Summary
The mammalian heart has generally been considered nonregenerative, but recent progress suggests that neonatal mouse hearts have a genuine capacity to regenerate following apex resection (AR). However, in this study, we performed AR or sham surgery on 400 neonatal mice from inbred and outbred strains and found no evidence of complete regeneration. Ideally, new functional cardiomyocytes, endothelial cells, and vascular smooth muscle cells should be formed in the necrotic area of the damaged heart. Here, damaged hearts were 9.8% shorter and weighed 14% less than sham controls. In addition, the resection border contained a massive fibrotic scar mainly composed of nonmyocytes and collagen disposition. Furthermore, there was a substantial reduction in the number of proliferating cardiomyocytes in AR hearts. Our results thus question the usefulness of the AR model for identifying molecular mechanisms underlying regeneration of the adult heart after damage.
Highlights
-
•
We examine the neonatal mouse heart regeneration model following apex resection
-
•
The apex-resected hearts do not hold the suggested complete regenerative potential
-
•
Instead, hearts heal by scarring and exhibit reduced cardiomyocyte proliferation
-
•
The challenging dogma that the neonatal heart is regenerative remains questionable
Neonatal mouse hearts have been reported to be regenerative following apex resection. However, in this study, Andersen, Sheikh, and colleagues were unable to reproduce this. Specifically, they found that the resected apex remains lost, cardiomyocyte proliferation is reduced, and that the wounded heart heals by a scarring response similar to that observed in the inert adult heart following damage.
Introduction
A key question in cardiovascular biology is to what degree the heart is able to regenerate after tissue damage from either cardiac stem cells or cardiomyocyte division. Cardiovascular disease including myocardial infarct is currently one of the leading causes of death worldwide, and the general view is that this is mainly caused by a genuine inability of the mammalian heart to regenerate upon damage (Vieira and Riley, 2011). Yet, this dogma was recently challenged by exciting data suggesting that the mouse heart retains regenerative ability up to 1 week after birth (Porrello et al., 2011), and without being reproduced by others, it has now been accepted as an established principle that neonatal mammalian hearts do enclose a true cardiac-regenerative potential following apex resection (AR) (Aguirre et al., 2013, Garbern and Lee, 2013). As a minimal requirement, complete cardiac regeneration should include the restoration of the functional continuity of cardiomyocytes, as well as blood supply in the necrotic area of the damaged heart with no sign of scar formation. Indeed, urodele amphibians and zebrafish have been shown to possess a high capacity to repair the heart following damage such as AR that meets these minimal criteria (Garbern et al., 2013). Accordingly, the zebrafish heart is regenerated in 60 days following AR, with full recovery of the myocardium (Poss et al., 2002). The mammal and zebrafish heart anatomy/physiology diverge substantially (Garbern et al., 2013). It was therefore a breakthrough in regenerative medicine of the mammalian heart when Porrello et al. in 2011 showed that the neonatal mouse heart (1 day old) holds an intrinsic capability to regenerate completely following resection of 10% of the heart apex (Porrello et al., 2011). As in the zebrafish heart (Jopling et al., 2010), the regenerative response in mice was primarily accomplished through reentry of cardiomyocytes into the cell cycle (Porrello et al., 2011). Interestingly, this ability was only transient and lost by postnatal day 7 (P7) (Porrello et al., 2011), a scenario the authors most recently suggested is caused by the homeobox transcription factor Meis1 inhibiting cardiomyocyte proliferation (Mahmoud et al., 2013). Remarkably, the repairing response seems to be faster in mice (21 days) (Porrello et al., 2011) than in teleost fish (60–180 days) (Lafontant et al., 2012, Poss et al., 2002). Furthermore, the regenerated neonatal mouse heart reportedly showed no signs of major scarring after 21 days (Porrello et al., 2011), which is in contrast to the mammalian adult heart that lacks substantial regenerative capacity (Garbern et al., 2013, Vieira and Riley, 2011). In addition, urodele amphibians and teleost fish show substantial scarring up until 60–180 days postinjury (Lafontant et al., 2012, Oberpriller and Oberpriller, 1974, Poss et al., 2002).
The reported availability of the neonatal mouse heart regeneration model is thus extremely valuable to researchers in order to identify factors that may be used for improving regeneration of the adult heart in the large group of patients suffering from cardiac infarcts. We thus originally set out to identify factors enabling regeneration of the heart. However, our data do not yield evidence of a complete regenerative response in the neonatal mouse heart following AR.
Results
Establishing the AR Model in Inbred C57Bl/6 Mice
The study by Porrello et al. was performed in the outbred ICR/CD-1 mouse strain (Porrello et al., 2011). However, many transgenic mouse models including ours use inbred mouse strains. We therefore set out to clarify and evaluate the regenerative potential of AR hearts in C57Bl/6 mice. We used the exact same surgery protocol (Figure 1A) as described previously by Porrello et al. (2011) for AR. The heart-to-body weight was reduced by 8.4% in AR hearts as compared with sham hearts immediately following resection (Figures 1B and 1C). The average resection size is thus similar to the approximate 10% accomplished by Porrello et al. (2011). Supportive of that, we achieved a 90% survival of AR animals following surgery, whereas maternal cannibalization further lowered this to 80%–85% (Figure 1D), which is moderately higher than the 70% survival reported for ICR/CD-1 mice (Porrello et al., 2011). Thus, our surgery setup for AR in C57BL/6 mice seems comparable to that reported for the ICR/CD1 mice.
Figure 1.
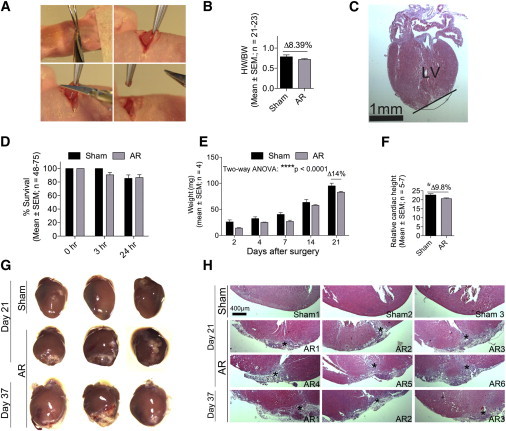
C57Bl/6 Neonatal Mouse Hearts Do Not Regenerate following AR
(A) Open thoracotomy was used to remove 5%–10% of the heart apex from hypothermia-anesthetized neonatal C57Bl/6 mice.
(B and C) Hearts from sham (n = 21) and apex-resected (n = 23) mice were removed immediately after AR and weighed to determine the amount of resected heart tissue. Hematoxylin staining of AR hearts (n = 10) confirmed localization of the resected area. Black line indicates resection border. LV, left ventricle.
(D) Animal survival was determined immediately (0 hr), then 3 and 24 hr after surgery.
(E) The cardiac weight in sham and AR animals was determined at different time points following surgery. Two-way ANOVA was used to test significance (∗∗∗∗p < 0.0001 for weight over time).
(F) Relative cardiac height at day 21 in sham (n = 5) and AR (n = 7) hearts. Student’s t test was used to test significance (∗p < 0.05).
(G) Three representative stereomicroscopic pictures of sham and AR hearts.
(H) Representative HE stainings from three to six sham or AR hearts at day 21 or 37 that enclose the AR damage. All hearts (n = 16) were sectioned throughout and examined (see Figure S1) to identify the area of apex lesion marked by an asterisk (∗).
See also Figure S1.
Neonatal C57Bl/6 Mice Do Not Regenerate Their Hearts following AR
Unexpectedly, however, we found that the AR heart weights were compromised throughout the study, with a persistent 14% reduction in AR hearts at day 21 (Figure 1E). Additionally, the resected hearts were 9.8% shorter at day 21 than sham controls (Figures 1F and 1G). These results thus indicated that the apex was healed but not regenerated with healthy tissue replenishing the removed apex. In accordance, stereomicroscopic (Figure 1G) and histological (Figure 1H; Figure S1 available online) examinations of AR and sham hearts showed an extensive remodeling of the damaged area in 100% of AR hearts at day 21, which was absent in sham specimens. Of note, scars were observed in 15.7% ± 4.1% of hematoxylin and eosin (HE)-stained sections (778 ± 54 per heart; n = 7). These data thus surprisingly suggested that C57BL/6 hearts were unable to regenerate.
Apex-Resected C57Bl/6 Hearts Are Healed with Accumulation of Many Noncardiomyocytes
An extensive quantitative real-time PCR study showed that mesenchymal, fat, hematopoietic, and vascular gene programs were significantly induced in AR hearts at day 21 as compared to sham hearts (Figure S2), whereas epicardial and cardiomyocyte gene programs overall remained unchanged at this endpoint (Figure S2). Hematopoietic cells such as macrophages are responsible for cleaning up necrotic constituents in areas of heart damage but are also main drivers of cardiac fibrosis (Santini and Rosenthal, 2012). We found increasing mRNA levels of Cd45, a hematopoietic marker (Figure 2A), already 3 hr after AR, reflecting an immediate influx of inflammatory cells as confirmed by immunohistochemistry (Figure 2B). The majority of these inflammatory cells disappeared between days 7 and 14 (Figures 2A and 2B), but some were still evident at day 21 in the lesioned area of AR hearts (Figure 2B). Heart fibrosis is characterized by deposition of collagen I and a transition of cardiac fibroblasts to myofibroblasts secreting large amounts of fibronectin. We found the inflammatory response in AR hearts to be clearly accompanied by an increase in both Pro-collagen I and Fibronectin (Figure 2A). This was confirmed by substantial amounts of collagen in the lesioned area as visualized by Sirius Red staining (Figure 2C) and immunohistochemistry (Figure 2D). In contrast to sham hearts, 100% of AR hearts contained numerous noncardiomyocytes (cardiac myosin−/desmin−/nonmuscle myosin+, collagen I+) in the apex (Figure 2D). Moreover, mRNAs for Atrial Natriuretic Peptide (ANP) and Brain Natriuretic Peptide (BNP), two markers of cardiac damage and hypertrophy, were increased in the AR hearts (Figure 2E) with a concomitant 7.5% increase in cardiac width (Figure 2F), the latter possibly indicating hypertrophic growth. Together, these data demonstrated that resection of heart muscle in neonates induced a massive fibrotic response resulting in substantial scarring at day 21.
Figure 2.
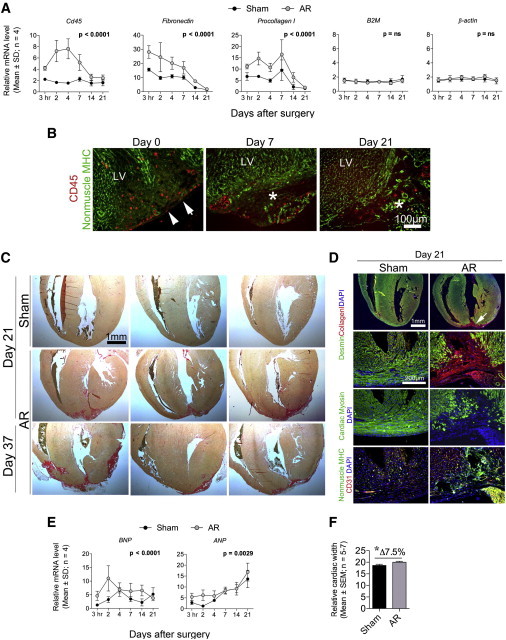
C57Bl/6 Hearts Become Substantially Fibrotic following AR and Show Signs of Hypertrophy
(A) Quantitative real-time PCR of sham and AR hearts (n = 4) at indicated time points. Statistical significance tested by two-way ANOVA is indicated.
(B and D) Paraffin- and cryo-embedded C57Bl/6 AR hearts (n = 4–8) were sectioned and immunostained for (B) CD45/nonmuscle myosin and (D) desmin/collagen; cardiac myosin; nonmuscle myosin/CD31. Representative images were processed (contrast/brightness and color balance) equally in Adobe Photoshop to enable merging. An asterisk (∗) indicates lesion area, whereas white arrows reflect the AR line.
(C) Sirius Red stainings of representative sham (n = 5) and AR (n = 11) heart sections from C57Bl/6 mice identify collagen disposition in the lesioned apex.
(E) Quantitative real-time PCR for hypertrophic markers (ANP and BNP) of sham and AR hearts (n = 4) at indicated time points.
(F) The relative width of C57Bl/6 hearts from sham and AR animals was measured at day 21. Significance was tested by a Student’s t test (∗p < 0.05).
For (A) and (E), quantitative real-time PCR raw data were normalized against B2M and β-actin, which were stably expressed as determined by the qBase+ platform (M:0.579 and CV:0.202) (Hellemans et al., 2007, Vandesompele et al., 2002).
See Figure S2 for quantitative real-time PCR profiling on gene programs at day 21 following AR.
The Damaged Apex Comprises Modest Vascularization, but Few Proliferating Cardiomyocytes
Ideally, cardiac regeneration should be able to restore the functional continuity of cardiomyocytes, as well as blood supply (endothelial and smooth muscle cells) in the necrotic area of the damaged heart. We did encounter blood vessels with CD31+ endothelia, and surrounding smooth muscle cells (aSMA+ and NG2+) in the lesioned area (Figures 2D and 3A). These vessels may either be preexisting vessels developed in the border zone prior to lesion, but they may also represent newly formed vascularity in the damaged area because both Cd31 (Figure 2B) and numerous other vascular genes (Figure S2) were increased in AR hearts as compared with sham controls. Yet, the vascularization was still incomplete as compared with normal vascularized myocardium (Figures 2D and 3A). Porrello et al. suggest that the regenerative heart response is due to an increased number of cardiomyocytes undergoing division 1–7 days following AR (Porrello et al., 2011). We therefore quantified the total number of proliferating cells (5-ethynyl-2-deoxyuridine positive [EdU+]) as well as the number of proliferating cardiomyocytes (EdU+/cardiac myosin+/collagen I−) 1–7 days postsurgery by pulse-chase labeling experiments (Figures 3C–3F). As such, there was a clear tendency (p = 0.06) for lower numbers of proliferating cells in the AR hearts compared to sham hearts, and a profound reduction in the overall number of dividing cardiomyocytes throughout the heart, with remarkably few being present in the apex area (Figures 3G and 3H). Instead, the proliferating cells in the apex region of AR hearts (Figures 3F and 3H) were fibroblastic in phenotype (EdU+/cardiac myosin−/collagen I+) and accounted for the majority of proliferating cells in this zone (Figure 3F). In addition, we did not observe any difference in AR and sham hearts of Meis1b transcripts (Figure 3G), and levels were decreasing with normal cardiac development (Figure 3H). This contradicts the suggested ability of increasing Meis1b levels to inhibit cardiomyocyte proliferation beyond P7 (Mahmoud et al., 2013). Therefore, our data do not support enhanced cardiomyocyte proliferation 1–7 days following AR as previously reported by Porrello et al. (2011). To exclude that the regeneration process merely was delayed in C57Bl/6 mice, we prolonged the examination period with 16 days, thus analyzing the hearts histologically at day 37. Yet, the hearts were still substantially compromised at day 37 with large visual fibrotic scars (Figures 1H, 1I, and 2C), numerous noncardiomyocytes in the lesion area (data not shown), and insufficient vascularization (data not shown), further suggesting that C57Bl/6 hearts heal by remodeling rather than by true regeneration.
Figure 3.
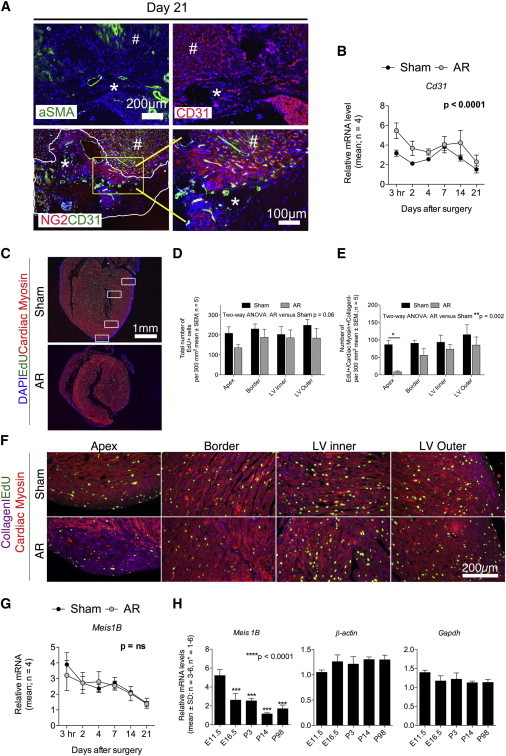
The Apex-Resected Zone in C57Bl/6 Hearts Reveals Limited Vascularization and Numbers of Proliferating Cardiomyocytes
(A) Cryosectioned C57Bl/6 AR hearts (n = 4–8) were immunostained for aSMA, CD31, and NG2/CD31. Representative images were processed (contrast/brightness and color balance) equally in Photoshop to enable merging. An asterisk (∗) indicates lesion area, whereas a number sign (#) refers to nondamaged myocardium.
(B) Quantitative real-time PCR for Cd31 of sham and AR hearts (n=4) at indicated time points. Quantitative real-time PCR raw data were normalized against the stably expressed B2M and β-actin genes (see Figure 2).
(C–F) Paraffin-embedded C57Bl/6 AR and sham hearts (n = 5) from EdU pulse-chase labeled animals were sectioned and immunostained for EdU/cardiac myosin/collagen I/DAPI to enable identification of proliferating cells in total and proliferating cardiomyocytes specifically. Images were taken in four different zones (white boxes in C and areas magnified in F) of each heart and used for cell number quantification (D and E) as described in Experimental Procedures.
(G) Quantitative real-time PCR for Meis1b of sham and AR hearts (n = 4) at indicated time points. Quantitative real-time PCR raw data were normalized against the stably expressed B2M and β-actin genes (see Figure 2).
(H) Meis1b is downregulated with cardiac development. Hearts were microdissected from C57Bl/6 mice at indicated time points and used for quantitative real-time PCR. Primers used were previously described by Mahmoud et al. (2013). Each biological experiment (n = 3–6) included between one and six hearts. qPCR raw data were normalized against Gapdh and β-actin, which were stably expressed as determined by the qBase+ platform (M:0.306, CV: 0.106) (Hellemans et al., 2007, Vandesompele et al., 2002). Statistical significance was tested using a one-way ANOVA with a Dunnett’s multiple comparison posttest. E11.5, embryonic day 11.5.
ICR/CD1 Mice Also Fail to Accomplish Complete Regeneration of Their Hearts
Porrello et al. showed a high regenerative capacity of apex-resected hearts in outbred ICR/CD1 mice (Porrello et al., 2011). Although no difference in regenerative capacity has been reported for C57Bl/6 and ICR/CD1 mouse strains, other regeneration schemes such as wound healing in the ear are highly dependent on the strain (Heber-Katz, 1999). We therefore next performed a new series of AR in 24 ICR/CD1 mice to reevaluate the cardiac-regenerative response recently described by Porrello et al. (2011). However, we did not find any evidence suggesting a complete cardiac-repairing response in the neonatal heart of ICR/CD1 mice either (Figure 4). As with the C57Bl/6 hearts, we found substantial fibrotic scars in 21.5% ± 4.7% of HE-stained sections (846 ± 60 per heart; n = 11), many noncardiomyocytes (cardiac myosin−/desmin−/nonmuscle myosin+/collagen I+), and an incomplete vascular network in 100% of resected ICR/CD1 hearts at day 21 (Figures 4 and S3). These data thus suggest that the absence of complete heart regeneration following heart muscle resection is mouse strain independent.
Figure 4.
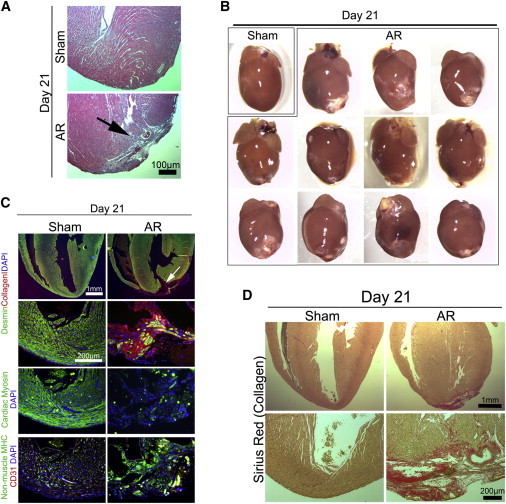
Apex-Resected ICR/CD1 Hearts Do Not Reveal Any Major Regenerative Response but Become Fibrotic
Representative sham (n = 3) and AR (n = 11) hearts from outbred ICR/CD-1 neonates (n = 24) were analyzed at day 21 by (A) HE staining (arrow identifies lesion), (B) stereomicroscopy, (C) immunofluorescence, and (D) Sirius Red staining. Representative sections are shown (see Figure S3). For (C), images were processed (contrast/brightness and color balance) equally in Photoshop to enable merging.
Discussion
Mortality after myocardial infarction is relatively high, and surviving patients are often severely compromised due to insufficient heart-pump function. At the cellular level, damage to the heart’s contractile constituents, the cardiomyocytes, is irreversible, and treatments merely serve to reduce symptoms, and only a minority of patients receives a heart transplant. Novel therapeutics to reestablish the cardiac tissue following infarcts are therefore needed, and factors improving heart regeneration will be of enormous value. The groundbreaking discovery of a mammalian regenerative heart model in mice (Porrello et al., 2011) has thus gained immense attention and hope for identifying factors that may improve cardiac regeneration. However, none of our results here suggests that the neonatal mammalian heart is able to accomplish complete regeneration following heart muscle resection as recently suggested by Porrello et al. (2011). Instead, we demonstrate that the resected heart apex remains lost and that the resection border is healed by a fibrotic scar composed of myofibroblasts, adipocytes, and sparse vessels. These nonmyogenic cells likely originate from the pericardium/epicardium (Smart et al., 2007, Zhou et al., 2008), the only structure in the apex that seemed completely regenerated. In contrast, we observed a lack of cardiac regeneration likely caused by a reduced number of proliferating cardiomyocytes within AR hearts as compared with noninjured hearts. Although the absolute number of proliferating cardiomyocytes would have been optimally determined by quantifying a proliferation marker in combination with a nuclear cardiomyocyte genetic label (Ang et al., 2010), our approach using EdU with cardiac myosin/collagen I seems valid for the relative measurements we performed, and importantly, Porrello et al. used a similar approach (Porrello et al., 2011). Our results are thus contradictory to those reported in the original study performed by the Sadek research group (Porrello et al., 2011).
We can only speculate on the reasons for the contradictions between our study and the Porrello study (Porrello et al., 2011). Importantly, we resected equal amounts of apex tissue, an obvious key parameter that otherwise could have influenced the outcome. Likewise, we used the same time schedule during surgery as previously described by Porrello et al. (2011), where the extent of hypothermia may be critical for the heart tissue survival and therefore also the regenerative ability. The use of different mouse strains may impose another problem to the results; yet, this seems less likely because we performed AR in both C57Bl/6 and ICR/CD1 mice and found no difference in scar formation at day 21. Supportive of that, the Sadek research group as well as others have reported a regenerative capacity of the heart in B6C3F1 (hybrid of C57Bl/6 × C3H/He) (Xin et al., 2013) and C57Bl/6 (Haubner et al., 2012) mice. Even so, ICR/CD1 mice from different breeding facilities have been shown to diverge (Aldinger et al., 2009), and unknown factors associated with the mice thus cannot be absolutely excluded for having an impact on the heart’s ability to regenerate. Finally, one may consider the analysis procedures used in our study and the ones performed by Porrello et al. Both studies base a substantial amount of the results on histological examinations. Porrello et al. report in their supplemental information that they observe a small fibrotic scar in 2 out of 140 (1.4%) sections from damaged hearts (Porrello et al., 2011). Yet, they do not describe if this accounts for each heart examined or just a single outlier. It is also unclear how many hearts were examined and if the 140 sections were randomly chosen throughout the heart or in one series. For comparison, we observed substantial scarring in 100% (31 out of 31) of AR hearts using histology, and the lesioned area was present in 19.5% ± 5.2% of sections, regardless of mouse strain. Notably, Porrello et al. sliced 140 sections (each 5 μm) from one, day 21 heart, whereas we obtained 820 ± 66 sections (each 5 μm). Thus, it may be that Porrello et al. overlooked the lesioned area, which often is located in the beginning or the end of sectioning (Figure S1).
Our results therefore contradict that the AR model may be used for identifying factors that enable cardiomyocyte replenishment and improve heart regeneration following damage. We cannot, however, eliminate the possibility that a few cardiomyocytes are being regenerated following AR, but the amplitude of this response, if present, may therefore be insufficient for a complete cardiac repair. One obvious limitation of the model seems to be the ongoing hyperplastic and hypertrophic growth present in the neonatal mouse hearts, which makes it difficult to distinguish an eventual small true regenerative response from simple developmental features. In that regard, it is important to note that the well-recognized zebrafish heart resection model is performed in adult animals (Jopling et al., 2010, Lafontant et al., 2012, Poss et al., 2002) free of immediate hyperplastic growth.
Yet, from our results, we cannot exclude that other cardiac regeneration models such as the left anterior-descending (LAD) artery ligation in the neonatal myocardium (Haubner et al., 2012, Mahmoud et al., 2013, Xin et al., 2013) do unravel a true and complete regenerative potential of the heart. Still, we find it questionable that a full cardiac regenerative response is accomplished in only 21 days (Mahmoud et al., 2013, Porrello et al., 2011), and in one study (Haubner et al., 2012), after only 7 days following LAD damage of the mouse heart. For comparison, Jesty et al. recently showed that cryo-injury to the neonatal heart is associated with scar formation even after 94 days (Jesty et al., 2012), a scenario that is also seen up until 60–180 days after cardiac injury in simpler organisms like teleost fish (Jopling et al., 2010, Lafontant et al., 2012). However, in a previous study, a genetic model of cardiomyocyte ablation showed that the cardiomyocyte pool is restored in the fetal mouse heart by enhanced proliferation of remaining cardiomyocytes (Drenckhahn et al., 2008). One may speculate if a similar scenario could take place after neonatal mouse heart LAD damage, whereas the AR model lacks an intact 3D heart architecture, a feature that likely is important to some extent for mammalian regeneration to be accomplished. In summary, we thus believe that additional clarifying data are required from the scientific community on this controversial matter to firmly establish whether the mammalian heart is regenerative, otherwise our data substantiate the view that it is not.
Experimental Procedures
A detailed version of our Experimental Procedures can be found in the Supplemental Experimental Procedures. All animal experiments were approved by the Danish Council for Supervision with Experimental Animals (#2011/561-1966). AR was performed at P1 as previously described by Porrello et al. (2011). Briefly, neonates were anesthetized by hypothermia, and the apex was resected until left ventricle chamber exposure, after which the thoracic wall and skin were sutured. Sham mice underwent the exact same procedure without resecting the apex of the heart. For pulse-chase labeling experiments, mice were injected 1 day after surgery with EdU, and the number of proliferating cardiomyocytes and cells in total was counted after 7 days. Relative quantitative PCR (qPCR) and histology were performed as previously described (Andersen et al., 2009). All analyses comprised at least four independent experiments, and statistical significance (p < 0.05) was tested as indicated.
Author Contributions
D.C.A. conceived and designed the study; performed collection, analysis, and interpretation of data; prepared and approved the manuscript; and provided funding. S.G. performed data collection and analysis. C.H.J. designed the study; performed data collection, analysis, and interpreted the data; and participated in the discussion and approval of the manuscript. S.P.S. performed interpretation of data, approved the manuscript, and provided funding.
Acknowledgments
We would like to thank Charlotte Nielsen, Tonja L. Jørgensen, and Anette Kliem (LMCC, Odense University Hospital) for technical assistance on this study and Dr. Bruce Conklin (Gladstone Institute, CA) for language editing. The work was supported by The Novo Nordisk Foundation, The Danish National Research Council (#09-073648), The Lundbeck Foundation (#R48-A4785), Lægeforeningen (#2011-3271/480853-109), Tømrermester Alfred Andersen og Hustru’s Fond, Hertha Christensens Foundation, Eva and Henry Frænkels Foundation, and Department of Clinical Biochemistry and Pharmacology/Odense University Hospital.
Published: April 3, 2014
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Supplemental Information includes Supplemental Experimental Procedures and three figures can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2014.02.008.
Contributor Information
Ditte Caroline Andersen, Email: dandersen@health.sdu.dk.
Søren Paludan Sheikh, Email: soeren.sheikh@rsyd.dk.
Supplemental Information
References
- Aguirre A., Sancho-Martinez I., Izpisua Belmonte J.C. Reprogramming toward heart regeneration: stem cells and beyond. Cell Stem Cell. 2013;12:275–284. doi: 10.1016/j.stem.2013.02.008. [DOI] [PubMed] [Google Scholar]
- Aldinger K.A., Sokoloff G., Rosenberg D.M., Palmer A.A., Millen K.J. Genetic variation and population substructure in outbred CD-1 mice: implications for genome-wide association studies. PLoS ONE. 2009;4:e4729. doi: 10.1371/journal.pone.0004729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen D.C., Petersson S.J., Jørgensen L.H., Bollen P., Jensen P.B., Teisner B., Schroeder H.D., Jensen C.H. Characterization of DLK1+ cells emerging during skeletal muscle remodeling in response to myositis, myopathies, and acute injury. Stem Cells. 2009;27:898–908. doi: 10.1634/stemcells.2008-0826. [DOI] [PubMed] [Google Scholar]
- Ang K.L., Shenje L.T., Reuter S., Soonpaa M.H., Rubart M., Field L.J., Galiñanes M. Limitations of conventional approaches to identify myocyte nuclei in histologic sections of the heart. Am. J. Physiol. Cell Physiol. 2010;298:C1603–C1609. doi: 10.1152/ajpcell.00435.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenckhahn J.D., Schwarz Q.P., Gray S., Laskowski A., Kiriazis H., Ming Z., Harvey R.P., Du X.J., Thorburn D.R., Cox T.C. Compensatory growth of healthy cardiac cells in the presence of diseased cells restores tissue homeostasis during heart development. Dev. Cell. 2008;15:521–533. doi: 10.1016/j.devcel.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Garbern J.C., Lee R.T. Cardiac stem cell therapy and the promise of heart regeneration. Cell Stem Cell. 2013;12:689–698. doi: 10.1016/j.stem.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbern J.C., Mummery C.L., Lee R.T. Model systems for cardiovascular regenerative biology. Cold Spring Harb Perspect Med. 2013;3:a014019. doi: 10.1101/cshperspect.a014019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubner B.J., Adamowicz-Brice M., Khadayate S., Tiefenthaler V., Metzler B., Aitman T., Penninger J.M. Complete cardiac regeneration in a mouse model of myocardial infarction. Aging (Albany, N.Y. Online) 2012;4:966–977. doi: 10.18632/aging.100526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heber-Katz E. The regenerating mouse ear. Semin. Cell Dev. Biol. 1999;10:415–419. doi: 10.1006/scdb.1999.0328. [DOI] [PubMed] [Google Scholar]
- Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jesty S.A., Steffey M.A., Lee F.K., Breitbach M., Hesse M., Reining S., Lee J.C., Doran R.M., Nikitin A.Y., Fleischmann B.K., Kotlikoff M.I. c-kit+ precursors support postinfarction myogenesis in the neonatal, but not adult, heart. Proc. Natl. Acad. Sci. USA. 2012;109:13380–13385. doi: 10.1073/pnas.1208114109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C., Sleep E., Raya M., Martí M., Raya A., Izpisúa Belmonte J.C. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontant P.J., Burns A.R., Grivas J.A., Lesch M.A., Lala T.D., Reuter S.P., Field L.J., Frounfelter T.D. The giant danio (D. aequipinnatus) as a model of cardiac remodeling and regeneration. Anat. Rec. (Hoboken) 2012;295:234–248. doi: 10.1002/ar.21492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud A.I., Kocabas F., Muralidhar S.A., Kimura W., Koura A.S., Thet S., Porrello E.R., Sadek H.A. Meis1 regulates postnatal cardiomyocyte cell cycle arrest. Nature. 2013;497:249–253. doi: 10.1038/nature12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberpriller J.O., Oberpriller J.C. Response of the adult newt ventricle to injury. J. Exp. Zool. 1974;187:249–253. doi: 10.1002/jez.1401870208. [DOI] [PubMed] [Google Scholar]
- Porrello E.R., Mahmoud A.I., Simpson E., Hill J.A., Richardson J.A., Olson E.N., Sadek H.A. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss K.D., Wilson L.G., Keating M.T. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- Santini M.P., Rosenthal N. Myocardial regenerative properties of macrophage populations and stem cells. J. Cardiovasc. Transl. Res. 2012;5:700–712. doi: 10.1007/s12265-012-9383-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart N., Risebro C.A., Melville A.A., Moses K., Schwartz R.J., Chien K.R., Riley P.R. Thymosin beta-4 is essential for coronary vessel development and promotes neovascularization via adult epicardium. Ann. N Y Acad. Sci. 2007;1112:171–188. doi: 10.1196/annals.1415.000. [DOI] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:H0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J.M., Riley P.R. Epicardium-derived cells: a new source of regenerative capacity. Heart. 2011;97:15–19. doi: 10.1136/hrt.2010.193292. [DOI] [PubMed] [Google Scholar]
- Xin M., Kim Y., Sutherland L.B., Murakami M., Qi X., McAnally J., Porrello E.R., Mahmoud A.I., Tan W., Shelton J.M. Hippo pathway effector Yap promotes cardiac regeneration. Proc. Natl. Acad. Sci. USA. 2013;110:13839–13844. doi: 10.1073/pnas.1313192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Ma Q., Rajagopal S., Wu S.M., Domian I., Rivera-Feliciano J., Jiang D., von Gise A., Ikeda S., Chien K.R., Pu W.T. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454:109–113. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


