Abstract
Although hydrolysis is known to be as important as synthesis in the growth and development of the bacterial cell wall, the coupling between these processes is not well understood. Bond cleavage can generate deleterious pores, but may also be required for the incorporation of new material and for the expansion of the wall, highlighting the importance of mechanical forces in interpreting the consequences of hydrolysis in models of growth. Critically, minimal essential subsets of hydrolases have now been identified in several model organisms, enabling the reduction of genetic complexity. Recent studies in Bacillus subtilis have provided evidence for both the presence and absence of coupling between synthesis and hydrolysis during sporulation and elongation, respectively. In this review, we discuss strategies for dissecting the relationship between synthesis and hydrolysis using time-lapse imaging, biophysical measurements of cell-wall architecture, and computational modeling.
Introduction
Across the bacterial kingdom, virtually all species possess a peptidoglycan (PG) cell wall, a macromolecular network of glycans cross-linked by peptides [1,2]. The shape of the cell is dictated physically by the outward force due to turgor pressure, which is balanced by the expansion of the cell-wall network. During growth and division, newly synthesized PG subunits are exported from the cytoplasm and then incorporated into the wall by a host of enzymes whose collective activity involves joining subunits into glycan strands and cross-linking them into the existing network [1,2]. While there has been a focus in recent years on the role of cytoskeletal elements and their association with PG synthesis [3–7], at a conceptual level, growth of the network has long been thought to also require the cleavage of pre-existing crosslinks in order to incorporate new material. On the generational time scale, some cleavage must occur to loosen the cell wall in some locations in order to intercalate the new material necessary to achieve micron-scale expansions. The genomes of Gram-negative and Gram-positive species all encode a wide variety of hydrolase enzymes responsible for the specific cleavage of different PG bonds; hydrolases are involved in several critical functions, including PG maturation, turnover, recycling, autolysis, and cleavage of the septum during cell division [8,9].
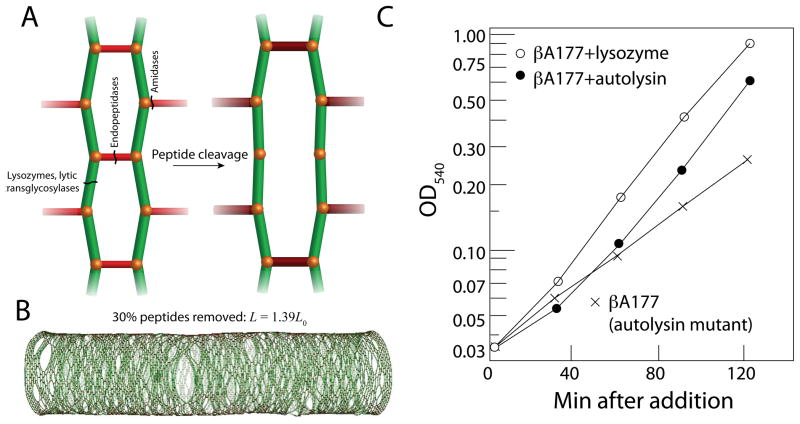
While a large number of hydrolases across many species have been characterized biochemically and structurally [10], there has been comparatively little investigation into their specific roles in vivo or the biophysical consequences of PG hydrolysis. The cleavage of a bond within the PG transfers the force that was borne by the crosslink onto the rest of the wall, resulting in expansion primarily in the local area of the cleaved bond [11] (Fig. 1A). The cleavage of a bond can also result in the generation of pores in the wall, which can represent a significant danger to the cell; during antibiotic treatment, cell lysis typically results from the formation of large pores that permit the cytoplasmic membrane to bleb into the extracellular environment [11]. This potential danger has stimulated the longstanding assumption that the insertion of new material must be coordinated in space and time with the hydrolysis of old material in order to avoid unregulated bond cleavage [9], and is supported indirectly by evidence that chemical inhibition of cell-wall synthesis often leads to cell lysis [12], though not in hydrolysis mutants [12–14]. In addition to qualitative models that have been proposed based on this concept [9], our recent biophysical modeling of cell-wall growth implemented the insertion of new strands into openings left by the cleavage of crosslinks [15,16]. While steady-state growth dictates that bond breaking should occur proportionally to bond formation, there is little direct evidence of spatially concerted hydrolysis and synthesis during rod-shaped growth. Moreover, computational simulations have demonstrated that as many as 30% of the crosslinks can be removed from the cell wall without disrupting the shape or integrity of the cell (although the wall does elongate due to the increase in stress borne by the remainder of the crosslinks), indicating that the cell wall may be highly robust to fluctuations in hydrolase activity [11] (Fig. 1B). Hence, it remains possible that a mechanism of growth without any coordination between synthesis and hydrolysis would result in the observed growth rate and maintenance of cell shape.
Figure 1. The role of hydrolases in cell-wall expansion.
(A) Specific hydrolases cleave crosslinks (red) between glycan strands (green), at the root of the peptide stem, or between glycan subunits. Cleavage of crosslinks transfers stress to the surrounding material (size and color of peptides indicate the amount of extension), leading to stretching of the PG network. (B) In silico, the removal of 30% of the crosslinks from a rod-shaped, Gram-negative PG network causes the cell to become longer, but does not affect the mechanical integrity of the cell wall or its shape, suggesting that organisms such as E. coli can tolerate large fluctuations in hydrolase activity. (C) A mutation in a B. subtilis gene encoding an autolysin results in lower levels of PG hydrolysis and a slower growth rate (crosses) relative to wild-type cells. The growth rate of the mutant can be increased by adding purified autolysin (filled circles) or lysozyme (open circles), indicating that hydrolysis is a major determinant of elongation and growth rate. (B) is modified from Ref. [11]; (C) is modified from Ref. [17].
Each type of peptide or glycosidic bond within the PG is targeted by a specific family of hydrolases. A number of excellent reviews address the biochemistry of these enzymes (e.g., [9,10]); this review focuses on the physical consequences of the hydrolysis of crosslinks (by endopeptidases and amidases) or glycans (by lytic transglycosylases), which promotes wall expansion by changing the distribution of forces across the cell surface [11]. An excellent example of the physical connection between hydrolase activity and growth occurs in the rod-shaped bacterium Bacillus subtilis, in which a temperature-sensitive mutant in an autolysin with impaired hydrolase activity exhibits a thicker wall than the wild-type strain [17,18], suggesting that synthesis rates are not significantly reduced in the face of reduced turnover. The growth rate of these mutant cells was substantially reduced, providing a clear example of the role of hydrolysis in wall expansion. Moreover, the addition of purified autolysin or lysozyme caused the growth rate to increase [17] (Fig. 1C) and reduced the wall thickness [18]. These data provide evidence that hydrolysis is required for growth, and suggest an intriguing physical connection between the molecular-scale activity of hydrolases and macroscopic, biophysical quantities such as growth rate and wall thickness that may be easier to measure in living cells.
The two best-studied rod-shaped bacterial species are the Gram-negative Escherichia coli and the Gram-positive B. subtilis. Although these organisms encode dozens of PG hydrolases [10,19], the regulation of these enzymes is virtually unknown. Genetic complexity and the possibility of multiple redundancies in a given growth environment have rendered genetic studies difficult, despite the wealth of information and tools available for studying growth. In addition, due to the difficulty of directly measuring hydrolase activity in vivo and the diversity of potential phenotypes resulting from the perturbation of hydrolase activity, it has been difficult to define mechanistic models of the role of hydrolase activity in cell-wall growth in vivo. Until the past year, there was no direct evidence that hydrolase activity was necessary for growth; there is no measureable phenotype when the Penicillin Binding Protein (PBP) endopeptidases in E. coli are deleted [20]. However, the recent identification of minimal, essential sets of hydrolases in both E. coli [21,22] and B. subtilis [23,24] has opened new avenues for study involving time-lapse microscopy, cell biology, and genetics.
In this review, we discuss recent experimental data that address the coordination between hydrolase activity and growth. In E. coli and B. subtilis, the recent discovery of minimal hydrolase subsets required for growth can be leveraged in the context of recent models of the cytoskeletal coordination of growth [3–5]. In B. subtilis, recent studies have highlighted the role of hydrolysis in two aspects of morphogenesis: the migration of the forespore membrane during engulfment [25,26] and the regulation of cell-wall thickness and its coupling to growth rate and mechanical strain [27]. In the latter case, a combination of biophysical experiments and modeling was used to support the existence of a coupling mechanism between hydrolysis and cell-wall synthesis. Here, we focus on the physical interpretations of these experiments in the context of our current mechanistic understanding of PG synthesis. In doing so, we hope to motivate new classes of experiments that utilize the existing biochemical knowledgebase and recent genetic advances in hydrolase biology, particularly emphasizing the quantitative dynamics of PG synthesis and mechanical stresses within the PG, to discover the ways in which cell-wall cleavage is integrated with the insertion of new material.
Hydrolases are required for growth
In the past year, studies separately identified small subsets of hydrolytic enzymes in several model organisms that are required for growth. In B. subtilis, LytE and CwlO are endopeptidase-like autolysins; a lytE cwlO double mutant is not viable and exhibits defects in cell-wall synthesis and elongation [23,24]. In Vibrio cholerae, a set of three genes required for elongation includes an endopeptidase [28]. In E. coli, Spr, YebA, and YdhO were identified as DD-endopeptidases, enzymes that cleave D-alanine-meso-diamino-pimelic acid crosslinks. The triple-deletion mutant is not viable and cell-wall synthesis is dramatically reduced [21]. This common behavior in widely divergent bacterial species is the first evidence that PG hydrolysis is required for rod-shaped growth. The set of required hydrolases in E. coli can be further winnowed in particular media, as growth rate appears to be correlated with the strengths of the phenotypes of hydrolase mutants. An spr yebA double mutant cannot grow on rich medium (LB) or on minimal medium supplemented with 1% tryptone, and an spr deletion is unable to grow on nutrient agar at high temperature [21]. In contrast, deletions in E. coli endopeptidases such as pbp4 and pbp7 have no discernible phenotype [20]. As might be expected from the behavior of strains lacking hydrolases, sacculi from an spr yebA ydhO mutant exhibited a higher degree of crosslinking via chromatographic analysis [21], implying a stiffer cell wall. However, phase-contrast microscopy revealed the formation of large ovoid cell shapes and extensive cell lysis [21], a nonintuitive result of the reduction in overall hydrolase activity due to the triple deletion. Future quantitative studies of the dynamics of growth at different expression levels of these hydrolases, along with the measurement of other cellular features such as membrane abundance and turgor pressure, should help to reveal the mechanism by which these essential hydrolases prevent lysis.
Spatial regulation of hydrolysis
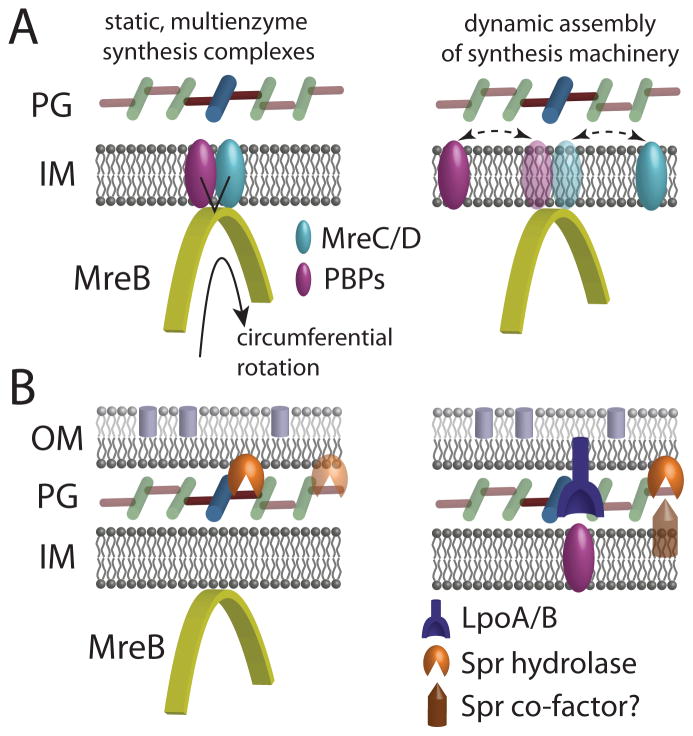
Recent fluorescence microscopy studies have reinforced the central role of the bacterial actin homolog MreB in rod-shaped elongation. In both E. coli [3] and B. subtilis [4,5], MreB was recently discovered to move in a circumferential manner that is dependent on cell-wall synthesis (Fig. 2A); in B. subtilis, MreB colocalizes with the wall synthetic enzyme PBP2H [4]. This rotation may distribute the insertion of new PG into the cell wall across the surface of the cell, ensuring that a relatively small number of MreB polymers can produce a robust shape insensitive to fluctuations in insertion pattern or environmental perturbations such as osmotic shock [3]. These data support the prevailing model that PG growth in rod-shaped organisms involves multienzyme complexes that each contain several PBPs, whose coordinated activities result in the processive incorporation of new strands into the existing PG network. It must be noted that such a complex has never been isolated, nor is there any direct evidence in E. coli of the colocalization of MreB with any of the PBPs. Moreover, from a mechanistic standpoint it has not been demonstrated that such spatiotemporal coordination is beneficial in any way for the robustness of growth. Directed, circumferential motion of these enzymes, as has been observed in B. subtilis [4,5], would appear to be a prerequisite for the existence of such a complex.
Figure 2. Hydrolase localization relative to cell-wall synthesis.
(A) The bacterial actin homolog MreB (yellow) binds to the cytoplasmic face of the inner membrane and moves in a circumferential direction. In E. coli, this motion is dependent on the activity of PBP2 during cell-wall synthesis, though it has yet to be determined whether MreB serves as a spatial organizing center for stable synthesis complexes (left) or as a transient rest stop for the dynamic association of PBPs (right). (B) The bifunctional enzymes PBP1A/B rely on the outer-membrane lipoprotein cofactors LpoA/B for their activity. The hydrolase Spr is an outer-membrane lipoprotein essential for growth on nutrient agar; it is unknown whether its activity is spatially coordinated with MreB (left), or whether other co-factors are required for its activity.
Notwithstanding the lack of evidence that coordination between synthesis and hydrolysis is necessary for robust bacterial growth, biochemical evidence suggests that certain hydrolases in E. coli can interact with the PBPs [29]. In B. subtilis, the fluorescence colocalization of MreBH with the lytic transglycosylase LytE and the phenotypic similarities between lytE and mreBH mutants support the existence of spatiotemporal coordination between synthesis and hydrolysis [30]. It has yet to be determined which molecular factors are responsible for the circumferential motion of MreB; hydrolases may contribute to this motion. The identification of minimal subsets of hydrolases required for cell growth and wall insertion [21,23,24] provides a framework for addressing this question. However, in B. subtilis, MreB and the PG synthetic machinery continues to move circumferentially during depletion of the essential hydrolases LytE and CwlO until the cells eventually lyse [31]. irIn E. coli, these hydrolases are required for the incorporation of new murein, indicating that hydrolysis may be coupled to insertion [21]. Interestingly, Spr is predicted to be an outer membrane lipoprotein [21]. The lipoproteins LpoA/B were found to regulate the synthesis of PG by PBP1A/B, respectively [7]; Spr exhibits low activity on isolated sacculi, suggesting that perhaps the hydrolysis of material in vivo requires association with factors in the inner or outer membranes (Fig. 2B).
Effect of hydrolysis on growth rate
In the “inside-out” model of Gram-positive elongation, the synthesis and insertion of new PG occurs adjacent to the membrane at the inner surface of the cell wall [32]. Through an unspecified mechanism, an increase in stress in the outer layers that forces them to stretch could result in the rupture of bonds within the PG; when the strain of extension becomes too large, the outer layers of the cell wall are ultimately removed. Although there is no evidence that PG material is only incorporated adjacent to the membrane, this model presents a conceptually simple mechanism for elongation in which the synthesis of new layers balances the removal of old layers. For this cycle to drive elongation rather than simply adding material to thicken the wall, two factors are required: positive turgor pressure to stretch the wall, and a mechanism for increasing the stress in layers after they are synthesized. Hydrolysis provides such a mechanism, weakening the oldest layers so that they are less load-bearing and redistributing stress to the newest layers. In this picture, hydrolase activity regulates growth rate by breaking apart the wall, while PG synthesis maintains the integrity of the wall by adding new, stiff layers.
How does the balance between synthesis and hydrolysis dictate the dynamics and structural properties of the wall in B. subtilis? We recently determined how the thickness of the wall and its extension due to turgor pressure depend on growth rate [27]. Using transmission electron and epifluorescence microscopy, we found that wall thickness and extension in exponentially growing cells were similar in rich and poor media, with a three-fold change in growth rate. To link these observations with potential mechanisms of elongation, we developed a biophysical model of a rod-shaped Gram-positive wall that balances the mechanical effects of synthesis of new material and removal of old material through hydrolysis. This model predicts that thickness will scale with the ratio of the rates of synthesis and hydrolysis; more PG synthesis will lead to a thicker wall, while more PG turnover will thin the wall. For a fixed turgor pressure, hydrolysis weakens the wall and synthesis strengthens it, and wall strain is thus predicted to increase inversely with the thickness. In contrast, growth rate is predicted to scale inversely with the synthesis rate, and quadratically with the hydrolysis rate because hydrolysis reduces both the number of layers and their strengths, both of which lead to more expansion. Thus, our model indicates that cells can vary their growth rate while maintaining constant cell-wall thickness and strain if the rates of synthesis and hydrolysis are coupled such that their ratio is fixed. These results are consistent with previous experiments analyzing a temperature-sensitive B. subtilis autolysin mutant with a large reduction in hydrolase activity. This mutant has a thicker wall [18] and a slower growth rate, which can be accelerated by increasing the rate of hydrolysis [17].
PG hydrolysis drives engulfment during sporulation
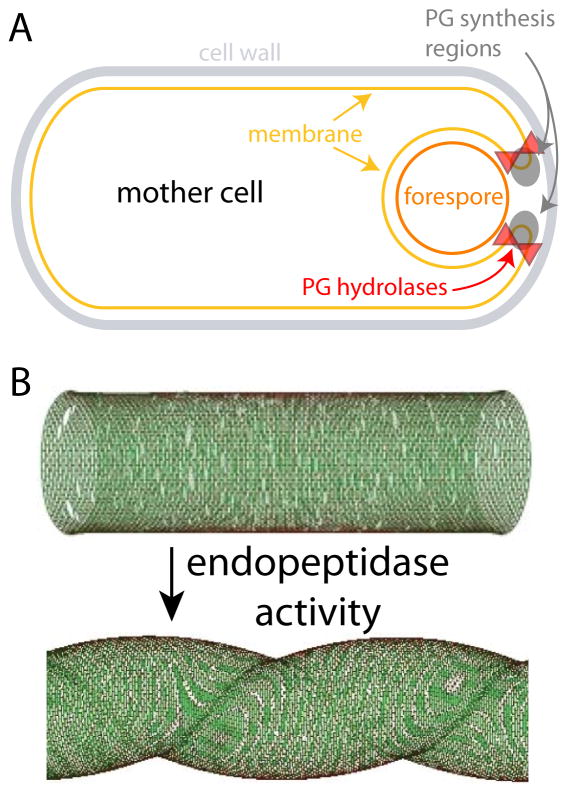
B. subtilis survives under conditions of nutrient deprivation or in harsh environments through sporulation, when a septum is formed near one of the poles and division occurs asymmetrically. The smaller cell (the forespore) eventually becomes the spore through a process called engulfment that involves a complex cascade of signaling, localization, and PG synthesis and hydrolysis. Engulfment requires spatiotemporal coordination between the synthesis of the cell wall and the cell membrane. Directly before engulfment, hydrolase-mediated PG degradation between the septal membranes allows the mother cell membrane to move around the forespore [33,34]. As engulfment continues, the membrane of the mother cell migrates around the forespore, forming a double-bilayer membrane that eventually surrounds the spore. When the migrating arms of the membrane meet, they fuse to release the forespore into the mother cell.
PG synthesis and hydrolysis are key components of the force-generation mechanism for engulfment. During the initial stages of engulfment, the sites of PG synthesis coincide with the leading edge of the engulfing membrane, and the inhibition of PG synthesis blocks membrane migration [26]. During the last stage of engulfment, PG synthesis localizes to the last site of attachment between the two cells; antibiotics that inhibit PG synthesis prevent cell separation [26]. The PG hydrolases SpoIID and SpoIIP first localize at the sporulation septum, where they dissolve the septal PG [35]. During membrane migration around the forespore, these enzymes localize at the leading edge of the membrane arms [25], where PG synthesis is also concentrated (Fig. 3A). In a mutant that displayed asymmetrical migration of the membrane arms, the speeds of the two arms were proportional to the fluorescence levels of SpoIIP [25]. As another example of the coupling between PG synthesis and hydrolysis, these studies suggest that the force generated by hydrolysis and synthesis can be transduced by the cell for migration of the leading edges of the engulfing membrane. This observation has inspired the idea that PG synthesis may be equivalent to a molecular motor that can drive not only proteins but also cellular-scale structures in a directed fashion.
Figure 3. Morphogenetic processes involving coordination between synthesis and hydrolysis.
(A) During B. subtilis sporulation, PG synthesis and hydrolysis are colocalized to the leading edge of the migrating arms of the engulfing membrane. (B) The spiral morphology of H. pylori requires endopeptidase activity and relaxation of PG crosslinking. (B) is modified from Ref. [11].
Discussion
Given the importance of hydrolysis in wall expansion, biophysical measurements of the differential movement of surface markers across the cell wall can shed light on where this activity is taking place and provide constraints for the evaluation of mechanistic models of wall growth [16]; such measurements of the pattern of outer membrane expansion motivated a model involving the incorporation of new material in large bursts [36]. Although it may be difficult to directly image hydrolase activity, novel fluorescent D-amino acid technologies for localizing the insertion of new PG enable the correlation of PG insertion with heterogeneity in cell-wall expansion, which would provide further evidence for the coupling of synthesis and hydrolysis [37,38]. Importantly, appreciation for the role of hydrolases in all bacterial morphogenetic processes has been revitalized by recent studies of specific enzymes involved in E. coli and B. subtilis elongation [21,22,24] and B. subtilis sporulation [25,26]. The potential for PG hydrolysis to directly impact morphogenetic processes is also illustrated by the role of endopeptidases in defining the helical shape of Helicobacter pylori cells [39] (Fig. 3B).
Although hydrolysis clearly plays a central role in many aspects of cell-wall growth, modification, and turnover, studies of this process remain stymied by difficulties in quantitatively measuring turnover in vivo and/or imaging hydrolysis directly. While the identification of essential hydrolases should motivate cell biological studies of localization and dynamics, it will be difficult to ascertain when and where these enzymes are active. Moreover, there are distinct gaps in our knowledge of how (if at all) hydrolases interact with PG synthesis enzymes; bioinformatic and structural biological tools for predicting such protein-protein interactions may shed light on the coordination of synthesis and hydrolysis. The efficacy of a model in which strands are cross-linked to the existing PG layer while hydrolytic activity cleaves old material randomly across the cell surface, rather than directly adjacent to the new strands (Fig. 4A,B), could be tested via straightforward modifications of our previous biophysical simulations. If growth rates and morphologies are similar between the two models, then hydrolysis need not be spatiotemporally coordinated with insertion. If hydrolases are indeed spatiotemporally coupled to synthesis in vivo, a further level of mechanical regulation has been proposed for regulating growth patterning involving autolysins that cleave only crosslinks that are stretched in order to stimulate the insertion of new material [40], though simulations involving tension-dependent insertion failed to maintain a rod-like shape [15]. If synthesis is regulated by such mechanically regulated hydrolase activity, then the rate of PG insertion may be dependent on turgor pressure; experiments carried out in environments with oscillating osmolarity would reveal such a relationship.
Figure 4. Potential models for the coordination of hydrolysis and synthesis.
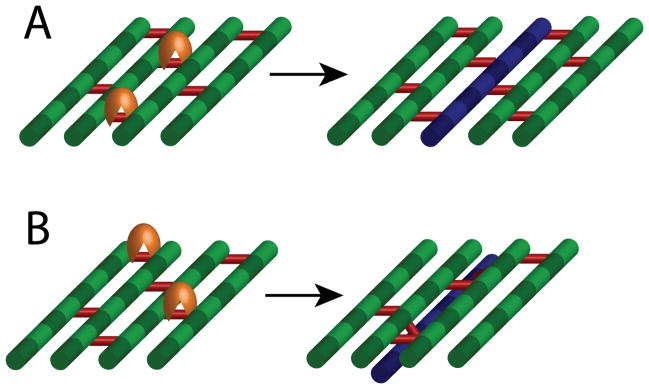
(A) The model of coupled synthesis and hydrolysis assumes that the insertion of a new glycan strand (blue) occurs concurrently with cleavage of adjacent peptides by hydrolases (orange). (B) An alternative model involves the incorporation of a new strand through crosslinking, while cleavage occurs randomly across the cell surface. A combination of biophysical simulations and quantitative measurements of cell growth will likely be necessary to distinguish between these models.
The connection between hydrolases and cell-wall expansion has important implications for the growth of densely packed cellular communities in which hydrolase molecules released by individual cells could rebind to neighbors rather than escape into the extracellular milieu, thereby effecting an increase in the hydrolase concentration and expansion rate of both the cells and the community. Moreover, antibiotics that disrupt the balance between synthesis and hydrolysis may actually increase the growth rate at sublethal concentrations, suggesting the potential for complex responses and the evolution of resistance to cell wall-targeting antibiotics [41]. A complete understanding of the regulation of hydrolase activity and its coupling to PG synthesis, wall architecture, and cell geometry will require computational techniques and physical models in concert with genetic tools and in vivo imaging. The recent surge of interest in hydrolases reflects a growing appreciation for the systems-level integration of biology, chemistry, and physics in all morphogenetic processes.
Acknowledgments
The authors acknowledge assistance from Samantha Desmarais and Manjula Reddy for careful readings of the manuscript. K.C.H. acknowledges support from NSF CAREER Award 114-9328.
References
- 1.Scheffers D-J, Pinho MG. Bacterial cell wall synthesis: new insights from localization studies. Microbiol Mol Biol Rev. 2005;69:585. doi: 10.1128/MMBR.69.4.585-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holtje JV. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *3.van Teeffelen S, Wang S, Furchtgott L, Huang KC, Wingreen NS, Shaevitz JW, Gitai Z. The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc Natl Acad Sci USA. 2011;108:15822–15827. doi: 10.1073/pnas.1108999108. van Teeffelen et al, Dominguez-Escober et al, and Garner et al demonstrated that MreB rotates around the long axis of E. coli and B. subtilis in a cell-wall synthesis-dependent manner. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *4.Dominguez-Escobar J, Chastanet A, Crevenna AH, Fromion V, Wedlich-Soldner R, Carballido-Lopez R. Processive Movement of MreB-Associated Cell Wall Biosynthetic Complexes in Bacteria. Science. 2011;333:225–228. doi: 10.1126/science.1203466. van Teeffelen et al, Dominguez-Escober et al, and Garner et al demonstrated that MreB rotates around the long axis of E. coli and B. subtilis in a cell-wall synthesis-dependent manner. [DOI] [PubMed] [Google Scholar]
- *5.Garner EC, Bernard R, Wang W, Zhuang X, Rudner DZ, Mitchison T. Coupled, Circumferential Motions of the Cell Wall Synthesis Machinery and MreB Filaments in B. subtilis. Science. 2011;333:222–225. doi: 10.1126/science.1203285. van Teeffelen et al, Dominguez-Escober et al, and Garner et al demonstrated that MreB rotates around the long axis of E. coli and B. subtilis in a cell-wall synthesis-dependent manner. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paradis-Bleau C, Markovski M, Uehara T, Lupoli TJ, Walker S, Kahne DE, Bernhardt TG. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell. 2010;143:1110–1120. doi: 10.1016/j.cell.2010.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Typas A, Banzhaf M, van den Berg van Saparoea B, Verheul J, Biboy J, Nichols RJ, Zietek M, Beilharz K, Kannenberg K, von Rechenberg M, et al. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell. 2010;143:1097–1109. doi: 10.1016/j.cell.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartmann R, Bock-Hennig SB, Schwarz U. Murein hydrolases in the envelope of Escherichia coli. Properties in situ and solubilization from the envelope. Eur J Biochem. 1974;41:203–208. doi: 10.1111/j.1432-1033.1974.tb03261.x. [DOI] [PubMed] [Google Scholar]
- 9.Holtje JV. From growth to autolysis: the murein hydrolases in Escherichia coli. Arch Microbiol. 1995;164:243–254. doi: 10.1007/BF02529958. [DOI] [PubMed] [Google Scholar]
- 10.Vollmer W, Joris B, Charlier P, Foster S. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev. 2008;32:259–286. doi: 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- 11.Huang KC, Mukhopadhyay R, Wen B, Gitai Z, Wingreen NS. Cell shape and cell-wall organization in Gram-negative bacteria. Proc Natl Acad Sci U S A. 2008;105:19282–19287. doi: 10.1073/pnas.0805309105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomasz A. The mechanism of the irreversible antimicrobial effects of penicillins: how the beta-lactam antibiotics kill and lyse bacteria. Annu Rev Microbiol. 1979;33:113–137. doi: 10.1146/annurev.mi.33.100179.000553. [DOI] [PubMed] [Google Scholar]
- 13.Goodell EW, Lopez R, Tomasz A. Suppression of lytic effect of beta lactams on Escherichia coli and other bacteria. Proc Natl Acad Sci U S A. 1976;73:3293–3297. doi: 10.1073/pnas.73.9.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitano K, Tomasz A. Escherichia coli mutants tolerant to beta-lactam antibiotics. J Bacteriol. 1979;140:955–963. doi: 10.1128/jb.140.3.955-963.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *15.Furchtgott L, Wingreen NS, Huang KC. Mechanisms for maintaining cell shape in rod-shaped Gram-negative bacteria. Mol Microbiol. 2011;81:340–353. doi: 10.1111/j.1365-2958.2011.07616.x. This study explored mechanisms of maintaining a rod-like shape in Gram-negative bacteria by developing a computational model of the cell wall based on realistic physical parameters. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang S, Furchtgott L, Huang KC, Shaevitz JW. Helical insertion of peptidoglycan produces chiral ordering of the bacterial cell wall. Proc Natl Acad Sci USA. 2012;109:E595–604. doi: 10.1073/pnas.1117132109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan DP, Beckman MM. Mutant of Bacillus subtilis demonstrating the requirement of lysis for growth. J Bacteriol. 1971;105:629–636. doi: 10.1128/jb.105.2.629-636.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan DP, Pelvit MC, Cunningham WP. Structural difference between walls from ends and sides of the rod-shaped bacterium Bacillus subtilis. J Bacteriol. 1972;109:1266–1272. doi: 10.1128/jb.109.3.1266-1272.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vollmer W, Bertsche U. Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochim Biophys Acta. 2008;1778:1714–1734. doi: 10.1016/j.bbamem.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 20.Meberg BM, Paulson AL, Priyadarshini R, Young KD. Endopeptidase penicillin-binding proteins 4 and 7 play auxiliary roles in determining uniform morphology of Escherichia coli. J Bacteriol. 2004;186:8326–8336. doi: 10.1128/JB.186.24.8326-8336.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **21.Singh SK, SaiSree L, Amrutha RN, Reddy M. Three redundant murein endopeptidases catalyse an essential cleavage step in peptidoglycan synthesis of Escherichia coli K12. Molecular Microbiology. 2012;86:1036–1051. doi: 10.1111/mmi.12058. This study identified a set of essential hydrolases in E. coli and showed that depletion of these enzymes inhibits cell-wall synthesis and leads to cell lysis. [DOI] [PubMed] [Google Scholar]
- 22.Vollmer W. Bacterial growth does require peptidoglycan hydrolases. Molecular Microbiology. 2012;86:1031–1035. doi: 10.1111/mmi.12059. [DOI] [PubMed] [Google Scholar]
- 23.Bisicchia P, Noone D, Lioliou E, Howell A, Quigley S, Jensen T, Jarmer H, Devine KM. The essential YycFG two-component system controls cell wall metabolism in Bacillus subtilis. Mol Microbiol. 2007;65:180–200. doi: 10.1111/j.1365-2958.2007.05782.x. [DOI] [PubMed] [Google Scholar]
- **24.Hashimoto M, Ooiwa S, Sekiguchi J. Synthetic lethality of the lytE cwlO genotype in Bacillus subtilis is caused by lack of D, L-endopeptidase activity at the lateral cell wall. J Bacteriol. 2012;194:796–803. doi: 10.1128/JB.05569-11. This study used fluorescence microscopy to show that endopeptidase activity of LytE and CwlO along the lateral cell wall is essential for growth. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gutierrez J, Smith R, Pogliano K. SpoIID-mediated peptidoglycan degradation is required throughout engulfment during Bacillus subtilis sporulation. J Bacteriol. 2010;192:3174–3186. doi: 10.1128/JB.00127-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer P, Gutierrez J, Pogliano K, Dworkin J. Cell wall synthesis is necessary for membrane dynamics during sporulation of Bacillus subtilis. Mol Microbiol. 2010;76:956–970. doi: 10.1111/j.1365-2958.2010.07155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **27.Misra G, Rojas ER, Gopinathan A, Huang KC. Mechanical consequences of cell-wall turnover in the elongation of a gram-positive bacterium. Biophys J. 2013;104:2342–2352. doi: 10.1016/j.bpj.2013.04.047. This study used electron microscopy and microfluidics to show that B. subtilis cells have the same cell-wall thickness and mechanical extension during growth in rich and poor media. The authors showed that this maintenance of cell wall structural properties can be explained by a biophysical model of Gram-positive elongation in which the rate of cell-wall hydrolysis is proportional to the rate of synthesis. This model provides a quantitative framework for understanding the acceleration in growth rate when B. subtilis cells are treated with purified autolysin enzymes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **28.Dorr T, Cava F, Lam H, Davis BM, Waldor MK. Substrate specificity of an elongation-specific peptidoglycan endopeptidase and its implications for cell wall architecture and growth of Vibrio cholerae. Mol Microbiol. 2013 doi: 10.1111/mmi.12323. This study identified a set of genes in V. cholerae that are essential for elongation. One of these genes is an endopeptidase, and its deletion decreases the average glycan strand length of the peptidoglycan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Romeis T, Holtje JV. Specific interaction of penicillin-binding proteins 3 and 7/8 with soluble lytic transglycosylase in Escherichia coli. J Biol Chem. 1994;269:21603–21607. [PubMed] [Google Scholar]
- 30.Carballido-Lopez R, Formstone A, Li Y, Ehrlich SD, Noirot P, Errington J. Actin homolog MreBH governs cell morphogenesis by localization of the cell wall hydrolase LytE. Dev Cell. 2006;11:399–409. doi: 10.1016/j.devcel.2006.07.017. [DOI] [PubMed] [Google Scholar]
- **31.Meisner J, Llopis PM, Sham LT, Garner E, Bernhardt TG, Rudner DZ. FtsEX is required for CwlO peptidoglycan hydrolase activity during cell wall elongation in Bacillus subtilis. Mol Microbiol. 2013 doi: 10.1111/mmi.12330. This study showed that the hydrolytic activity of CwlO is regulated by FtsEX, and that the peptidoglycan synthetic machinery continues to move circumferentially during depletion of CwlO and LytE until lysis occurs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koch AL, Doyle RJ. Inside-to-outside growth and turnover of the wall of gram-positive rods. J Theor Biol. 1985;117:137–157. doi: 10.1016/s0022-5193(85)80169-7. [DOI] [PubMed] [Google Scholar]
- 33.Illing N, Errington J. Genetic regulation of morphogenesis in Bacillus subtilis: roles of sigma E and sigma F in prespore engulfment. J Bacteriol. 1991;173:3159–3169. doi: 10.1128/jb.173.10.3159-3169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piggot PJ, Coote JG. Genetic aspects of bacterial endospore formation. Bacteriol Rev. 1976;40:908–962. doi: 10.1128/br.40.4.908-962.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morlot C, Uehara T, Marquis KA, Bernhardt TG, Rudner DZ. A highly coordinated cell wall degradation machine governs spore morphogenesis in Bacillus subtilis. Genes Dev. 2010;24:411–422. doi: 10.1101/gad.1878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *36.Ursell TS, Trepagnier EH, Huang KC, Theriot JA. Analysis of surface protein expression reveals the growth pattern of the gram-negative outer membrane. PLoS Comput Biol. 2012;8:e1002680. doi: 10.1371/journal.pcbi.1002680. This study used pulse-chase labeling of an abundant outer membrane and time-lapse microscopy, coupled with biophysical modeling, to demonstrate that growth of the E. coli outer membrane occurs in large bursts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *37.Kuru E, Hughes HV, Brown PJ, Hall E, Tekkam S, Cava F, de Pedro MA, Brun YV, VanNieuwenhze MS. In Situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angew Chem Int Ed Engl. 2012;51:12519–12523. doi: 10.1002/anie.201206749. Kuru et al and Siegrist et al developed fluorescent D-amino acid probes for reporting cell-wall synthesis, enabling future studies elucidating the spatiotemporal coordination between cell-wall synthesis and expansion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *38.Siegrist MS, Whiteside S, Jewett JC, Aditham A, Cava F, Bertozzi CR. (D)-amino acid chemical reporters reveal peptidoglycan dynamics of an intracellular pathogen. ACS Chem Biol. 2013;8:500–505. doi: 10.1021/cb3004995. Kuru et al and Siegrist et al developed fluorescent D-amino acid probes for reporting cell-wall synthesis, enabling future studies elucidating the spatiotemporal coordination between cell-wall synthesis and expansion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sycuro LK, Pincus Z, Gutierrez KD, Biboy J, Stern CA, Vollmer W, Salama NR. Peptidoglycan crosslinking relaxation promotes Helicobacter pylori’s helical shape and stomach colonization. Cell. 2010;141:822–833. doi: 10.1016/j.cell.2010.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koch AL. Additional arguments for the key role of “smart” autolysins in the enlargement of the wall of gram-negative bacteria. Res Microbiol. 1990;141:529–541. doi: 10.1016/0923-2508(90)90017-k. [DOI] [PubMed] [Google Scholar]
- 41.Bollenbach T, Quan S, Chait R, Kishony R. Nonoptimal microbial response to antibiotics underlies suppressive drug interactions. Cell. 2009;139:707–718. doi: 10.1016/j.cell.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]