Abstract
Objective
To analyze the association between −1082A/G polymorphism in interleukin-10 (IL-10) gene and ischemic stroke (IS) risk by meta-analysis.
Methods
We carried out a systematic electronic search in PubMed, BIOSIS Previews, Science Direct, Chinese National Knowledge Infrastructure, Chinese Biomedical Database, Weipu database and WANGFANG Database. Pooled odds ratios (ORs) with 95% confidence intervals (95%CIs) were calculated to assess the strength of the association.
Results
7 studies were included. There was no significant association between IL-10 −1082A/G polymorphism and IS risk under all genetic models in overall estimates (A vs. G: OR = 1.23,95%CI = 0.85–1.79;AA vs. GG: OR = 1.01,95%CI = 0.47–2.19; AG vs. GG: OR = 0.76, 95%CI = 0.38–1.55; AA+AG vs. GG: OR = 0.89,95%CI = 0.46–1.73; AA vs. AG+GG: OR = 1.39, 95%CI = 0.91–2.13). Similarly, no associations were found in subgroup analysis based on ethnicity and source of controls. However, removing the study deviating from Hardy–Weinberg equilibrium (HWE) produced statistically significant associations for overall estimates under recessive model(AA VS. AG+GG OR 1.58, 95% CI 1.04–2.42) and among Asians in all genetic models (A VS.G OR 1.64, 95% CI 1.07–2.53; AA vs. GG OR1.91, 95% CI 1.31–2.80; AG vs. GG OR1.44, 95% CI 1.09–1.91; AA+AG vs. GG OR 1.54, 95% CI 1.18–2.01;AA VS. AG+GG OR 1.79, 95% CI 1.07–3.00). Even after Bonferroni correction, the associations were observed still significantly in Asians under the two models (AA vs. GG OR1.91, 95% CI 1.31–2.80, P = 0.0008; AA+AG vs. GG OR 1.54, 95% CI 1.18–2.01, P = 0.001).
Conclusion
This meta-analysis indicates that IL10 −1082 A/G polymorphism is associated with IS susceptibility in Asians and the −1082 A allele may increase risk of IS in Asians. Considering the sample size is small and between-study heterogeneity is remarkable, more studies with subtle design are warranted in future.
Introduction
Ischemic stroke (IS) is a major cause of adult disability and death in the world[1], which is a heterogeneous multifactorial disease associated with genetic and environmental factors[2]. During the past few years, more and more evidence showed that inflammatory molecules and the genetic variation of the genes which encoded these inflammatory cytokines might take part in the pathogenesis of stroke [3]. Inflammatory mechanisms may not only play important roles in the manifestation and development of IS, but also may be vulnerable to IS in time via accumulation of atherosclerotic disease and maintain of atrial fibrillation [4]. Several candidate genes of inflammatory cytokines are implicated in the pathogenesis of IS, one of which is interleukin −10 (IL-10).
IL-10(Gene ID: 3586) is a multifunctional cytokine with anti-inflammatory properties, which has been showed involving in the inflammatory process of IS[5]. The human IL-10 gene is located on chromosome 1q31-32, in which some polymorphisms have been found in the promoter region, such as, −1082A/G (rs1800896), −592C/A (rs1800872) and −829C/T (rs1800871) [6]. The −1082A/G (also named as −1087A/G in some studies) polymorphism could affect IL-10 production [7]. And it is believed that the A/G substitution is relevant to low/high amount of IL-10 secretion, respectively [7].
Emerging studies have reported the associations between −1082G/A polymorphism in IL-10 gene and IS risk[3], [8], [9], [10], [11], [12], [13], but the results are inconclusive. Given that a single study may be too underpowered to provide reliable conclusion owing to relatively small sample size, we performed this meta-analysis to estimate the association between IL10 −1082A/G polymorphism and IS susceptibility more precisely.
Materials and Methods
Search Strategy
This meta-analysis conformed to the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) criteria [14]. We carried out systematic literature searches in PubMed, BIOSIS Previews, Science Direct, Chinese Biomedical Database (http://sinomed.imicams.ac.cn/index.jsp), Weipu database (http://cstj.cqvip.com/), Chinese National Knowledge Infrastructure (http://dlib3.cnki.net/kns50/),and WANFANG Database (http://g.wanfangdata.com.cn/) up to 10th October 2013 to identify relevant studies, using the following key words: (“interleukin-10” OR “interleukin 10” OR “IL-10” OR “IL10”) and (stroke OR “cerebrovascular accident” OR “cerebral ischemia” OR “cerebral infarction”) and (“polymorphism” OR “mutation” OR “genotype” OR “allele” OR “variation” OR “variant”). In addition, hand searching of the references in selected literatures and the abstracts presented at relevant conferences were performed for other potential related studies. Languages were limited to English and Chinese.
Selection Criteria
Studies meeting the following criteria were included:(1) the study should evaluate the relationship between IL-10 gene −1082A/G polymorphism and IS risk;(2) the study had to be a case-control design; (3) genotype distributions in both cases and controls were available for calculating an odds ratio (OR) with 95% confidence interval (CI); (4)Computed tomographic(CT) or magnetic resonance imaging(MRI) were used to assess the diagnosis of IS. The following were exclusion criteria: (1) reviews, abstracts or animal studies; (2)studies were not relevant to IL-10 gene −1082A/G polymorphism or IS; (3) studies did not report genotype frequencies;(4) the design were based on sibling pairs or family. If studies were repeated or overlapped publications, the most complete one was included. If studies did not report detailed data, we would get in touch with authors to obtain the relevant information.
Data Extraction
Two investigators (Jin and Li) reviewed and extracted data independently in accordance with the inclusion criteria. The results were compared, and if any disagreement appeared, a third investigator (Peng) was invited to evaluate such studies, then the discrepancy was resolved by discussion. The following information were extracted: the name of first author, year of publication, country (ethnicity), diagnostic criteria of IS, study design, sample size, allele numbers and genotype distributions in cases and controls.
Quality Assessment
The quality of included studies were assessed by 2 investigators (Jin and Li) independently on the basis of Newcastle-Ottawa Scale (NOS)[15] which consisted of three aspects: selection, comparability, and exposure, and each satisfactory answer received one point. Studies with a score equal to or higher than five were regarded as of high quality.
Statistical Analysis
Pooled ORs with corresponding 95% CIs were calculated to evaluate the strength of relationship between IL 10 gene −1082 A/G polymorphism and IS risk under the following five genetic models: the allele model (A vs. G), the homozygote model (AA vs. GG), the heterozygote model (AG vs. GG), the dominant model (AA+AG vs. GG), and the recessive model (AA vs. AG+GG). Z-test was used to assess the significance of the pooled OR, in which P<0.05 was considered as statistically significant. The Q-test and I2-statistics were employed for evaluating the between-study heterogeneity, which was considered as significant when PQ≤0.10 or I2>50%[16]. Then, the overall or pooled OR was obtained by a random-effect (DerSimonian-Laird method)[17] or a fixed-effect model (Mantel- Haenszel method)[18] in the presence (PQ≤0.10 or I2>50%) or absence (PQ>0.10 or I2≤50%) of heterogeneity, respectively. Furthermore,to explore the sources of heterogeneity, we conducted subgroup analysis based on ethnicity, source of controls,respectively. Moreover, Bonferroni method, controlling for false positive error rate, was utilized to adjust for multiple comparisons. As we performed multiple comparisons in this meta-analysis for 25 times, the P value which was less than 0.05/25 (0.002) indicated statistical significance after Bonferroni correction. To validate the reliability of the results, sensitivity analysis was performed though omitting one case–control study each time, as well as limiting this meta-analysis to studies which were conformed to HWE. HWE of genotype distribution in the controls of included studies was conducted using an online program (http://ihg2.helmholtz-muenchen.de/cgi-bin/hw/hwa1.pl), and P<0.05 was considered significantly deviating from HWE. Publication bias was evaluated by visual inspection of symmetry of Begg's funnel plot and assessment of Egger's test[19] (P<0.05 was regarded as representative of statistical significance). All statistical analyses were performed using software RevMan 5.1 and STATA 11.0.
Results
Study Characteristics
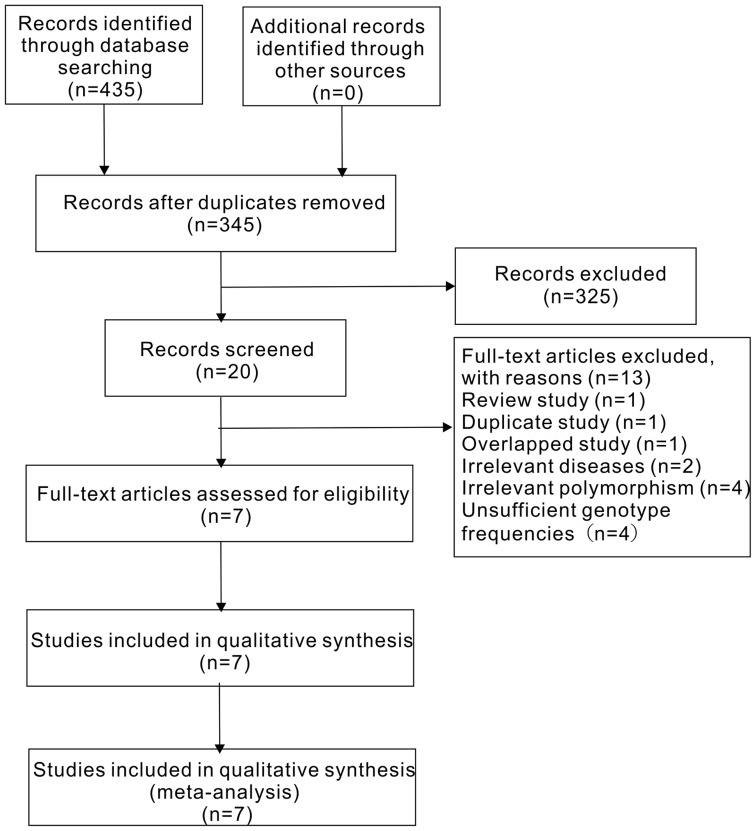
Figure 1 displayed the selection process of this study. A total of 435 literatures were identified after an initial search. Of these studies, the first screening excluded 415 citations based on inclusion criteria, leaving 20 articles for further selection. Among the remaining 20 literatures, 2 explored other diseases instead of IS[20], [21]; 4 researched other polymorphisms of IL-10 gene[22], [23], [24], [25]; 2 reported overlapped data [4], [8], then the one with more complete information was included[8]; 1 was a duplicate study[26];1 was a review study[27],4 did not have sufficient genotype frequencies[28], [29], [30], [31].Finally, a total of 7 studies were included in our meta-analysis [3], [8], [9], [10], [11], [12], [13],consisting of 1533 cases and 1227 controls.
Figure 1. Flow diagram of the study selection process.
The detailed characteristics of the included studies are listed in Table 1.
Table 1. Characteristics of studies included in the meta-analysis.
| Study | Ethnicity | Study design | Control source | Genotype distribution(case/control) | HWE | NOS | ||
| AA | AG | GG | (P) | |||||
| Marousi2011[8] | Greece | Case-control | Hospital based | 47/53 | 71/71 | 27/21 | 0.72 | 6 |
| Munshi2010[3] | Indian | Case-control | Population based | 92/63 | 241/218 | 147/189 | 0.99 | 6 |
| Tuttolomondo2012[9] | Caucasian | Case-control | Hospital based | 58/20 | 14/17 | 24/11 | 0.07 | 6 |
| Jin 2011[11] | Chinese | Case-control | Hospital based | 161/78 | 27/12 | 1/2 | 0.09 | 7 |
| Zhang2007[12] | Chinses | Case-control | Hospital based | 202/120 | 2/11 | 0/0 | 0.61 | 6 |
| Lin 2009[13] | Chinese | Case-control | Hospital based | 153/83 | 28/32 | 0/0 | 0.08 | 6 |
| Sultana 2011[10] | Indian | Case-control | Population based | 154/163 | 44/47 | 40/16 | 0.000 | 7 |
All included studies were of high quality as the NOS score of each one was higher than 5 points and the genotype distributions in all controls were consistent with HWE except 1 study[10].All the 7 eligible studies were case-control studies, 2 of them were in a population-based design[3], [10], the remaining were hospital-based[8], [9], [11], [12], [13]. All studies used stroke-free people as controls except 1 study[12] which recruited healthy people as controls. All the cases were recruited from hospitalized patients and had a brain CT or MRI to assess the diagnosis of IS. 1 study only involved first-ever stroke patients[13], 2 studies included first-ever and recurrent strokes[3], [8], others did not describe the detailed information[9], [10], [11], [12]. All studies did not have age limitations for cases. Moreover, ethnic groups in these studies were as following: 2 were Caucasians[8], [9],2 were Indians[3], [10], and 3 were Chinese[11], [12], [13] (n = 3).
Quantitative Synthesis: Overall study
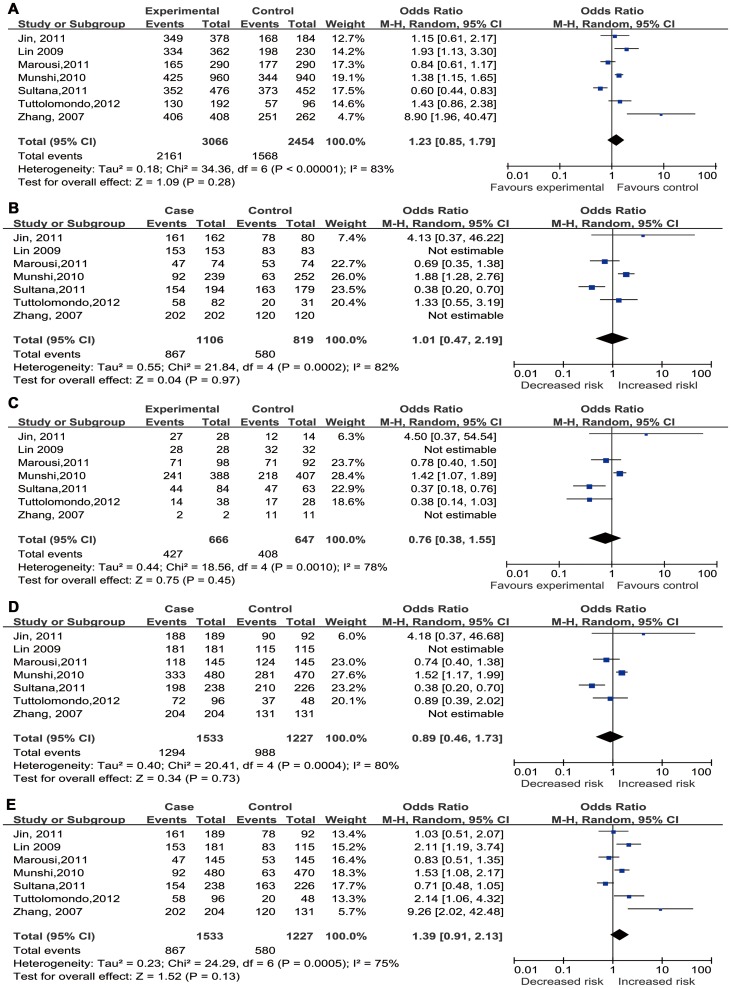
As shown in Table 2 and Figure 2,significant between-study heterogeneity appeared under all genetic models for overall analysis,thus, a random-effect model was utilized to calculate the pooled estimates. Overall, no significant relationship between IL-10 −1082 A/G polymorphism and IS risk was found in all genetic models (A vs. G: OR = 1.23,95%CI = 0.85–1.79; AA vs. GG: OR = 1.01,95%CI = 0.47–2.19; AG vs. GG: OR = 0.76,95%CI = 0.38–1.55; AA+AG vs. GG: OR = 0.89,95%CI = 0.46–1.73; AA vs. AG+GG: OR = 1.39,95%CI = 0.91–2.13).
Table 2. Summary risk estimates for association between IL-10-1082A/G polymorphism and IS.
| Comparisons | Stratifications | Studies(n) | M | Pooled estimate | Heterogeneity | ||
| OR(95%CI) | PZ | I2 (%) | PH | ||||
| A vs. G | Overall | 7 | R | 1.23(0.85, 1.79) | 0.35 | 83 | <0.0001 |
| Asians | 5 | R | 1.37(0.80, 2.34) | 0.26 | 87 | <0.0001 | |
| Non-Asians | 2 | R | 1.06(0.63, 1.78) | 0.82 | 66 | 0.09 | |
| Hospital-based | 5 | R | 1.47(0.90, 2.40) | 0.13 | 73 | 0.006 | |
| Population-based | 2 | R | 0.92(0.41, 2.07) | 0.84 | 95 | <0.0001 | |
| AA vs. GG | Overall | 5 | R | 1.01(0.47, 2.19) | 0.97 | 82 | 0.0002 |
| Asians | 3 | R | 1.15(0.29, 5.54) | 0.84 | 90 | <0.0001 | |
| Non-Asians | 2 | F | 0.88(0.51, 1.52) | 0.66 | 25 | 0.25 | |
| Hospital -based | 3 | F | 0.96(0.57, 1.62) | 0.87 | 29 | 0.25 | |
| Population-based | 2 | R | 0.86(0.18, 4.14) | 0.94 | 95 | <0.0001 | |
| AG vs. GG | Overall | 5 | R | 0.76 (0.38, 1.55) | 0.45 | 78 | 0.001 |
| Asians | 3 | R | 0.99 (0.31, 3.19) | 0.99 | 84 | 0.002 | |
| Non-Asians | 2 | F | 0.63 (0.36, 1.08) | 0.09 | 28 | 0.24 | |
| Hospital -based | 3 | F | 0.69(0.41, 1.18) | 0.17 | 46 | 0.16 | |
| Population-based | 2 | R | 0.76(0.21, 2.80) | 0.69 | 91 | 0.0006 | |
| AA+AG vs.GG | Overall | 5 | R | 0.89(0.46, 1.73) | 0.73 | 80 | 0.0004 |
| Asians | 3 | R | 1.02(0.30, 3.46) | 0.97 | 89 | 0.0001 | |
| Non-Asians | 2 | F | 0.79(0.48, 1.30) | 0.36 | 0 | 0.72 | |
| Hospital -based | 3 | F | 0.85(0.53, 1.38) | 0.52 | 0 | 0.39 | |
| Population-based | 2 | R | 0.78(0.20, 3.06) | 0.72 | 80 | 0.0004 | |
| AA vs.AG+GG | Overall | 7 | R | 1.39(0.91, 2.13) | 0.13 | 75 | 0.0005 |
| Asians | 5 | R | 1.48(0.85, 2.57) | 0.17 | 79 | 0.0007 | |
| Non-Asians | 2 | R | 1.29(0.51, 3.23) | 0.59 | 79 | 0.03 | |
| Hospital -based | 5 | R | 1.68(0.93, 3.04) | 0.09 | 72 | 0.006 | |
| Population-based | 2 | R | 1.05(0.49, 2.23) | 0.90 | 88 | 0.004 | |
NA, data not available;
M, Statistical model.
R, random-effects model; F, fixed-effects model.
PZ, P value for Z test; PH, P value for heterogeneity.
Figure 2. Forest plots for association between IL-10 −1082 A/G polymorphism and IS risk in different genetic models.
((A) Allele model(A vs. G); (B) Homozygote model (AA vs. GG); (C) Heterozygote model (AG vs. GG); (D) Dominant model (AA+AG vs. GG); (E) Recessive model (AA vs. AG+GG)).
Notably, there was no GG genotype in either case or control group of both the Lin's study and Zhang's study[12], [13], resulting in that the two studies did not contribute to the pooled ORs in homozygote model, heterozygote model,and dominant model. In other words, in the three models, there were only 5 substantially valuable studies for overall analysis. The main results of this meta-analysis were shown in Table 2 and Figure 2.
Subgroup analysis
To explore the sources of heterogeneity, we performed further subgroup analyses by ethnicity and source of controls respectively. Similarly, there were no significant associations in the subgroup analyses, and significant heterogeneity in most of the comparison models still existed. Table 2 showed the detailed results.
Sensitivity Analysis
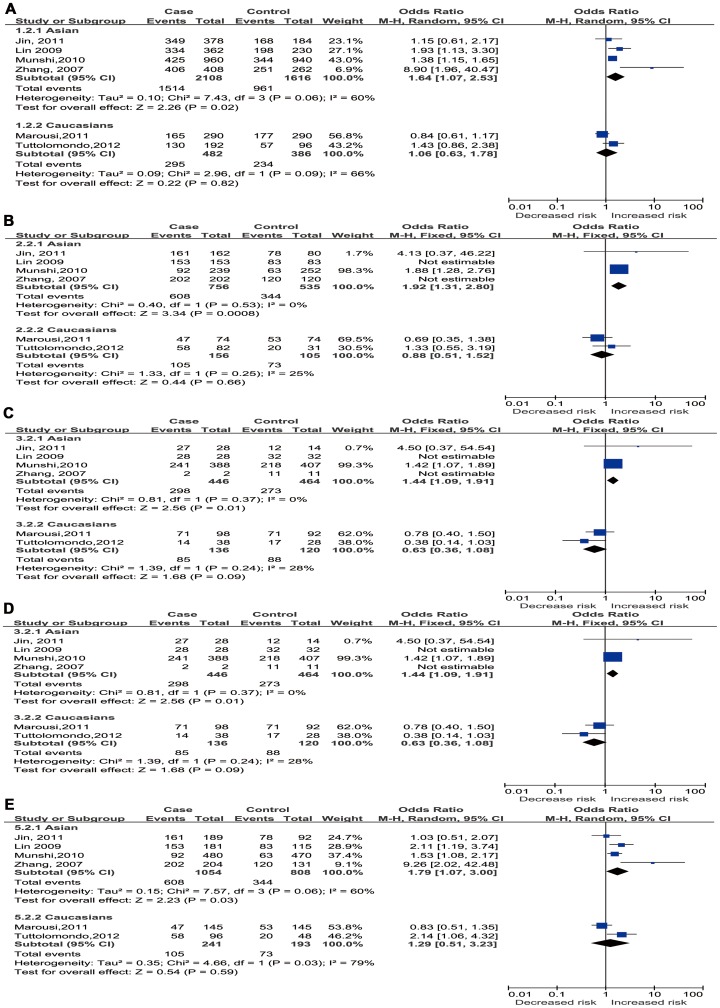
Sensitivity analysis was conducted to evaluate the stability of the results. After excluding the study deviating from HWE[10], statistically significant associations were observed for Asians under all the genetic models (A VS.G: OR 1.64, 95% CI 1.07–2.53, P = 0.02; AA vs. GG: OR 1.91, 95% CI 1.31–2.80,P = 0.0008; AG vs. GG: OR1.44, 95% CI 1.09–1.91,P = 0.01 AA+AG vs. GG: OR1.54, 95% CI 1.18–2.01,P = 0.001,AA VS. AG+GG: OR 1.79, 95% CI 1.07–3.00, P = 0.03, whereas the corresponding pooled ORs of total available studies which meant overall analysis for all ethnicities were not materially altered (data not shown), except in recessive model(AA VS. AG+GG OR 1.58, 95% CI 1.04–2.42, P = 0.03). However, after Bonferronic correction for multiple testing, the associations revealed still significantly in Asian subgroup under homozygote and dominant models (AA vs. GG: OR 1.91, 95% CI 1.31–2.80, P = 0.0008; AA+AG vs. GG: OR1.54, P = 0.001). The results indicated that the homozygote AA and A allele carriers (AA+AG) had nearly a 91% and 54% increased risk of IS respectively, when compared with the homozygote GG in Asians. In particular, removing the study which was deviated from HWE eliminated the heterogeneity in subgroup analysis of Asians in homozygote, heterozygote, and dominant models, but not in the allele and recessive models and the pooling analysis (The results were shown in Table 3 and Figure 3.).Moreover, omitting the other 6 eligible studies one by one, the corresponding pooled ORs in overall comparisons and subgroup analysis were not significantly changed and the significant heterogeneity between studies still existed (data not shown).
Table 3. Sensitivity analysis: Study deviated from HWE were excluded in Asians under all models and for overall studies in recessive model.
| Comparisons | Stratifications | Studies(n) | M | Pooled estimate | Heterogeneity | ||
| OR(95%CI) | PZ | I2 (%) | PH | ||||
| A vs. G | Asians | 5 | R | 1.37(0.80, 2.34) | 0.26 | 87 | <0.0001 |
| SA-A | 4 | R | 1.64(1.07, 2.34) | 0.02 | 60 | 0.02 | |
| AA vs. GG | Asians | 5 | R | 1.15(0.29, 5.54) | 0.84 | 90 | <0.0001 |
| SA-A | 4 | F | 1.91(1.31, 2.80) | 0.0008* | 0 | 0.53 | |
| AG vs. GG | Asians | 5 | R | 0.99 (0.31, 3.19) | 0.99 | 84 | 0.002 |
| SA-A | 4 | F | 1.44 (1.09, 1.91) | 0.01 | 0 | 0.37 | |
| AA+AG vs.GG | Asians | 5 | R | 1.02 (0.30, 3.46) | 0.97 | 89 | 0.0001 |
| SA-A | 4 | F | 1.54(1.18, 2.01) | 0.001* | 0 | 0.42 | |
| AA vs.AG+GG | Overall | 7 | R | 1.39(0.91, 2.13) | 0.13 | 75 | 0.0005 |
| SA-O | 6 | R | 1.58(1.04, 2.42) | 0.03 | 66 | 0.01 | |
| Asians | 5 | R | 1.48(0.85, 2.57) | 0.17 | 79 | 0.0007 | |
| SA-A | 4 | R | 1.79(1.07, 3.00) | 0.03 | 60 | 0.03 | |
M, Statistical model.
R, random-effects model; F, fixed-effects model.
PZ, P value for Z test; PH, P value for heterogeneity.
SA-A: Sensitivity analysis (Study deviated from HWE were exclude).in Asians.
SA-O: Sensitivity analysis (Study deviated from HWE were exclude).in overall studies.
*:the association is sill significant after Bonferronic correction for multiple testing.
Figure 3. Forest plots for association between IL-10 −1082 A/G polymorphism and IS risk based on ethnicity for studies in Hardy-Weinberg equilibrium.
((A) Allele model (A vs. G); (B) Homozygote model (AA vs. GG); (C) Heterozygote model (AG vs. GG); (D) Dominant model (AA+AG vs. GG); (E) Recessive model (AA vs. AG+GG)).
Publication Bias
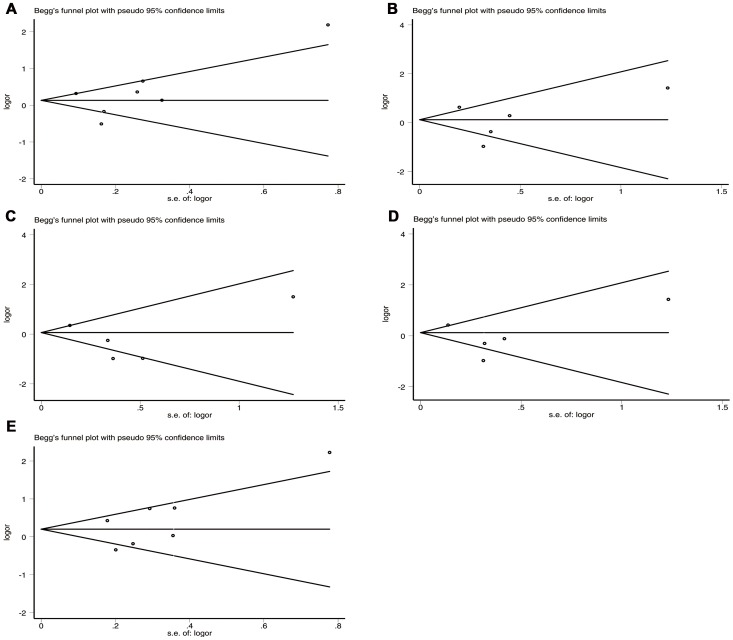
No obvious visual asymmetry was observed in Begg's funnel plots, and the results of Egger's test revealed no statistical evidence for publication bias among studies (P = 0.632 for allele model; P = 0.773 for homozygote model; P = 0.384 for heterozygote model; P = 0.503 for dominant model; P = 0.202 for recessive model) (Table 4 and Figure 4).
Table 4. Publication bias tests for association between IL-10-1082A/G polymorphism and IS.
| Comparisons | Egger test | Begg test | |
| P value | 95%CI | P value | |
| A vs. G | 0.632 | (−4.045, 6.047) | 0.548 |
| AA vs. GG | 0.773 | (−9.428, 7.727) | 0.806 |
| AG vs. GG | 0.384 | (−7.216, 3.719) | 0.806 |
| AA+AG vs. GG | 0.503 | (−7.828, 4.812) | 0.806 |
| GG vs. AG+AA | 0.202 | (−2.163, 7.922) | 0.133 |
Figure 4. Begg's funnel plots for IL-10 −1082 A/G polymorphism and IS risk.
((A) Allele model (A vs. G); (B) Homozygote model (AA vs. GG); (C) Heterozygote model(AG vs. GG); (D) Dominant model (AA+AG vs. GG); (E) Recessive model (AA vs. AG+GG)).
Discussion
It is now accepted that genetics and environmental factors contribute to IS susceptibility and outcome. Similarly, the inflammation reaction is also relevant to IS. IL-10 is a potent anti-inflammatory cytokine with multiple functions taking part in inflammation reaction as well as the development of IS. Recently, the associations between IL-10 gene −1082 A/G polymorphism and the risk of IS have been intensively investigated, however, the results are inconsistent. Therefore, we designed this meta-analysis to draw a more precise conclusion for the association between IL-10 −1082 A/G polymorphism and IS risk.
In this meta-analysis, no association of the IL-10 −1082A/G polymorphism with IS risk was found under all comparisons, and in subgroup analysis by ethnicity or source of controls. However, for Asians, after excluding the study deviating from HWE[10], the data indicated that IL-10 A allele was associated with increased risk of IS in Asians. The inconsistent outcome between Asians on subgroup analysis with overall estimates partly caused by genetic diversity in different ethnicities. In addition, the different subtypes of ischemic stroke may contribute to the conflicting results,for stroke in Asians is more often due to intracranial atherosclerosis than in other populations. Furthermore, as IS is a multifactorial disease, except genetic factors, environmental factors also take important parts in IS etiology. Thus, this discrepancy may also attribute to other environmental factors, such as different geographic distribution economic status, climate, lifestyle, diet,and so on. Importantly, there is currently no consensus for whether to include studies deviating from HWE. But if the results are different between including or excluding studies deviating from HWE, it is suggested that the analysis without studies departed from HWE may be more valid [32].
Significant between-study heterogeneity displayed among all comparison models. Considering that the diversity in design, difference of ethnicity, sample sizes, and measurement errors may contribute to common sources of heterogeneity [33], we conducted the subgroup analysis by ethnicity and control of sources trying to clarify the sources of heterogeneity. Unluckily, we did not effectively eliminate the heterogeneity, indicating us that all above factors should be taken into consideration. In addition, other factors such as subtype of IS, gender distribution, past medical history, personal history and so on, might also be responsible for the heterogeneity. Notably, after removing the study deviating from HWE, the heterogeneity were removed for subgroup analysis in Asians under homozygote, heterozygote, and dominant models, suggesting that the study deviating from HWE[10] was the main source of heterogeneity in the three models for Asians.
As far as we know, this is the first comprehensive meta-analysis exploring the association between IL-10 −1082 A/G polymorphism and IS risk up to now, which involved Caucasian, Indian and Chinese populations. In addition, more studies were included in our study than a recently published meta-analysis concerning −1082A/G polymorphism and IS risk only in South Asians[27]. Our meta-analysis also has some advantages. Firstly, the search and selection studies were conducted strictly. Secondly, the results of NOS indicated that the included studies were credible. Thirdly, no evidence of publication bias was found by Begg's funnel plot and Egger's test. Fourthly, multiple testing to adjust for multiple comparisons was performed which could reduce the type I error rate. In addition, we performed sensitive analysis by excluding studies deviating from HWE, considering that deviations from HWE in healthy populations may be a sign of selection bias or population stratification[34].
Despite of the advantages mentioned above, the current study has some inevitable limitations that should be acknowledged. First, there was significant heterogeneity among included studies. Even though we used the random-effects model to calculate pool ORs, the precision of outcome would be affected. Second, owing to limiting detailed information such as lacking of subtyping of the ischemic strokes in individual study, we failed to perform further subgroup analysis to adjust these possible confounders. Third, the sample size in individual included studies was respectively small, especially in homozygote, heterozygote and dominant models. Forth, only English and Chinese language studies were included in this meta-analysis which might have led to bias. Despite of conducting an exhaustive search for eligible studies, some relevant studies so called “grey literatures” might be still missed. Fifth, similar to a case–control study, meta-analysis was a retrospective study, which might lead to recall bias. Last but not the least, this study only explored one variation in IL-10 gene, which ignored such gene–gene and gene-environmental interactions.
In conclusion,this meta-analysis indicates that IL- 10 gene −1082 A/G polymorphism is associated with IS susceptibility in Asians and the −1082 A allele may increase risk of IS in Asian populations. However, considering the limitations mentioned above, more well designed studies with adequately sized populations are needed in future.
Supporting Information
PRISMA 2009 Checklist.
(DOC)
Funding Statement
The authors have no support or funding to report.
References
- 1. Amarenco P, Bogousslavsky J, Caplan LR, Donnan GA, Hennerici MG (2009) Classification of stroke subtypes. Cerebrovasc Dis 27: 493–501. [DOI] [PubMed] [Google Scholar]
- 2. Dichgans M (2007) Genetics of ischaemic stroke. Lancet Neurol 6: 149–161. [DOI] [PubMed] [Google Scholar]
- 3. Munshi A, Rajeshwar K, Kaul S, Al-Hazzani A, Alshatwi AA, et al. (2010) Interleukin-10-1082 promoter polymorphism and ischemic stroke risk in a South Indian population. Cytokine 52: 221–224. [DOI] [PubMed] [Google Scholar]
- 4. Marousi S, Antonacopoulou A, Kalofonos H, Papathanasopoulos P, Karakantza M, et al. (2011) Functional inflammatory genotypes in ischemic stroke: could we use them to predict age of onset and long-term outcome? Stroke Res Treat 2011: 792923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liesz A, Suri-Payer E, Veltkamp C, Doerr H, Sommer C, et al. (2009) Regulatory T cells are key cerebroprotective immunomodulators in acute experimental stroke. Nat Med 15: 192–199. [DOI] [PubMed] [Google Scholar]
- 6. Eskdale J, Gallagher G, Verweij CL, Keijsers V, Westendorp RG, et al. (1998) Interleukin 10 secretion in relation to human IL-10 locus haplotypes. Proc Natl Acad Sci U S A 95: 9465–9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kilpinen S, Huhtala H, Hurme M (2002) The combination of the interleukin-1alpha (IL-1alpha-889) genotype and the interleukin-10 (IL-10 ATA) haplotype is associated with increased interleukin-10 (IL-10) plasma levels in healthy individuals. Eur Cytokine Netw 13: 66–71. [PubMed] [Google Scholar]
- 8. Marousi S, Ellul J, Antonacopoulou A, Gogos C, Papathanasopoulos P, et al. (2011) Functional polymorphisms of interleukin 4 and interleukin 10 may predict evolution and functional outcome of an ischaemic stroke. Eur J Neurol 18: 637–643. [DOI] [PubMed] [Google Scholar]
- 9. Tuttolomondo A, Di Raimondo D, Forte GI, Casuccio A, Vaccarino L, et al. (2012) Single nucleotide polymorphisms (SNPs) of pro-inflammatory/anti-inflammatory and thrombotic/fibrinolytic genes in patients with acute ischemic stroke in relation to TOAST subtype. Cytokine 58: 398–405. [DOI] [PubMed] [Google Scholar]
- 10. Sultana S, Kolla VK, Jeedigunta Y, Penagaluru PK, Joshi S, et al. (2011) Tumour necrosis factor alpha and interleukin 10 gene polymorphisms and the risk of ischemic stroke in south Indian population. J Genet 90: 361–364. [DOI] [PubMed] [Google Scholar]
- 11. Jin L, Ni PH, Wu JM, Fu Yi, Ge HL (2011) The correlation between gene polymorphism of IL-10-819C/T and -1082G/A and cerebral infarction (In Chinese). Laboratory Medicine 26: 717–721. [Google Scholar]
- 12. Zhang GZ, Pan SY, Du R, lu BX, Li W (2007) The relationship between interleukin-10 gene polymorphisms and cerebral infarction (In Chinese). Chin J Cerebrovasc Dis 4: 294–297. [Google Scholar]
- 13.Lin KX (2009) Study on The Relationship between Interleukin–10 Gene Polymorphisms and Cerebral Infarction. Unpublished MsD dissertation [MsD]: Fujian Medical University.
- 14. Moher D, Liberati A, Tetzlaff J, Altman DG (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 6: e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stang A (2010) Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 25: 603–605. [DOI] [PubMed] [Google Scholar]
- 16. Attia J, Thakkinstian A, D'Este C (2003) Meta-analyses of molecular association studies: methodologic lessons for genetic epidemiology. J Clin Epidemiol 56: 297–303. [DOI] [PubMed] [Google Scholar]
- 17.DerSimonian R (1996) Meta-analysis in the design and monitoring of clinical trials. Stat Med 15: : 1237–1248, 1249–1252. [DOI] [PubMed] [Google Scholar]
- 18. MANTEL N, HAENSZEL W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22: 719–748. [PubMed] [Google Scholar]
- 19. Egger M, Davey SG, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Timasheva YR, Nasibullin TR, Zakirova AN, Mustafina OE (2008) Association of interleukin-6, interleukin-12, and interleukin-10 gene polymorphisms with essential hypertension in Tatars from Russia. Biochem Genet 46: 64–74. [DOI] [PubMed] [Google Scholar]
- 21. Xie GQ, Zhao LC, Li Y, Wang H, Liang LR, et al. (2008) The Relationship Between Interleukin-10 Gene -592A/C Polymorphism and Carotid Atherosclerosis (In Chinese). Molecular Cardiology of China 8: 305–309. [Google Scholar]
- 22. Park HK, Kim DH, Yun DH, Ban JY (2013) Association between IL10, IL10RA, and IL10RB SNPs and ischemic stroke with hypertension in Korean population. Mol Biol Rep 40: 1785–1790. [DOI] [PubMed] [Google Scholar]
- 23. Balding J, Livingstone WJ, Pittock SJ, Mynett-Johnson L, Ahern T, et al. (2004) The IL-6 G-174C polymorphism may be associated with ischaemic stroke in patients without a history of hypertension. Ir J Med Sci 173: 200–203. [DOI] [PubMed] [Google Scholar]
- 24.Li H (2008) Study on Relationship among Polymorphisms of three Inflammatory Cytokines,Serum Level of Lipoprotein and Cerebral Infarction. Unpublished MsD dissertation [MsD]: Southern Medical University.
- 25. Xie G, Myint PK, Zaman MJ, Li Y, Zhao L, et al. (2013) Relationship of serum interleukin-10 and its genetic variations with ischemic stroke in a chinese general population. PLoS One 8: e74126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang GZ (2006) Correlation between Interleukin-1, 10,TNF-a,TNF-βGene Polymorphisms and Cerebral Infarction. Unpublished MsD dissertation [MsD]: First Military Medical University Southern Medical University.
- 27. Yadav S, Hasan N, Marjot T, Khan MS, Prasad K, et al. (2013) Detailed analysis of gene polymorphisms associated with ischemic stroke in South Asians. PLoS One 8: e57305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stoica AL, Stoica E, Constantinescu I, Uscatescu V, Ginghina C (2010) Interleukin-6 and interleukin-10 gene polymorphism, endothelial dysfunction, and postoperative prognosis in patients with peripheral arterial disease. J Vasc Surg 52: 103–109. [DOI] [PubMed] [Google Scholar]
- 29. Trompet S, Pons D, DE Craen AJ, Slagboom P, Shepherd J, et al. (2007) Genetic variation in the interleukin-10 gene promoter and risk of coronary and cerebrovascular events: the PROSPER study. Ann N Y Acad Sci 1100: 189–198. [DOI] [PubMed] [Google Scholar]
- 30. Fei GZ, Svenungsson E, Frostegard J, Padyukov L (2004) The A-1087IL-10 allele is associated with cardiovascular disease in SLE. Atherosclerosis 177: 409–414. [DOI] [PubMed] [Google Scholar]
- 31. Bis JC, Heckbert SR, Smith NL, Reiner AP, Rice K, et al. (2008) Variation in inflammation-related genes and risk of incident nonfatal myocardial infarction or ischemic stroke. Atherosclerosis 198: 166–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Thakkinstian A, McElduff P, D'Este C, Duffy D, Attia J (2005) A method for meta-analysis of molecular association studies. Stat Med 24: 1291–1306. [DOI] [PubMed] [Google Scholar]
- 33. Thompson SG (1994) Why sources of heterogeneity in meta-analysis should be investigated. BMJ 309: 1351–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Attia J, Thakkinstian A, D'Este C (2003) Meta-analyses of molecular association studies: methodologic lessons for genetic epidemiology. J Clin Epidemiol 56: 297–303. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PRISMA 2009 Checklist.
(DOC)