF, a Fur repressed sRNA of Neisseria gonorrhoeae . Microb Pathog 46: 166–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Payne SM (1994) Detection, isolation, and characterization of siderophores. Meth Enz 235: 329–344. [DOI] [PubMed] [Google Scholar]
- 23. Xiao Y, Gerth K, Muller R, Wall D (2012) Myxobacterium-produced antibiotic TA (myxovirescin) inhibits type II signal peptidase. Antimicro Agents Chemo 56: 2014–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xiao Y, Wei X, Ebright R, Wall D (2011) Antibiotic production by myxobacteria plays a role in predation. J Bacteriol 193: 4626–4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Raman R, Rajanikanth V, Palaniappan RU, Lin YP, He H, et al. (2010) Big domains are novel Ca(2)+-binding modules: evidences from big domains of Leptospira immunoglobulin-like (Lig) proteins. PLoS One 5: e14377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bowden MG, Kaplan HB (1996) The Myxococcus xanthus developmentally expressed asgB-dependent genes can be targets of the A signal-generating or A signal-responding pathway. J Bacteriol 178: 6628–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Konovalova A, Petters T, Sogaard-Andersen L (2010) Extracellular biology of Myxococcus xanthus . FEMS Microbiol Rev 34: 89–106. [DOI] [PubMed] [Google Scholar]
- 28. Munoz-Dorado J, Inouye S, Inouye M (1993) Eukaryotic-like protein serine/threonine kinases in Myxococcus xanthus, a developmental bacterium exhibiting social behavior. J Cell Biochem 51: 29–33. [DOI] [PubMed] [Google Scholar]
- 29. Nariya H, Inouye S (2006) A protein Ser/Thr kinase cascade negatively regulates the DNA-binding activity of MrpC, a smaller form of which may be necessary for the Myxococcus xanthus development. Mol Microbiol 60: 1205–1217. [DOI] [PubMed] [Google Scholar]
- 30. Inouye S, Jain R, Ueki T, Nariya H, Xu CY, et al. (2000) A large family of eukaryotic-like protein Ser/Thr kinases of Myxococcus xanthus, a developmental bacterium. Microb Comp Genomics 5: 103–120. [DOI] [PubMed] [Google Scholar]
- 31. Wall D, Kolenbrander PE, Kaiser D (1999) The Myxococcus xanthus pilQ (sglA) gene encodes a secretin homolog required for Type IV pilus biogenesis, social motility and development. J Bacteriol 181: 24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shi W, Zusman DR (1993) The two motility systems of Myxococcus xanthus show different selective advantages on various surfaces. Proc. Natl Acad Sci USA 90: 3378–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schwyn B, Neilands JB (1987) Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160: 47–56. [DOI] [PubMed] [Google Scholar]
- 34.Team RDC (2010) R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing.
- 35.Smyth GK (2005) Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer. pp. 397–420.
- 36. Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193. [DOI] [PubMed] [Google Scholar]
- 37. Smyth GK (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statist Appl Gen Mol Biol 3: Article3. [DOI] [PubMed] [Google Scholar]
- 38. Benjamini Y, Hochberg Y (1995) Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J Royal Stat Soc Series B 57: 289–300. [Google Scholar]
- 39. Krug D, Zurek G, Revermann O, Vos M, Velicer GJ, et al. (2008) Discovering the hidden secondary metabolome of Myxococcus xanthus: a study of intraspecific diversity. Appl Environ Micro 74: 3058–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kunze B, Bedorf N, Kohl W, Hofle G, Reichenbach H (1989) Myxochelin A, a new iron-chelating compound from Angiococcus disciformis (Myxobacterales). Production, isolation, physico-chemical and biological properties. J Antibio 42: 14–17. [DOI] [PubMed] [Google Scholar]
- 41. Bhat S, Zhu X, Patel RP, Orlando R, Shimkets LJ (2011) Identification and localization of Myxococcus xanthus porins and lipoproteins. PLoS One 6: e27475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, et al. (2011) The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucl Acids Res 39: D561–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Goldman BS, Nierman WC, Kaiser D, Slater SC, Durkin AS, et al. (2006) Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl Acad Sci USA 103: 15200–15205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cell cohesion (aka agglutination) is a phenotype that can be used to distinguish yellow and tan variants. The cohesion assay was performed as described in Methods. Tan strains (WT-T, asgB, xre228, and dkxG) were compared with the WT-Y variant; data are presented for three independent assays.
(TIF)
The WT-T (right) strain produces a larger siderophore halo than the WT-Y (left) strain. Cultures were grown to 5×108 cells ml−1 (Klett 100) and concentrated 10-fold in TPM buffer. 5 µl aliquots were spotted on CTPM blue agar (containing CAS-Fe) and incubated at 32°C for 4 hr.
(TIF)
DNA microarray data supplement to Tables 1 and 2. Additional data are listed in Tables 3 and 4. Supporting array table legend: Genes showing significant expression changes in at least one of the M. xanthus tan strains (12 datasets: four biological replicates with triplicate technical replicates) in DNA tiling array studies relative to the WT-Y variant. Column headings: fold increase represents the change in expression and is derived from coefficient value; ID = MXAN # and description refers to annotation; both are derived from the M. xanthus genome data at NCBI http://www.ncbi.nlm.nih.gov/genome/1120.
(DOCX)
Strains and molecular reagents.
(DOCX)