Summary
Throughout their journey to forming new individuals, germline stem cells must remain totipotent, particularly by maintaining a specific chromatin structure. However, the place epigenetic factors occupy in this process remains elusive. So far, “sensitization” of chromatin by modulation of histone arrangement and/or content was believed to facilitate transcription-factor-induced germ cell reprogramming. Here, we demonstrate that the combined reduction of two epigenetic factors suffices to reprogram C. elegans germ cells. The histone H3K4 demethylase SPR-5/LSD1 and the chromatin remodeler LET-418/Mi2 function together in an early process to maintain germ cell status and act as a barrier to block precocious differentiation. This epigenetic barrier is capable of limiting COMPASS-mediated H3K4 methylation, because elevated H3K4me3 levels correlate with germ cell reprogramming in spr-5; let-418 mutants. Interestingly, germ cells deficient for spr-5 and let-418 mainly reprogram as neurons, suggesting that neuronal fate might be the first to be derepressed in early embryogenesis.
Graphical Abstract

Highlights
-
•
SPR-5/LSD1 and LET-418/Mi2 interact to jointly control germ cell status
-
•
C. elegans germ cells reprogram as neurons in spr-5 let-418 mutants
-
•
SPR-5 and LET-418 counteract COMPASS-dependent H3K4 methylation in the germline
-
•
High H3K4me3 levels in germ cells correlate with somatic reprogramming
Two chromatin factors, the chromatin remodeler LET-418/Mi2 and the H3K4 demethylase SPR-5/LSD1, interact physically and genetically to form an epigenetic barrier against germ cell reprogramming toward somatic fates in C. elegans. Käser-Pébernard and colleagues demonstrate that the main role for this interaction is to counteract the COMPASS-mediated H3K4 methylation, and they propose that the balance of H3K4 methylation defines germ cell status in C. elegans.
Introduction
To ensure that all lineages will develop after fertilization, germ cells must proceed through gametogenesis while maintaining totipotency and resisting somatic differentiation. After their induction, mammalian primordial germ cells (PGCs) express the transcription factors sufficient to not only maintain their pluripotency, such as Oct4, Sox2, or Nanog, but also activate the epigenetic changes essential to PGC specification, including chromosome X inactivation, histone H3K9 demethylation, and genome-wide erasure of methylated DNA (reviewed in Magnúsdóttir et al., 2012). The use of nonvertebrate systems such as C. elegans or D. melanogaster to study germ cell specification revealed that combinations of genetic and epigenetic events were the key to somatic fate repression. To maintain their unique status, C. elegans PGCs globally repress mRNA transcription and establish a specific chromatin structure and composition to tightly control gene expression (Wang and Seydoux, 2013). Recently, germline reprogramming was “artificially” obtained by the simultaneous ectopic expression of master somatic fate inducers (“terminal selector genes”) and the downregulation of chromatin repressors such as LIN-53/RbAP46-48 and the H3K27 methyl-transferase Polycomb (Patel et al., 2012, Tursun et al., 2011), implying that specific combinations of transcriptional and epigenetic factors were capable of controlling the germ cell program.
The ATP-dependent nucleosome remodeler Mi2 is the core component of the nucleosome remodeling and deacetylase complex (NuRD), a multisubunit transcriptional repressor complex known to play a major role in mammalian cell fate determination and capable of different scopes of activities depending on its subunit content (reviewed in Bowen et al., 2004). Embryonic stem cells (ESCs) deficient for the NuRD subunit MBD3 are unable to undertake lineage commitment (Kaji et al., 2006). Conditional knockout mice models showed that Mi2/NuRD was essential in terminal differentiation programs, including T cell maturation (Williams et al., 2004) and Schwann cell-directed peripheral nerve myelination (Hung et al., 2012). In addition, recent findings propose that the NuRD repressive activity is required to limit pluripotency gene expression, thereby permitting ESC differentiation (Reynolds et al., 2012a).
Recently, histone H3 lysine 4 (H3K4) demethylase LSD1/KMD1A was identified as a de novo member of the NuRD complex in HeLa cell extracts (Wang et al., 2009) and in ESCs (Whyte et al., 2012), independently of the chromatin repressor complex CoREST, of which it is the core component (Lee et al., 2005). LSD1 carries differentiation-licensing functions in common with the NuRD complex. lsd1−/− ESCs fail to fully deactivate pluripotency gene enhancers to complete differentiation programs (Whyte et al., 2012). LSD1 silences the bivalent promoter of developmental genes, which combine activating H3K4me2/3 and repressing H3K27me3 histone marks, to maintain ESC pluripotency (Adamo et al., 2011). LSD1 is also involved in multiple developmental programs, including myoblast differentiation (Choi et al., 2010) and neuronal development (Ceballos-Chávez et al., 2012, Fuentes et al., 2012), and is a putative metastatic breast cancer suppressor (Wang et al., 2009). In Drosophila, loss of LSD1 leads to ovarian germline tumorigenesis, because somatic gonadal cells become unable to produce the lineage specification signals required for germline stem cell differentiation (Eliazer et al., 2011).
In C. elegans, homologs for both the NuRD complex and LSD1 were identified. C. elegans LET-418/Mi2 is a subunit of a NuRD-like complex, together with Rb-binding protein LIN-53/RbAp48, histone deacetylase HDA-1/HDAC1, metastasis-associated protein homolog LIN-40/MTA1, and DCP-66/p66(α/β) (Passannante et al., 2010, Unhavaithaya et al., 2002; our unpublished data). The C. elegans NuRD-like complex was previously involved in controlling the vulval cell fate (von Zelewsky et al., 2000). In addition, similar to its Drosophila dMi2 homolog (Kunert et al., 2009), LET-418 interacts tightly with the zinc finger protein MEP-1 and HDA-1/HDAC1 in a distinct MEP-1-interacting complex (MEC) involved in repressing germline gene expression in somatic cells (Passannante et al., 2010, Unhavaithaya et al., 2002).
Three C. elegans genes encode putative LSD1 homologs: Suppressor of Presenilin 5 (spr-5), T08D10.2/lsd-1 and R13G10.2/amx-1. The SPR-5 protein displays a demonstrated biochemical H3K4 demethylase activity and functions in promoting fertility (Katz et al., 2009). SPR-5 is partially responsible for the specific erasure of H3K4me2 marks in the PGCs at their birth. spr-5 mutants progressively accumulate H3K4me2 in PGCs throughout generations, correlating with the progressive “mortal germline” sterile phenotype peaking at 28–30 generations (Katz et al., 2009).
All these observations suggest that the functions of LSD1 and Mi2/NuRD in controlling cell lineage specification are ancient and well conserved across species. In order to decipher the molecular mechanisms by which LSD1 and NuRD determine cell fate in vivo, we set up to analyze their common functions in the developmental model organism C. elegans. Here, we describe an interaction between the C. elegans LET-418/Mi2-containing complexes and SPR-5/LSD1. In addition to the physical interaction between SPR-5, LET-418, and associated complexes, spr-5 and let-418 interact genetically to promote the normal development of germline stem cells. Concomitant loss of SPR-5 and LET-418 leads to immediate sterility, aberrant gonad development, and germline teratoma incidence. SPR-5 and LET-418 together maintain the germline stem cell status and form an epigenetic barrier to reprogramming. This infers the existence of a conserved link between LSD1 and Mi2-related complexes and shows that specific epigenetic regulators collaborate intricately to control cell fate during germ cell development.
Results
LET-418 and SPR-5 Physically Interact In Vivo
To first test whether the C. elegans homologs of LSD1 and Mi2 interacted physically, coimmunoprecipitation (co-IP) assays of SPR-5 and LET-418 were performed in embryonic extracts of wild-type and spr-5 null (by134 allele) strains (Figure 1A). Although anti-LET-418 antibodies did not pull down visible amounts of SPR-5, anti-SPR-5 antibodies recovered detectable levels of the LET-418 protein in wild-type, but not spr-5-null-derived, samples (Figure 1A), demonstrating that SPR-5 and LET-418 interact in vivo.
Figure 1.

SPR-5 Interacts with LET-418 and MEP-1 In Vivo
(A) Coimmunoprecipitation of SPR-5 and LET-418 in embryonic extracts using anti-LET-418 or anti-SPR-5 antibodies. wt, wild-type; spr-5, spr-5(by134) null allele.
(B) Coimmunoprecipitation of LET-418 using anti-SPR-5 (SPR-5 IP) antibodies with (+) or without (−) DNaseI/ethidium bromide (DNase+EtBr) pretreatment.
(C) Coimmunoprecipitation of LET-418 and FLAG-tagged MEP-1, using anti-SPR-5 and anti-FLAG antibodies, in embryonic extracts of a control wild-type strain (−) or a strain stably expressing MEP-1::3xFLAG::GFP (+).
See also Figure S1.
To determine whether this interaction was bridged by DNA, the anti-SPR-5 immunoprecipitation was repeated using wild-type embryonic extract pretreated with DNaseI and ethidium bromide (EtBr), which separate all proteins from DNA (Figure 1B). LET-418 was still detectable in the DNase/EtBr-treated anti-SPR-5 eluate, ruling out DNA bridging as a cause for the interaction (Figure 1B).
An interaction was thereafter detected in embryonic extracts between 3×Flag-tagged MEP-1 and SPR-5 (Figure 1C) and between HDA-1 and SPR-5 (Figure S1A available online). Provided that HDA-1 is potentially a member of both the NuRD and MEC complexes, SPR-5 might be interacting preferentially with the MEC complex or with both. We therefore tested the interaction of SPR-5 with other NuRD complex members. A weak but reproducible interaction was detected between SPR-5 and GFP-tagged LIN-53, a NuRD member homologous to mammalian RbAp48 (Harrison et al., 2006) (Figure S1B). Hence, SPR-5 interacts with both LET-418-containing NuRD and MEC complex subunits in vivo.
Simultaneous Downregulation of SPR-5- and LET-418-Containing Complexes Leads to Synthetic Sterility
To understand the genetic relationship between spr-5 and let-418, double mutants were generated. Similar to the spr-5(101) mutant allele phenotype reported previously (Katz et al., 2009), the spr-5(by134) null strain started to lose fertility after seven generations (Figure 2A). Conversely, let-418(n3536) temperature-sensitive (let418ts) hypomorphic worms maintain fertility over generations at 20°C, whereas they produce an L1-arrested progeny at 25°C (von Zelewsky et al., 2000; Figure 2A). Interestingly, combining spr-5(by134) with let-418(n3536) mutations at 20°C led to a maternal effect sterile phenotype (Figure 2A). The first generation of spr-5(by134); let-418(n3536) double homozygotes was fertile, whereas 97.8% (312/319) second-generation worms were sterile, with the remaining seven subfertile worms producing a total of 38 sterile third-generation progeny (Figure 2A). To determine whether this synergistic effect on fertility could also be obtained using RNAi-mediated gene targeting, spr-5(by134) L4 larvae were transferred onto let-418(RNAi) feeding plates at the semipermissive temperature of 20°C and their progeny observed. spr-5(by134); let-418(RNAi) mutants were synthetic sterile in the next generation, causing 100% sterility in spr-5(by134); let-418(RNAi) versus 27% in let-418(RNAi) animals at 20°C (Figure S2C). This effect was also observed at 15°C, but to a lower extent (Figure S2B). Simultaneous loss of spr-5 and let-418 function therefore leads to synthetic sterility in the next generation.
Figure 2.

Simultaneous Downregulation of SPR-5 and LET-418 Causes Sterility and Germline Tumor Formation
(A) Double spr-5(by134); let-418(n3536) mutants are maternal-effect sterile. Percentage of fertile adults, at 20°C, over the indicated generations; ≥100 adults were counted per strain per generation.
(B) Nomarski pictures of the indicated genotypes 4 days postbirth.
(C) DAPI staining of the indicated mutant germlines 6 days postbirth. Double spr-5; let-418 mutants develop a germline tumor. Plain arrows, oocytes; empty arrows, sperm; gray arrows, embryos; striped arrows, endomitotic oocytes. Scale bar = 20 μm.
(D) Enlarged view of the squared zones from (C), showing DAPI-stained nuclei for the indicated genotypes. Fine dashed circles, larger nuclei with decondensed chromatin; long dashed circles, smaller nuclei different from wild-type germ cells. Scale bar = 20 μm.
All strains in (B)–(D) displayed the unc-46(e177) allele in their background. See also Figure S2.
This phenotype did not involve the other C. elegans LSD1 homologs, because let-418ts worms exposed to lsd-1 and amx-1(RNAi) produced a 100% fertile progeny (Figure S2D).
Synthetic sterility was also observed in spr-5(by134) mutants grown on mep-1(RNAi) at 15°C and 20°C, on hda-1(RNAi) at 15°C, and on dcp-66(RNAi) at 15°C and 20°C (Figures S2B and S2C). No effect was observed on lin-53(RNAi) or on hda-1(RNAi) (at 20°C), due to embryonic lethality (Figures S2B and S2C; our unpublished data). These results imply that SPR-5 interacts with both the LET-418-containing NuRD and MEC complexes to promote fertility.
SPR-5 was first identified as a physical and functional member of the C. elegans CoREST complex (Eimer et al., 2002), a chromatin-remodeling complex with repressive transcriptional activities (Andrés et al., 1999, Lakowski et al., 2006). Three subunits were identified as putative CoREST complex members: spr-1, spr-3, and spr-4 (Smialowska and Baumeister, 2006). let-418ts worms produced a 100% fertile progeny when fed on spr-3 or spr-4 (RNAi) and 97.3% fertile progeny when fed on spr-1(RNAi) (Figure S2E), indicating that the CoREST complex was probably not involved in SPR-5/LET-418 function.
Overall, we found that spr-5 interacts physically and genetically with NuRD and MEC complex members in a CoREST-independent manner to promote germline immortality over generations.
The Double spr-5; let-418 Mutation Triggers Germline Tumor Formation
Morphologically, the sterile spr-5(by134); let-418ts sterile progeny was slow growing, shorter and thinner than wild-type, and displayed abnormally developed gonads (data not shown). DAPI (4′,6-diamidino-2-phenylindole dihydrochloride) staining showed that the double-mutant gonads did not extend in the classical “U” shape but instead formed a large oval shape invading the whole center of the worm, occasionally with a small gonadal protrusion on one side (Figure S2A). To confirm these observations, double-null spr-5 let-418 mutants were generated from spr-5(by134); let-418(s1617)/+ heterozygote parents. let-418(1617) M+Z− single mutants are sterile, most likely due to precocious oocyte endomitosis (Figure 2C; Table 1). Double-null spr-5(by134); let-418(s1617) M+Z− mutants are also sterile, smaller than single mutants, and grow abnormally shaped oval gonads (Figures 2B and 2C). Similar to what we observed with the previous genotypes (Figure S2A), the sterility in double spr-5(by134); let-418(s1617) worm was due to abnormal germ cell progression and incomplete gametogenesis (Figure 2C). Their oval gonads retained normal germ cell nuclei at their extremities, whereas the central region contained both small condensed and large decondensed nuclei, which are normally not found in wild-type germlines (Figures 2C and 2D). Sperm was present in the central gonad, but oocytes were never observed, demonstrating a strong defect in completing gametogenesis (Figures 2C and 2D; Table 1). Seven days postbirth, 100% of the spr-5(by134); let-418(s1617) worms displayed a large disorganized germline (Figure 2C; Table 1). We are qualifying these abnormal spr-5; let-418 germlines of “germline tumors,” although they are different from the proximal or distal proliferative germline tumors previously observed in C. elegans (Francis et al., 1995, Subramaniam and Seydoux, 2003). Germline tumors were observed for all the combinations of spr-5 and let-418 alleles or RNAi tested; in addition, the progeny of spr-5(by134); let-418(RNAi) or spr-5(by134); dcp-66(RNAi) also developed abnormal germline tumors (Table S1), suggesting that the LET-418-containing NuRD and MEC complexes are involved in this abnormal germ cell progression.
Table 1.
Gonads of the spr-5 let-418 Double Mutants Are Tumorous
| Strain | % Sperm | % Oocyte | % Embryos | % Endomitoses | % Tumors | n |
|---|---|---|---|---|---|---|
| unc-46(e177) | 100 | 100 | 100 | 0 | 0 | 28 |
| spr-5(by134); unc-46(e177) | 97 | 92 | 92 | 0 | 0 | 39 |
| let-418(s1617) unc-46(e177) | 100 | 94 | 0 | 91 | 0 | 32 |
| spr-5(by134); let-418(s1617) unc-46(e177) | 17 | 0 | 0 | 0 | 100 | 36 |
Quantification of events occurring in the germlines of DAPI-stained 7-day-old adults of the indicated genotypes. % sperm, percentage of worms producing sperm; % oocyte, percentage of worms producing mature oocytes; % embryos, percentage of worms producing fertilized embryos; % endomitoses, percentage of worms containing endomitotic, nonfertilized oocytes; % tumors, percentage of worms presenting a tumorous germline; n, total number of worms counted.
spr-5 let-418 Germ Cells Lose Pluripotency while Maintaining Cell Division
To find out whether the tumorous germlines in spr-5; let-418 worms were hyperproliferative, replication activity was monitored using a BrdU incorporation assay (Figure 3; Table S2). In parallel, the germ cell status was checked by immunostaining PGL-1, a component of the cytosolic P granules (Kawasaki et al., 1998) cosegregating specifically with germ cells (Hird et al., 1996). Wild-type and spr-5(by134) or let-418(s1617) single-null mutants displayed comparable amounts of BrdU-positive cells in the distal mitotic region of the gonads at 4 and 7 days after birth (Figure 3). All their germ cells were P granule positive and progressed normally toward gametogenesis (Figure 3; Table S2). On the contrary, double spr-5(by134); let-418(s1617) mutants held disorganized, BrdU-positive cells in every part of their abnormally oval gonad (Figure 3). Strikingly, 78% of the BrdU-positive cells were P granule negative (Figure 3; Table S2), indicating that the double-mutant germ cells lost pluripotency while maintaining an active replication.
Figure 3.

spr-5 let-418 Germline Tumors Replicate Rapidly while Losing Pluripotency
BrdU incorporation assay in single or double spr-5(by134) let-418(s1617) mutants 4 or 7 days postbirth at 20°C. Gonad immunostaining against BrdU (green) and the P granule component PGL-1 (red) plus DAPI staining of the DNA content (blue). Empty arrows, PGL-1-positive, BrdU-positive cells; plain arrows, PGL-1-negative, BrdU-positive cells; right panel squares, enlargements of the boxed areas on their left. All these strains contained the unc-46(e177) mutation in their genetic background. Scale bar = 20 μm. See also Figure S3 and Table S2.
The mitotic potential of gonadal cells was also assessed by immunostaining phosphorylated histone H3 serine 10 (PH3) (Hendzel et al., 1997) in 7-day-old worms (Figure S3; Table S3). Wild-type and single spr-5 or let-418 mutant gonads were entirely P granule positive, with a few PH3-positive cells, reflecting the low mitotic rate at this age (Figure S3; Table S3). Strangely, germlines of spr-5(by134); let-418(s1617) double mutants contained two distinct populations of cells, separable by the intensity of their PH3 staining. The first population of “high-PH3-signal” cells was defined as cells displaying PH3 levels comparable to the control and single mutants; these cells were all devoid of P granules (Figure S3; Table S3). On the other hand, a second population of “low-PH3-signal” cells, never observed in the control strains, was composed essentially of P-granule-positive cells and might indicate the presence of a slowly dividing population (Figure S3; Table S3). Altogether, our data suggest that spr-5; let-418 germlines are tumoral, because replication and mitosis are maintained in the whole gonad and are not restricted to a mitotic zone. In addition, cells in the central tumors keep dividing but fail to maintain their germ cell status, which is evocative of cells undertaking a somatic fate.
spr-5 let-418 Germ Cells Reprogram into Neurons
To detect the likely activation of somatic differentiation pathways, reporter constructs expressing GFP or red fluorescent protein under the control of tissue-specific promoters were introduced in the spr-5(by134) background and ectopic expression was examined in let-418(RNAi)-treated worms. Strikingly, strong ectopic expression of pan-neuronal reporter unc-119p::gfp was detected specifically within the spr-5(by134); let-418(RNAi) worm gonads starting at the young-adult stage (Figure 4A). This ectopic expression increased progressively between 4 and 7 days after birth and was detected in a large majority of worms (73%–75% at day 7; Figure 4A). Moreover, the pan-cellular expression of the transgene allowed us to observe that the GFP-positive cells adopted a neuron-like morphology and developed cellular projections similar to neuronal axons (Figure 4A). Nomarski pictures confirmed that the unc-119p::GFP-positive cells had lost the “pan-fried egg” shape of normal germ cells, became flatter, and projected axon-like structures (Figure 4B). Neuronal transformation was further confirmed by testing the expression of two other pan-neuronal transgenes, rab-3p::nls::rfp and unc-33p::gfp, in the spr-5(by134); let-418(RNAi) mutant background. In both cases, these transgenes were found ectopically expressed in the mutant tumorous germlines (Figure S4A). Strikingly, this phenomenon was also observed when the unc-119p::gfp transgene was expressed into spr-5(by134) worms exposed to dcp-66(RNAi) or mep-1(RNAi) worms (Figure S4B; data not shown). Thus, downregulating one member of a LET-418-containing complex is therefore sufficient to produce synthetic cell-fate catastrophes in spr-5 worm germlines.
Figure 4.

spr-5 let-418 Germ Cells Reprogram toward a Neuronal Fate
(A) Ectopic expression of the pan-neuronal reporter unc-119p::gfp within the spr-5(by134) let-418(RNAi) germinal gonad. spr-5(by134) or control worms (wild-type) expressing the unc-119p::gfp reporter were grown on let-418(RNAi) at 20°C and monitored for ectopic GFP expression in their gonads at 5, 7, 11 and 13 days postbirth. The percentage of worms ectopically expressing unc-119p::GFP in their gonads is indicated within each representative picture. Dashed lines, gonad limits; n.d., not determined; ∗, distal tip (when observable). Scale bar = 20 μm.
(B) Nomarski pictures of unc-119p-driven GFP-positive cells in the spr-5(by134) let-418(RNAi) germlines 7 days postbirth. Scale bar = 10 μm.
See also Figure S4.
SPR-5 and LET-418 Protect the Germline against Multiple Somatic Fates
To determine whether other cell fates were induced in the spr-5; let-418 mutants, muscle- and intestinal-specific transgenic reporters were tested in our system. Among them, only the muscular unc-97::gfp reporter was found to be ectopically expressed in a subset of spr-5(by134); let-418(RNAi) germlines (54%; Figure S4A; data not shown). To confirm this, ectopic expression of the MYO-3 muscular marker expression (Miller et al., 1983) was followed in early and late adult tumors (Figure S5A). Cytoplasmic expression of MYO-3, which is distinct from the signal generally observed in sheath cells around wild-type gonads, was detected in PGL-1-negative cells in a small amount of old-adult tumorous germlines (Figure S5A). These cells formed distinct “clonal” groups (Figure S5A), substantiating our previous finding that cells kept dividing after losing P granules (Figure 3). Because of its late occurrence, this germ-to-muscle reprogramming might be general of germline tumors in aging worms, although this phenomenon was not documented in the literature. To rule out this possibility, worms with mutations causing proliferative tumors (mog-6(q465); gld-3(q730) (Belfiore et al., 2004) and glp-1(ar202) gain of function (Pepper et al., 2003) mutations, respectively) were grown to old age and ectopic expression of MYO-3 was assessed by immunostaining (Figure S5B). Although the tumorous germlines of both mutants were filled with proliferative germ cells, there was no P granule loss or ectopic MYO-3 expression, confirming that muscular differentiation was not simply an age-related process but was specific to the spr-5; let-418 tumorous germlines (Figures S5A and S5B).
Altogether, our data infer that the spr-5; let-418 germline forms a type of teratoma, in which germ cells keep dividing but fail to complete meiosis, lose their pluripotent status, and reprogram into neurons, muscles, and possibly other undetermined cell types.
Germline Tumor Formation in spr-5 let-418 Mutants Is Linked to a COMPASS-Complex-Dependent Increase in H3K4 Methylation Levels in Chromatin
In spr-5 mutants, the loss of H3K4 demethylase activity is accompanied by an increase in H3K4me2/3 levels in the PGCs (Katz et al., 2009). To determine whether germline defects in the double spr-5; let-418 mutants were caused by ectopic accumulation of methylated H3K4 on chromatin, we first undertook a genetic approach that consisted of decreasing H3K4 methyltransferase activity in those mutants. In C. elegans, most of the H3K4 methylation is ensured by the SET1 homolog SET-2 (Greer et al., 2010, Xiao et al., 2011, Xu and Strome, 2001), in association with homologs of the yeast SET1/COMPASS complex components such as WDR-5.1 (Li and Kelly, 2011), DPY-30 (Pferdehirt et al., 2011), ASH-2 (Greer et al., 2010), and RbBP5 (Li and Kelly, 2011). Endogenous levels of SET-2 or WDR-5.1 were therefore downregulated by RNAi in the double-null mutants spr-5(by134); let-418(s1617), and germline tumor formation was monitored (Figure 5A). control(RNAi)-treated, spr-5(by134); let-418(S1617) worms developed germline tumors in a majority (91%) of individuals, whereas set-2(RNAi)-treated worms were partially rescued, with 36% worms harboring two normally shaped gonads, and 64% developed only one unilateral tumor (Figure 5A; Table S4). wdr-5.1(RNAi)-treated worms were also partially rescued, but to a lesser extent (Figure 5A; Table S4). The rescued gonads, although smaller, presented a normal mitosis-through-meiosis progression, up to spermatogenesis; however, there was no oogenesis (Figure 5A), indicating that gametogenesis was only partially rescued.
Figure 5.

The Cooperative Control of COMPASS-Dependent H3K4 Methylation Levels by SPR-5 and LET-418 Is Linked to Germ Cell Status
(A) Germline tumorigenesis of spr-5 let-418 mutants is partially rescued by set-2 and wdr-5.1(RNAi). DAPI staining of wild-type and double spr-5(by134) let-418(s1617) null mutants grown on control, set-2, or wdr-5.1(RNAi) 7 days postbirth at 20°C. Right panels show an enlarged view of the right arm of spr-5; let-418 mutant gonads. Dashed lines show the position of the somatic gonad. ∗, distal tip (when visible). Scale bar = 20 μm.
(B) P-granule-negative cells of the spr-5(by134); let-418(s1617) germlines show elevated H3K4me3 levels, which are partially rescued by set-2(RNAi) and wdr-5.1(RNAi). Immunostaining of control or spr-5(by134); let-418(s1617) germlines after exposure to control, set-2, or wdr-5.1(RNAi) 7 days postbirth at 20°C. Full arrows show P-granule (GLH-2)-positive germ cells, whereas empty arrows point to P-granule-negative cells. Scale bar = 20 μm.
(C) Quantification of (A), showing the percentage of cells with elevated H3K4me3 signal in P-granule-positive (light gray bars) or P-granule-negative (dark gray bars) cells. Results are represented as the mean percentage of cells with high H3K4me3 signal, counted manually using the ImageJ cell counting tool on immunostaining pictures, between two biological replicates. Error bars = SD; p values: t test (1), p = 0.00025; t test (2), p = 0.00012. All the strains present in this figure contained the unc-46(e177) mutation in their genetic background.
To confirm these results, we generated a double spr-5(by134); set-2(ok952) mutant in which the SET-2 enzymatic activity is mildly compromised (Simonet et al., 2007, Xiao et al., 2011). Strikingly, 96% of the spr-5(by134); set-2(ok952); let-418(RNAi) progeny showed no germline tumor and contained two normally shaped gonads in which spermatogenesis took place; among these, 9% produced fertilized embryos (Figure S6, two first rows; Table S5).
In summary, reduction of the COMPASS H3K4 methyltransferase activity partially rescues the germline program in spr-5; let-418 animals, allowing for the maintenance of germ cell status, progression through meiosis, and suppression of somatic differentiation.
To verify whether the observed tumoral phenotypes correlated with a COMPASS-dependent increase in H3K4 methylation levels, germlines of spr-5(by134); let-418(s1617) double-null mutants treated with set-2 or wdr-5.1(RNAi) were immunostained for trimethylated H3K4 (H3K4me3) (Figure 5B). Strikingly, elevated levels of H3K4me3 were specifically observed in the P-granule-negative cells of spr-5(by134); let-418(s1617) mutants (Figures 5B and 5C). Coherent with the previous observations that downregulation of COMPASS activity rescued the spr-5(by134); let-418(s1617) mutant phenotypes, set-2 and wdr-5.1(RNAi) treatment resulted in a reduction not only of the number of P-granule-negative cells but also of the global H3K4me3 levels in those cells (Figures 5B and 5C; data not shown). Hence, simultaneous targeting of SPR-5 and LET-418 functions leads to an abnormal, COMPASS-dependent, increase in H3K4me3 levels in germ cells, which tightly correlates with the loss of germ cell status.
Germ Cell Pluripotency Is Maintained via H3K4 Methylation Control
Our results strongly support the hypothesis that a strong increase in H3K4 methylation can lead to germ cell reprogramming. Sterile spr-5(by134) late-generation worms, which display a high H3K4 methylation level in their germ cell chromatin, should therefore show neuronal differentiation of their germline (Katz et al., 2009). To test this hypothesis, sterile spr-5(by134) worms, which stochastically appear at each generation, were analyzed for ectopic expression of the pan-neuronal unc-119p::gfp reporter and compared to their fertile counterparts (Figure 6A). Strikingly, ectopic unc-119p::gfp expression was detected in the germline of more than three-quarters of the sterile spr-5(by134) population, although it was never observed in fertile worms (Figure 6). This ectopic expression was accompanied by loss of germ cell shape toward a neuron-like morphology, including axonal extensions (Figure 6A; data not shown). Side-by-side immunostaining of fertile and sterile spr-5(by134); unc-119p::gfp dissected gonads confirmed that the H3K4me3 level in sterile, P-granule-negative spr-5(by134) germlines (Figure 6B, right gonad) is visibly higher than in the P-granule-positive, fertile spr-5(by134) germline (Figure 6B, left gonad). Hence, the progressive accumulation of H3K4 methyl marks on germ cell chromatin over generations directly leads to loss of pluripotency, germ cell reprogramming, and sterility in spr-5(by134) null worms.
Figure 6.
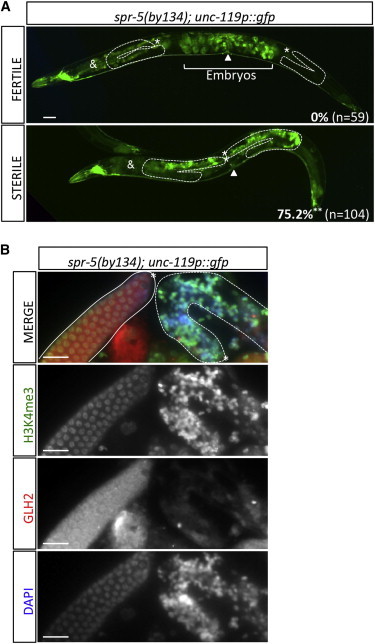
Loss of Fertility in spr-5(by134) Single Mutants Is Also Associated with Germ Cell Reprogramming and Elevated H3K4me3 Levels
(A) Percentage of worms ectopically expressing the pan-neuronal reporter unc-119p::gfp in the germline of spr-5(by134) fertile and sterile individuals of a same generation 7 days postbirth at 20°C. Two representative pictures are presented. n, total number counted. t test p value = 0.0025 (∗∗). White dashed lines show gonad borders. ∗, distal tip; &, background fluorescence of the intestine; white arrow, position of the vulva. Scale bar = 20 μm.
(B) Side-by-side immunostaining showing H3K4me3 (green) an GLH-2(red) levels in dissected gonads from fertile and sterile spr-5(by134) individuals from (A). Left gonad (white full line), fertile worm; right gonad (white dashed line), sterile worm. Scale bar = 20 μm.
To confirm these results, we studied the impact of depleting other H3K4 demethylases on germ cell development in the absence of LET-418. H3K4 histone demethylases (KDM) can be organized in two groups relative to their functional domain (reviewed in Rotili and Mai, 2011). In C. elegans, only four H3K4 KDMs were identified. The amine-oxidase family includes the three LSD1 homologs spr-5, lsd-1, and amx-1 (Katz et al., 2009, Maures et al., 2011), whereas the JumonjiC (JmJC)/JARID family is only represented by the RBP2 homolog rbr-2 (Christensen et al., 2007). As mentioned above, depletion of lsd-1 or amx-1 via RNAi did not reduce the fertility of let-418ts worms at semipermissive temperature (Figure S2D), implying that none of these LSD1 homologs were involved in the SPR-5/LET-418-dependent mechanisms of germ cell protection. Similarly, the rbr-2(tm1231) mutation did not lead to increased sterility when combined to let-418(RNAi) (Figure S6). However, double spr-5(by134); rbr-2(tm1231) mutants were synthetic lethal when exposed to let-418(RNAi) (Figure S6; Table S5), suggesting that spr-5 and rbr-2 might function principally in separate and complementary pathways. This synthetic lethality was largely rescued when downregulating SET-2 activity, to an higher extent with respect to fertility than in spr-5(by134); let-418(RNAi) worms (Figure S6; Table S5). Thus, the combined loss of two H3K4 KDMs, SPR-5 and RBR-2, in combination with LET-418 might generate a global, COMPASS-mediated increase in H3K4 methylation levels incompatible with embryonic development.
Altogether, we believe that the LET-418-containing NuRD and MEC complexes specifically interact with the H3K4 demethylase SPR-5, but not with RBR-2, AMX-1, or LSD-1, to form an epigenetic barrier to germ cell reprogramming. A deficiency in SPR-5 activity, accompanied by loss of NuRD or MEC function, leads to inappropriate levels of H3K4 methylation on germ cell chromatin due to uncontrolled COMPASS activity and triggers loss of germ cell status and somatic differentiation.
Discussion
Here, we describe the identification of an epigenetic mechanism necessary and sufficient to maintain pluripotency and/or avoid precocious differentiation of the germ cell lineage in C. elegans. Histone H3K4 demethylase SPR-5/LDS1 physically interacts with the LET-418-containing chromatin-remodeling NuRD and MEC complexes. This is accompanied by a genetic interaction, leading to a synthetic sterile phenotype in double mutants and indicating that SPR-5 and LET-418 have a collaborative role in promoting fertility. We then discovered that spr-5; let-418 germ cells progressed anarchically, eventually lost their pluripotent status, and undertook somatic differentiation, leading to teratoma formation. Most of the reprogramming germlines contained a large number of neuron-shaped cells, suggesting that the neuronal fate is a major target for combined SPR-5 and LET-418-complex regulation.
The LET-418/SPR-5 Physical Interaction Might Potentiate Their Biochemical Function
Our finding that NuRD and SPR-5 work together to control germ cell fate suggests that the state of the chromatin linked to germ cell differentiation relies principally on the three biochemical activities of these complexes, namely histone deacetylation, histone demethylation, and nucleosome remodeling. Identifying a physical LET-418/SPR-5 interaction was surprising considering the synthetic phenotypes of the double spr-5; let-418 mutants, which would not be expected if those factors worked together in a single complex. A first, simple interpretation is that the SPR-5 and LET-418 complexes interact in embryonic and adult somatic cells, but not in the germline. Arguing against this idea, immunostaining experiments show that both proteins colocalize around the adult germ cell chromatin (data not shown). More excitingly, we postulate that the LET-418/SPR-5 interaction has a synergistic effect on their biochemical activities. As an illustration, Reynolds et al. recently demonstrated that NuRD-mediated deacetylation of histone H3K27 was necessary for Polycomb/PRC2 access and subsequent methylation of this residue to regulate the expression of poised developmental genes in ESCs (Reynolds et al., 2012b). Furthermore, the activity of human LSD1 depends on the histone code surrounding the H3K4 residue and is inhibited by H3K9 acetylation (Forneris et al., 2005). The NuRD complex’s histone deacetylase activity could be necessary to deacetylate H3K9 and promote LSD1-dependent H3K4 demethylation. In our hands, concomitant loss of LET-418 and SPR-5/LSD1 increased the amount of methylated H3K4 on chromatin in cells that had lost their germline status. Downregulating members of the COMPASS H3K4 methyltransferase complex partially rescued this H3K4me3 increase, comforting the hypothesis that SPR-5 and LET-418 are synergistically limiting the COMPASS-dependent H3K4 methylation in germ cells.
Surprisingly, we did not observe any genetic interaction impairing germline development between let-418 and the three other C. elegans H3K4 demethylases rbr-2, lsd-1, and amx-1, re-enforcing the idea that the let-418/spr-5 interaction is specific and involved in promoting proper germ cell progression toward gametogenesis. However, spr-5; rbr-2 double mutants, which grow normally and are fertile at low generation count, were synthetic lethal when exposed to let-418(RNAi). This implies that in the absence of spr-5 and let-418 functions, rbr-2 is essential to the worms’ development and that SPR-5 and RBR-2 function primarily on different targets and possibly in different tissues. The fact that COMPASS downregulation rescues spr-5; rbr-2 mutants on let-418(RNAi) confirms that all these phenotypes were generated by a deregulation of H3K4 methylation on chromatin, but the functions and tissue specificity harbored by rbr-2 during development remain elusive. rbr-2 was proposed to act mainly in somatic cells during early embryogenesis, spr-5 being then considered as the main germline-specific H3K4 KDM (Wang et al., 2011). rbr-2 also counteracts the effects of ash-2 in the germline of aging worms (Greer et al., 2010). Finally, extensive studies on two rbr-2 null alleles demonstrated that RBR-2 and SPR-5 are required to maintain germline immortality at high temperatures (Alvares et al., 2014). In our hands, the germline of rbr-2(tm1231) mutants did not display higher-than-normal H3K4me3 levels (data not shown). However, more extensive studies investigating the intricate relationship among SPR-5, RBR-2, and SET-2 will be necessary to understand their unique function in controlling germ cell status.
LET-418 and SPR-5: An Epigenetic Barrier to Somatic Programs in Germ Cells
In our model, SPR-5- and LET-418-containing complexes are the prime inhibitors of COMPASS-mediated H3K4 methylation, forming a strong epigenetic barrier against germ cell differentiation, for which the mechanistics of action start to become unraveled. The observation that late-generation spr-5 sterile mutants also exhibit reprogrammed germlines directly links H3K4 methylation to somatic fate. In wild-type animals, LET-418 complexes might target SPR-5 to chromatin and potentiate SPR-5 activity by deacetylating histones but also maintain a certain level of competition with COMPASS for specific sites on chromatin. In the absence of SPR-5, this steric competition is at least partially acting to protect chromatin, explaining why only a small subpopulation of spr-5 mutant worms, versus all the spr-5; let-418 mutants, undergo germline reprogramming. An overdose of H3K4me2/3 marks on germ cell chromatin, above a defined threshold, would therefore represent an irreversible “somatic signal,” condemning the germline to mortality.
Noticeably, among every single let-418 mutant allele already studied in our laboratory, germ cell reprogramming was never obtained (our unpublished data). This might indicate that SPR-5 might be coupled to additional chromatin factors to protect germ cell chromatin against COMPASS activity.
Neuronal Differentiation: The “Default” Program?
A majority of sterile spr-5 and spr-5; let-418 worms developed neurons in their reprogrammed germline. Interestingly, the first asymmetric divisions of the C. elegans embryo produces one germ cell progenitor (P) and one somatic cell progenitor (AB), from which most (254/259) neurons originate (Sulston et al., 1983). SPR-5- and LET-418-containing complexes might constitute important guardians of the neuronal differentiation program within P-granule-positive germ cell progenitors. It would be of upmost interest to determine whether the LET-418/SPR-5 interaction is specific to germ cells and/or occurs specifically at the promoter of differentiation genes and gets disrupted once cells exit the “P” lineage, lose pluripotency, and enter somatic differentiation.
Experimental Procedures
C. elegans strains, expression vectors, and antibodies used in these experiments are described in detail in the Supplemental Information.
C. elegans Strains and Cultures
The Bristol N2 strain was used as the wild-type strain of reference in all experiments, which were performed at 20°C under standard worm culture conditions (Brenner, 1974), unless otherwise stated.
RNAi
RNAi constructs were all amplified from the Ahringer RNAi library (Kamath and Ahringer, 2003), except for the control RNAi control (pPD129.36, Fire lab L4440), LET-418, and MEP-1 RNAi vectors (pFG98 and pMP167 RNAi vectors; information available upon request). RNAi experiments were performed by feeding as described in Kamath et al. (2001). Briefly, L4 mothers of the indicated genotype were fed on RNAi plates at 20°C (unless otherwise stated) and allowed to lay fertilized embryos for 24–36 hr. Their F1 progeny were then analyzed at the indicated time of growth postbirth on the RNAi plates at 20°C (where “birth” = egg laying time). F1 worms were transferred to fresh plates every 2–3 days when necessary.
Coimmunoprecipitations
Co-IP assays were performed following standard protocols described in detail in the Supplemental Information.
Transgenerational Fertility Assay
To start the transgenerational fertility assays at generation count zero, the spr-5(by134) BR3417 strain was outcrossed with N2 and homozygote spr-5(by134) mutants were selected by PCR. At each generation, six nonstarved, fertile adults were transferred to a fresh plate at 20°C, allowed to lay eggs for 24 hr, and eliminated. At least 100 worms of the next generation were scored per strain per generation. “Fertile” worms contained visible embryos in their uterus 4 days postbirth at 20°C.
DAPI Staining
Rapid DAPI staining protocol was applied as follows. Briefly, worms were harvested and washed in M9, fixed for 10 minutes in methanol at −20°C, washed in M9, stained with 2 μg/ml DAPI (Sigma-Aldrich), washed extensively in M9, and mounted in Vectashield mounting medium (Vector Laboratories) before being examined under a UV-light microscope (Zeiss Axioplan 2 microscope, Zeiss AxioCam Color camera, AxioVision 4.6 software).
BrdU Assays
BrdU assays were performed as described in Biedermann et al. (2009) with modifications, as described in the Supplemental Information.
Cell Counting in Germline Tumors
Cells positive for PGL-1, GLH-2, BrdU, PH3, or H3K4me3 signal were counted manually using the Cell Counter application of the ImageJ software (National Institutes of Health). In the BrdU and PH3 experiments, the amounts of counted cells are indicated in the corresponding tables. For the H3K4me3 experiment (Figure 6), a minimum of 500 DAPI-positive nuclei were counted in each category, totalized from at least five different pictures for each of the indicated strains.
Acknowledgments
This work was funded by Swiss National Science Foundation (SNF) grant 31003A_125577 (to F.M. and C.W.) and SNF Marie Heim-Vögtlin subsidy PMPDP3_129019 (to S.K.-P.). We thank S. Strome, K. Bennet, D. Miller, and A. Puoti for sharing strains and Y. Molleyres and L. Bulliard for technical assistance. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440).
Published: March 27, 2014
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Supplemental Information includes Supplemental Experimental Procedures, six figures, and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2014.02.007.
Supplemental Information
References
- Adamo A., Sesé B., Boue S., Castaño J., Paramonov I., Barrero M.J., Izpisua Belmonte J.C. LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nat. Cell Biol. 2011;13:652–659. doi: 10.1038/ncb2246. [DOI] [PubMed] [Google Scholar]
- Alvares S.M., Mayberry G.A., Joyner E.Y., Lakowski B., Ahmed S. H3K4 demethylase activities repress proliferative and postmitotic aging. Aging Cell. 2014;13:245–253. doi: 10.1111/acel.12166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés M.E., Burger C., Peral-Rubio M.J., Battaglioli E., Anderson M.E., Grimes J., Dallman J., Ballas N., Mandel G. CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc. Natl. Acad. Sci. USA. 1999;96:9873–9878. doi: 10.1073/pnas.96.17.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfiore M., Pugnale P., Saudan Z., Puoti A. Roles of the C. elegans cyclophilin-like protein MOG-6 in MEP-1 binding and germline fates. Development. 2004;131:2935–2945. doi: 10.1242/dev.01154. [DOI] [PubMed] [Google Scholar]
- Biedermann B., Wright J., Senften M., Kalchhauser I., Sarathy G., Lee M.H., Ciosk R. Translational repression of cyclin E prevents precocious mitosis and embryonic gene activation during C. elegans meiosis. Dev. Cell. 2009;17:355–364. doi: 10.1016/j.devcel.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Bowen N.J., Fujita N., Kajita M., Wade P.A. Mi-2/NuRD: multiple complexes for many purposes. Biochim. Biophys. Acta. 2004;1677:52–57. doi: 10.1016/j.bbaexp.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceballos-Chávez M., Rivero S., García-Gutiérrez P., Rodríguez-Paredes M., García-Domínguez M., Bhattacharya S., Reyes J.C. Control of neuronal differentiation by sumoylation of BRAF35, a subunit of the LSD1-CoREST histone demethylase complex. Proc. Natl. Acad. Sci. USA. 2012;109:8085–8090. doi: 10.1073/pnas.1121522109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J., Jang H., Kim H., Kim S.T., Cho E.J., Youn H.D. Histone demethylase LSD1 is required to induce skeletal muscle differentiation by regulating myogenic factors. Biochem. Biophys. Res. Commun. 2010;401:327–332. doi: 10.1016/j.bbrc.2010.09.014. [DOI] [PubMed] [Google Scholar]
- Christensen J., Agger K., Cloos P.A., Pasini D., Rose S., Sennels L., Rappsilber J., Hansen K.H., Salcini A.E., Helin K. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007;128:1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Eimer S., Lakowski B., Donhauser R., Baumeister R. Loss of spr-5 bypasses the requirement for the C.elegans presenilin sel-12 by derepressing hop-1. EMBO J. 2002;21:5787–5796. doi: 10.1093/emboj/cdf561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliazer S., Shalaby N.A., Buszczak M. Loss of lysine-specific demethylase 1 nonautonomously causes stem cell tumors in the Drosophila ovary. Proc. Natl. Acad. Sci. USA. 2011;108:7064–7069. doi: 10.1073/pnas.1015874108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forneris F., Binda C., Vanoni M.A., Battaglioli E., Mattevi A. Human histone demethylase LSD1 reads the histone code. J. Biol. Chem. 2005;280:41360–41365. doi: 10.1074/jbc.M509549200. [DOI] [PubMed] [Google Scholar]
- Francis R., Barton M.K., Kimble J., Schedl T. gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans. Genetics. 1995;139:579–606. doi: 10.1093/genetics/139.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes P., Cánovas J., Berndt F.A., Noctor S.C., Kukuljan M. CoREST/LSD1 control the development of pyramidal cortical neurons. Cereb. Cortex. 2012;22:1431–1441. doi: 10.1093/cercor/bhr218. [DOI] [PubMed] [Google Scholar]
- Greer E.L., Maures T.J., Hauswirth A.G., Green E.M., Leeman D.S., Maro G.S., Han S., Banko M.R., Gozani O., Brunet A. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature. 2010;466:383–387. doi: 10.1038/nature09195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison M.M., Ceol C.J., Lu X., Horvitz H.R. Some C. elegans class B synthetic multivulva proteins encode a conserved LIN-35 Rb-containing complex distinct from a NuRD-like complex. Proc. Natl. Acad. Sci. USA. 2006;103:16782–16787. doi: 10.1073/pnas.0608461103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendzel M.J., Wei Y., Mancini M.A., Van Hooser A., Ranalli T., Brinkley B.R., Bazett-Jones D.P., Allis C.D. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- Hird S.N., Paulsen J.E., Strome S. Segregation of germ granules in living Caenorhabditis elegans embryos: cell-type-specific mechanisms for cytoplasmic localisation. Development. 1996;122:1303–1312. doi: 10.1242/dev.122.4.1303. [DOI] [PubMed] [Google Scholar]
- Hung H., Kohnken R., Svaren J. The nucleosome remodeling and deacetylase chromatin remodeling (NuRD) complex is required for peripheral nerve myelination. J. Neurosci. 2012;32:1517–1527. doi: 10.1523/JNEUROSCI.2895-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji K., Caballero I.M., MacLeod R., Nichols J., Wilson V.A., Hendrich B. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat. Cell Biol. 2006;8:285–292. doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- Kamath R.S., Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Kamath R.S., Martinez-Campos M., Zipperlen P., Fraser A.G., Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2:H0002. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz D.J., Edwards T.M., Reinke V., Kelly W.G. A C. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell. 2009;137:308–320. doi: 10.1016/j.cell.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki I., Shim Y.H., Kirchner J., Kaminker J., Wood W.B., Strome S. PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell. 1998;94:635–645. doi: 10.1016/s0092-8674(00)81605-0. [DOI] [PubMed] [Google Scholar]
- Kunert N., Wagner E., Murawska M., Klinker H., Kremmer E., Brehm A. dMec: a novel Mi-2 chromatin remodelling complex involved in transcriptional repression. EMBO J. 2009;28:533–544. doi: 10.1038/emboj.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowski B., Roelens I., Jacob S. CoREST-like complexes regulate chromatin modification and neuronal gene expression. J. Mol. Neurosci. 2006;29:227–239. doi: 10.1385/JMN:29:3:227. [DOI] [PubMed] [Google Scholar]
- Lee M.G., Wynder C., Cooch N., Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- Li T., Kelly W.G. A role for Set1/MLL-related components in epigenetic regulation of the Caenorhabditis elegans germ line. PLoS Genet. 2011;7:e1001349. doi: 10.1371/journal.pgen.1001349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnúsdóttir E., Gillich A., Grabole N., Surani M.A. Combinatorial control of cell fate and reprogramming in the mammalian germline. Curr. Opin. Genet. Dev. 2012;22:466–474. doi: 10.1016/j.gde.2012.06.002. [DOI] [PubMed] [Google Scholar]
- Maures T.J., Greer E.L., Hauswirth A.G., Brunet A. The H3K27 demethylase UTX-1 regulates C. elegans lifespan in a germline-independent, insulin-dependent manner. Aging Cell. 2011;10:980–990. doi: 10.1111/j.1474-9726.2011.00738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D.M., 3rd, Ortiz I., Berliner G.C., Epstein H.F. Differential localization of two myosins within nematode thick filaments. Cell. 1983;34:477–490. doi: 10.1016/0092-8674(83)90381-1. [DOI] [PubMed] [Google Scholar]
- Passannante M., Marti C.O., Pfefferli C., Moroni P.S., Kaeser-Pebernard S., Puoti A., Hunziker P., Wicky C., Müller F. Different Mi-2 complexes for various developmental functions in Caenorhabditis elegans. PLoS ONE. 2010;5:e13681. doi: 10.1371/journal.pone.0013681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel T., Tursun B., Rahe D.P., Hobert O. Removal of Polycomb repressive complex 2 makes C. elegans germ cells susceptible to direct conversion into specific somatic cell types. Cell Rep. 2012;2:1178–1186. doi: 10.1016/j.celrep.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper A.S., Killian D.J., Hubbard E.J. Genetic analysis of Caenorhabditis elegans glp-1 mutants suggests receptor interaction or competition. Genetics. 2003;163:115–132. doi: 10.1093/genetics/163.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pferdehirt R.R., Kruesi W.S., Meyer B.J. An MLL/COMPASS subunit functions in the C. elegans dosage compensation complex to target X chromosomes for transcriptional regulation of gene expression. Genes Dev. 2011;25:499–515. doi: 10.1101/gad.2016011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds N., Latos P., Hynes-Allen A., Loos R., Leaford D., O’Shaughnessy A., Mosaku O., Signolet J., Brennecke P., Kalkan T. NuRD suppresses pluripotency gene expression to promote transcriptional heterogeneity and lineage commitment. Cell Stem Cell. 2012;10:583–594. doi: 10.1016/j.stem.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds N., Salmon-Divon M., Dvinge H., Hynes-Allen A., Balasooriya G., Leaford D., Behrens A., Bertone P., Hendrich B. NuRD-mediated deacetylation of H3K27 facilitates recruitment of Polycomb Repressive Complex 2 to direct gene repression. EMBO J. 2012;31:593–605. doi: 10.1038/emboj.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotili D., Mai A. Targeting histone demethylases: a new avenue for the fight against cancer. Genes Cancer. 2011;2:663–679. doi: 10.1177/1947601911417976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonet T., Dulermo R., Schott S., Palladino F. Antagonistic functions of SET-2/SET1 and HPL/HP1 proteins in C. elegans development. Dev. Biol. 2007;312:367–383. doi: 10.1016/j.ydbio.2007.09.035. [DOI] [PubMed] [Google Scholar]
- Smialowska A., Baumeister R. Presenilin function in Caenorhabditis elegans. Neurodegener. Dis. 2006;3:227–232. doi: 10.1159/000095260. [DOI] [PubMed] [Google Scholar]
- Subramaniam K., Seydoux G. Dedifferentiation of primary spermatocytes into germ cell tumors in C. elegans lacking the pumilio-like protein PUF-8. Curr. Biol. 2003;13:134–139. doi: 10.1016/s0960-9822(03)00005-8. [DOI] [PubMed] [Google Scholar]
- Sulston J.E., Schierenberg E., White J.G., Thomson J.N. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Tursun B., Patel T., Kratsios P., Hobert O. Direct conversion of C. elegans germ cells into specific neuron types. Science. 2011;331:304–308. doi: 10.1126/science.1199082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unhavaithaya Y., Shin T.H., Miliaras N., Lee J., Oyama T., Mello C.C. MEP-1 and a homolog of the NURD complex component Mi-2 act together to maintain germline-soma distinctions in C. elegans. Cell. 2002;111:991–1002. doi: 10.1016/s0092-8674(02)01202-3. [DOI] [PubMed] [Google Scholar]
- von Zelewsky T., Palladino F., Brunschwig K., Tobler H., Hajnal A., Müller F. The C. elegans Mi-2 chromatin-remodelling proteins function in vulval cell fate determination. Development. 2000;127:5277–5284. doi: 10.1242/dev.127.24.5277. [DOI] [PubMed] [Google Scholar]
- Wang J.T., Seydoux G. Germ cell specification. Adv. Exp. Med. Biol. 2013;757:17–39. doi: 10.1007/978-1-4614-4015-4_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang H., Chen Y., Sun Y., Yang F., Yu W., Liang J., Sun L., Yang X., Shi L. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009;138:660–672. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- Wang S., Fisher K., Poulin G.B. Lineage specific trimethylation of H3 on lysine 4 during C. elegans early embryogenesis. Dev. Biol. 2011;355:227–238. doi: 10.1016/j.ydbio.2011.04.010. [DOI] [PubMed] [Google Scholar]
- Whyte W.A., Bilodeau S., Orlando D.A., Hoke H.A., Frampton G.M., Foster C.T., Cowley S.M., Young R.A. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482:221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C.J., Naito T., Arco P.G., Seavitt J.R., Cashman S.M., De Souza B., Qi X., Keables P., Von Andrian U.H., Georgopoulos K. The chromatin remodeler Mi-2beta is required for CD4 expression and T cell development. Immunity. 2004;20:719–733. doi: 10.1016/j.immuni.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Xiao Y., Bedet C., Robert V.J., Simonet T., Dunkelbarger S., Rakotomalala C., Soete G., Korswagen H.C., Strome S., Palladino F. Caenorhabditis elegans chromatin-associated proteins SET-2 and ASH-2 are differentially required for histone H3 Lys 4 methylation in embryos and adult germ cells. Proc. Natl. Acad. Sci. USA. 2011;108:8305–8310. doi: 10.1073/pnas.1019290108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Strome S. Depletion of a novel SET-domain protein enhances the sterility of mes-3 and mes-4 mutants of Caenorhabditis elegans. Genetics. 2001;159:1019–1029. doi: 10.1093/genetics/159.3.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


