Summary
The bone marrow failure syndromes (BMFS) are a heterogeneous group of rare blood disorders characterized by inadequate haematopoiesis, clonal evolution, and increased risk of leukaemia. Single nucleotide polymorphism arrays (SNP-A) have been proposed as a tool for surveillance of clonal evolution in BMFS. To better understand the natural history of BMFS and to assess the clinical utility of SNP-A in these disorders, we analysed 124 SNP-A from a comprehensively characterized cohort of 91 patients at our BMFS centre. SNP-A were correlated with medical histories, haematopathology, cytogenetic and molecular data. To assess clonal evolution, longitudinal analysis of SNP-A was performed in 25 patients. We found that acquired copy number-neutral loss of heterozygosity (CN-LOH) was significantly more frequent in acquired aplastic anaemia (aAA) than in other BMFS (odds ratio 12.2, p<0.01). Homozygosity by descent was most common in congenital BMFS, frequently unmasking autosomal recessive mutations. Copy number variants (CNVs) were frequently polymorphic, and we identified CNVs enriched in neutropenia and aAA. Our results suggest that acquired CN-LOH is a general phenomenon in aAA that is probably mechanistically and prognostically distinct from typical CN-LOH of myeloid malignancies. Our analysis of clinical utility of SNP-A shows the highest yield of detecting new clonal haematopoiesis at diagnosis and at relapse.
Keywords: bone marrow failure, aplastic anaemia, chromosomal rearrangements, clonal evolution, cytogenetic diagnosis
Introduction
The bone marrow failure syndromes (BMFS) are a diverse group of life-threatening blood disorders characterized by inadequate haematopoiesis, clonal evolution, and increased risk of haematological malignancies. BMF can be acquired or inherited. The prototypical inherited BMFS are frequently caused by inherited defects in key housekeeping pathways, such as DNA repair in Fanconi Anaemia (FA), telomere maintenance in Dyskeratosis Congenita (DC), and ribosomal biogenesis in Diamond Blackfan Anaemia (DBA) and Shwachman Diamond Syndrome (SDS) (Shimamura and Alter 2010). In acquired BMFS, such as acquired aplastic anaemia (aAA), a specific trigger is frequently not identified; in aAA the final pathogenic insult is thought to be immune-mediated destruction of haematopoietic stem and progenitor cells (Young, et al 2006). Despite recent advances in the understanding of the molecular pathogenesis of BMFS, the ability to diagnose, risk-stratify and treat patients with these rare disorders remains limited. Up to a quarter of patients with an apparent inherited BMFS cannot be given a specific diagnosis despite extensive testing (Alter, et al 2010, Teo, et al 2008). A subset of patients with a clinical diagnosis of a prototypical inherited BMFS, such as DBA, lack a mutation in genes that are known to be linked to that disorder. Conversely, patients with the same genetic defect can differ greatly in disease severity (Shimamura and Alter 2010).
In both the acquired and the inherited BMFS, the major contributors to mortality are complications of progressive cytopenias, and - albeit to a lesser extent - transformation to myelodysplastic syndrome (MDS) and acute myeloid leukaemia (AML). The main predictor of malignant transformation is acquisition of clonal cytogenetic abnormalities. Several nonrandom chromosomal abnormalities in BMFS have been described. Recurrent monosomy 7, trisomy 8, deletion of 13q, trisomy 6, and copy number-neutral loss of heterozygosity (CN-LOH) of 6p have been reported in aAA (Afable, et al 2011, Katagiri, et al 2011, Maciejewski and Selleri 2004). Monosomy 7, isochromosome 7q, and deletion 20q were reported in SDS (Donadieu, et al 2012, Dror, et al 2002), and the gain of 1q, monosomy 7, gain of 3q, and deletion of 11q were linked to poor prognosis in FA (Mehta, et al 2010, Quentin, et al 2011, Tonnies, et al 2003). While annual surveillance with bone marrow biopsies has been the standard of care for many BMFS, beyond a handful of ominous abnormalities (e.g. monosomy 7), the extent and significance of genetic changes in BMFS is largely uncertain.
Recently, single nucleotide polymorphism arrays (SNP-A) were proposed as a promising tool for high resolution cytogenetic analysis and surveillance of early clonal changes in BMFS (Afable, et al 2011, Katagiri, et al 2011, Kojima, et al 2011, Quentin, et al 2011), however, their clinical utility still remains to be established (Kojima, et al 2011). In 2009, the Comprehensive Bone Marrow Failure Center (CBMFC) at the Children’s Hospital of Philadelphia (CHOP) and the Hospital of the University of Pennsylvania (Penn) incorporated high-density SNP-A as an adjunct to conventional cytogenetics in the evaluation of BMFS patients. Here we present a comprehensive analysis of genetic changes in BMFS using 124 SNP-A from 91 patients, who were referred for evaluation of bone marrow failure. SNP-A genotyping was correlated with medical histories, haematopathology, cytogenetic, and molecular data. To assess the potential role of SNP-A in screening for early clonal evolution, longitudinal analysis of SNP-A was performed in 25 patients. Our analysis revealed distinct patterns of genomic abnormalities in BMFS, with acquired CN-LOH being significantly more frequent in aAA compared to non-aAA BMFS, and showed that clonal haematopoiesis in BMFS is most frequently detected at diagnosis and upon relapse.
Methods
Patients and Controls
The Penn-CHOP BMFS cohort is an open prospective/retrospective cohort for the investigation of molecular mechanisms of BMFS established in accordance with the procedures approved by the Institutional Review Boards of CHOP and of the University of Pennsylvania. Informed consent was obtained in accordance with the Declaration of Helsinki from all study participants or their legal guardians before participation. All paediatric and adult patients who were referred to CBMFC between 2009 and 2012 for an evaluation of BMFS and had SNP-A genotyping available were eligible for the current study. For all patients, race was self-reported. Complete medical histories, blood counts, bone marrow biopsy and metaphase cytogenetics were available for 90 patients. One patient with a genetically confirmed diagnosis of DC, who is an affected parent of a child also enrolled in this study, did not have a bone marrow biopsy or cytogenetics available for review.
Normal controls used for the association analysis were recruited from well-child visits within the CHOP healthcare network as a part of ongoing studies at the CHOP Center for Applied Genomics, in accordance with the procedures approved by the CHOP Institutional Review Board. Controls were screened for the presence of chronic health conditions and genetic abnormalities using electronic health records. Genomic DNA extracted from whole blood was used for SNP-A genotyping. Overall, 1,618 healthy controls were genotyped with Illumina Quad610 Beadchip (Illumina, San Diego, CA, USA). The data for the 268 normal controls, genotyped using Illumina Omni1-Quad Beadchip, was obtained from Illumina.
Diagnostic Group Assignment
Patients were assigned to diagnostic categories using standard criteria (Alter 2003), Kaufman, et al 1991). For FA, DC, SDS, DBA and severe congenital neutropenia (SCN) patients, diagnoses were confirmed with syndrome-specific testing (Bessler, et al 2008). Diagnosis of aAA required exclusion of other entities that could mimic its presentation. Patients with aAA who had > 1% of CD55-negative, CD59-negative cells (white blood cells or red blood cells) were analysed separately as aAA with Paroxysmal Nocturnal Haemoglobinuria (AAPNH). Patients with MDS were classified according to the 2008 World Health Organization (WHO) Guidelines (Swerdlow, et al 2008). Patients who could not be classified as a specific BMFS were grouped according to the primary cell line affected. Patients with an isolated congenital anaemia were grouped as Anaemia Not Otherwise Specified (Anaemia NOS); this group included patients who fulfilled the diagnostic criteria for DBA but did not have an identifiable molecular defect. Patients with an isolated neutropenia were grouped as Neutropenia NOS; autoimmune, cyclic, and benign ethnic neutropenia were excluded. Patients with cytopenias and physical malformations were grouped as BMF NOS. Eight additional patients who were referred to the CBMFC for a clinical suspicion of an occult inherited BMFS but whose evaluation revealed an alternative diagnosis were included for completeness. These patients were analysed separately as Other; their diagnoses were juvenile myelomonocytic leukaemia (JMML), X-linked agammaglobulinaemia, haemophagocytic lymphohistiocytosis, iron-refractory iron deficiency anaemia, paediatric AML with unusually severe complications of chemotherapy, primary combined immunodeficiency syndrome (DOCK8 mutation), autoimmune disease-associated cytopenias, and Rothmund- Thompson Syndrome.
SNP-A Analysis
Illumina Infinium SNP-A genotyping was performed on DNA extracted from bone marrow aspirates at the CHOP Center for Applied Genomics according to the manufacturer’s protocol. From 2009 to 2011, genotyping was performed with the Illumina Quad610 Beadchip. Due to the manufacturer’s technology upgrade, starting in the autumn of 2011, genotyping was undertaken with the Illumina Omni1-Quad Beadchip. Overall, 67 samples from 51 patients were genotyped with the Quad610, and 57 samples from 54 patients were genotyped with the Omni1-Quad. Fourteen patients were genotyped with both Beadchips. SNP-A data have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO), accession number GSE48484.
The majority of patients did not have a paired constitutional DNA sample. In the absence of constitutional DNA, genomic abnormalities were classified as acquired when they were clonal within the patient sample as determined by the B-allele frequency (BAF) and/or represented classic recurrent cytogenetic abnormalities (i.e. monosomy 7 or trisomy 8). All other abnormalities, such as long tracts of homozygosity with the BAF of 1 or 0 were conservatively assumed to be constitutional; this approach is in agreement with published data (Afable, et al 2011).
All SNP-A were analysed in GenomeStudio (Illumina), which allows direct visualization of the BAF and log R ratio (LRR). For detection of inherited CN-LOH, we used a CN-LOH prediction algorithm requiring a minimum of 100 homozygous SNPs and 1 MB size; analysis was then focused on CN-LOH exceeding the 99th percentile of CN-LOH size in the control population (Supporting Methods). All CN-LOH regions in excess of the 99th percentile of controls were visually confirmed in GenomeStudio. For evaluation of inherited copy number variants (CNVs), SNP-A from patients and controls were analysed with CNV Workshop (Gai, et al 2010), with a minimum thresholds of 5 SNPs for deletions and 10 SNPs for duplications. Predicted CNVs with over 50% overlap with telomeric, centromeric and immunoglobulin variable regions were excluded. For enrichment analysis, frequencies of CNVs within known genes in patients were compared to frequencies in controls; patients and controls were matched for ethnicity and SNP-A Beadchip. Because of small numbers of patients in some of the diagnostic groups, enrichment analysis was limited to the two largest subgroups: Caucasian patients with aAA and Neutropenia NOS. All CNVs significantly enriched in the patient population were visually confirmed in GenomeStudio. All chromosome coordinates were based on hg19 (NCBI build 37).
Cytogenetics and Haematopathology
Cytogenetic analysis and fluorescence in situ hybridization (FISH) of marrow aspirates were performed according to standard methods. Haematopathology of bone marrow biopsies was performed by a clinical haematopathologist in a blinded fashion, as this study was conceived after the completion of haematopathology review. In accordance with the department policy, all controversial cases were subject to a clinical consensus conference.
Statistical Analysis
Frequencies of genetic abnormalities or other clinical features between patient groups were compared using Fisher’s exact test with a significant p-value <0.05 (two-tailed). For enrichment analysis of CNVs, Bonferroni correction for multiple testing was performed, and significance was estimated with one-tailed p-value < 0.05.
Results
Patient Characteristics
Patient characteristics are shown in Table I; a detailed breakdown including array information is presented in Supporting Table 1. This was a diverse, predominantly paediatric cohort with 69 (76%) paediatric and 22 (24%) adult patients, representative of the patient population seen at a tertiary bone marrow failure referral centre. The largest diagnostic category was aAA with 31 patients, eight of whom had a detectable PNH clone. Seventeen patients had a classical inherited BMFS - FA, DBA, DC, SDS, or SCN. Four paediatric patients had MDS. In agreement with prior series (Alter, et al 2010, Teo, et al 2008), 32 patients with BMFS could not be assigned to a specific diagnostic entity after extensive testing. The majority of patients (86 of 91) had a normal karyotype. Sixty-six patients had a single SNP-A, while 25 patients had two or more SNP-A obtained over a median follow-up of 13 months.
Table I.
Patient Characteristics.
| Diagnostic Category | Number of Patients (n=91) |
Female | Male | Median Age, years (Range) |
Family History of Haematological Disorder (%) |
Abnormal Karyotype (%) |
||
|---|---|---|---|---|---|---|---|---|
| BMFS | Aplastic | No PNH Clone | 23 | 9 | 14 | 12(<1 – 46) | 0.0 | 0.0 |
| Anaemia | PNH Clone | 8 | 7 | 1 | 17 (5 – 61) | 12.5 | 0.0 | |
| Diamond-Blackfan Anaemia | 3 | 0 | 3 | 12 (9 –14) | 33.3 | 0.0 | ||
| Dyskeratosis Congenita | 6 | 2 | 4 | 13.5 (7 – 40) | 66.7 | 0.0 | ||
| Fanconi Anaemia | 3 | 0 | 3 | 6 (1 – 9) | 100.0 | 0.0 | ||
| Shwachman Diamond Syndrome | 4 | 3 | 1 | 3.5 (1 – 20) | 50.0 | 0.0 | ||
| Severe Congenital Neutropenia | 1 | 1 | 0 | 20 | 0.0 | 0.0 | ||
| Myelodysplastic Syndrome* | 4 | 3 | 1 | 11 (<1 – 17) | 50.0 | 75.0 | ||
| Anaemia NOS | 5 | 3 | 2 | 2 (1 – 16) | 20.0 | 0.0 | ||
| Neutropenia NOS | 14 | 7 | 7 | 11 (1 – 30) | 7.1 | 0.0 | ||
| Bone Marrow Failure NOS | 12 | 5 | 7 | 5 (1 – 22) | 41.7 | 16.7 | ||
| Other** | 8 | 1 | 7 | 6 (<1 – 18) | 0.0 | 25 | ||
| Total | 91 | 41 | 50 | |||||
Three patients with Refractory Cytopenia of Childhood (RCC), one patient with Refractory Anaemia with Excess Blasts-2 (RAEB-2) (WHO Classification).
See Methods.
BMFS, bone marrow failure syndrome; PNH, paroxysmal nocturnal haemoglobinuria; NOS, not otherwise specified
Acquired CN-LOH is Significantly More Frequent in aAA Compared to Other BMFS and Shows Dynamic Changes Over Disease Course
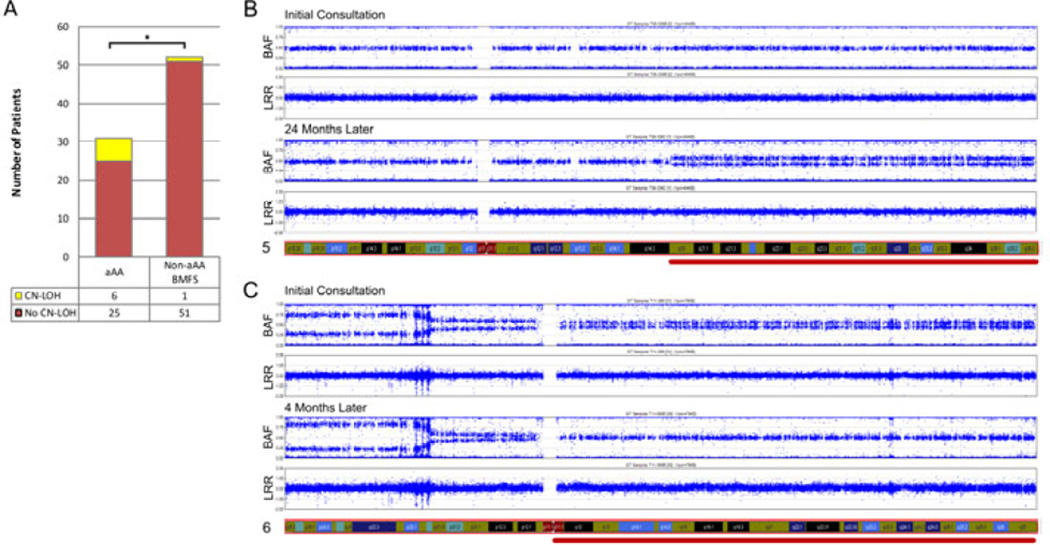
Acquired CN-LOH was found in 7 patients with BMFS: six patients with aAA and one with SDS (Table II). Six of the 31 aAA patients (19%) had at least one region of acquired CN-LOH; all had a normal karyotype. The most common abnormality in aAA was CN-LOH involving the short arm of chromosome 6, seen in 4 patients. All patients with 6p CN-LOH had multiple breakpoints of the CN-LOH region, indicating the presence of multiple clones. Additionally, there were three other regions of acquired CN-LOH, at 5q15qter, 6q12qter and 15q12qter. 6q12qter was seen in conjunction with a 6p CN-LOH abnormality. Compared to 6p CN-LOH clones, these clones were relatively small, estimated at 5–15% clone size. The patient with SDS who had a CN-LOH abnormality was a compound heterozygote for two germline SBDS mutations and acquired CNLOH favouring the hypomorphic allele; this patient is reported in detail elsewhere (Parikh, et al 2012). There was no statistically significant association between acquired CN-LOH and the diagnostic categories of BMFS when tested for heterogeneity across the 10 individual diagnostic groups listed in Table I (Fisher-Freeman-Halton exact test of 10×2 table, p=0.3885). However, acquired CN-LOH was significantly more frequent in patients with aAA than in the combined category of non-aAA BMFS patients in our cohort, with an Odds Ratio (OR) of 12.240 (95% confidence interval [CI] 1.333–573.696, p<0.01) (Figure 1A). To evaluate the stability of acquired CN-LOH in aAA, we analysed serial SNP-As in nine aAA patients who had SNP-A performed at multiple time points. Of 9 patients in this subset, 4 were found to have acquired CN-LOH, and 2 of the 4 were found to have dynamic changes - acquisition or loss - of the CN-LOH clones (Figure 1B–C, Supporting Text 1A).
Table II.
Acquired Copy Number-Neutral Loss of Heterozygosity (Acquired CN-LOH)
| Abnormality | Patient Number | Diagnosis | CN-LOH Region | Clone Size | Karyotype | Driving Gene |
|---|---|---|---|---|---|---|
| BMFS Patients | ||||||
| 5q CN-LOH | 1 | AA | 5q15qter | 10–15% | Normal | Unknown |
| 6p CN-LOH | 2 | AA | 6pterp12.1 | Multiple | Normal | Probably HLA gene family |
| 3 | AA | 6pterp11.2 | Multiple | Normal | ||
| 4 | AA | 6pterp21.2 | 30% | Normal | ||
| 5 | AA | 6pterp11.1 | Multiple | Normal | ||
| 6q CN-LOH | 3 | AA | 6q12qter | 5–10% | Normal | Unknown |
| 15q CN-LOH | 6 | AAPNH | 15q12qter | Multiple | Normal | Unknown |
| 7q CN-LOH | 7 | SDS | 7q11.21qter | 10–15% | Normal | SBDS mutation |
| Non-BMFS Patients | ||||||
| 11q CN-LOH | 8 | Other (JMML) | 11q13.4qter | 95–100% | Normal | CBL mutation |
| 13q CN-LOH | 9 | Other (AML) | 13q12.11qter | 95–100% | 47,XY,+8 | FLT3 ITD |
BMFS, bone marrow failure syndrome; AA, aplastic anaemia; PNH, paroxysmal nocturnal haemoglobinuria; SDS, Shwachman Diamond Syndrome; JMML, juvenile myelomoncytic leukaemia; AML, acute myeloid leukaemia; ITD, internal tandem duplication.
Figure 1. Acquired CN-LOH is Significantly More Frequent in aAA Compared to Other BMFS, and Exhibits Dynamic Changes.
A. Histogram depicting relative frequencies of acquired copy number-neutral loss of heterozygosity (CN-LOH) in acquired aplastic anaemia (aAA) compared to other bone marrow failure syndromes (BMFS). The number of patients with acquired CNLOH is shown in yellow, and without acquired CN-LOH—in red; the numbers are tabulated below the graph. The difference between aAA and other BMFS is statistically significant (*). B and C. SNP-A genotyping at two different time points for two aAA patients are shown in B and C respectively. Genotyping results at each time point are shown as two scatter plots. The top plot of each pair shows B-allele Frequency (BAF, a relative frequency of the minor allele) on the Y-axis, and the chromosomal location on the X-axis. The bottom plot of each pair shows Log R Ratio (LRR, a measure of normalized total signal intensity for both alleles) on the Y-axis, and the chromosomal location on the X-axis. In a region with acquired CN-LOH, the copy number (indicated by the LRR) remains constant, while there is a decreased frequency of the heterozygous alleles (indicated by the change of BAF plot). Both patients exhibit dynamic changes in acquired CN-LOH clones. The patient in Panel B developed a new acquired CN-LOH clone on chromosome 5q during aAA relapse (red line). The patient in Panel C had multiple acquired CN-LOH clones involving both 6p and 6q at diagnosis of aAA. Single nucleotide polymorphism arrays (SNP-A) performed after one course immunosuppressive therapy showed a loss of the 6q CN-LOH (red line) and an expansion of the 6p CN-LOH clone.
Acquired Copy Number Alterations (CNAs) Are Frequent in MDS and Unclassified BMFS
Acquired CNA were found in 5 patients with BMFS (3 in MDS and 2 in BMF NOS) (Table III). Three patients with MDS had regions of acquired CNA characteristic of MDS (abnormalities of chromosome 7 and trisomy 8). Of the 13 patients with unclassified BMFS (BMF NOS) in our cohort, two had regions of acquired CNA.
Table III.
Acquired Copy Number Abnormalities (Acquired CNAs)
| Abnormality | Patient | Diagnosis | DNA | Copy Number Abnormality | Clone Size | Karyotype |
|---|---|---|---|---|---|---|
| Isochromosome7q | 10 | MDS | BM | 7pterp11.1 loss 7q11.21qter gain | >95% | 46,XX,i(7)(q10)[16]/ 46,XX[1] |
| Monosomy 7 | 11 | MDS | BM | Chr 7 loss | 50% | 45,XX,-7[13]/ 46,XX[7] |
| Trisomy 8 | 12 | MDS | BM | Chr 8 gain | 20–30% | 47,XX,+8[5]/ 46,XX[15] |
| 9 | Other (AML) | BM | Chr 8 gain | >95% | 47,XY,+8[17] | |
| Ring chromosome 21, Monosomy 21 | 13 | BMF NOS | BM | 21q11.2q22.12 loss, 21q22.13qter loss | 85% monosomy 21; 15% r(21)(q11.2q22.12) | 46,XY,r(21)[1]/ 45,XY,-21[19] |
| Skin | 21q11.2q22.12 loss, 21q22.13q22.3 loss | 15% monosomy 21, 85% r(21)(q11.2q22.12) | 46,XY,r(21)[17]/ 45,XY,-21[3] | |||
| Trisomy 13 | 14 | BMF NOS | BM | Chr 13 gain | 5–10% | Normal |
MDS, myelodysplastic syndrome; AML, acute myeloid leukaemia; BMF NOS, bone marrow failure not otherwise specified; BM, bone marrow
Patient 14 was a 6-year-old female with Shwachman-like BMFS. SBDS gene testing was normal, and metaphase cytogenetics failed to detect a cytogenetic abnormality. SNP-A revealed a 5- 10% clone with a gain of chromosome 13 (Supporting Text 1B). Patient 13 was a 1-year-old male with congenital thrombocytopenia and multiple anomalies at birth. Constitutional karyotype and SNP-A performed on a skin biopsy at birth revealed constitutional mosaicism for ring chromosome 21 (85%) and monosomy 21 (15%). SNP-A of the bone marrow aspirate, performed to evaluate congenital thrombocytopenia, showed an expansion of the monosomy 21 clone in the bone marrow compared to the skin, resulting in haploinsufficiency for RUNX1. RUNX1 haploinsufficiency was previously linked to congenital thrombocytopenia (van der Crabben, et al 2010, Walker, et al 2002) (Supporting Text 1C, Supporting Figure 1).
Inherited Regions of Homozygosity Are Common in BMFS, and Can Unmask Recessive Mutations
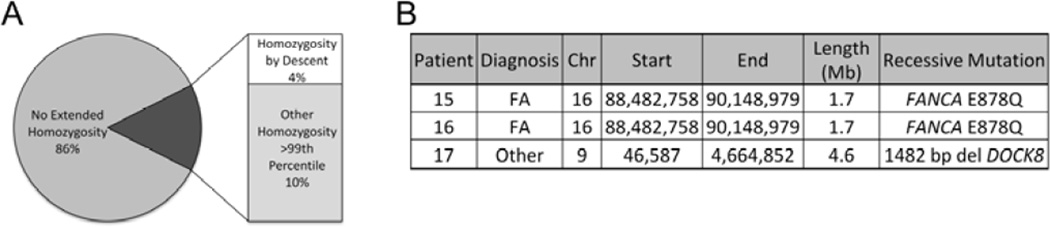
Fourteen percent of the patient cohort (13 of 91 patients) had inherited regions of CN-LOH exceeding the 99th percentile of CN-LOH size seen in the normal population (Figure 2A). Four of 13 had known consanguinity; these patients had multiple regions of homozygosity due to identity by descent. Three of the 4 patients had a pathogenic autosomal recessive mutation within the region of homozygosity (Figure 2B). Two brothers with FA shared a region of homozygosity encompassing a FANCA mutation. A third patient, referred for cytopenias and frequent infections, had CN-LOH in the region of chromosome 9 encompassing a DOCK8 mutation, known to cause a primary combined immunodeficiency syndrome (Su 2010). Nine patients without consanguinity had a single tract of extended homozygosity (Supporting Table 2), probably reflecting an ancestral haplotype with high linkage disequilibrium or a region of cryptic relatedness within the family (Gibson, et al 2006). Seven patients had homozygosity at distinct autosomal sites, while two had an overlapping region of homozygosity on the Xchromosome. The median number of genes contained within these tracts of homozygosity was 66, ranging from 23 to 357.
Figure 2. Inherited Regions of Extended Homozygosity Are Common, and Frequently Uncover Recessive Mutations in Inherited BMFS.
A. Pie chart showing the distribution of inherited regions of homozygosity in excess of 99th percentile of what is seen in the normal population. Fourteen percent of the patient cohort (13 of 91 patients) had abnormally large regions of homozygosity; of these, four patients had known consanguinity. B. Three of the four patients with known consanguinity harboured pathogenic autosomal recessive mutation within the homozygosity region. FA, Fanconi Anaemia.
Polymorphic Copy Number Variants (CNVs) May Contribute to Pathogenesis of BMFS
While the nature of our study precluded a genome-wide association analysis, we were able to identify several CNVs that were enriched in the two largest diagnostic subgroups in our cohort (Neutropenia NOS and aAA) compared to ethnically-matched controls. In the Neutropenia NOS subgroup, we found a significant enrichment of predicted deletions within the complex inverted duplication region on chromosome 5q13.2, which includes the genes General transcription factor IIH, polypeptide 2 (GTF2H2), NLR family apoptosis inhibitory protein (NAIP), and LOC647859, as well as an enrichment of deletions in the intronic regions of strawberry notch homolog (SBNO2) and zinc finger protein 516 (ZNF516). In the aAA subgroup, we found a significant enrichment of predicted deletions in SBNO2. Further studies, which are beyond the scope of this retrospective clinical analysis, are needed to determine the significance of these findings. The complete dataset is presented in Supporting Tables 3A–3D.
A targeted analysis of candidate genes implicated in bone marrow failure and haematological malignancies confirmed abnormalities detected previously, and identified 13 additional abnormalities of unknown significance (Supporting Figure 2, Supporting Table 4). Six candidate genes were included within the long tracts of homozygosity in patients with a history of consanguinity. Five genes were located within larger regions of acquired cytogenetic abnormalities (e.g. monosomy 7). Two genes had small CNVs (a duplication of exons 30–35 of RTEL1, and an intronic deletion of ETV1).
Clinical Utility of SNP Genotyping is Highest at Diagnosis and at Relapse
To better define the role of clinical SNP-A testing in the BMFS patients, we compared clinical findings of conventional cytogenetics and SNP-A genotyping. From a total of 109 SNP-A performed on 86 patients with a normal karyotype, 86.2% of SNP-A were reported as abnormal using the clinical reporting thresholds of > 20 SNPs for CNV and >5 Mb for CN-LOH. When stratified by SNP-A platform, 75.4% of the lower density Quad610 SNP-A and 98% of the higher density Omni1-Quad SNP-A were reported as abnormal (Supporting Table 5). While the majority of variants identified by SNP-A represented polymorphisms, a number of abnormalities were pathogenic and were not identified by conventional karyotyping. These included CN-LOH, a small clone of trisomy 13, and the delineation of the ring chromosome 21 breakpoint in the case of RUNX1 haploinsufficiency.
Because many BMFS patients undergo annual surveillance with bone marrow biopsies and cytogenetics, we were interested in the value of serial SNP-A genotyping as an adjunctive test in longitudinal follow-up of this patient population. Among 25 patients who had multiple SNP-A performed in the course of their disease (median interval between genotyping of 13 months), the three most common reasons for repeating SNP-A genotyping were: 1) routine testing at the time of an annual bone marrow biopsy (n=20, 62.5%), 2) follow-up of a known acquired CN-LOH clone in a stable patient (n=7, 21%), and 3) disease relapse (n=4, 12%). Notably, the yield of detecting a newly acquired abnormality on repeat SNP-A genotyping was significantly higher at the time of relapse (2 of 4) than when performed routinely in a stable patient without a known CN-LOH abnormality (0 of 20); this difference was statistically significant (OR 41, 95% CI 1.50- 1120.65, p=0.022) (Figure 3). When compared solely based on clinical stability at the time of repeat SNP-A genotyping, the likelihood of detecting a new acquired abnormality was significantly higher in the setting of relapse (2 of 4) than in the setting of stable disease (1 of 28) (OR 27, 95% CI 1.23 to 808.54, p=0.035).
Figure 3. Clinical Utility of SNP Array Genotyping in the Longitudinal Follow-Up of BMFS.
Histogram comparison showing the relative yield of detecting a new acquired abnormality on repeat single nucleotide polymorphism arrays (SNP-A), stratified by the clinical setting when repeated SNP-A genotyping was obtained. While the most common reason for repeating SNP-A genotyping in this series was routine testing at the time of an annual bone marrow biopsy, the yield of detecting a newly acquired abnormality was significantly higher when SNP genotyping was repeated at clinical progression. This difference was statistically significant (*). LOH, loss of heterozygosity.
Discussion
In this genome-wide SNP-A analysis of a comprehensively characterized BMFS patient cohort, we have found that BMFS have distinct patterns of genetic abnormalities. Acquired CN-LOH is significantly more frequent in aAA than in other BMFS (OR 12.2, p<0.01). In contrast, acquired CNAs are more typical of MDS and unclassified BMFS. Extended tracts of homozygosity are common in this patient population and can contribute to the pathogenesis of inherited BMFS by unmasking recessive loci. We found several polymorphic CNVs that are enriched in patients with aAA and neutropenia and may serve as disease modifiers. From a clinical utilization standpoint, we have shown that SNP-A genotyping is a useful adjunct in the diagnostic evaluation of BMFS, and repeat SNP-A may be helpful in the follow-up of patients at the time of relapse.
Interestingly, while acquired 6p CN-LOH was previously noted to be a recurrent abnormality in aAA and linked to the loss of particular HLA alleles (Afable, et al 2011, Katagiri, et al 2011), the greater prevalence of acquired CN-LOH in aAA compared to other BMFS has not been previously appreciated. The paucity of acquired CN-LOH in non-aAA BMFS in our study is consistent with prior analyses of hypoplastic MDS and FA (Afable, et al 2011, Quentin, et al 2011). Expanding our comparative analysis to include the published SNP-A studies in individual BMFS (Afable, et al 2011, Katagiri, et al 2011, Quentin, et al 2011) (Supporting Table 6) supports our conclusion that acquired CN-LOH is significantly more frequent in aAA (OR 6.5, CI 1.653–56.053, p=0.002), occurring in an estimated 13% of aAA cases. This difference probably reflects the distinct pathogenesis of aAA, and may be helpful diagnostically.
Intriguingly, while roughly 75% of acquired CN-LOH in aAA involve 6p, over 25% of acquired CNLOH involve other, non-6p regions, of which none were recurrent in the present study as well as the two previously published studies (Afable, et al 2011, Katagiri, et al 2011). Our data suggest that these CN-LOH clones tend to be smaller, non-dominant, and can vary over time. Despite representing clonal haematopoiesis, acquired CN-LOH clones in aAA appear to be distinct from the typical oncogenic mutation-driven acquired CN-LOH seen in myeloid malignancies (e.g. those associated with mutations in CBL, JAK2, TET2, FLT3) where dominant CN-LOH clones recur at specific regions due to a strong proliferative advantage conferred by specific oncogenic mutations. Instead, we hypothesize that the high frequency of diverse, non-dominant clones of acquired CN-LOH in aAA may emerge as a result of relative resistance to immune destruction, and may represent a general mechanism of immune escape in this disease. While 6p CN-LOH is thought to be caused by an escape of immune recognition through the loss of selected HLA subtypes (Afable, et al 2011, Katagiri, et al 2011), the other CN-LOH clones may arise due to decreased immune recognition or greater immune tolerance conferred by one of the parental alleles within the affected region. Follow-up studies are needed to determine the prognostic significance of CN-LOH clones in aAA, and to answer whether acquired CN-LOH can be predictive of response to immunosuppressive therapy. An important immediate clinical implication of the recurrent acquired 6p CN-LOH in aAA is the potential for inaccurate or ambiguous human leucocyte antigen (HLA) typing in this patient population when using peripheral blood for HLA typing (Smith, et al 2012). Given the frequency of 6p CN-LOH in aAA, in the absence of SNP-A information, HLA typing using buccal cells instead of peripheral blood should be considered in this patient population.
The potential utility of SNP-A in the clinical evaluation of BMFS was discussed at the Third Consensus Conference on the Treatment of Aplastic anaemia (Kojima, et al 2011), with the expert panel concluding that additional studies are needed to determine the clinical role of SNP-A in the evaluation and follow-up of patients with aplastic anaemia. Our analysis investigated the clinical utility of SNP-A in the evaluation of an unselected cohort of patients with BMFS. Our results show that SNP-A were particularly helpful as an adjunct to conventional cytogenetics at the time of initial diagnosis (e.g. to identify regions of acquired CN-LOH and inherited homozygosity, acquired CNAs with a small clone size, and CNVs). Our longitudinal analysis indicates that repeating SNP-A genotyping in the short-to-intermediate term follow-up is unlikely to reveal clonal evolution in a clinically stable patient. In contrast, repeating SNP-A at the time of relapse is significantly more likely to reveal a newly acquired abnormality. Additional studies are needed to assess the role of longitudinal SNP-A follow-up over a longer timeframe, and to clarify possible differences between the individual BMFS in a subgroup analysis.
This study is largely a retrospective analysis of a clinical dataset, and, as such, has limitations. We did not have access to paired constitutional DNA for the majority of patients. In accordance with the published characteristics of acquired and inherited CNVs (Afable, et al 2011), we used the B-allele frequencies to conservatively classify clonal abnormalities (e.g. trisomy 8, 6p CNLOH) as acquired, and non-clonal abnormalities as inherited. Inherited variants in our study were smaller and predominantly interstitial, in agreement with prior studies (Afable, et al 2011); however, it is possible that our study slightly underestimated the number of acquired variants due to classifying them as inherited. Due to the heterogeneous nature of this patient cohort, we were limited in our ability to perform a full association analysis. Further studies are needed to validate candidate CNVs found in our study, and also to identify additional modifier loci in BMFS.
In conclusion, we present a genome-wide SNP-A analysis of a comprehensively characterized patient cohort with congenital and acquired BMFS. Our results show distinct patterns of genomic abnormalities in BMFS, and demonstrate that acquired CN-LOH is significantly more frequent in aAA compared to other BMFS. Additional longitudinal studies are needed to better characterize the significance of acquired CN-LOH clones in aAA, as well as to validate and further characterize the role of modifier genes in BMFS.
Supplementary Material
Acknowledgements
We thank all patients and their referring physicians for participation in our studies. We are grateful to Kelly Thomas and Cecilia Kim for help with SNP-A genotyping, and to Donna Wilmoth and Laura Tooke for technical assistance. This work was supported by the NIH 5-T32-HL-07439-34 grant to D.B., NIH/NIGMS T32-GM008638 grant to J.R., and by the NCI NIH R01 CA105312 and Buck Family Endowed Chair in Hematology to MB.
Footnotes
Author Contributions
D.B. and M.B. designed the study, G.P. designed the human subjects research aspects of the project, T.O, J.C. and M.B. enrolled patients, D.B., T.O., J.C., and M.B. performed clinical record review, D.B., J.R., N.P., J.B. analysed SNP arrays, H.X., X.G. and J.P. performed computational CNV and LOH predictions, D.B. and H.X. performed bioinformatic analysis, D.B., H.X. and Y.L. performed statistical analysis, H.H. provided genotyping of normal controls and performed SNP array genotyping, D.B., H.X., J.R., N.P., M.P., P.M., J.B., and M.B. analysed and interpreted the data, D.B. wrote the manuscript, D.B., H.X., J.R., N.P., T.O., Y.L., G.P., P.M., J.B., and M.B. edited the manuscript. All authors approved the manuscript. The authors declare no competing financial interests.
References
- Afable MG, 2nd, Wlodarski M, Makishima H, Shaik M, Sekeres MA, Tiu RV, Kalaycio M, O'Keefe CL, Maciejewski JP. SNP array-based karyotyping: differences and similarities between aplastic anemia and hypocellular myelodysplastic syndromes. Blood. 2011;117:6876–6884. doi: 10.1182/blood-2010-11-314393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter BP. Inherited bone marrow failure syndromes. In: Nathan DG, Orkin SH, Look AT, Ginsburg D, editors. Nathan and Oski’s Hematology of Infancy and Childhood. Philadelphia, PA: WB Saunders; 2003. pp. 280–365. [Google Scholar]
- Alter BP, Giri N, Savage SA, Peters JA, Loud JT, Leathwood L, Carr AG, Greene MH, Rosenberg PS. Malignancies and survival patterns in the National Cancer Institute inherited bone marrow failure syndromes cohort study. British journal of haematology. 2010;150:179–188. doi: 10.1111/j.1365-2141.2010.08212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessler M, Mason PJ, Link DC, Wilson DB. Inherited bone marrrow failure syndromes. In: Nathan DG, Orkin SH, Ginsburg D, Look AT, Fisher DE, Lux S, editors. Nathans and Oski's Hematology of Infancy and Childhood. Saunders; 2008. [Google Scholar]
- Donadieu J, Fenneteau O, Beaupain B, Beaufils S, Bellanger F, Mahlaoui N, Lambilliotte A, Aladjidi N, Bertrand Y, Mialou V, Perot C, Michel G, Fouyssac F, Paillard C, Gandemer V, Boutard P, Schmitz J, Morali A, Leblanc T, Bellanne-Chantelot C. Classification of and risk factors for hematologic complications in a French national cohort of 102 patients with Shwachman-Diamond syndrome. Haematologica. 2012;97:1312–1319. doi: 10.3324/haematol.2011.057489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror Y, Durie P, Ginzberg H, Herman R, Banerjee A, Champagne M, Shannon K, Malkin D, Freedman MH. Clonal evolution in marrows of patients with Shwachman-Diamond syndrome: a prospective 5-year follow-up study. Experimental hematology. 2002;30:659–669. doi: 10.1016/s0301-472x(02)00815-9. [DOI] [PubMed] [Google Scholar]
- Gai X, Perin JC, Murphy K, O'Hara R, D'Arcy M, Wenocur A, Xie HM, Rappaport EF, Shaikh TH, White PS. CNV Workshop: an integrated platform for highthroughput copy number variation discovery and clinical diagnostics. BMC Bioinformatics. 2010;11:74. doi: 10.1186/1471-2105-11-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson J, Morton NE, Collins A. Extended tracts of homozygosity in outbred human populations. Human molecular genetics. 2006;15:789–795. doi: 10.1093/hmg/ddi493. [DOI] [PubMed] [Google Scholar]
- Katagiri T, Sato-Otsubo A, Kashiwase K, Morishima S, Sato Y, Mori Y, Kato M, Sanada M, Morishima Y, Hosokawa K, Sasaki Y, Ohtake S, Ogawa S, Nakao S. Frequent loss of HLA alleles associated with copy number-neutral 6pLOH in acquired aplastic anemia. Blood. 2011;118:6601–6609. doi: 10.1182/blood-2011-07-365189. [DOI] [PubMed] [Google Scholar]
- Kaufman DW, Kelly JP, Levy M, Shapiro S. The Drug Etiology of Agranulocytosis and Aplastic Anemia. New York: Oxford; 1991. [Google Scholar]
- Kojima S, Nakao S, Young N, Bacigalupo A, Gerard G, Hirano N, Maciejewski J, Deeg J, Marsh J, Zhang FK, Lee JW, Ozawa K. The Third Consensus Conference on the treatment of aplastic anemia. International journal of hematology. 2011;93:832–837. doi: 10.1007/s12185-011-0873-0. [DOI] [PubMed] [Google Scholar]
- Maciejewski JP, Selleri C. Evolution of clonal cytogenetic abnormalities in aplastic anemia. Leukemia & lymphoma. 2004;45:433–440. doi: 10.1080/10428190310001602363. [DOI] [PubMed] [Google Scholar]
- Mehta PA, Harris RE, Davies SM, Kim MO, Mueller R, Lampkin B, Mo J, Myers K, Smolarek TA. Numerical chromosomal changes and risk of development of myelodysplastic syndrome--acute myeloid leukemia in patients with Fanconi anemia. Cancer genetics and cytogenetics. 2010;203:180–186. doi: 10.1016/j.cancergencyto.2010.07.127. [DOI] [PubMed] [Google Scholar]
- Parikh S, Perdigones N, Paessler M, Greenbaum B, Tooke LS, Biegel JA, Mason PJ, Bessler M. Acquired copy number neutral loss of heterozygosity of chromosome 7 associated with clonal haematopoiesis in a patient with Shwachman-Diamond syndrome. British journal of haematology. 2012;159:480–482. doi: 10.1111/bjh.12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quentin S, Cuccuini W, Ceccaldi R, Nibourel O, Pondarre C, Pages MP, Vasquez N, Dubois d'Enghien C, Larghero J, Peffault de Latour R, Rocha V, Dalle JH, Schneider P, Michallet M, Michel G, Baruchel A, Sigaux F, Gluckman E, Leblanc T, Stoppa-Lyonnet D, Preudhomme C, Socie G, Soulier J. Myelodysplasia and leukemia of Fanconi anemia are associated with a specific pattern of genomic abnormalities that includes cryptic RUNX1/AML1 lesions. Blood. 2011;117:e161–e170. doi: 10.1182/blood-2010-09-308726. [DOI] [PubMed] [Google Scholar]
- Shimamura A, Alter BP. Pathophysiology and management of inherited bone marrow failure syndromes. Blood Reviews. 2010;24:101–122. doi: 10.1016/j.blre.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AG, Fan W, Regen L, Warnock S, Sprague M, Williams R, Nisperos B, Zhao LP, Loken MR, Hansen JA, Pereira S. Somatic mutations in the HLA genes of patients with hematological malignancy. Tissue Antigens. 2012;79:359–366. doi: 10.1111/j.1399-0039.2012.01868.x. [DOI] [PubMed] [Google Scholar]
- Su HC. Dedicator of cytokinesis 8 (DOCK8) deficiency. Current opinion in allergy and clinical immunology. 2010;10:515–520. doi: 10.1097/ACI.0b013e32833fd718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow S, Campo E, Harris NL, editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissue. Geneva, Switzerland: World Health Organization; 2008. [Google Scholar]
- Teo JT, Klaassen R, Fernandez CV, Yanofsky R, Wu J, Champagne J, Silva M, Lipton JH, Brossard J, Samson Y, Abish S, Steele M, Ali K, Athale U, Jardine L, Hand JP, Tsangaris E, Odame I, Beyene J, Dror Y. Clinical and genetic analysis of unclassifiable inherited bone marrow failure syndromes. Pediatrics. 2008;122:e139–e148. doi: 10.1542/peds.2007-3415. [DOI] [PubMed] [Google Scholar]
- Tonnies H, Huber S, Kuhl JS, Gerlach A, Ebell W, Neitzel H. Clonal chromosomal aberrations in bone marrow cells of Fanconi anemia patients: gains of the chromosomal segment 3q26q29 as an adverse risk factor. Blood. 2003;101:3872–3874. doi: 10.1182/blood-2002-10-3243. [DOI] [PubMed] [Google Scholar]
- van der Crabben S, van Binsbergen E, Ausems M, Poot M, Bierings M, Buijs A. Constitutional RUNX1 deletion presenting as non-syndromic thrombocytopenia with myelodysplasia: 21q22 ITSN1 as a candidate gene in mental retardation. Leukemia Research. 2010;34:e8–e12. doi: 10.1016/j.leukres.2009.06.030. [DOI] [PubMed] [Google Scholar]
- Walker LC, Stevens J, Campbell H, Corbett R, Spearing R, Heaton D, Macdonald DH, Morris CM, Ganly P. A novel inherited mutation of the transcription factor RUNX1 causes thrombocytopenia and may predispose to acute myeloid leukaemia. British journal of haematology. 2002;117:878–881. doi: 10.1046/j.1365-2141.2002.03512.x. [DOI] [PubMed] [Google Scholar]
- Young NS, Calado RT, Scheinberg P. Current concepts in the pathophysiology and treatment of aplastic anemia. Blood. 2006;108:2509–2519. doi: 10.1182/blood-2006-03-010777. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.